ABSTRACT
Branchio-oto-renal syndrome (BOR) is a disorder characterized by hearing loss, and craniofacial and/or renal defects. Variants in the transcription factor Six1 and its co-factor Eya1, both of which are required for otic development, are linked to BOR. We previously identified Sobp as a potential Six1 co-factor, and SOBP variants in mouse and humans cause otic phenotypes; therefore, we asked whether Sobp interacts with Six1 and thereby may contribute to BOR. Co-immunoprecipitation and immunofluorescence experiments demonstrate that Sobp binds to and colocalizes with Six1 in the cell nucleus. Luciferase assays show that Sobp interferes with the transcriptional activation of Six1+Eya1 target genes. Experiments in Xenopus embryos that either knock down or increase expression of Sobp show that it is required for formation of ectodermal domains at neural plate stages. In addition, altering Sobp levels disrupts otic vesicle development and causes craniofacial cartilage defects. Expression of Xenopus Sobp containing the human variant disrupts the pre-placodal ectoderm similar to full-length Sobp, but other changes are distinct. These results indicate that Sobp modifies Six1 function and is required for vertebrate craniofacial development, and identify Sobp as a potential candidate gene for BOR.
KEY WORDS: Sobp, Cranial placodes, Neural crest, Pre-placodal ectoderm, Otic vesicle, Branchio-oto-renal syndrome
Summary: Sobp interacts with Six1 in the cell nucleus and represses the transcriptional activation of Six1+Eya1 target genes. In Xenopus embryos, Sobp functions during early stages of inner ear development.
INTRODUCTION
Hearing is an important component of human communication and hearing loss disorders negatively affect quality of life. Around 32 million children worldwide have disabling hearing loss, 40% of which have a genetic cause (Krug, 2016). Although hundreds of genes are associated with syndromic and non-syndromic congenital hearing loss, many cases still have an unknown cause (Shearer et al., 2017). Branchio-otic and branchio-oto-renal syndromes (BOR) are autosomal dominant disorders in which affected individuals present with variable degrees of hearing loss due to defects in inner, middle and outer ears, as well as branchial arch-associated dysmorphologies, including branchial fistulas and cysts (Moody et al., 2015; Smith, 2018). Variants in EYA1 or SIX1 have been identified in ∼50% of affected individuals, but the underlying genetic causes of remaining cases are unknown (Ruf et al., 2003, 2004; Sanggaard et al., 2007; Lee et al., 2007; Klingbeil et al., 2017). Thus, identifying genes that interact with SIX1 may uncover novel genes linked to BOR or other deafness syndromes.
The Six1 transcription factor, which is homologous to Drosophila Sine oculis (So) (Cheyette et al., 1994), plays a role in many cellular processes and in the embryo regulates the development of craniofacial tissues affected in BOR (Kawakami et al., 2000; Kumar, 2009; Xu, 2013; Moody and LaMantia, 2015). Loss of Six1 in mouse and Xenopus causes several craniofacial defects, including disruption of inner, middle and outer ear development (Zheng et al., 2003; Li et al., 2003; Laclef et al., 2003; Ozaki et al., 2004; Brugmann et al., 2004; Schlosser et al., 2008; Guo et al., 2011; Tavares et al., 2017; Sullivan et al., 2019). A hallmark of Six1 transcriptional activity is that it can be modulated by co-factors that influence it to either activate or repress target genes (Ohto et al., 1999; Li et al., 2003; Silver et al., 2003; Brugmann et al., 2004). Transcriptional activation is mediated by the Eya family of co-activators. Upon binding to Six1, Eya factors are translocated into the nucleus, where Eya phosphatase activity triggers the recruitment of co-activators that switch Six1 function from repression to activation (Ohto et al., 1999; Li et al., 2003).
A screen to identify proteins that interact with Drosophila So identified So-binding protein (Sobp), which is co-expressed with so in the anterior region of the eye disc (Kenyon et al., 2005a). In mouse, Sobp was identified as the spontaneous recessive variant causing deafness and vestibular-mediated circling behavior in the Jackson circler (jc) mouse (Calderon et al., 2006; Chen et al., 2008). In humans, SOBP was linked to mental retardation, anterior maxillary protrusion, strabismus and mild hearing loss (MRAMS; OMIM #613671) in seven individuals of the same family. The homozygous variant inserts an early stop codon at arginine 661 (p.R661X), causing a 212 amino acid truncation in the C terminus (Basel-Vanagaite et al., 2007; Birk et al., 2010).
As Sobp variants cause otic phenotypes in mouse and humans, and are expressed in the pre-placodal ectoderm (PPE) and otic vesicle in Xenopus embryos (Neilson et al., 2010), we speculated that Sobp might function by associating with Six1 and thereby contributing to BOR (Moody et al., 2015). Here, we address the developmental function of Sobp. We find that, although sobp is expressed in the same tissues as six1, higher levels of Sobp are detected where Six1 expression is lower, suggesting that it may repress Six1 activity. We show that Sobp binds to Six1, competes with Eya1 binding to the complex and significantly interferes with the transcriptional activation of Six1+Eya1 target genes. Structural and functional analyses show that Sobp constructs either lacking a conserved C-terminal nuclear localization signal (NLS) or containing a variant similar to human p.R661X (Xenopus Sobp p.R651X) can still interact with Six1, indicating that the domain that interacts with Six1 is not in the C terminus. In vivo studies establish Sobp as a crucial factor for PPE and neural crest (NC) gene expression, and for later otic vesicle development. These findings also show that, while the p.R651X variant disrupts gene expression in the PPE in the same manner as full-length Sobp, changes in other domains and otic vesicle patterning are distinct. Together, these results demonstrate that Sobp plays a crucial role in Six1 transcriptional function during several aspects of vertebrate craniofacial development, and suggest it may be a candidate gene for BOR and other deafness syndromes.
RESULTS
Sobp expression in comparison to that of Six1
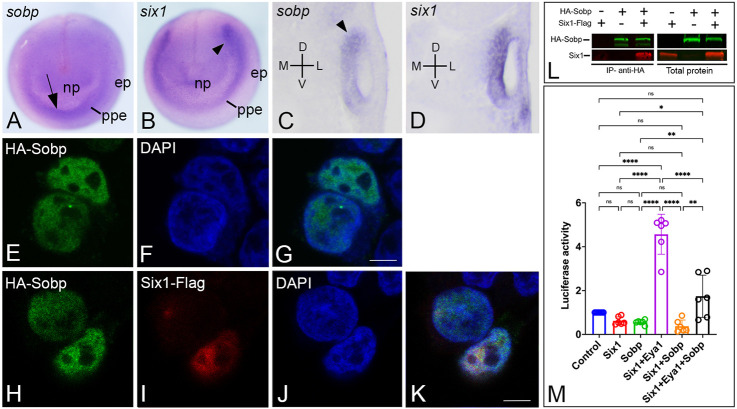
We reported previously that sobp is expressed in several of the embryonic tissues that also express six1, including the PPE and otic vesicle (Neilson et al., 2010). A closer comparison at the neural plate stage showed that sobp expression was stronger in the anterior PPE and weaker in the posterior PPE, whereas six1 expression was the reverse (Fig. 1A,B). At larval stages, both six1 and sobp were expressed in the medial wall of the otic vesicle, but sobp expression appeared more intense dorsally compared with its ventral domain (Fig. 1C,D; Fig. S1A,B). Co-transfection assays in HEK293T cells showed that Sobp protein was mainly nuclear (Fig. 1E-G), as previously described in mouse (Chen et al., 2008), where it colocalized with Six1 in most cells expressing both constructs (Fig. 1H-K). These findings are the first direct evidence in vertebrates of Sobp and Six1 co-expression in tissues and cellular compartments that would allow their interaction.
Fig. 1.
Sobp is expressed with Six1 in the cell nucleus and represses the transcriptional activation of Six1+Eya1 target genes. (A-D) In situ hybridization for sobp (A,C) and six1 (B,D). At neural plate stages (A,B), although sobp expression in the PPE overlaps with that of six1, its expression is more intense in the anterior domain (arrow), whereas six1 expression is more intense in the posterior domain (arrowhead). In transverse sections through the larval otic vesicle (C,D), sobp is expressed with six1 in the ventral-medial wall. sobp expression is more intense at the dorsal pole (arrowhead). D, dorsal; ep, epidermis; L, lateral; M, medial; np, neural plate; ppe, pre-placodal ectoderm; V, ventral. (E-K) Confocal images of HEK293T cells expressing HA-Sobp (green, E-G) and cells co-expressing HA-Sobp (green) and Six1-Flag (red) (H-K). Sobp is localized in the cell nuclei in both the absence and presence of Six1-Flag. Cell nuclei are stained with DAPI (blue, F,G,J,K). Scale bars: 5 μm. (L) HEK293T cells were co-transfected with combinations of HA-Sobp and/or Six1-Flag followed by multiplex fluorescence western blot detection for HA-Sobp (green) and Six1-Flag (red). Six1 was detected after HA-Sobp was immunoprecipitated with anti-HA magnetic beads (IP, left two rows). Right two rows show expression of the constructs prior to immunoprecipitation. (M) Graph depicting the luciferase activity of the pGL3-6xMEF3-luciferase reporter in HEK293T cells transfected with different combinations of constructs expressing Six1, Eya1 and/or Sobp. Data are normalized to Renilla expressed with a constitutive promoter. Luciferase activity is significantly induced (P<0.0001) by Six1+Eya1, whereas Sobp reduces this induction to levels indistinguishable from control (Six1+Eya1 versus Six1+Eya1+Sobp, P<0.0001; control versus Six1+Eya1+Sobp, P=0.2226). Six1 (P=0.9984), Sobp (P=0.9184) and Six1+Sobp (P=0.9184) did not cause any significant changes in luciferase activity compared with control. ns, not significant; *P<0.05, **P<0.01, ****P<0.0001. Experiments were repeated in duplicate at least three independent times. Error bars represent s.d. with circles depicting individual data points.
Sobp binds to Six1 and modulates the transcriptional activation of Six1+Eya1 target genes
Drosophila Sobp has been shown to bind to So by yeast two hybrid and GST-pulldown assays (Kenyon et al., 2005a). To test whether this interaction occurred with vertebrate proteins, HEK293T cells were co-transfected with constructs driving expression of Six1-Flag and/or HA-Sobp followed by immunoprecipitation (IP). Western blot analysis detected Six1 after HA-Sobp IP (Fig. 1L); reverse IP confirmed this finding (data not shown).
To assess whether Sobp modulates Six1 transcriptional activity, HEK293T cells were co-transfected with a Six1-inducible reporter (Ford et al., 2000) and with different combinations of Six1, Eya1 and/or Sobp (Fig. 1M; Fig. S2). As demonstrated previously (Patrick et al., 2009), in the presence of Eya1, Six1 induced a significant increase in luciferase activity over control (∼5 fold increase, P<0.0001). In contrast, Sobp by itself (P=0.8899) or in the presence of Six1 (P=0.9184) did not cause a significant change in luciferase activity. However, co-transfection of Sobp with both Six1 and Eya1 significantly repressed luciferase activity compared with Six1+Eya1 levels (P<0.0001), returning luciferase activity to levels statistically indistinguishable from control (P=0.2226). These data show that Sobp can be classified as a bona fide Six1 co-factor because it binds to Six1 and is able to significantly interfere with the transcriptional activation of the Six1+Eya1 reporter.
