Abstract
The novel NKG2C*03 allele encodes a hybrid of the NKG2C*01 and NKG2C*02 primary structures.
Keywords: alleles, genetic polymorphism, HLA‐E, human genetics, natural killer cell lectin‐like receptors, NKG2C receptor
The CD94:NKG2 family of heterodimeric receptors, expressed on subsets of human natural killer (NK) and T lymphocytes, monitor the expression of HLA‐E, presenting peptides mostly derived from the signal sequence of other HLA class I alpha chains.1, 2 CD94:NKG2 heterodimers deliver inhibitory or activating signals, depending on the NKG2 subunit (mainly NKG2A and NKG2C, respectively). CD94 and all NKG2 subunits are type II membrane‐integral glycoproteins of the C‐type‐lectin‐like superfamily, with 49%–94% identity in their coding sequence. Their genes are located in the Natural Killer gene Complex (NKC) on chromosome 12, which encompasses nearly 2 Mbp, and encodes additional homologs of the same family.
The activating NKG2C (or CD159c) subunit is encoded by the KLRC2 gene (more often referred to as NKG2C), with a length of ca. 6 kbp comprising a 696‐bp coding sequence segmented into six exons.1, 3, 4 Homo‐ and heterozygous complete KLRC2/NKG2C gene deletions are seen in up to 8% and 32%, respectively, of Spanish Caucasoids and other populations.3, 5, 6 Several studies have addressed the distribution, functional effect, and importance of NKG2C deletion in different populations and health conditions, particularly cytomegalovirus infection, which specifically triggers differentiation and expansion of NK cells that express high NKG2C levels, and display a characteristic phenotype and function.3, 5, 6 In contrast, studies on NKG2C sequence polymorphism are scarce,1, 3, 4 possibly because the receptor family is often deemed, albeit inexactly, non‐polymorphic. A common complementary DNA sequence was designated NKG2C*01 by Shum et al,1, 3, 4 who also named NKG2C*02 a second sequence, found in individuals of different ethnicities, and diverging from the former by two single‐nucleotide nonsynonymous polymorphisms (c. 5G > A, Ser2Asn, and c.305C > T, Ser102Phe). The first change affects the cytoplasmic tail, whilst the second is located in the stem that connects the transmembrane region with the ligand‐binding domain.
We report here a novel allele, identified in two unrelated Spanish Caucasoids, that encodes asparagine 2, like NKG2C*02; and serine 102, like NKG2C*01; being the rest of their primary structures identical (Figure 1). The complete NKG2C coding region was amplified from an NKG2C‐hemizygous donor in two overlapping genomic segments, using a proof‐reading DNA polymerase and two pairs of oligonucleotide primers in separate reactions—KLRC2F‐383/KLRR+485b (5′–ctattttatcttatggcacacaatcc–3′/5′–ctggatagctttattgaagtgtca–3′), and KLRFg669/KLRR+623 (5′–cagtgtggatcttcaatg–3′/5′–gtcataaacaatcccatcag–3′) (PCR conditions available upon request). Sequence analysis revealed the new allele to be most similar to the NKG2C*01 sequence in NKC clone with accession number AC277791.1–5815 identities in 5820 nucleotides, the most relevant difference being the codon 2 substitution AGT > AAT (Ser2Asn). Following the previously used format,4 we have designated the new allele with the name NKG2C*03 (GenBank MW291142). The limited number of nucleotide changes that separate the few genomic NKG2C*01/*02 sequences available in public databases does not allow for asserting whether NKG2C*03 is an intermediate evolutionary step between previously known alleles or evolved from these by interallelic recombination or point mutation. The structural and functional importance of CD94/NKG2 sequence polymorphisms, as well as the detailed three‐dimensional configuration of the involved stem and cytoplasmic domains, remain unexplored.
FIGURE 1.
