Abstract
Mutations in the oncogene PARK7, which codes for DJ-1, have been associated with early-onset autosomal recessive Parkinson’s disease (PD); however, the exact role of DJ-1 in PD remains elusive. Fibroblasts from a PD patient with a uniparental disomy, 1 bp deletion in PARK7 were reprogrammed into the induced pluripotent stem cell (iPSC) line: NIHTVBi015-A. For control purposes, CRISPR-Cas9 editing was used to mimic the mutation in the Gibco Human Episomal iPSC line: TMOi001-A is the control line (A18945) and TMOi001-A-3 is the control-edited line (2B10). All 3 lines exhibit normal karyotyping and expression of pluripotent markers: OCT4, SOX2, and NANOG. These lines provide a translational environment to study DJ-1-related function in PD.
1. Resource table
| Unique stem cell lines identifier | 1. NIHTVBi015-A |
| 2. TMOi001-A-3 | |
| 3. TMOi001-A | |
| Alternative names of stem cell lines | 4. HT188 (NIHTVBi015-A) |
| 5. 2B10 (TMOi001-A-3) | |
| 6. A18945 (TMOi001-A) | |
| Institution | 1. NIH National Heart, Lung, and Blood Institute IPSC Core |
| 2. National Institutes of Health | |
| 3. Commercially available | |
| Contact information of distributor | Mark Cookson, cookson@mail.nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | 1. Fibroblasts |
| 2. CD34 + cord blood | |
| 3. CD34 + cord blood | |
| Clonality | Clonal |
| Method of reprogramming | 1. Sendai virus |
| 2. Three-plasmid, seven-factor EBNA-based episomal system | |
| 3. Three-plasmid, seven-factor EBNA-based episomal system | |
| Multiline rationale | Control and disease pair |
| Gene modification | 1. No |
| 2. Yes | |
| 3. No | |
| Type of modification | Induced mutation |
| Associated disease | Parkinson’s disease |
| Gene/locus | NM:001123377.1 (PARK7): Chr1(GRCh37): g.8037720del; p.Ala111Leufs*7 |
| Method of modification | CRISPR |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | N/A |
| Cell line repository/bank | N/A |
| Ethical approval | 1. NIH Undiagnosed Diseases Program, 76-HG-0238 |
2. Resource utility
The PARK7 gene encodes for DJ-1 and causes an autosomal recessive early-onset form of Parkinson’s disease (Bonifati et al., 2003). The iPSC line from a PD patient with a PARK7 loss-of-function mutation, and the line mimicking the mutation in the A18945 control line, are valuable tools in which to study DJ-1-related mechanisms of neurodegeneration.
3. Resource details
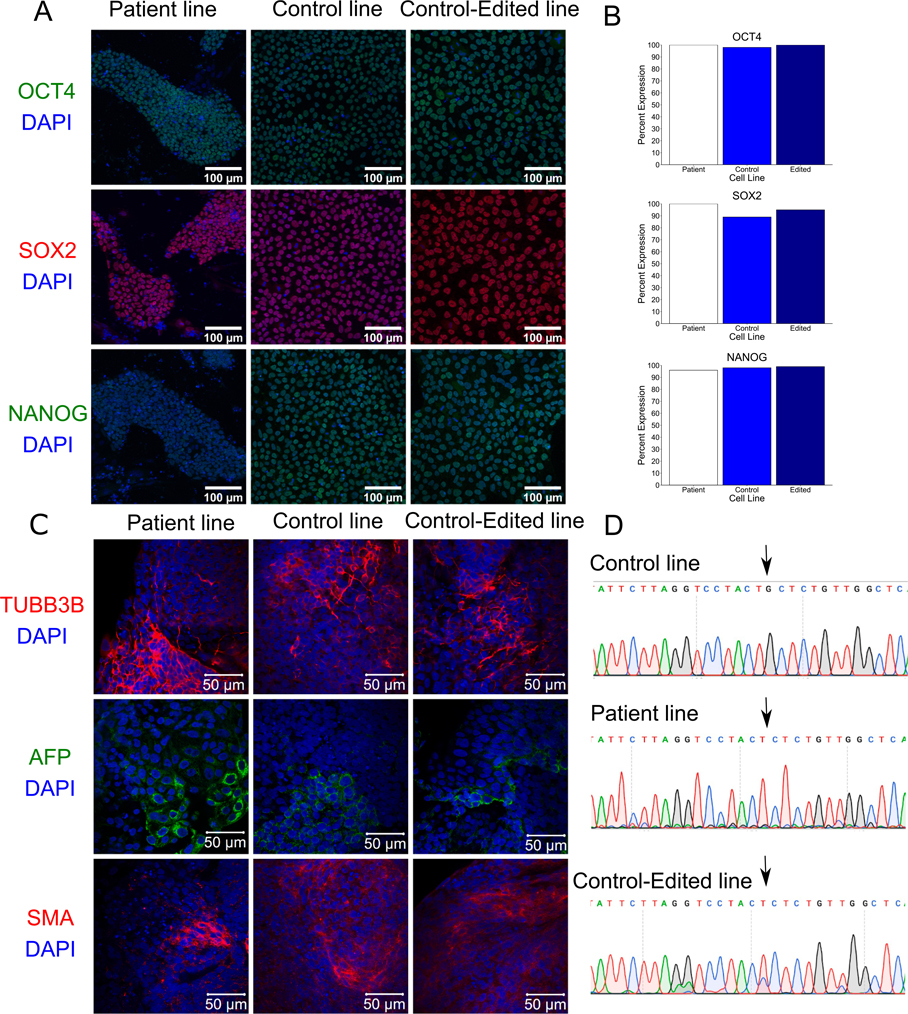