Pa2g4 is a recently identified Six1 co-factor that reduces the transcriptional activation of Six1+Eya1 target genes by competing with the ability of Eya1 to bind to Six1 (Neilson et al., 2017). To determine whether Sobp also competes for binding to Six1, HEK293T cells were co-transfected with Six1-Flag, Myc-Eya1 and increasing amounts of HA-Sobp. Co-IP experiments (Fig. 2A) showed that equimolar amounts (1.0×) of Six1, Eya1 and Sobp did not disrupt Six1 binding to Eya1, whereas the amount of Six1 bound to Eya1 was diminished at a five- or 10-fold increase in Sobp. Interestingly, low (0.5×) and equimolar (1.0×) levels of Sobp resulted in higher levels of Six1 bound to Eya1 relative to when Six1 and Eya1 were co-transfected without Sobp. Because Eya factors are localized in the cytosol (Fig. 2C-F) and become nuclear when bound to Six factors (Fig. 2G-J) (Ohto et al., 1999; Li et al., 2003), we tested whether the competition for binding to Six1 would disturb the ability of Six1 to translocate Eya1 to the nucleus (Fig. 2O-C′). Cells co-transfected with combinations of Six1-Flag, Myc-Eya1 and increasing concentration of HA-Sobp showed that Eya1 was translocated to the nucleus by Six1 (Fig. 2O,S,X,Z) at equimolar amounts of Six1, Eya1 and Sobp, with most transfected cells also expressing Sobp in the nucleus (Fig. 2P,T,W,AA). Remarkably, Sobp was able to bind to (Fig. 2B) and partially translocate Eya1 to the nucleus (Fig. 2K-N) without Six1 co-expression. However, and in agreement with the competition co-IP results, a fivefold increase in Sobp led to detection of Eya1 in the cytosol (Fig. 2Q-R,BB,CC) even though both Six1 (Fig. 2U,BB) and Sobp (Fig. 2V,CC) were nuclear. Together, these data indicate that Sobp interference with the transcriptional activation of Six1+Eya1 target genes is achieved through a dose-dependent competition mechanism.
Fig. 2.
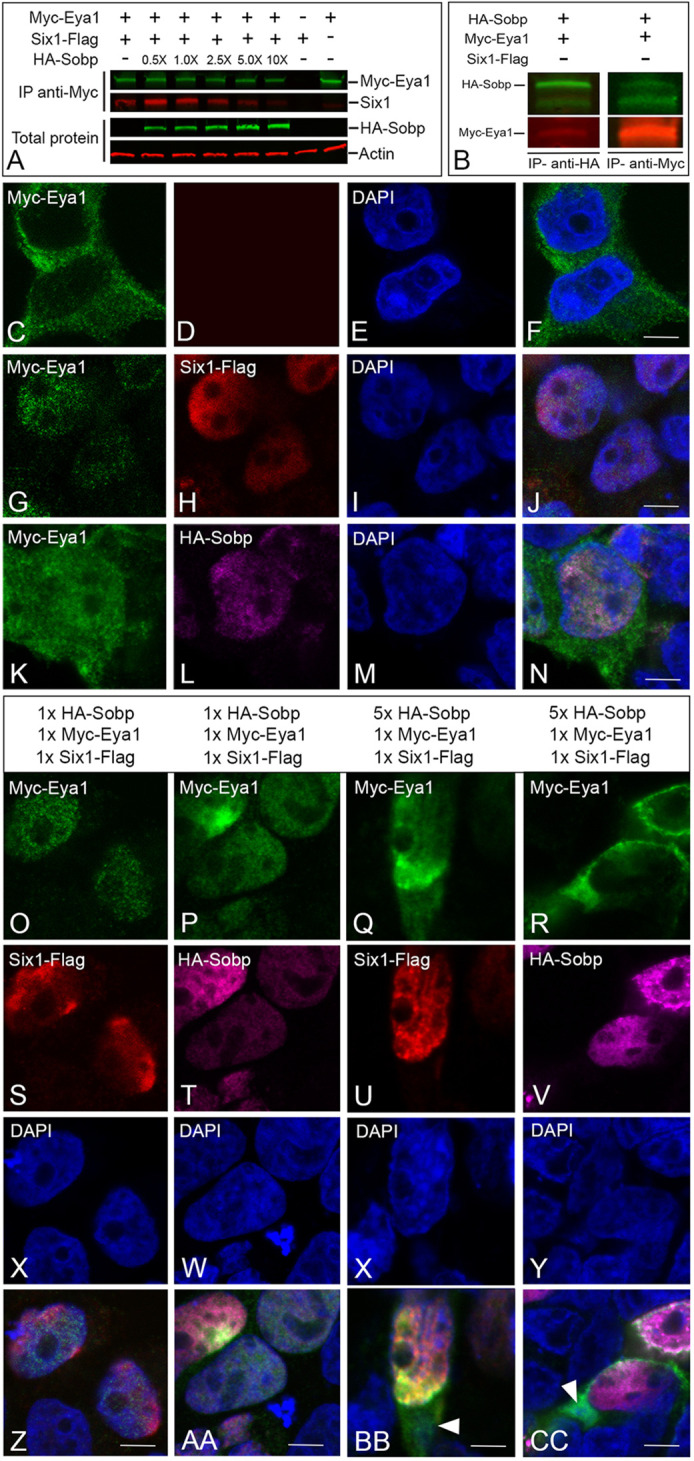
Sobp reduces the transcriptional activation of Six1+Eya1 target genes by disrupting the Six1/Eya1 interaction. (A) HEK293T cells co-transfected with equimolar amounts of Six1-Flag and/or Myc-Eya1 were additionally transfected with increasing amounts of HA-Sobp. Although low (0.5×) and equimolar (1.0×) levels of Sobp resulted in higher levels of Six1 bound to Eya1 relative to when Six1 and Eya1 were co-transfected without Sobp, the amount of Six1 bound to Myc-Eya1 decreased with increasing levels of HA-Sobp (2.5×, 5.0× and 10×). The bottom two rows show expression before immunoprecipitation of increasing levels of HA-Sobp with β-actin as loading control. (B) HEK293T cells were co-transfected with HA-Sobp and Myc-Eya1 followed by multiplex fluorescence western blot detection for HA-Sobp (green) and Myc-Eya1 (red). Myc-Eya1 is detected when HA-Sobp is immunoprecipitated (IP, anti-HA, left column). The reverse immunoprecipitation (anti-Myc, right column) confirmed this interaction. (C-N) Confocal images of HEK293T cells expressing Myc-Eya1 (green), Six1-Flag (red) and/or HA-Sobp (magenta). Myc-Eya1 is located exclusively in the cytosol (C,F) and is completely translocated to the cell nucleus by Six1-Flag in the majority of the cells (G,H,J). Surprisingly, HA-Sobp also partially translocates Myc-Eya1 to the cell nucleus (K,L,N) in the absence of Six1. Cell nuclei are stained with DAPI (blue in E,F,I,J,M,N). Scale bars: 5 μm. (O-CC) Confocal images of HEK293T cells expressing Myc-Eya1 (green, O-R,Z-CC), Six1-Flag (red, S,U,Z,BB) and HA-Sobp (magenta, T,V,AA,CC). Myc-Eya1 was completely translocated into the cell nucleus by Six1 when cells received equimolar amounts (1×) of Six1-Flag, Myc-Eya1 and HA-Sobp (O,P,S,T,Z,AA), whereas cytosolic Myc-Eya1 (arrowheads in BB and CC) was detected in many cells when there was a fivefold increase in HA-Sobp. Nuclear DAPI staining, blue (X-CC). Scale bars: 5 μm.
Six1 can transport Sobp to the nucleus
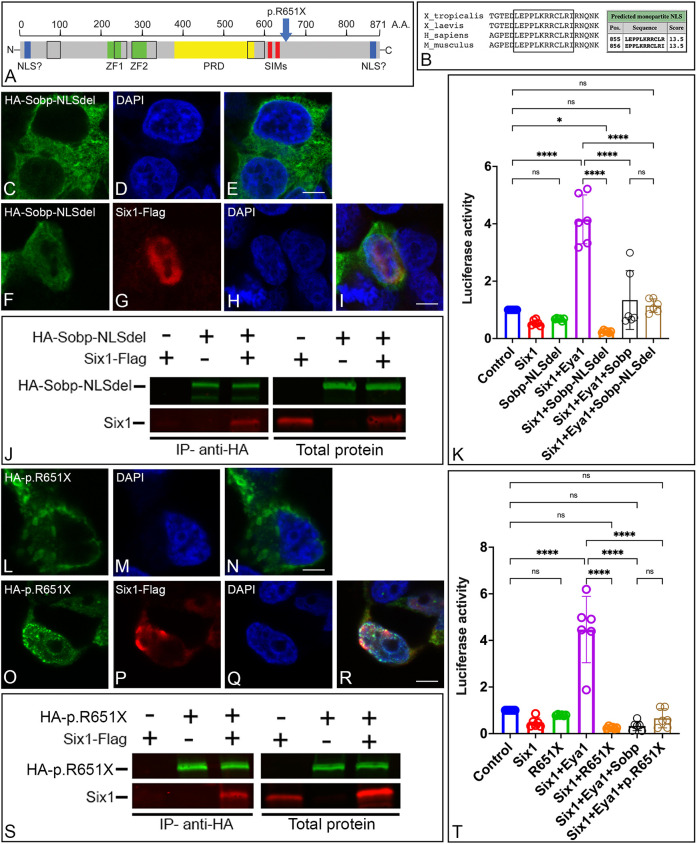
Because Sobp is located in the nucleus in the absence of Six1 co-transfection (Fig. 1E-G) (Chen et al., 2008), it must contain a NLS. A bioinformatic analysis (Kosugi et al., 2009) identified in Xenopus Sobp N-terminal and C-terminal putative NLSs, similar to that detected in the mouse (Chen et al., 2008) and human (Birk et al., 2010) sequences. However, only the putative C-terminal NLS had a cutoff score above 8, which indicates protein localization exclusively in the nucleus (Fig. 3A-B) (Kosugi et al., 2009). Deletion of this C-terminal domain caused HA-Sobp-NLSdel to be detected mainly in the cytosol (Fig. 3C-E). Interestingly, when cells additionally expressed Flag-tagged Six1, Sobp-NLSdel was translocated to the nucleus (Fig. 3F-I). Removal of the NLS did not disrupt binding of Sobp to Six1 (Fig. 3J) or diminish the ability of Sobp to reduce the transcriptional activation of the Six1+Eya1 reporter (P<0.0001; Fig. 3K). These findings identify a single C-terminal NLS in Sobp and demonstrate that, in its absence, Sobp can bind to Six1 in the cytosol and be translocated to the nucleus via this interaction.
Fig. 3.
