Abstract
Rattus norvegicus (Norway rat) is the main reservoir host of pathogenic Leptospira, the causative agent of leptospirosis, in urban environments. Pathogenic Leptospira forms biofilms in the environment, possibly contributing for bacterial survival and maintenance. Nonetheless, biofilms have not yet been studied in natural animal reservoirs presenting leptospiral renal carriage. Here, we described biofilm formation by pathogenic Leptospira inside the renal tubules of R. norvegicus naturally infected and captured in an urban slum endemic for leptospirosis. From the 65 rats carrying Leptospira in their kidneys, 24 (37%) presented biofilms inside the renal tubules. The intensity of leptospiral colonization in the renal tubules (OR: 1.00; 95% CI 1.05–1.1) and the type of occlusion pattern of the colonized renal tubules (OR: 3.46; 95% CI 1.20–9.98) were independently associated with the presence of Leptospira biofilm. Our data showed that Leptospira interrogans produce biofilms during renal chronic colonization in rat reservoirs, suggesting a possible role for leptospiral biofilms in the pathogenesis of leptospirosis and bacterial carriage in host reservoirs.
Author summary
Leptospirosis is an infectious disease caused by pathogenic Leptospira bacteria. The main reservoir hosts of Leptospira are the brown rats (Rattus norvegicus), which are chronically colonized in the kidneys. Leptospires form biofilms, which are communities of microorganisms embedded in an extracellular polysaccharidic matrix. Leptospira pathogenesis in reservoir hosts is poorly understood. We captured 87 brown rats from an impoverished urban community that is endemic for leptospirosis. To investigate the biofilm in the rats’ kidneys, we co-localized leptospires and saccharides of the biofilm extracellular matrix in the renal tubules, using immunohistochemistry anti-Leptospira and carbohydrate staining, respectively. We quantified Leptospira using molecular tools and characterized the biofilm using electron microscopy. We analysed demographic data to identify variables correlated with renal carriage. We found that Leptospira infected 78 rats. From those, 65 were positive for immunohistochemistry in the kidneys and 24 (37%) were biofilm-positive. We found significant positive correlation between the intensity of colonization and the presence of biofilm in the kidneys. The intensity of colonization was also associated with the rats’ gender and age. Biofilm formation by Leptospira in the kidneys of natural reservoir rats fills a gap into the knowledge of leptospirosis pathogenesis.
Introduction
Leptospirosis is an infectious disease of public health global importance [1]. It is estimated that more than one million cases and nearly 60 thousand deaths occur yearly in the world due to leptospirosis [2]. The incidence of this zoonotic disease is higher in developing and tropical countries, especially during rainy seasons [3,4]. Leptospirosis is caused by pathogenic spirochetes from the genus Leptospira, which is composed of 66 pathogenic and saprophytic species [5,6]. The development of leptospirosis depends, among others, on epidemiologic factors such as sanitation and flooding, on the pathogenicity and infecting dose of Leptospira, and on host susceptibility to infection [7].
Rattus norvegicus (Norway rats) are the main reservoir hosts of leptospires in urban settings worldwide. It is estimated that naturally infected Norway rats daily excrete approximately 9.0e+10 leptospires in the urine, strongly contaminating the environment [8]. Pathogenic Leptospira densely colonize the kidneys of rat reservoir hosts, where they form aggregates in the proximal renal tubules [9,10]. Experimentally infected R. norvegicus with a high dose of pathogenic leptospires remain asymptomatic and with stable renal colonization, what is observed at the first week post infection, persevering for more than four months [11]. Histopathological analysis of the kidneys of naturally infected R. norvegicus revealed minor alterations as interstitial nephritis and tubular epithelial hyaline droplets. However, it was not possible to detect if those alterations were related to Leptospira infection, once numerous environmental exposures could cause similar alterations [9,12]. During infection in chronic animal models, leptospires form biofilm-like structures during renal tubular colonization [13], what may explain the antibiotic tolerance observed when the treatment occurs during the disease chronic phase [14].