To date, all PARK7 mutations related to PD are loss-of-function mutations (Hauser et al., 2016). Fibroblasts were taken from an early-onset PD patient carrying a homozygous single base pair deletion in the PARK7 gene (c.331del (p.A111L); Table 1). Fibroblasts were converted to the pluripotent state (patient iPSC line HT188 (NIHTVBi015-A)) using the Sendai method of reprogramming (NIH National Heart, Lung, and Blood Institute IPSC Core). We tried to correct the mutation using CRISPR-editing techniques, but low transfection rates prevented this. Therefore, we inserted the mutation into the commercially available control iPSC line A18945 (Gibco; (TMOi001-A)) using CRISPR-editing (edited iPSC line 2B10 (TMOi001-A-3)) so to have a genetically matched control line. All lines have normal morphology, but the patient iPSC line needs to be seeded onto mouse embryonic fibroblasts (MEFS) to survive (Supplementary Fig. 1A). Pluripotency was confirmed through the detection of OCT4, SOX2, and NANOG at the protein level using immunocyctochemistry (Fluorescent Human ES/iPS Cell Characterization Kit, Sigma-Aldrich, SCR078; Fig. 1A–B). All three lines were able to undergo embryoid body differentiation and express markers for the three layers including beta-tubulin (TUBB3B; ectoderm), alpha-feto protein (AFP; endoderm), and muscle actin (SMA, mesoderm; Fig. 1C). All three lines have normal karyotyping with no detectable clonal abnormalities at the stated level of band resolution (Table 2, Supplemental Fig. 1B; WiCell). According to short tandem repeat (STR) analysis (WiCell), all lines were unique and not contaminated with other cell lines. However, while the patient iPSC line did not display allelic imbalances, the control and edited iPSC lines did express allelic imbalances which may be due to chromosomal gains, losses, and/or amplifications (see journal archive). The control iPSC line was matched with four other cell lines that were generated from it, including the edited iPSC line (Table 2). All lines were sequenced to verify that the guanine at c.331 locus was either present (control line, arrow) or absent (patient and edited lines) in a homozygous fashion (Fig. 1D). To detect potential off-target effects of the deletion and CRISPR-editing, all lines were submitted for whole genome sequencing (WGS, Psomagen). No off-target effects were identified in the top seven predicted sites (Supplementary Fig. 1D). Sequences for all three lines have been made available (Table 2). Finally, all lines tested negative for mycoplasma (Supplementary Fig. 1C) (see Table 3).
Table 1.
Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
|
| ||||||
| HT188 (NIHTVBi015-A) | HT188 | Female | 35 | Northern European | Homozygous PARK7 c.331del (p.A111L) | Parkinson’s disease |
| 2B10 (TMOi001-A-3) | 2B10 | Female | Unknown | Unknown | Homozygous PARK7 c.331del (p.A111L) | Mimics mutation |
| A18945 (TMOi001-A-3) | A18945 | Female | Unknown | Unknown | Commercially available control line | |
Fig. 1.