Deletion of the nuclear localization signal and the p.R651X variant of sobp do not disrupt interaction with Six1. (A) Schematic representation of the Sopb protein structure showing its different domains and the location of the p.R651X variant identified in individuals with MRAMS. Black outline indicates protein domains that are highly conserved between different species, including D. melanogaster, M. musculus, G. gallus and X. laevis/tropicalis; blue boxes/NLS denote putative nuclear localization signals; green boxes/ZF1/ZF2 denote FCS zinc-finger domains; yellow box/PRD denotes proline-rich domain; red boxes/SIMs denote SUMO-interacting motifs. (B) Comparison of the C-terminal region of Sobp between species showing a highly conserved domain that is predicted to be a NLS with a high cutoff score according to cNLS mapper. (C-I) Confocal images of HEK293T cells expressing a construct lacking the C-terminal NLS (HA-Sobp-NLSdel, green, C,E,F,I). This construct is cytosolic in the majority of the transfected cells (C,E). Cells co-expressing HA-Sobp-NLSdel (green, F,I) with Six1-Flag (red, G,I) show partial translocation to the nucleus. DAPI (blue, D,E,H,I). Scale bars: 5 μm. (J) HEK293T cells were co-transfected with combinations of HA-Sobp-NLSdel and/or Six1-Flag followed by multiplex fluorescent western blot detection for HA-Sobp-NLSdel (green) and Six1 (red). Six1 was detected after HA-Sobp-NLSdel was immunoprecipitated with anti-HA magnetic beads (IP, left two rows). Right two rows show expression of the constructs before immunoprecipitation. (K) Graph depicting the luciferase activity of the pGL3-6xMEF3-luciferase reporter in HEK293T cells transfected with different combinations of constructs expressing Six1, Eya1, Sobp and/or Sobp-NLSdel. The C-terminal NLS is not required for repression of the transcriptional activation of the Six1+Eya1 reporter (control versus Six1+Eya1+Sobp-NLSdel, P=0.9959; Six1+Eya1+Sobp versus Six1+Eya1+Sobp-NLSdel, P=0.9912). ns, not significant; *P<0.05, ****P<0.0001. Experiments were repeated in duplicate at least three independent times. Error bars represent s.d. with circles depicting individual data points. (L-R) Confocal images of HEK293T cells expressing the p.R651X variant of sobp (HA-p.R651X, green, L,N,O,R). The variant is cytosolic in the majority of transfected cells (L,N). Cells co-expressing p.R651X (green, O,R) and Six1-Flag (red, P,R) show partial translocation of HA-p.R651X to the cell nucleus. Cell nuclei are stained with DAPI (M,N,Q,R). Scale bars: 5 μm. (S) HEK293T cells were co-transfected with combinations of HA-p.R651X and/or Six1-Flag followed by multiplex fluorescence western blot detection for HA-p.R651X (green) and Six1 (red). Six1 was detected after HA-p.R651X was immunoprecipitated with anti-HA magnetic beads (IP, left two rows). Right two rows show expression of the constructs before immunoprecipitation. (T) Graph depicting the luciferase activity of the pGL3-6xMEF3-luciferase reporter in HEK293T cells transfected with different combinations of constructs expressing Six1, Eya1, Sobp and/or p.R651X. Truncated Sobp (p.R651X) is still able to repress the transcriptional activation of the Six1+Eya1 reporter (control versus Six1+Eya1+p.R651X, P=0.8603; Six1+Eya1+Sobp versus Six1+Eya1+p.R651X, P=0.9362). Experiments were repeated in duplicate at least three independent times. Error bars represent s.d. with circles depicting individual data points. ns, not significant; ****P<0.0001.
The human p.R661X variant does not disrupt its interaction with Six1
We introduced the human p.R661X variant found in a MRAMS family (Birk et al., 2010) in Xenopus Sobp by replacing the arginine located at amino acid 651 with a stop codon (c.1951 A>T; p.R651X) generating a truncated Sobp lacking part of the C-terminus, including the NLS (Fig. 3A). As expected, HA-p.R651X is located primarily in the cytosol (Fig. 3L-N) as the NLS was missing. Cytosolic HA-p.R651X was translocated to the nucleus in most cells also expressing Six1-Flag (Fig. 3O-R). Despite the large deletion, p.R651X still bound to Six1 (Fig. 3S) and significantly reduced the transcriptional activation of the Six1+Eya1 reporter (P<0.0001; Fig. 3T). These data demonstrate that, although the p.R651X variant of sobp lacks a large region of the C terminus, the domain for Six1 interaction remains intact.
Sobp is required for development of neural crest and placode domains
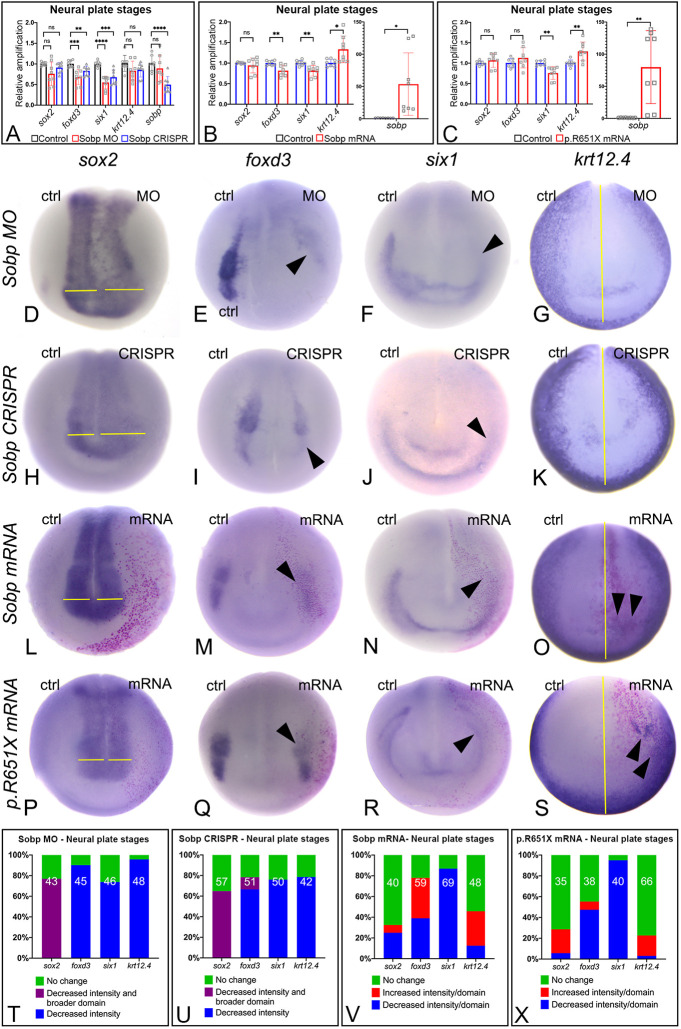
To determine whether Sobp has a role in NC and PPE development, we performed two types of knockdown (KD) experiments: a translation-blocking antisense morpholino oligonucleotide (MO) against sobp; and an F0 analysis after CRISPR/Cas9-mediated genome editing (Fig. 4; Fig. S3). First, we assessed by qPCR whether KD in the whole embryo caused changes in the expression of genes characteristic of neural plate (sox2), NC (foxd3), PPE (six1) and epidermis (krt12.4) at neural plate stages (Fig. 4A). KD using either method showed decreased mRNA levels for foxd3 (NC) (MO, P<0.001/q=0.000602; CRISPR, P<0.01/q=0.002520) and six1 (PPE) (MO, P<0.0001/q=0.000007; CRISPR, P<0.001/q=0.000274), but not for sox2 (MO, P=0.033437/q=0.033771; CRISPR, P=0.071790/q=0.036254) or krt12.4 (MO, P=0.114167/q=0.086482; CRISPR, P=0.104590/q=0.042254). We also monitored levels of sobp mRNA (Fig. 4A) and confirmed that, as expected, it was decreased by CRISPR/Cas9 editing likely followed by nonsense-mediated mRNA decay (P<0.0001/q=0.000162), but not by a translation-blocking MO (P=0.359946/q=0.218127). To assess whether these whole-embryo gene expression changes were also observed as changes in the sizes of specific expression domains, we monitored similar KD on one side of embryos by in situ hybridization (ISH). Sobp KD by MO or by CRISPR each caused a broader sox2 domain that was fainter in intensity (MO, 77.0%, Fig. 4D,T; CRISPR, 64.9%, Fig. 4H,U) and reduction of the foxd3 (MO, 90.2%, Fig. 4E,T; CRISPR, 66.6%, Fig. 4I,U), six1 (MO, 73.9%, Fig. 4F,T; CRISPR, 76%, Fig. 4J,U) and krt12.4 (MO, 95.8%, Fig. 4G,T; CRISPR, 78.6%, Fig. 4K,U) domains on the injected side of the embryos. The results from the two different assays are summarized in Table S1. The qPCR and ISH approaches both showed reductions in foxd3 and six1 expression, but the ISH assay was more sensitive to changes in the spatial domains of sox2 and krt12.4. Whereas qPCR did not detect a change in sox2 levels in the whole embryo, the sox2 domain was broader and fainter, which would likely lead to no significant overall change as assessed by qPCR. Whereas qPCR did not detect a change in krt12.4 levels in the whole embryo, the krt12.4 domain was obviously reduced along the anterior neural plate (Fig. 4G,K); this may be too subtle a change to be detected by qPCR of a whole embryo covered by epidermis.
Fig. 4.
Sobp is required for proper formation of the embryonic ectodermal domains at neural plate stages. (A) qPCR analysis of whole neural plate embryos injected with MO or after CRISPR shows that Sobp KD caused a significant decrease in the mRNA levels for foxd3 (MO, ∼33%; CRISPR, ∼20%) and six1 (MO, ∼40%; CRISPR, ∼32%) relative to uninjected control embryos, whereas changes in sox2 or krt12.4 were not significant. Levels of sobp mRNA verified reduced transcripts after CRISPR (MO, not significant; CRISPR, ∼50% decrease). (B) qPCR analysis of whole embryos injected with sobp mRNA shows that increasing Sobp (∼50-fold increase) significantly reduced mRNA levels for foxd3 (∼20%) and six1 (∼20%), and increased that of krt12.4 (∼1.3 fold), whereas sox2 levels were not significantly affected. (C) qPCR analysis of whole embryos injected with p.R651X mRNA shows that increased expression (∼80-fold increase) significantly reduced mRNA levels for six1 (∼25%) and increased that of krt12.4 (∼1.3 fold), whereas foxd3 and sox2 levels were not significantly affected. ns, not significant; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. qPCR experiments were repeated at least four independent times. Error bars represent s.d. with symbols depicting individual data points. (D-S) In situ hybridization for sox2 (neural plate, D,H,L,P), foxd3 (neural crest, E,I,M,Q), six1 (PPE, F,J,N,R) and krt12.4 (epidermis, G,K,O,S). Images are representative of the most frequent phenotype, except for krt12.4 in O and S. Knockdown of Sobp on one side in morphants (MO, D-G) or F0 crispants (CRISPR, H-K) reduced the intensity of sox2 expression concomitant with expansion of its domain, indicated by yellow lines (D,H). They also reduced expression of foxd3 (arrowheads, E,I), six1 (arrowheads, F,J) and krt12.4 (indicated by distance from the midline, yellow line, G,K). Increased expression of Sobp (L-O) or the p.R651X variant of sobp (P-S) on one side caused a decrease in the expression of foxd3 (arrowheads, M, Q) and six1 (arrowheads, N,R), whereas sox2 expression was unchanged (L,P). Although krt12.4 expression was unchanged in most embryos, we detected ectopic expression overlapping the lineage tracer (arrowheads, O,S). (T-X) Frequencies of changes in gene expression illustrated in D-S. The number in each bar denotes sample sizes.
To determine whether increasing Sobp protein above endogenous levels altered gene expression, we increased Sobp expression by mRNA injections. First, we assessed by qPCR whether increasing Sobp throughout the embryo caused overall changes in gene expression (Fig. 4B). Increasing Sobp significantly reduced whole-embryo mRNA levels of foxd3 (P<0.01/q=0.002780) and six1 (P<0.01/q=0.002780), and increased levels of krt12.4 (P<0.05/q=0.009927); sox2 levels were not significantly altered (P=0.397516/q=0.200746). To assess whether these whole embryo gene expression changes also were observed as changes in the sizes of specific expression domains, we targeted sobp mRNA to the dorsal-animal and ventral-animal blastomeres of eight-cell embryos that are the major precursors of NC and PPE (Moody and Kline, 1990) and assessed changes in expression domains by ISH. Similar to the qPCR data, the majority of embryos showed no changes in the sox2 domain on the injected side (67.5%, Fig. 4L,V). However, we also detected a decrease in the sox2 domain in some embryos (25.0%). The effects on the foxd3 domain were pleiotropic: it was smaller in 39.0% (Fig. 4M,V) and broader in 39.0% (Fig. 4V) of embryos. Similar to the qPCR data, the six1 domain was smaller in the majority of the embryos (86.9%, Fig. 4N,V). Even though the krt12.4 domain was increased in some embryos with ectopic expression of this gene overlapping the lineage tracer (33.3%, Fig. 4O,V), consistent with the qPCR results, for most embryos the domain of this gene was not detectably changed (54.2%, Fig. 4V). Overall, these data, which are summarized in Table S1, demonstrate that increasing the levels of Sobp disrupts the relative sizes of the gene expression domains of foxd3 and six1 and can induce ectopic expression of krt12.4.