Bacteria of the genus Leptospira form biofilms in vitro [15] and in rural environment [16]. Biofilms are communities of microorganisms attached to a surface and involved by a self-produced exopolymeric matrix [17,18]. The transcriptome of saprophytic Leptospira biflexa revealed important insights into Leptospira biology during biofilm formation. Of note, there was a great shift in gene regulation during biofilm, as the diminishment of alginate gene expression, upregulation of a putative gene for a virulence-associated protein, and upregulation of outer membrane proteins (OMPs) encoding genes [19]. A recent study analyzed in vitro several aspects of the biofilm formed by pathogenic Leptospira and showed that inside the biofilm leptospires are protected against stressful environmental conditions as pH and temperature [20].
The biofilm lifestyle is ubiquitous in Bacteria. This ubiquity is related to the protection offered by biofilms against harsh environmental and host conditions [18,21]. Biofilms are described as colonization and virulence factors [22], participating in the pathogenesis of several diseases caused by pathogenic or opportunistic bacteria [23,24]. Bacterial biofilms are associated with many medical chronic conditions as cystic fibrosis pneumonia, dental plaque, and catheter contamination, among others [24–27]. The spirochete Borrelia form biofilms in vitro and in vivo in the skin of infected patients diagnosed with borrelial lymphocytoma [28,29]. Biofilms were also described as crucial for the transmission of Yersinia pestis and host-pathogen interactions in plague, due to biofilm formation in the midgut of fleas [30].
Knowledge about pathogenic Leptospira biology, host-pathogen interactions, and survival inside the hosts, as well as their transmission mechanisms to other hosts are of outermost importance to understand and counteract the infection in susceptible hosts [31,32]. In this study we demonstrated Leptospira biofilms in the kidneys of naturally infected Rattus norvegicus captured in an urban slum endemic for leptospirosis. We identified the factors associated with renal biofilm formation and characterized the histopathological kidney alterations in natural reservoir rats.
Methods
Ethics statement
This work was approved by the Institutional Animal Care and Use Committee at the Oswaldo Cruz Foundation (Salvador, Brazil; protocol number 003/2012).
Study sites and animals
Animal captures followed a previously described methodology with few modifications (COSTA et al., 2015a). Briefly, from May 2013 to August 2014, we captured 86 Norway rats (Rattus norvegicus) in Pau da Lima slum, in Salvador, Bahia. Pau da Lima is an urban community with 0.46 Km2 area and four valleys [33], and was selected for this study given its high incidence of severe human leptospirosis [34]. We systematically sampled the study site by setting two Tomahawk live traps at each of 108 sampling points [35], and recorded the capture site and entered/validated demographic data in Redcap database. We euthanized the rats, recorded the site of collection, sex, and weight, and used mass/weight as a proxy for estimating rat’s age, dividing them into juveniles (≤ 200 g), sub-adults (201–400 g) and adults (≥ 401 g). We collected the rats’ urine directly from the bladder using a 1mL syringe and froze the urine in -80° C until qPCR analysis [8]. During necropsies, we collected and preserved the right kidney in 10% buffered formalin and further processed for histology. We also collected and divided the left kidney; half of it we preserved in -80°C [8] and the other half was processed for scanning electron microscopy (SEM), as described below.
Quantitative real-time PCR (qPCR) of Leptospira load in kidneys
We performed quantitative real-time polymerase-chain reaction (qPCR) strictly as described by COSTA et al., 2015b. Briefly, we extracted DNA from 200 μL of urine and 25 mg of frozen kidney using Maxwell 16 System DNA Purification Kits (Promega Corp., USA). We performed qPCR for pathogenic leptospires using 5’ nuclease (Taq-Man) assay, and primers for lipL32, a gene solely present in pathogenic leptospires [36]. We performed the quantitative amplification using an ABI 7500 Real-Time PCR System (Applied Biosystems, USA).
Histological processing, immunofluorescence (IF), and immunohistochemistry (IHC)
We screened the kidney sections for L. interrogans infection using immunofluorescence (IF) qualitative method imprint technique, following the protocol described by CHAGAS-JUNIOR et al. [37]. Next, we embedded the kidneys in paraffin wax, cut the blocks in 2-μm serial sections and processed for histopathology, in the following order: (1) hematoxylin-eosin (HE); (2) immunohistochemistry (IHC) anti-Leptospira interrogans; (3) periodic acid-Schiff stain (PAS); (4) PAS Silver Methenamine (PAS-M); (5) Alcian blue pH 2.5 (AB); (6) Mayer’s Mucicarmine (MM); (7) AZAN; (8) Picrosirius Red (PIFIG). We used HE, PAS, PAS-M, AZAN and PIFIG to analyze pathologic alterations, according to routine protocols. We then applied a questionnaire for histopathological analysis.