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Photography | Normal | Supplementary Fig. 1A |
| Phenotype | Qualitative analysis | Verified expression of OCT4, SOX2, and NANOG | Fig. 1A |
| Quantitative analysis | Assess % of positive cells for antigen markers: OCT4: all above 98%, SOX2: all above 89%, NANOG: all above 96% | Fig. 1B | |
| Genotype | Karyotype (G-banding) and resolution | 1. 46,XX Resolution 500–550 2. 46,XX Resolution 450–575 3. 46,XX Resolution 425–500 |
Supplementary Fig. 1B |
| Identity | Microsatellite PCR (mPCR) OR STR analysis |
Not performed 16 loci tested: 1. No matches 2. No matches 3. Four matches to other cell lines derived from this commercial line |
N/A Submitted in archive with journal |
| Mutation analysis (IF APPLICABLE) | Sequencing | Homozygous, c.331del | Fig. 1D |
| WGS | No off-target effects. | Sequencing deposited at Sequence Read Archive: SRA accession PRJNA622304. Release date: 2020-09-01. https://www.ncbi.nlm.nih.gov/sra/PRJNA622304 Supplementary Fig. 1D | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by RT-PCR. All negative | Supplementary Fig. 1C |
| Differentiation potential | Embryoid body formation | Expression of muscle actin, β-III tubulin and α-feto protein. | Fig. 1C |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | N/A |
| HLA tissue typing | Not performed | N/A | |
Table 3.
Reagents details.
| Antibodies used for immunocytochemistry/flow-citometry | |||
| Antibody | Dilution | Company Cat # and RRID | |
|
| |||
| Pluripotency Marker | Mouse anti-Oct-4, Alexa Fluor® 488 conjugate | 1:100 | Millipore Sigma, MAB4419A4, RRID:AB_2847875 |
| Pluripotency Marker | Mouse anti-Sox-2, Cy3 conjugate | 1:100 | Millipore Sigma, MAB4423C3, RRID:AB_2847876 |
| Pluripotency Marker | Mouse anti-Nanog, Alexa Fluor® 488 conjugate | 1:100 | Millipore Sigma, SCR078, MABD24A4, RRID:AB_2847877 |
| Mesoderm marker | smooth muscle actin | 1:200 | Invitrogen, A25531, RRID:AB_2651005 |
| Endoderm marker | α-fetoprotein | 1:200 | Invitrogen, A25530, RRID:AB_2651004 |
| Ectoderm marker | β-III tubulin | 1:200 | Invitrogen, A25532, RRID:AB_2651003 |
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
|
| |||
| Mycoplasma testing | Mycoplasma | Forward primers: CGCCTGAGTAGTACGTTCGC; CGCCTGAGTAGTACGTACGC; TGCCTGAGTAGTACATTCGC; TGCCTGGGTAGTACATTCGC; CGCCTGAGTAGTATGCTCGC/ Reverse primers: GCGGTGTGTACAAGACCCGA; GCGGTGTGTACAAAACCCGA; GCGGTGTGTACAAACCCCGA |
|
| Mutation analysis for HT188 | PARK7 | TTCTGTGCTTTTGCCAGATG/TCTTTAGCAAGAGGGTGTGTTG | |
| CRISPR diagnostic primers | Donor oligo | TGGGCTCAAGCAATTTTTCTA/ATGGAGCGAGACTCCATCT | |
| gRNA for A18945 | PARK7 mutation | caccgCTTAGGTCCTACTGCTCTGT/ aaacACAGAGCAGTAGGACCTAAGc | |
| Donor oligo | TGCGATTTTTTAAACATGGGCTTTTCTATATCTGCACTTAGAT | ||
| CTTTTTATTTTTATTCTTAGGTCCTACTCTCTGTTGGCTCATGA | |||
| AATAGGTTTTGGAAGTAAAGTTACAACACACCCTCTTGCTAA | |||
| AGACAAAATGATGAATGGAGGTAAGTATATGCTTGTTTTTGT | |||
| TTGTTTGTTTGTTTTTTGAGATGGAGTC | |||
4. Materials and methods
4.1. Reprogramming
For the patient HT188 line, fibroblasts were reprogrammed following the Sendai Cytotune 1 (A1378001) and Cytotune 2 (A16517) virus kit (Life Technologies) instructions by NIH National Heart, Lung, and Blood Institute IPSC Core as previously described (Beers et al., 2015). The Gibco Human Episomal iPSC Line (A18945; ThermoFisher Scientific) was commercially purchased and derived from CD34+ cord blood using the three-plasmid, seven-factor EBNA-based episomal system.