Because the p.R651X variant of sobp does not disrupt the interaction of this protein with Six1, we assessed whether p.R651X would cause the same changes in gene expression as full-length Sobp. First, qPCR analysis of whole embryos (Fig. 4C) showed a significant decrease in six1 (P<0.01/q=0.002134) and significant increase in krt12.4 (P<0.01/q=0.004706), but no significant changes in foxd3 (P=0.211730/q=0.142565) or sox2 (P=0.307810/q=0.155444) mRNA levels. The only difference in comparison with full-length Sobp was a lack of effect on foxd3 levels (Table S1). By ISH, we found that the sox2 domain was not affected in the majority (71.4%, Fig. 4P,X) of embryos, similar to the qPCR results and similar to, but slightly more frequently than, the full-length Sobp ISH data (P<0.05). The foxd3 domain was reduced in about half (47.4%, Fig. 4Q,X) and increased in a small number (7.9%, Fig. 4X) of embryos, even though qPCR did not detect a change. In comparison with full-length Sobp, the foxd3 domain showed a significantly different distribution in phenotypes (P<0.01). Consistent with the qPCR results and indistinguishable from full-length Sobp (P=0.3772), the six1 domain was decreased at a high frequency (95.0%, Fig. 4R,X). Although whole embryo levels of krt12.4 were increased, its domain was not detectably changed in most embryos (77.3%, Fig. 4X). However, a low percentage of embryos showed ectopic expression of krt12.4 in the area where the lineage tracer was present (19.7%, Fig. 4S,X), albeit at a lower frequency than full-length sobp (P<0.05). These data demonstrate that introduction of the p.R651X variant in Sobp caused similar changes in six1 expression compared with full-length Sobp, but its effects on sox2, foxd3 and krt12.4 expression were subtly different. Importantly, in spite of its inability to translocate to the nucleus in the absence of Six1 (Fig. 3L,N), p.R651X is sufficiently functional to alter NC and PPE gene expression in whole embryos.
Proper levels of Sobp are required for otic vesicle development
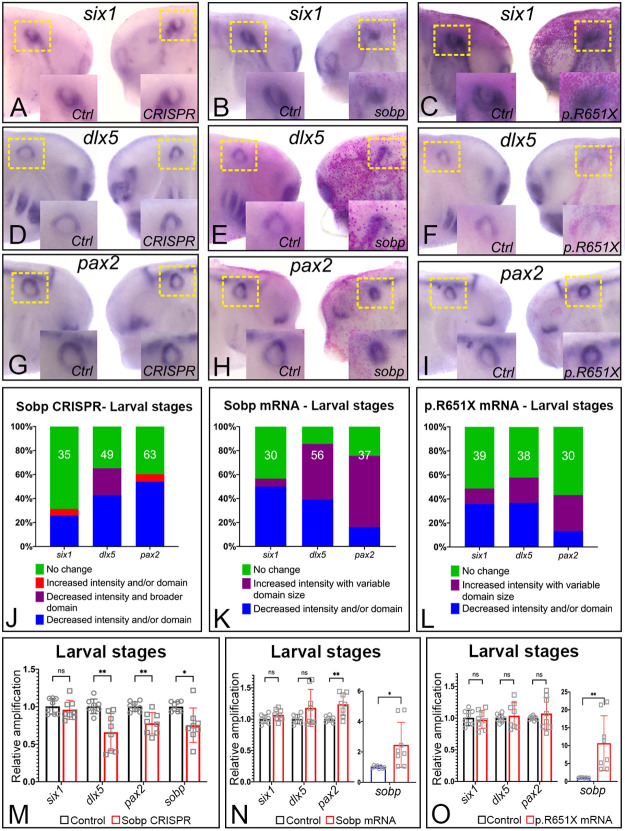
As both KD and increased expression of Sobp altered PPE gene expression, we analyzed whether these changes led to disruptions in the otic vesicle at larval stages (Fig. 5, Table S2). KD of sobp by CRISPR (Fig. 5A,D,G,J) revealed a decrease in the intensity and domain size of the otic expression of dlx5 (42.9%, Fig. 5D,J) and pax2 (54.0%, Fig. 5G,J); interestingly, six1 was not altered in most sobp crispants (68.6%, Fig. 5A,J) as only 25.7% of embryos showed a slight decrease in expression (Fig. 5J). Similar findings were shown by qPCR (Fig. 5M): decreased dlx5 (P<0.01/q=0.002700) and pax2 (P<0.01/q=0.002668), and no significant change in six1 (P=0.450307/q=0.227405).
Fig. 5.
Dorsal-ventral patterning of the otic vesicle requires proper expression levels of Sobp. (A-I) In situ hybridization for six1 (A-C), dlx5 (D-F) and pax2 (G-I) at larval stages. Images in each box are the control and injected sides of the same embryo, and are representative of the most frequent phenotype for CRISPR (A,D,G) and sobp mRNA (B,E,H). Images for p.R651X mRNA represent the less frequently observed phenotype (C,F,I). Insets show a higher magnification of the otic vesicle. Yellow dotted boxes denote the areas contained in the insets. Sobp knockdown leads to decreased otic expression of dlx5 (D) and pax2 (G), whereas six1 expression is unchanged (A). Increased Sobp causes a decrease in six1 expression (B) and increased expression with a variable domain size of dlx5 (E) and pax2 (H). Less frequently, increased p.R651X Sobp expression caused a decrease in six1 (C) and dlx5 (F) and increased expression of pax2 (I). (J-L) Frequencies of changes in gene expression illustrated in A-I. The number in each bar denotes the sample size. (M) qPCR analysis of whole larval heads after CRISPR (∼25% decrease in sobp mRNA) shows significant decrease in dlx5 (∼34%) and pax2 (∼22%) mRNAs, whereas changes in six1 are not significant. (N) qPCR analysis of whole larval heads injected with sobp mRNA (∼2.5-fold increase) shows a significant increase in pax2 (∼1.4 fold); changes in six1 and dlx5 are not significant. (O) qPCR analysis of whole larval heads injected with p.R651X mRNA (∼10-fold increase) shows no significant changes in six1, dlx5 or pax2. ns, not significant; *P<0.05, **P<0.01. qPCR experiments were repeated at least four independent times. Error bars represent s.d. with symbols depicting individual data points.
Increased Sobp had variable effects on otic vesicle genes (Fig. 5B,E,H,K; Table S2). six1 expression was decreased in 50.0%, increased in 6.7% and unchanged in 43.3% (Fig. 5B,K) of larvae. dlx5 showed increased expression with a variable domain size in 46.4% (Fig. 5E,K) and decreased expression and domain size in 39.3% (Fig. 5K) of larvae. pax2 showed increased expression with a variable domain size in 59.5% (Fig. 5H,K) and decreased expression in 16.2% (Fig. 5K) of larvae. qPCR of whole heads (Fig. 5N) confirmed the effects were variable because there were no significant changes in the mRNA levels for six1 (P=0.106946/q=0.079244) or dlx5 (P=0.117690/q=0.079244). Only pax2 showed a significant increase in mRNA levels (P<0.01/q=0.005414) after increased Sobp expression. These findings show that altering Sobp levels that change PPE gene expression are followed by disruptions of otic vesicle development.
We tested whether the p.R651X variant of sobp also altered otic gene expression (Fig. 5C,F,I,L; Table S2). Effects on otic gene expression were variable with most of the analyzed larvae showing no changes in six1 (51.3%), dlx5 (42.1%) or pax2 (56.7%; Fig. 5L). Less frequently, we observed a decrease in the expression and domain size for six1 (35.9%, Fig. 5C,L) and dlx5 (36.8%, Fig. 5F,L), whereas pax2 showed increased expression (30.0%, Fig. 5I,L). Comparing the frequencies of gene expression changes between embryos injected with wild-type sobp versus p.R651X mRNAs showed effects on six1 were similar (P=0.742), whereas dlx5 and pax2 were affected by p.R651X significantly less frequently (dlx5, P<0.0001; pax2, P<0.05). qPCR of whole heads (Fig. 5O) confirmed the effects were variable because there were no significant changes in six1 (P=0.671981/q=0.678701), dlx5 (P=0.621603/q=0.678701) or pax2 (P=0.435624/q=0.678701) mRNA levels. These data demonstrate that the p.R651X variant of sobp mildly disrupts six1 otic vesicle expression similar to full-length Sobp, but has diminished effects on dlx5 and pax2.
Sobp is required for craniofacial cartilage development
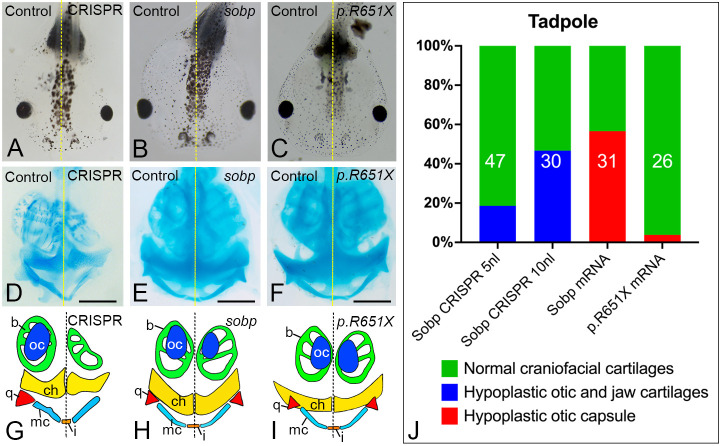
Because altered Sobp levels affected NC gene expression (Fig. 4), we examined whether this leads to defects in cranial cartilages at tadpole stages (Fig. 6). Gross analysis of surviving tadpole heads revealed that sobp KD by CRISPR (Fig. 6A) caused mild to severe cranial cartilage hypoplasia on the injected side in a subset (5 nl, 18.6%; 10 nl, 46.7%) of F0 tadpoles, whereas injection of sobp mRNA (Fig. 6B) caused only mild hypoplasia. Tadpoles injected with the p.R651X variant of sobp (Fig. 6C) did not have apparent cranial cartilage defects. Staining tadpoles with Alcian Blue demonstrated that the observed hypoplasia in Sobp F0 crispants was the consequence of deformed Meckel's and ceratohyal cartilages, hypoplastic branchial arch cartilages, and absent quadrate and otic cartilages (Fig. 6D,G,J). Most of the cartilaginous elements of the sobp mRNA-injected tadpole heads were normal; only the otic capsule was mildly hypoplastic in 56.6% of tadpoles (Fig. 6E,H,J). p.R651X mRNA-injected tadpole cranial cartilages, including the otic capsule, were minimally affected; only 3.8% of tadpoles showed a mildly hypoplastic otic capsule (Fig. 6F,I,J). These findings indicate that early changes in gene expression caused by loss or increased expression of Sobp lead to craniofacial skeletal defects, particularly disruption of the otic capsule.
Fig. 6.
Sobp is required for craniofacial cartilage development. (A-C) Gross morphology of tadpoles after unilateral CRISPR knockdown (5 nl, A), increased sobp (B) or increased p.R651X variant of sobp (C). Survival rates were: CRISPR 5 nl, ∼65.9%; 10 nl, ∼40.0%; sobp mRNA, ∼96.3%; p.R651X mRNA, ∼87.2%. Hypoplasia of head structures on the injected side is noticeable from a dorsal view in a subset of Sobp crispants (A) and increased Sobp (B), but not of increased p.R651X (C). (D-I) Ventral views of Alcian Blue staining of tadpoles (D-F) and drawings of the stained cartilages (G-I) show severe cranial cartilage defects of crispants (5 nl, 18.6%, D,G; 10 nl, 46.7%), including deformed Meckel's (mc) and ceratohyal (ch) cartilages, hypoplastic branchial arch cartilages (b), absent quadrate (q) and absent otic capsule (oc) cartilages. Increased sobp (E,H) resulted in hypoplasia of the otic capsule (56.6%), whereas the majority of the p.R651X mRNA-injected tadpoles (96.2%, F,I) did not have apparent defects. i, infrarostral cartilage. Scale bars: 500 μm. (J) Frequencies of defects in the cranial cartilages depicted in D-I. The number in each bar denotes the sample size.