For biofilm detection, we used AB pH 2.5 and MM (special stains for carbohydrates), according to routine protocols. For MM staining, we deparaffinized the samples in xylol baths twice for 10 min each; immersed in absolute alcohol baths twice for 30 s each; hydrated in tap water; and placed the slides in Weigert’s iron hematoxylin for 7 min. Then, we washed the samples under tap water for 10 min; added mucicarmine solution for 1 h; washed quickly in non-pyrogenic distilled water, added metanil yellow for 1 min, and washed again in non-pyrogenic distilled water. We then dehydrated the samples in 95% alcohol; placed 2x in absolute alcohol baths for 30 s each; clarified in xylol baths twice for 30 s each; and assembled the slides. For AB staining, we deparaffinized the samples in xylol baths twice for 10 min each; immersed in absolute alcohol baths twice for 30 s each and hydrated under tap water. Then, we immersed the slides in a 3% acetic acid solution for 10 min, followed by immersion in 2% alcian blue solution for 30 min and washed in non-pyrogenic distilled water to remove the excess dye. We immersed the slides into Harris hematoxylin for 1 min, rinsed in tap water for 5 min, dehydrated in absolute alcohol baths three times for 3 min each, clarified in two xylol baths, and assembled the slides. We obtained the microscopy images using an Olympus BX51 optical microscope with objectives of 20x and 40x and the Image-Pro Plus software (Media Cybernetics, USA). We analyzed biofilm formation only in the kidneys of positive rats for IHC anti-L. interrogans.
We performed IHC analysis according to CRODA et al. (2008) [38], with the following modifications: we blocked the slides with 10% skimmed milk in 1× phosphate buffered saline (NaCl 140 mM; 2.7 mM KCl; 10 mM Na2HPO4; 1.8 mM KH2PO4; pH 7.4) and incubated with rabbit primary antibodies anti-Leptospira interrogans serovar Icterohaemorrhagiae strain RGA diluted 1:1.000 at room temperature for 1 h. As negative controls, we used kidneys of negative rats for L. interrogans infection. Once we identified renal tubules positive to IHC anti-L. interrogans, we performed co-localization of these tubules in serial renal sections with tubules concomitantly positive for AB staining.
Patterns of leptospiral colonization in rat kidneys
We analyzed different patterns of leptospiral colonization in R. norvegicus kidneys using a previous methodology described by SANTOS et al., 2015. We inferred colonization intensity by counting the number of IHC positive colonized tubules (CTs) in a half-kidney cortex. This information generated the variable “number of colonized tubules”, in this work denominated CT count, considered to be a quantitative measurement of infection. We analyzed the distribution of CTs by performing a qualitative evaluation, registering if CTs were observed isolated in the cortex (one CT) or agglomerated (two or more CTs), and with focal or multifocal distribution. Finally, we qualitatively evaluated the tubular intraluminal IHC, considering two patterns: kidney tubules with IHC marking restricted to the renal epithelial membrane (considered as partial occlusion of the CT) or kidney tubules with IHC marking both restricted to the renal epithelial membrane and completely occluding the lumen of CT (considered as partial/complete occlusion of the CT).
Electron microscopy using ruthenium red probe
Using a sterile scalpel, we cut fresh half kidneys of captured rats in 1–2 mm cubes, fixed in 2% glutaraldehyde/0.1 M sodium cacodylate, and kept at 4°C until processing. A non-colonized rat kidney was processed as negative control. We then transferred the samples to new tubes containing the same fixative with or without 0.2% ruthenium red (RR), washed in 0.1 M sodium cacodylate and post-fixed in 1% osmium tetroxide/0.2 M sodium cacodylate for one hour. We washed and dehydrated the samples in graded series of ethanol (from 30–100%), followed by critical point dry (Leica EM CPD030, Austria), sputter-coated with gold (DESCK IV; Denton Vacuum, USA), and examined using a scanning electron microscope (JSM6394LV; JEOL, Japan) operated at 10 kV. For this analysis, we used a subsampling of four positive rats for Leptospira infection with renal biofilm and one negative rat as a control.