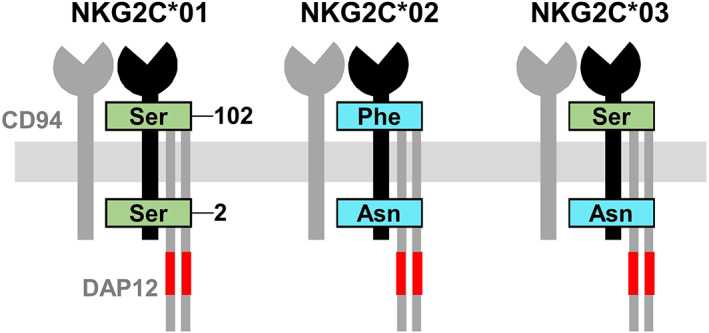
Representation of the CD94:NKG2C receptor, including the signaling adaptor DAP12 and its immunoreceptor tyrosine‐based activation motifs (ITAMs, in red). Well‐characterized polymorphisms of the NKG2C primary structure are shown
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Judit Asenjo designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. Aura Muntasell and Miguel López‐Botet designed the study and revised the manuscript. Manuela Moraru designed the study and directed research. Carlos Vilches designed the study, directed research and wrote the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Elvira Ramil, from the DNA sequencing core facility of Instituto de Investigación Sanitaria Puerta de Hierro—Segovia de Arana, for continued support. This work was partially funded by grants SAF2016‐80363‐C2‐1/2‐R from the Spanish Ministerio de Ciencia e Innovación (MCINN, Agencia Estatal de Investigación‐AEI/FEDER, EU), and PCIN‐2015‐191‐C02‐01/02 (FP7‐MINECO ERA‐NET Infect‐ERA program, EU) to C.V. and M.L.B. M.M. is hired by grant GCB15152947MELE from the Asociación Española contra el Cáncer Foundation; J.A. is hired by grant PID2019‐110609RB‐C22/AEI/10.13039/501100011033 (AEI/FEDER, EU) to C.V. Publication costs are defrayed by a nominative grant to support research from Consejería de Sanidad de la Comunidad de Madrid in 2021 to Fundación de Investigación Biomédica Puerta de Hierro.
Asenjo J, Muntasell A, López‐Botet M, Moraru M, Vilches C. Complete genomic characterization of a new KLRC2 allele, NKG2C*03 . HLA. 2021;98(3):259-261. 10.1111/tan.14231
Manuela Moraru and Carlos Vilches share last authorship.
Funding information Consejería de Sanidad de la Comunidad de Madrid, Grant/Award Number: Nominative grant to support research in FIBHUPH; FP7‐MINECO Infect‐ERA program, EU, Grant/Award Number: PCIN‐2015‐191‐C02‐01/02; Fundación Científica Asociación Española Contra el Cáncer, Grant/Award Number: GCB15152947MELE; Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación, FEDER, EU, Grant/Award Number: PID2019‐110609RB‐C22/AEI/10.13039/501100011033
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Cantoni C, Biassoni R, Pende D, et al. The activating form of CD94 receptor complex: CD94 covalently associates with the Kp39 protein that represents the product of the NKG2‐C gene. Eur J Immunol. 1998;28(1):327‐338. [DOI] [PubMed] [Google Scholar]
- 2.Lopez‐Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non‐classical HLA class I molecules. Semin Immunol. 2000;12(2):109‐119. [DOI] [PubMed] [Google Scholar]
- 3.Hikami K, Tsuchiya N, Yabe T, Tokunaga K. Variations of human killer cell lectin‐like receptors: common occurrence of NKG2‐C deletion in the general population. Genes Immun. 2003;4(2):160‐167. [DOI] [PubMed] [Google Scholar]
- 4.Shum BP, Flodin LR, Muir DG, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168(1):240‐252. [DOI] [PubMed] [Google Scholar]
- 5.Lopez‐Botet M, Muntasell A, Vilches C. The CD94/NKG2C+NK‐cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin Immunol. 2014;26(2):145‐151. [DOI] [PubMed] [Google Scholar]
- 6.Muntasell A, Lopez‐Montanes M, Vera A, et al. NKG2C zygosity influences CD94/NKG2C receptor function and the NK‐cell compartment redistribution in response to human cytomegalovirus. Eur J Immunol. 2013;43(12):3268‐3278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