DISCUSSION
Development of the cranial sensory placodes is a complex process that requires different signaling pathways and expression of transcription factors that induce and/or repress gene expression (Saint-Jeannet and Moody, 2014; Moody and LaMantia, 2015). Previous work from our lab and others showed that Six1 functions at early stages of placode formation (Christophorou et al., 2009; Brugmann et al., 2004), during otic vesicle patterning (Zheng et al., 2003; Ozaki et al., 2004; Laclef et al., 2003) and hair cell formation in the inner ear (Zhang et al., 2017; Li et al., 2020; Ahmed et al., 2012). One important characteristic of Six1 factors that allows them to act in different contexts is their interaction with co-factors that modulate their transcriptional activities (Neilson et al., 2020, 2017; Li et al., 2003, 2020; Kenyon et al., 2005a,b; Heanue et al., 1999; Guo et al., 2011; Brugmann et al., 2004; Ahmed et al., 2012). We identified Sobp as a potential Six1 co-factor based on findings in Drosophila (Neilson et al., 2010; Kenyon et al., 2005a), but little is known about its function in vertebrate development or the mechanisms by which it interacts with Six1. Here, we demonstrate that Sobp is a bona fide Six1 co-factor that can repress the transcriptional activation of Six1+Eya1 target genes. In addition, we show that altering the levels of Sobp disrupts the patterning of embryonic ectodermal domains and otic vesicle genes (Fig. 7).
Fig. 7.
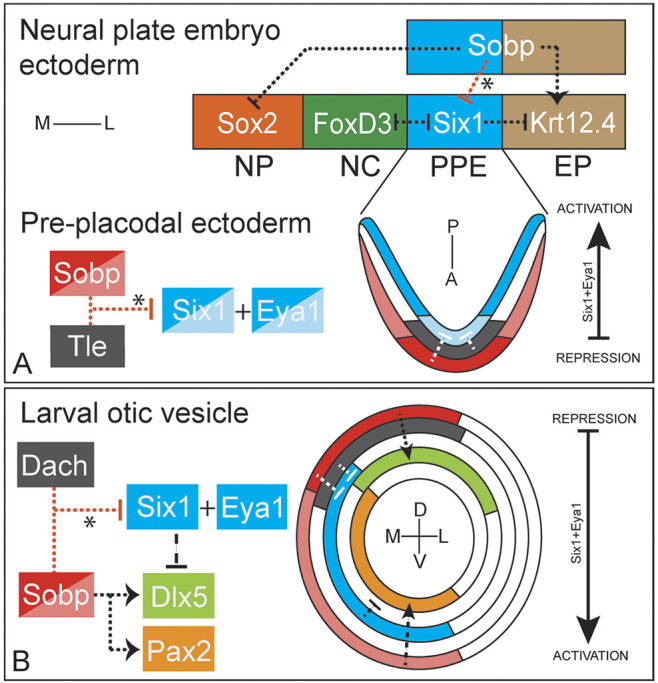
Model for Sobp interactions during craniofacial development. (A) Sobp expression in two ectodermal domains (PPE and EP) directly or indirectly induces epidermal genes (krt12.4), represses neural plate genes (sox2) and represses the Six1+Eya1 transcriptional activation (asterisk) in the PPE. Sobp effects outside the PPE are likely independent of Six1. Repression of the transcriptional activation of Six1+Eya1 target genes (asterisk) is likely achieved by higher levels of Sobp and lower levels of Six1, and the additional expression of the known Six1 co-repressors (e.g. Groucho/Tle) in the anterior PPE. Conversely, in the posterior PPE, lower levels of Sobp, higher levels of Six1 and lower/no Groucho/Tle expression leads to transcriptional activation. Eya1 is co-expressed with Six1 in this domain. A, anterior; EP, epidermis; L, lateral; M, medial; NP, neural plate; NC, neural crest; P, posterior; PPE, pre-placodal ectoderm. (B) Sobp expression in the medial wall of the otic vesicle is higher in the dorsal region, a domain where Dach co-repressors are also expressed. This expression pattern leads to repression of the transcriptional activation of Six1+Eya1 target genes (asterisk), thus allowing expression of dorsal otic genes (e.g. Dlx5). Ventrally, Sobp induces expression of Pax2 independent of Six1. Six1 and Eya1 have overlapping expression. D, dorsal; L, lateral; M, medial; V, ventral.
Sobp is a Six1 co-repressor
Sobp was first characterized in Drosophila as part of the retinal gene determination network based on its co-expression in the eye disc with So (Silver et al., 2003; Kenyon et al., 2005a). Our work is the first demonstration that in vertebrates Sobp interacts with Six1 and Eya1. Binding to Six1 was expected as Drosophila Sobp binds to So (Kenyon et al., 2005a), but binding to Eya1 was not expected; this interaction is reminiscent of fly Eya binding to the So co-repressor Dachshund (Chen et al., 1997). Similar to Eya factors, Sobp does not have a DNA-binding domain (Birk et al., 2010) and therefore relies on interaction with transcription factors to modulate transcription. However, unlike Eya1, a cytosolic protein that requires binding to Six factors for nuclear translocation (Ohto et al., 1999; Buller et al., 2001), Sobp is a nuclear protein (Chen et al., 2008; Birk et al., 2010). Our findings demonstrate that Sobp has a functional C-terminal NLS and after deletion of this crucial domain or a larger C-terminal domain containing the NLS, Sobp is located almost exclusively in the cytosol. Interestingly, variants of Sobp containing a deletion of this NLS or of the 220 C-terminal amino acids, including the NLS (p.R651X) still interfere with the transcriptional activation of Six1+Eya1 target genes after binding to Six1 and translocation to the nucleus. These data indicate that an interaction between Six1 and cytosolic proteins is not exclusive to Eya factors (Ohto et al., 1999).
Upon binding to the Six1+ Eya1 complex, Sobp reduces the ability of this complex to activate the transcription of target genes. Our findings indicate that this repressive function occurs through a dose-dependent competition mechanism: low levels of Sobp increase the interaction between Six1 and Eya1, whereas high levels of Sobp disrupt this interaction, leading to at least some Eya1 remaining in the cytosol. As Six1 protein stability is higher in the presence of Eya factors (Patrick et al., 2009), it is possible that low levels of Sobp similarly increase Six1 protein stability without inhibiting its transcriptional function. In addition, as Sobp can partially translocate Eya1 into the nucleus by itself, the increased amount of Six1 bound to Eya1 at lower Sobp concentrations may be a consequence of a higher level of Eya1 in the nucleus.
Because deletion of the C-terminal region of Sobp did not affect its ability to interfere with the transcriptional activation of Six1+Eya1 target genes, we speculate that other remaining regions of the protein are responsible for its activity. For example, the FCS zinc-finger domains (ZF1 and ZF2) (Chen et al., 2008) are similar to those in constituents of the polycomb repressive complex 1 (PRC1), which modifies histones in chromatin leading to transcriptional repression (Chen et al., 2008; Blackledge and Klose, 2021). The SUMO-interacting motifs (SIMs) may also be necessary as SUMOylation also causes transcriptional repression by modifying transcription factors and the assembly of nuclear protein complexes (Sun and Hunter, 2012), and SUMOylation of PRC1 is required for repression (Gill, 2010). It will be important to test the function of these domains in future experiments. It is interesting that individuals with BOR and some with MRAMS present with hearing loss but there are no reports of defects in balance caused by vestibular perturbations (Smith, 2018; Birk et al., 2010; Basel-Vanagaite et al., 2007), whereas the jc mouse presents with both hearing and balance defects (Chen et al., 2008; Calderon et al., 2006). A comparison between the MRAMS p.R661X and the jc S449fsX490 shows that the latter loses additional domains, including both SIMs and a region of the proline rich domain (PRD). As PRDs can be involved in protein/protein interactions (Yu et al., 1994), the jc variant could interrupt binding to Six1 or other factors. A full understanding of the Sobp functional domains and sites of protein-protein interactions is an important next goal.
Sobp is required to establish embryonic ectodermal domains
The PPE gives rise to all cranial sensory placodes and is characterized by the expression of six1 and eya1 (Schlosser and Ahrens, 2004; Pandur and Moody, 2000). sobp also is expressed in the PPE (Neilson et al., 2010), but we noticed by ISH that its expression is weaker in the posterior placodal region that will give rise to the otic and epibranchial placodes. This regional difference in expression is supported by a single cell RNA-seq (scRNA-seq) study that allowed us to compare the anterior and posterior placode regions of neural plate stage embryos (Briggs et al., 2018; Peshkin and Kirschner, 2020). sobp enrichment occurs in the anterior placodal cells that co-express tle1 and tle2, two members of the Groucho family of known Six1 co-repressors (Roth et al., 2010). Together, these findings suggest that at PPE stages, Six1 may induce gene expression in the posterior PPE by transcriptional activation and its activity may be limited in the anterior PPE by the presence of multiple co-repressors, including Sobp (Fig. 7A). This is consistent with the proposed role of Sobp in the fly eye field (Kenyon et al., 2005a).
The changes in gene expression after loss or increased Sobp expression that we observed also show that Sobp is required for the proper balance in size of the other embryonic ectodermal domains: neural plate, neural crest and epidermis. Our data indicate that Sobp appears to have a role in inducing directly or indirectly epidermal genes and repressing neural ectodermal genes (Fig. 7A). It is likely that NC and PPE were disrupted after loss of Sobp because the neural plate domain expanded, and that they were disrupted after increasing Sobp levels because the epidermis expanded. As Sobp and Six1 are co-expressed in the PPE, which is intercalated between the neural plate and epidermis, it is possible that, within the PPE, Sobp in combination with Six1 inhibits a neural plate fate while promoting an epidermal one. Alternatively, as Six1 is exclusively expressed in the PPE (Pandur and Moody, 2000; Briggs et al., 2018; Peshkin and Kirschner, 2020), Sobp function in the other ectodermal domains could be independent of its ability to modulate Six1+Eya1-mediated transcriptional activity. scRNA-seq analyses show that, at neural plate stages, sobp is not expressed in cells expressing sox2 but is co-expressed in krt19+ cells (Briggs et al., 2018). Because increased expression of the truncated p.R651X caused similar changes to full-length Sobp in the PPE, but not to genes in the other domains, we posit that the truncation in Xenopus p.R651X/human p.R661X does not disrupt the interaction with Six1. It is, however, possible that the truncation hampers interactions with other nuclear factors because p.R651X cannot translocate into the nucleus in domains where Six1 is not present. It will be important to determine with which other proteins Sobp interacts in different embryonic ectodermal domains.
Sobp contributes to patterning the otic vesicle
Our in vivo KD studies show that Sobp does not appear to be required for six1 expression in the otic vesicle. The decrease in otic six1 after increasing either Sobp or p.R651X is likely due to a reduction in the interaction between Six1 and Eya1 because, in mouse, Eya1 is required for otic vesicle expression of Six1 (Zheng et al., 2003). Although changes in gene expression in the otic vesicle may be influenced by earlier changes in the PPE, because six1 is still expressed in the majority of Sobp crispants, changes in PPE gene expression do not completely account for otic vesicle changes. Instead, we propose that the Sobp-Six1 relationship in the otic vesicle likely impacts dorsal-ventral (D-V) patterning.