Statistical analysis
We considered the rats positive to renal infection when they were positive for IF and/or IHC and/or qPCR anti-Leptospira. Rats were considered to have biofilms when they were positive in the co-localization technique (AB and IHC anti-Leptospira). Sixty-seven rats were analyzed for patterns of leptospiral colonization and 64 rats with positive IHC anti-L. interrogans were analyzed for biofilm presence. We assessed the association of Leptospira infection, biofilm formation and patterns of renal tubules colonization with demographic data (sex, weight category and site of collection) and Leptospira load in rats’ urine. We transformed leptospiral load (GEq of leptospiral DNA/mL) into log for all the analyses. We used Chi-square test to investigate Leptospira infection and biofilm formation occurrence in the rats’ population, and Kruskal-Wallis to evaluate leptospiral load in urine and CT count. To evaluate Leptospira infection and renal biofilm association with demographic characteristics and renal tubules pattern of colonization, we used Pearson Chi-square test or Fisher’s Exact test (n < 5). We used generalized linear model (GLM) to analyze the risk factors associated with Leptospira infection occurrence and renal biofilm formation in the chronically infected host, R. norvegicus. GLM was also used to analyze demographic data associated with histological characteristics of infected rats. We used R Version 1.3.1093 [39] and considered differences with p<0.05 as significant. All results are available in S1 Table.
Results
Reservoir Rattus norvegicus present renal biofilm formed by pathogenic Leptospira
We captured 87 R. norvegicus from May 2013 to August 2014, at Pau da Lima neighborhood (S1 Fig). From those, 78 (90%) rats were positive for Leptospira infection (p = 1.38–13) using one of the methods previously described. From the 78 rats, 65 were positive for immunohistochemistry (IHC) anti-Leptospira and were analyzed for the presence of renal biofilm. Twenty-four (37%) rats were positive (p = 0.04) (Table 1) for renal biofilm according to co-localization of Alcian Blue (AB) staining (Fig 1A and 1B) and IHC anti-Leptospira (Fig 1C and 1D), confirming the presence of polysaccharidic matrix and Leptospira biofilms inside the proximal renal tubules of infected rats. None of the kidneys were positive for Mayer mucicarmine (MM) staining (S2 Table). Negative controls did not stain for either AB or MM (S2 Table). We used as positive controls standard sections of dog intestine, which were positive for both AB and MM (S2 Table).
Table 1. Demographic characteristics, patterns of renal colonization and Leptospira shedding in wild Rattus norvegicus (Norway rats) positive for Leptospira infection and renal biofilm.
| Characteristics | Leptospira positive rats No. ¥ | Leptospira biofilm positive rats No. (%⁂) | OR (CI) |
|---|---|---|---|
| Total | 65 | 24 (37) * | |
| Sex | |||
| Intercept | 0.57 (0.29–1.12) | ||
| Female | 36 | 13 (36) | Ref. |
| Male | 29 | 11 (38) | 1.08 (0.39–2.98) |
| Weight categories | |||
| Intercept | 0.40 (0.13–1.28) | ||
| Adult | 14 | 4 (29) * | Ref. |
| Sub-adult | 38 | 15 (39) | 1.56 (0.31–7.82) |
| Juvenile | 13 | 5 (38) | 1.63 (0.43–6.16) |
| Site of capture | |||
| Intercept | 0.22 (0.05–1.03) | ||
| Valley 1 | 11 | 2 (18) * | Ref. |
| Valley 2 | 6 | 1 (17) | 0.90 (0.06–12.58) |
| Valley 4 | 33 | 17 (52) | 4.78 (0.89–25.59) |
| Leptospiral shedding in urine | |||
| Intercept | 0.14 (0.01–1.29) | ||
| Mean log10 qPCR urine1 | NA | 6.34 * | 1.24 (0.94–1.63) |
| CT count | |||
| Intercept | 0.28 (0.13–0.61) | ||
| Mean of CT2 | NA | 229 | 0.99 (1.0–1.01) ** |
| CT occlusion pattern | |||
| Intercept | 0.32 (0.15–0.68) | ||
| Partial | 37 | 9 (24) * | Ref. |
| Partial/complete | 28 | 15 (53) | 3.59 (1.25–10.32) ** |
| CT distribution | |||
| Intercept | 0.62 (0.36–1.06) | ||
| No. agglomerated CT | 55 | 21 (39) | Ref. |
| No. isolated CT | 10 | 3 (30) | 0.69 (0.16–2.98) |
* Significant differences in descriptive statistical analysis (P<0.05);
** Bold items indicating significant OR in generalized linear model analysis (P<0.05);
¥ Sample with 65 animals positive for IHC anti-Leptospira and analyzed for the presence of renal biofilm.