4.2. CRISPR editing
CRISPR editing was used to create the edited 2B10 line by inserting the single base pair mutation into the commercially available control A18945 line to mimic the patient-derived HT188 line. gRNA was cloned into the pSP-Ef1α-Cas9–2A-GFP plasmid backbone. Cells were transfected with the Cas9/gRNA plasmid (pSP-Ef1α-Cas9–2A-GFP; Integrated DNA Technologies) and donor oligo using lipofectamine™ stem transfection reagent (Thermo Fisher Scientific). Cells were prepped for fluorescence-activated cell sorting (FACS) sorting for GFP-positive cells then expanded until recovered. Cells were then treated with Accutase (Thermo Fisher Scientific) for a single-cell suspension and seeded to generate colonies for monoclonal lines. Once colonies had recovered, the Guide-it Mutation Detection kit (Takara) was used to prep samples for PCR amplification and sequencing analysis by Psomagen for verification of the mutation. gRNA, donor oligo and diagnostic primers to verify mutation insertion are listed in Table 2.
4.3. Karyotyping
Karyotyping was performed by the WiCell Characterization Lab (Madison, WI). Cells were seeded in a T25 flask and shipped 3 days after passaging (HT188 at p8, A18945 at p4, 2B10 at p5) when at ~50% confluency.
4.4. Short tandem repeat (STR) analysis
STR analysis was carried out by the WiCell Characterization Lab (Madison, WI). Cells were seeded in a T25 flask and shipped 3 days after passaging (HT188 at p4, A18945 at p4, 2B10 at p5) when at ~50% confluency.
4.5. Whole genome sequencing (WGS)
Cells (passage 4–5) underwent DNA extraction using the 25:24:1 Phenol:Chloroform:Isoamyl alcohol protocol. The resulting DNA pellet was resuspended in autoclaved water then sent to Psomagen for WGS and off-target analysis.
4.6. Mutation analysis
To verify the presence and absence of the mutation, all three lines were sequenced using the Sanger sequencing method, respectively. The DNeasy blood and tissue kit (Qiagen) was used to extract gDNA for PCR amplification with Terra PCR polymerase mix (Takara). AMPure purification reagents (Beckman) and sequencing using BigDye on a 3730 DNA Analyzer (Applied Biosystems) followed. Primers to verify mutation insertion are listed in Table 2.
4.7. Immunofluorescence staining
Immunofluorescence staining was carried out following the Fluorescent Human ES/iPS Cell Characterization Kit (Millipore Sigma, SCR078) protocol. Antibody information is listed in Table 2. Images taken on a Zeiss 780 confocal microscope.
4.8. Embryoid body (EB) formation
iPSC lines were dissociated into single cells with TrypLE™ Express (Life Technologies), and 10,000 cells/well were plated into low-cell-adhesion 96-well culture plates with V-bottomed conical wells (Sumitomo Bakelite) to form uniform EBs cultured in: DMEM media, 20% FBS, 1 mM L-glutamine, 0.1 mM β-mercaptoethanol, 1% nonessential amino acids, P/S. After 20 days, EBs were fixed with 4%PFA and stained for three markers: smooth muscle actin (mesoderm), beta-III Tubulin (ectoderm), alpha-fetoprotein (endoderm) by Molecular Probes® 3-Germ Layer Immunocytochemistry Kit (Cat. No. A25538).
4.9. Mycoplasma detection
Mycoplasma testing was performed by PCR (Uphoff and Drexler, 2004). Cell lines (passage 4–5) were scraped in PBS and spun down to remove supernatant. Pellets were lysed using proteinase K (1 mg/mL) and DirectPCR lysis reagent (Viagen Biotech). Detection primers is listed in Table 2.
Supplementary Material
Acknowledgements
This research was supported in part by the Intramural research Program of the National Institutes of Health (NIH; Bethesda, MD, USA), National institute on Aging. This work was supported in part by the Common Fund, Office of the Director, NIH and the Intramural Research Program of the National Human Genome Research Institute, NIH. The HT-188 iPSC line was generated/reprogrammed by the National Heart, Lung and Blood Institute iPSC Core at the NIH.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102506.
References
- Bonifati V, Rizzu P, van Baren M, Schaap O, Breedveld G, Krieger E, et al. , 2003. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259. [DOI] [PubMed] [Google Scholar]
- Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G, 2015. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep. 5 (1) 10.1038/srep11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser D, Primiani C, Cookson M, 2016. The effects of variants in the PARK2 (parkin), PINK1, and PARK7 (DJ-1) genes along with evidence for their pathogenicity. Curr. Protein Pept. Sci. 18, 702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff CC, Drexler HG, 2004. Detecting Mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol. Med. 88, 319–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.