D-V patterning of the otic vesicle is crucial for the development of ventral auditory structures and dorsal vestibular structures (Ohta and Schoenwolf, 2018; Nakajima, 2015; Bever et al., 2003). Published data and data presented here show that Six1, Eya1 and Sobp are co-expressed in the medial wall of the otic vesicle (Durruthy-Durruthy et al., 2014; David et al., 2001). Unlike the apparent uniform expression of six1, sobp expression is more intense dorsally, a region that, in mouse, express two known Six1 co-repressors: Dach1 and Dach2 (Ozaki et al., 2004; Li et al., 2002, 2003; Durruthy-Durruthy et al., 2014). These expression data suggest that the transcriptional activation of Six1+Eya1 target genes is repressed in the dorsal-medial domain of the otic vesicle, in agreement with previous findings in mouse that Six1 and Eya1 induce expression of ventral genes, such as Otx1, while repressing dorsal genes, such as Dlx5 (Zheng et al., 2003; Ozaki et al., 2004). We suggest that, in the otic vesicle, there is likely a dose-dependent mechanism of activation and repression of Six1+Eya1-mediated transcriptional activity involving co-factors such as Eya1, Sobp and Dach that contributes to D-V patterning (Fig. 7B). Dorso-medially, higher levels of Sobp and Dach would repress this transcriptional activity, allowing localized expression of dorsal genes such as Dlx5. Ventro-medially, Sobp induces Pax2 expression independently of Six1 expression, as Pax2 expression does not depend on Six1 in mouse (Ozaki et al., 2004); concurrently, Six1 and Eya1 are transcriptionally active in this domain because of lower levels of Sobp and the absence of Dach. Sobp likely interacts with other factors to accomplish otic D-V patterning because, compared with full-length Sobp, changes in gene expression were less frequent after increased p.R651X, which required increased expression of Six1 for nuclear translocation.
Sobp-mediated effects on cranial cartilage
Although we have focused our analyses on early stages of otic development, the changes after altering Sobp levels in the embryonic ectodermal domains ultimately led to severe cranial cartilage defects. The differences in severity between loss and increased Sobp expression might be explained by the targeting approach: for CRISPR/Cas9-mediated KD, embryos were injected at the two-cell stage, whereas mRNA injections were performed at the eight-cell stage only in precursors of the PPE and NC. Because Sobp appears to not be expressed in Xenopus branchial arches (Neilson et al., 2010), the defects in the cranial cartilages likely are secondary to changes in the early ectodermal domains rather than disruption of NC patterning in the branchial arches. Although additional work is required to understand the function of Sobp during formation of these cartilages, it is notable that the otic cartilage was consistently hypoplastic when levels of Sobp were altered. Perhaps this defect contributes to the hearing and vestibular deficits observed in jc mice.
In summary, we show that Sobp is a Six1 co-repressor that interacts with Six1 and most likely other factors during otic and craniofacial development. Although Sobp does not have an identified DNA-binding domain (Birk et al., 2010), it functionally modulates the Six1+Eya1 transcriptional complex. The variants found in individuals with MRAMS (Birk et al., 2010) and jc mice (Chen et al., 2008) that have overlapping but also very distinct phenotypes suggest that disruption of different domains in Sobp may perturb interactions with different partners. To better understand the function of Sobp during normal development and in congenital syndromes, it will be essential to assess the role of its various domains and identify any additional binding partners.
MATERIALS AND METHODS
Many of the methods were supported by Xenbase (http://www.xenbase.org/, RRID: SCR_003280) and the National Xenopus Resource (http://mbl.edu/xenopus/, RRID:SCR_013731).
Plasmid constructs
A full-length Xenopus tropicalis sobp plasmid was purchased from Open Biosystems (BC154687); subsequent sequence analysis identified it as having ∼95% homology to the predicted sequences for Xenopus laevis L-homeolog sobp (XM_018263336.1; XM_018263335.1) and S-homeolog sobp (XM_018265405.1). The ORF was subcloned into the BamHI site of pCS2+ (pCS2+-sobp) using the Clone EZ PCR cloning kit (GenScript). pCS2+-5′HA-sobp and pCS2+-sobp-3′HA were generated using the QuikChange lightning Site-directed mutagenesis kit (Agilent). The same kit was used to sequentially remove three nucleotides located at the 5′ end of the pCS2+-3′HA-sobp ORF to generate a construct whose transcribed mRNA does not bind to the designed translation-blocking antisense morpholino oligonucleotide (pCS2+-sobpMOI-3′HA; morpholino insensitive). To generate a plasmid containing the human p.R661X variant, the mutagenesis kit was used to introduce into the ORF of pCS2+-sobp and of pCS2+-5′HA-sobp a stop codon at amino acid 651 (c.1951A<T; p.R651X; pCS2+-p.R651X and pCS2+-5′HA-p.R651X). Finally, a conserved putative C-terminal nuclear localization signal (NLS) was identified by analyzing the amino acid sequences for Xenopus tropicalis Sobp (NP_001096678.1), Xenopus laevis Sopb (XP_018118825.1; XP_018120894.1), Homo sapiens SOBP (NP_060483.3) and Mus musculus Sobp (NP_780616.4) using cNLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). The putative NLS was deleted using the mutagenesis kit (pCS2+-5′HA-sobp-NLSdel). All plasmids were confirmed by full-length sequencing in both directions.
Cell transfection
HEK293T/17 cells (ATCC CRL-11268) were grown in Dulbecco's modified Eagle medium (Hyclone) supplemented with 10% fetal bovine serum (Lonza) and penicillin-streptomycin (Gibco). Cells were plated into 24-well plates (Thermo Fisher Scientific) for luciferase assays; into six-well plates (Fisher) for co-immunoprecipitation experiments (co-IP); and into one-well Nunc Lab-Tek Chamber slides (Thermo Fisher Scientific) for immunofluorescence (IF). Cells were transfected according to the manufacturer's protocol using X-tremeGENE 9 DNA (Sigma) for luciferase assays and IF; and lipofectamine 3000 (Thermo Fisher Scientific) for co-IP. Cells were processed for each assay 48 h after transfection.
Co-immunoprecipitation
HEK293T cells were transfected with combinations of pCS2+-3′Flag-Six1, pCS2+-5′HA-Sobp, pCS2+-5′HA-sobp-NLSdel, pCS2+-5′HA-p.R651X and/or pCS2+-5′Myc-Eya1. 1 µg of each plasmid was used unless noted in the figure legend. Cells were extracted after 48 h using 500 µl of ice-cold Pierce IP lysis buffer (Thermo Fisher Scientific) plus Halt Protease Inhibitor Cocktail with EDTA (Thermo Fisher Scientific). Cell debris was pelleted at 13,000 g for 10 min at 4°C. The supernatant was subjected to immunoprecipitation using Pierce anti-HA magnetic beads (Thermo Fisher Scientific), Pierce anti-c-Myc magnetic beads (Thermo Fisher Scientific) or Pierce anti-DYKDDDDK magnetic agarose (Thermo Fisher Scientific). Proteins were washed five times and eluted using the Pierce lane marker non-reducing sample buffer (Thermo Fisher Scientific). The eluted proteins were reduced using 100 mM DTT at 100°C for 5 min prior to SDS-PAGE and Western blot. For control experiments, 10 µl of each sample in IP lysis buffer was diluted with Laemmli sample buffer with 2% BME, incubated at 100°C for 5 min prior to SDS-PAGE and Western blot. Immobilon-FL PVDF membranes (Fisher) were probed with mouse anti-HA (6E2, Cell Signaling, 1:1000) to detect Sobp, mouse anti-Myc (9B11, Cell Signaling, 1:1000) to detect Eya1, rabbit anti-Six1 (D5S2S, Cell Signaling, 1:1000) and rabbit anti-β-actin (13E5, Cell Signaling, 1:1000). Secondary antibodies were IRDye 680RD donkey anti-rabbit IgG (92568073, Licor, 1:5000) and IRDye 800CW goat anti-mouse IgG (92532210, Licor, 1:5000). Experiments were repeated at least three independent times. Blots were scanned using the Licor Odyssey infrared imaging system.
Luciferase assay
Transfected HEK293T cells were harvested and analyzed using the Dual Luciferase Assay kit (Promega) following the manufacturer's directions. Each transfection included 200 ng of pGL3-6XMEF3-Firefly luciferase reporter (Ford et al., 2000) and 20 ng of Renilla luciferase reporter (pRL-TK), in addition to different combinations of pCS2+ (control), pCS2+-3′Flag-Six1, pCS2+-5′Myc-Eya1, pCS2+-5′HA-Sobp, pCS2+-5′HA-sobp-NLSdel and/or pCS2+-5′HA-p.R651X (400 ng each). At 48 h post-transfection, cells were resuspended directly in 100 µl of passive lysis buffer (PLB) and 20 µl of lysate was used in the analysis. Experiments were repeated at least five times. ANOVA with Tukey's post-hoc multiple comparisons test was performed using GraphPad Prism 9 software. Expression of exogenous proteins from the transfected plasmids was confirmed by standard western blotting using antibodies described for co-IP (Fig. S2).
Immunofluorescence
HEK293T cells were transfected with different combinations of pCS2+-3′Flag-Six1, pCS2+-5′HA-Sobp, pCS2+-5′HA-sobp-NLSdel, pCS2+-5′HA-p.R651X and/or pCS2+-5′Myc-Eya1. 2 µg of each plasmid was used unless noted in the figure legend. Cells were processed as described previously (Shah et al., 2020). Briefly, 48 h after transfection, cells were fixed in 4% paraformaldehyde and processed for immunostaining by standard methods using mouse anti-Flag (9A3, Cell Signaling, 1:400), rabbit anti-Myc (71D10, Cell Signaling, 1:250), rabbit anti-HA (C29F4, Cell Signaling) or mouse anti-HA (6E2, Cell Signaling, 1:800) followed by Alexa Fluor 488-conjugated anti-rabbit (4412, 1:1000) and Alexa Fluor 568-conjugated anti-mouse (A1104, 1:1000) secondary antibodies, and DAPI nuclear counterstain (R37605, Thermo Fisher Scientific). Experiments were repeated at least three independent times, and at least five fields per slide were analyzed using a Zeiss LSM 710 confocal microscope.
In vitro synthesis of mRNAs and antisense RNA probes
mRNAs encoding Xenopus tropicalis sobp, Xenopus tropicalis 5′HA-sobp, Xenopus tropicalis sobpMOI-3′HA, Xenopus tropicalis p.R651X and a nuclear-localized β-galactosidase (nβgal) lineage tracer were synthesized in vitro according to manufacturer's protocols (mMessage mMachine kit, Ambion). Antisense RNA probes for in situ hybridization (ISH) were synthesized in vitro (MEGAscript kit; Ambion), as previously described (Yan et al., 2009b).
Morpholino oligonucleotide knockdown
To knock down endogenous levels of Sobp protein in embryos, a 3′-carboxyfluorescein-labelled translation-blocking antisense morpholino oligonucleotide (MO) was purchased (GeneTools): TCCCCTCTTTTTCCATTTCTGCCAT. The MO binds at the ATG start site for both the predicted L- and S-homeologs for Xenopus laevis (Fig. S3A) and to Xenopus tropicalis sobp. To verify the ability of the MO to block sobp translation (Fig. S3B), Xenopus stage VI oocytes were injected with 9 ng of MO and then injected with either 2 ng of 5′HA-Sobp (MO-insensitive), Sobp-3′HA (MO-sensitive) or sobpMOI-3′HA (MO-insensitive) mRNA. Oocytes were cultured overnight at 18°C. Lysates were prepared and western blotting performed with rabbit anti-HA antibody (C29F4, Cell Signaling) as previously described (Neilson et al., 2017).The specificity of the MO was tested by injecting embryos with 9 ng of MO followed by the immediate microinjection of 50 pg of the MO-insensitive sobp mRNA (sobpMOI-3′HA), and processing embryos for foxd3 expression by ISH (Fig. S3C,D), as described below. Only 29.4% of these embryos had reduced foxd3 expression (Fig. S3C,D) compared with 90.2% for MO alone (Fig. 4E,T), demonstrating significant rescue of the foxd3 phenotype.
CRISPR/Cas9 gene editing
Based on the Xenopus laevis sobp genomic sequence obtained from Xenbase (http:/xenbase.org), a 20 bp target was designed by CRISPRscan (http://www.crisprscan.org) during the Xenopus Genome Editing Workshop at the National Xenopus Resource at the Marine Biological Laboratory (Woods Hole, MA, USA). Potential off-targets were predicted by GGGgenome (https://gggenome.dbcls.jp) using the most up-to-date version of the Xenopus laevis genome. The 5′ dinucleotides were converted to GG (Gagnon et al., 2014). The sequence is GGTTCTTGGATGGTACGGTA and targets the L- and S-homeologs for sobp in its second exon that encodes the first conserved region (Fig. S3A) (Chen et al., 2008; Kenyon et al., 2005a). A DNA template was produced by a PCR-based method using a universal reverse primer with a gene-specific forward primer containing a T7 polymerase promoter. The MEGAshortscript T7 transcription kit (Thermo Fisher Scientific) was used for sgRNA synthesis. sgRNA was mixed with Cas9 Protein (PNA Bio) and Texas Red Dextran, Lysine Fixable (Thermo Fisher Scientific) prior to injections. Injected embryos were incubated at 21°C and at least eight embryos were processed for sequencing to confirm DNA editing for the L- and S-homeologs for sobp each time injections were performed. Genotyping primer sequences are listed in Table S3. Insertion/deletion frequencies were calculated with the TIDE software package (http://shinyapps.datacurators.nl/tide/; Fig. S3E,F).
Embryo microinjections
Fertilized Xenopus laevis embryos were obtained by natural mating and in vitro fertilization (Moody, 2000). Injections for ISH and cartilage staining were performed unilaterally with 1 nl of MO (9 ng/nl) or 1nl of mRNA (100 pg mixed with 100 pg nβgal mRNA) in the dorsal-animal and ventral-animal blastomeres of eight-cell stage embryos; these blastomeres predominantly give rise to the neural crest and cranial placodes (Moody and Kline, 1990); or with 5 nl or 10 nl of 75 pg/nl of sgRNA mixed with 0.2 ng/nl of Cas9 protein (PNA Bio) and 0.1% Texas Red Dextran in one cell of two-cell stage embryos. Injections for quantitative real time PCR (qPCR) were performed with 1 nl of MO (2 ng/nl) or 2 nl of mRNA (100 pg mixed with 100 pg nβgal) in each cell of two-cell stage embryos; or with 10 nl of sgRNA/Cas9 solution in one-cell stage embryos. Microinjections were performed according to standard methods (Moody, 2018). Embryos were cultured in diluted Steinberg's solution until fixation or harvest.
Histochemistry and in situ hybridization
Embryos were cultured to neural plate (stage 16-18) and larval (stage 28-34) stages (Nieuwkoop and Faber, 1994), fixed in 4% paraformaldehyde (PFA), stained for β-Gal histochemistry and processed for in situ hybridization (ISH) as described previously (Yan et al., 2009a). In embryos in which the fluorescent label/dextran or nβgal lineage tracer were located in the appropriate tissue domains, the position, intensity and size of the expression domains of sox2, foxd3, six1, krt12.4, dlx5 and pax2 were compared between the injected, lineage-labeled side to the control, uninjected side of the same embryo, thus accounting for inter-embryo variation. Embryos for each assay were derived from a minimum of three different sets of outbred, wild-type parents. Differences in the frequency of gene expression changes were assessed for significance (P<0.05) by the Chi-square test using GraphPad Prism 9. A set of control larvae that was processed for six1 and sobp ISH was embedded in a gelatin-based medium [0.5% gelatin, 30% bovine serum albumin and 20% sucrose, hardened with glutaraldehyde (75 μl/ml)], and vibratome sectioned at 50 µm in the transverse plane. Whole-mount and serial section ISH images were collected with an Olympus SZX16 stereomicroscope coupled to an Olympus UC90 camera and cellSens Entry software.
RNA collection and qPCR
Three embryos at neural plate (stage 16-18) or three dissected heads at larval (stage 28-34) stages were collected in TRI-reagent (Zymo) and processed for RNA extraction with DNAse I treatment using the Direct-zol RNA Miniprep kit (Zymo). cDNA was synthesized using the iScript Advanced cDNA Synthesis kit (Bio-Rad). qPCR was performed using 5 ng cDNA with the SsoAdvanced Universal SYBR Green Mix (Bio-Rad). Primer sequences are listed in Table S4. qPCR of four biological replicates was performed in duplicate. PCR and data analysis were performed using a CFX Connect thermocycler (Bio-Rad). Statistical analysis was performed with GraphPad Prism 9, with significance calculated by unpaired t-tests followed by a False Discovery Rate approach using the two-stage step-up method (Benjamini et al., 2006) (FDR=1%).
Cartilage staining
Embryos were grown to tadpole stages and the ones that remained alive were photographed and counted for quantification of survival rates. They were subsequently processed as described previously (Young et al., 2017): fixed in 4% PFA for 1 h at room temperature then incubated in a solution of acid/alcohol containing 0.1% Alcian Blue. When staining was complete, tadpoles were washed in the acid/alcohol solution without Alcian Blue, bleached with a solution containing 1.2% hydrogen peroxide and 5% formamide, and cleared in 2% KOH with increasing concentrations of glycerol.
Supplementary Material
Acknowledgements
This work was made possible with the support of Xenbase (http://www.xenbase.org/, RRID: SCR_003280) and the National Xenopus Resource (http://mbl.edu/xenopus/, RRID:SCR_013731). We thank the Xenopus Genome Editing Workshop at the National Xenopus Resource that helped with sgRNA design and training on CRISPR/Cas technology. We also thank Francesca Pignoni, Kristy Kenyon and Dominique Alfandari for helpful discussions during the execution of this work. Finally, we thank many members of the Xenopus community for providing plasmids.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.L.P.T., K.M.N., S.A.M.; Methodology: A.L.P.T., K.M.N.; Validation: A.L.P.T.; Formal analysis: A.L.P.T.; Investigation: A.L.P.T.; Resources: H.D.M.; Data curation: A.L.P.T.; Writing - original draft: A.L.P.T.; Writing - review & editing: A.L.P.T., K.J., K.M.N., S.A.M.; Visualization: A.L.P.T., K.J.; Supervision: A.L.P.T., S.A.M.; Project administration: A.L.P.T., S.A.M.; Funding acquisition: S.A.M.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199684
References
- Ahmed, M., Wong, E. Y. M., Sun, J., Xu, J., Wang, F. and Xu, P.-X. (2012). Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377-390. 10.1016/j.devcel.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite, L., Rainshtein, L., Inbar, D., Gothelf, D., Hennekam, R. and Straussberg, R. (2007). Autosomal recessive mental retardation syndrome with anterior maxillary protrusion and strabismus: MRAMS syndrome. Am. J. Med. Genet. A 143A, 1687-1691. 10.1002/ajmg.a.31810 [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., Krieger, A. M. and Yekutieli, D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491-507. 10.1093/biomet/93.3.491 [DOI] [Google Scholar]
- Bever, M. M., Jean, Y. Y. and Fekete, D. M. (2003). Three-dimensional morphology of inner ear development in Xenopus laevis. Dev. Dyn. 227, 422-430. 10.1002/dvdy.10316 [DOI] [PubMed] [Google Scholar]
- Birk, E., Har-Zahav, A., Manzini, C. M., Pasmanik-Chor, M., Kornreich, L., Walsh, C. A., Noben-Trauth, K., Albin, A., Simon, A. J., Colleaux, L.et al. (2010). SOBP is mutated in syndromic and nonsyndromic intellectual disability and is highly expressed in the brain limbic system. Am. J. Hum. Genet. 87, 694-700. 10.1016/j.ajhg.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge, N. P. and Klose, R. J. (2021). Getting under the skin of Polycomb-dependent gene regulation. Genes Dev. 35, 301-303. 10.1101/gad.348257.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, J. A., Weinreb, C., Wagner, D. E., Megason, S., Peshkin, L., Kirschner, M. W. and Klein, A. M. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780. 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann, S. A., Pandur, P. D., Kenyon, K. L., Pignoni, F. and Moody, S. A. (2004). Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 131, 5871-5881. 10.1242/dev.01516 [DOI] [PubMed] [Google Scholar]
- Buller, C., Xu, X., Marquis, V., Schwanke, R. and Xu, P.-X. (2001). Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum. Mol. Genet. 10, 2775-2781. 10.1093/hmg/10.24.2775 [DOI] [PubMed] [Google Scholar]
- Calderon, A., Derr, A., Stagner, B. B., Johnson, K. R., Martin, G. and Noben-Trauth, K. (2006). Cochlear developmental defect and background-dependent hearing thresholds in the Jackson circler (jc) mutant mouse. Hear. Res. 221, 44-58. 10.1016/j.heares.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Chen, R., Amoui, M., Zhang, Z. and Mardon, G. (1997). Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91, 893-903. 10.1016/S0092-8674(00)80481-X [DOI] [PubMed] [Google Scholar]
- Chen, Z., Montcouquiol, M., Calderon, R., Jenkins, N. A., Copeland, N. G., Kelley, M. W. and Noben-Trauth, K. (2008). Jxc1/Sobp, encoding a nuclear zinc finger protein, is critical for cochlear growth, cell fate, and patterning of the organ of corti. J. Neurosci. 28, 6633-6641. 10.1523/JNEUROSCI.1280-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette, B. N. R., Green, P. J., Martin, K., Garren, H., Hartenstein, V. and Zipursky, S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977-996. 10.1016/0896-6273(94)90308-5 [DOI] [PubMed] [Google Scholar]
- Christophorou, N. A. D., Bailey, A. P., Hanson, S. and Streit, A. (2009). Activation of Six1 target genes is required for sensory placode formation. Dev. Biol. 336, 327-336. 10.1016/j.ydbio.2009.09.025 [DOI] [PubMed] [Google Scholar]
- David, R., Ahrens, K., Wedlich, D. and Schlosser, G. (2001). Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech. Dev. 103, 189-192. 10.1016/S0925-4773(01)00355-0 [DOI] [PubMed] [Google Scholar]
- Durruthy-Durruthy, R., Gottlieb, A., Hartman, B. H., Waldhaus, J., Laske, R. D., Altman, R. and Heller, S. (2014). Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell 157, 964-978. 10.1016/j.cell.2014.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, H. L., Landesman-Bollag, E., Dacwag, C. S., Stukenberg, P. T., Pardee, A. B. and Seldin, D. C. (2000). Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 275, 22245-22254. 10.1074/jbc.M002446200 [DOI] [PubMed] [Google Scholar]
- Gagnon, J. A., Valen, E., Thyme, S. B., Huang, P., Ahkmetova, L., Pauli, A., Montague, T. G., Zimmerman, S., Richter, C. and Schier, A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186. 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, G. (2010). SUMO weighs in on polycomb-dependent gene repression. Mol. Cell 38, 157-159. 10.1016/j.molcel.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Guo, C., Sun, Y., Zhou, B., Adam, R. M., Li, X., Pu, W. T., Morrow, B. E., Moon, A. and Li, X. (2011). A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J. Clin. Invest. 121, 1585-1595. 10.1172/JCI44630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heanue, T. A., Reshef, R., Davis, R. J., Mardon, G., Oliver, G., Tomarev, S., Lassar, A. B. and Tabin, C. J. (1999). Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13, 3231-3243. 10.1101/gad.13.24.3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., Sato, S., Ozaki, H. and Ikeda, K. (2000). Six family genes--structure and function as transcription factors and their roles in development. BioEssays 22, 616-626. [DOI] [PubMed] [Google Scholar]
- Kenyon, K. L., Li, D. J., Clouser, C., Tran, S. and Pignoni, F. (2005a). Fly SIX-type homeodomain proteins sine oculis and optix partner with different cofactors during eye development. Dev. Dyn. 234, 497-504. 10.1002/dvdy.20442 [DOI] [PubMed] [Google Scholar]
- Kenyon, K. L., Yang-Zhou, D., Cai, C. Q., Tran, S., Clouser, C., Decene, G., Ranade, S. and Pignoni, F. (2005b). Partner specificity is essential for proper function of the SIX-type homeodomain proteins sine oculis and optix during fly eye development. Dev. Biol. 286, 158-168. 10.1016/j.ydbio.2005.07.017 [DOI] [PubMed] [Google Scholar]
- Klingbeil, K. D., Greenland, C. M., Arslan, S., Llamos Paneque, A., Gurkan, H., Demir Ulusal, S., Maroofian, R., Carrera-Gonzalez, A., Montufar-Armendariz, S., Paredes, R.et al. (2017). Novel EYA1 variants causing Branchio-oto-renal syndrome. Int. J. Pediatr. Otorhinolaryngol. 98, 59-63. 10.1016/j.ijporl.2017.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., Hasebe, M., Tomita, M. and Yanagawa, H. (2009). Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 106, 10171-10176. 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug, E. G. E. A. (2016). Childhood Hearing Loss: Strategies for Prevention and Care. Geneva: World Health Organization. [Google Scholar]
- Kumar, J. P. (2009). The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol. Life Sci. 66, 565-583. 10.1007/s00018-008-8335-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef, C., Souil, E., Demignon, J. and Maire, P. (2003). Thymus, kidney and craniofacial abnormalities in Six1 deficient mice. Mech. Dev. 120, 669-679. 10.1016/S0925-4773(03)00065-0 [DOI] [PubMed] [Google Scholar]
- Lee, K. Y., Kim, S., Kim, U. K., Ki, C.-S. and Lee, S. H. (2007). Novel EYA1 mutation in a Korean branchio-oto-renal syndrome family. Int. J. Pediatr. Otorhinolaryngol. 71, 169-174. 10.1016/j.ijporl.2006.08.023 [DOI] [PubMed] [Google Scholar]
- Li, X., Perissi, V., Liu, F., Rose, D. W. and Rosenfeld, M. G. (2002). Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297, 1180-1183. 10.1126/science.1073263 [DOI] [PubMed] [Google Scholar]
- Li, X., Ohgi, K. A., Zhang, J., Krones, A., Bush, K. T., Glass, C. K., Nigam, S. K., Aggarwal, A. K., Maas, R., Rose, D. W.et al. (2003). Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426, 247-254. 10.1038/nature02083 [DOI] [PubMed] [Google Scholar]
- Li, J., Zhang, T., Ramakrishnan, A., Fritzsch, B., Xu, J., Wong, E. Y. M., Loh, Y.-H. E., Ding, J., Shen, L. and Xu, P.-X. (2020). Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 48, 2880-2896. 10.1093/nar/gkaa012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody, S. A. (2000). Cell lineage analysis in Xenopus embryos. Methods Mol. Biol. 135, 331-347. [DOI] [PubMed] [Google Scholar]
- Moody, S. A. (2018). Lineage tracing and fate mapping in xenopus embryos. Cold Spring Harb. Protoc. 2018, pdb.prot097253. 10.1101/pdb.prot097253 [DOI] [PubMed] [Google Scholar]
- Moody, S. A. and Kline, M. J. (1990). Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 182, 347-362. 10.1007/BF02433495 [DOI] [PubMed] [Google Scholar]
- Moody, S. A. and Lamantia, A. S. (2015). Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 111, 301-350. 10.1016/bs.ctdb.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody, S. A., Neilson, K. M., Kenyon, K. L., Alfandari, D. and Pignoni, F. (2015). Using Xenopus to discover new genes involved in branchiootorenal spectrum disorders. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 178, 16-24. 10.1016/j.cbpc.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, Y. (2015). Signaling regulating inner ear development: cell fate determination, patterning, morphogenesis, and defects. Congenit. Anom. 55, 17-25. 10.1111/cga.12072 [DOI] [PubMed] [Google Scholar]
- Neilson, K. M., Pignoni, F., Yan, B. and Moody, S. A. (2010). Developmental expression patterns of candidate cofactors for vertebrate six family transcription factors. Dev. Dyn. 239, 3446-3466. 10.1002/dvdy.22484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson, K. M., Abbruzzesse, G., Kenyon, K., Bartolo, V., Krohn, P., Alfandari, D. and Moody, S. A. (2017). Pa2G4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev. Biol. 421, 171-182. 10.1016/j.ydbio.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson, K. M., Keer, S., Bousquet, N., Macrorie, O., Majumdar, H. D., Kenyon, K. L., Alfandari, D. and Moody, S. A. (2020). Mcrs1 interacts with Six1 to influence early craniofacial and otic development. Dev. Biol. 467, 39-50. 10.1016/j.ydbio.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop, P. D. and Faber, J. (1994). Normal Table of Xenopus Laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the end of Metamorphosis. New York: Garland Pub. [Google Scholar]
- Ohta, S. and Schoenwolf, G. C. (2018). Hearing crosstalk: the molecular conversation orchestrating inner ear dorsoventral patterning. Wiley Interdiscip. Rev. Dev. Biol. 7, e302. 10.1002/wdev.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, H., Kamada, S., Tago, K., Tominaga, S.-I., Ozaki, H., Sato, S. and Kawakami, K. (1999). Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell Biol. 19, 6815-6824. 10.1128/MCB.19.10.6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, H., Nakamura, K., Funahashi, J.-I., Ikeda, K., Yamada, G., Tokano, H., Okamura, H.-O., Kitamura, K., Muto, S., Kotaki, H.et al. (2004). Six1 controls patterning of the mouse otic vesicle. Development 131, 551-562. 10.1242/dev.00943 [DOI] [PubMed] [Google Scholar]
- Pandur, P. D. and Moody, S. A. (2000). Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech. Dev. 96, 253-257. 10.1016/S0925-4773(00)00396-8 [DOI] [PubMed] [Google Scholar]
- Patrick, A. N., Schiemann, B. J., Yang, K., Zhao, R. and Ford, H. L. (2009). Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J Biol Chem, 284, 20781-20790. 10.1074/jbc.M109.016832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin, L. and Kirschner, M. W. (2020). A cell type annotation Jamboree—Revival of a communal science forum. Genesis 58, e23383. 10.1002/dvg.23383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, M., Bonev, B., Lindsay, J., Lea, R., Panagiotaki, N., Houart, C. and Papalopulu, N. (2010). FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development 137, 1553-1562. 10.1242/dev.044909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf, R. G., Berkman, J., Wolf, M. T. F., Nurnberg, P., Gattas, M., Ruf, E.-M., Hyland, V., Kromberg, J., Glass, I., Macmillan, J.et al. (2003). A gene locus for branchio-otic syndrome maps to chromosome 14q21.3-q24.3. J. Med. Genet. 40, 515-519. 10.1136/jmg.40.7.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf, R. G., Xu, P.-X., Silvius, D., Otto, E. A., Beekmann, F., Muerb, U. T., Kumar, S., Neuhaus, T. J., Kemper, M. J., Raymond, R. M.Jr.. et al. (2004). SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. USA 101, 8090-8095. 10.1073/pnas.0308475101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet, J.-P. and Moody, S. A. (2014). Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev. Biol. 389, 13-27. 10.1016/j.ydbio.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanggaard, K. M., Rendtorff, N. D., Kjaer, K. W., Eiberg, H., Johnsen, T., Gimsing, S., Dyrmose, J., Nielsen, K. O., Lage, K. and Tranebjærg, L. (2007). Branchio-oto-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur. J. Hum. Genet. 15, 1121-1131. 10.1038/sj.ejhg.5201900 [DOI] [PubMed] [Google Scholar]
- Schlosser, G. and Ahrens, K. (2004). Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 271, 439-466. 10.1016/j.ydbio.2004.04.013 [DOI] [PubMed] [Google Scholar]
- Schlosser, G., Awtry, T., Brugmann, S. A., Jensen, E. D., Neilson, K., Ruan, G., Stammler, A., Voelker, D., Yan, B., Zhang, C.et al. (2008). Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 320, 199-214. 10.1016/j.ydbio.2008.05.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A. M., Krohn, P., Baxi, A. B., Tavares, A. L. P., Sullivan, C. H., Chillakuru, Y. R., Majumdar, H. D., Neilson, K. M. and Moody, S. A. (2020). Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis. Model Mech. 13, dmm043489. 10.1242/dmm.043489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer, A. E., Hildebrand, M. S. and Smith, R. J. H. (2017). Hereditary hearing loss and deafness overview. In GeneReviews (ed. Adam M. P., Ardinger H. H., Pagon R. A., Wallace S. E., Bean L. J. H., Mirzaa G. and Amemiya A.). https://www.ncbi.nlm.nih.gov/books/NBK1434/ [Google Scholar]
- Silver, S. J., Davies, E. L., Doyon, L. and Rebay, I. (2003). Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell Biol. 23, 5989-5999. 10.1128/MCB.23.17.5989-5999.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. J. H. (2018). Branchiootorenal spectrum disorders. In GeneReviews (ed. Adam M. P., Ardinger H. H., Pagon R. A., Wallace S. E., Bean L. J. H., Stephens K. and Amemiya A.). https://www.ncbi.nlm.nih.gov/books/NBK1380/ [Google Scholar]
- Sullivan, C. H., Majumdar, H. D., Neilson, K. M. and Moody, S. A. (2019). Six1 and Irx1 have reciprocal interactions during cranial placode and otic vesicle formation. Dev. Biol. 446, 68-79. 10.1016/j.ydbio.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. and Hunter, T. (2012). Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J. Biol. Chem. 287, 42071-42083. 10.1074/jbc.M112.410985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares, A. L. P., Cox, T. C., Maxson, R. M., Ford, H. L. and Clouthier, D. E. (2017). Negative regulation of endothelin signaling by SIX1 is required for proper maxillary development. Development 144, 2021-2031. 10.1242/dev.145144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P.-X. (2013). The EYA-SO/SIX complex in development and disease. Pediatr. Nephrol. 28, 843-854. 10.1007/s00467-012-2246-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, B., Neilson, K. M. and Moody, S. A. (2009a). foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev. Biol. 329, 80-95. 10.1016/j.ydbio.2009.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, B., Neilson, K. M. and Moody, S. A. (2009b). Notch signaling downstream of foxD5 promotes neural ectodermal transcription factors that inhibit neural differentiation. Dev. Dyn. 238, 1358-1365. 10.1002/dvdy.21885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. J., Kjolby, R. A. S., Wu, G., Wong, D., Hsu, S.-W. and Harland, R. M. (2017). Noggin is required for first pharyngeal arch differentiation in the frog Xenopus tropicalis. Dev. Biol. 426, 245-254. 10.1016/j.ydbio.2016.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer, A. W. and Schrelber, S. L. (1994). Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76, 933-945. 10.1016/0092-8674(94)90367-0 [DOI] [PubMed] [Google Scholar]
- Zhang, T., Xu, J., Maire, P. and Xu, P.-X. (2017). Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 13, e1006967. 10.1371/journal.pgen.1006967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, W., Huang, L., Wei, Z.-B., Silvius, D., Tang, B. and Xu, P.-X. (2003). The role of Six1 in mammalian auditory system development. Development 130, 3989-4000. 10.1242/dev.00628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.