⁂ Percentage relative to the number of Leptospira positive rats;
1The mean log10 qPCR urine of negative rats for renal biofilm was 7.34.
2The mean of CT count of negative rats for renal biofilm was 113. Ref.: reference category; refers to the default chosen category against which other categories are compared to in regression models when we use categorical dependent variables. Intercept: regression constant; predicts a linear value when all predictor variables are set to zero. Abbreviation: OR, Odds Ratios; CI, Confidence Interval.
Fig 1. Histopathological investigation of leptospiral infection and biofilm formation in kidney serial sections of wild Rattus norvegicus naturally infected with pathogenic Leptospira interrogans.
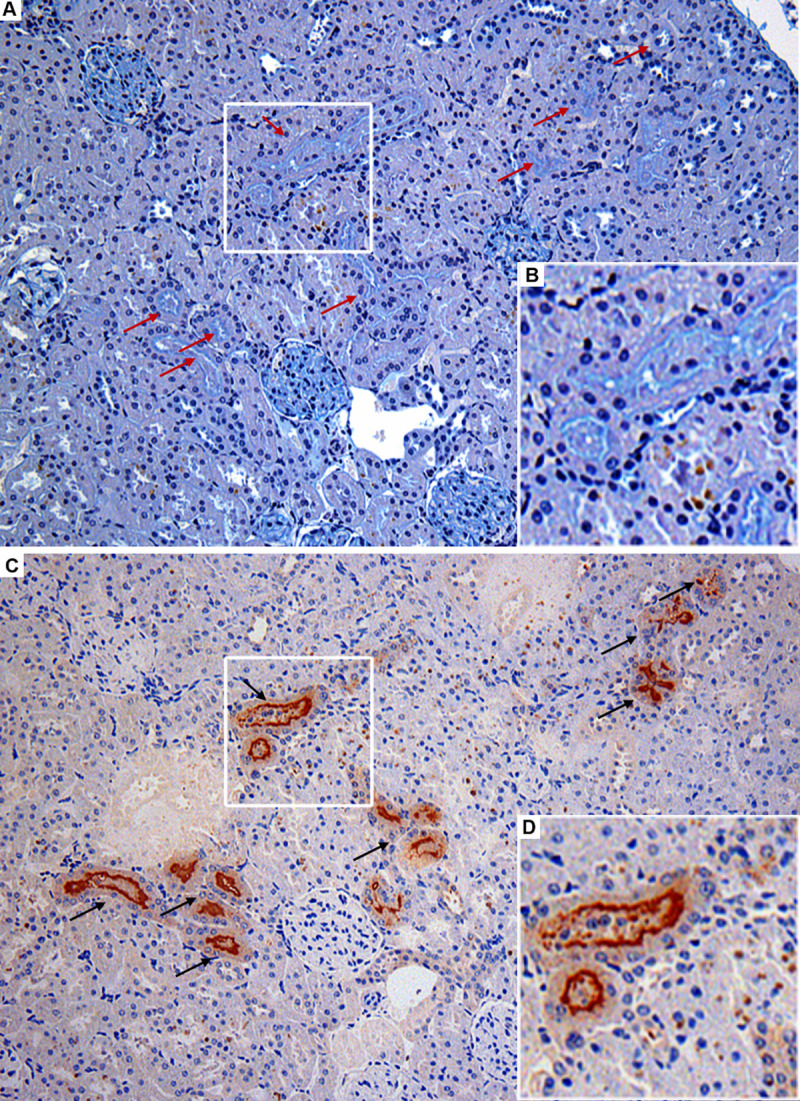
(A) Alcian Blue (AB) positively stained renal tubules (red arrows) observed in light turquoise blue, indicating the presence of biofilm matrix; insert B with detail of biofilm staining. (C) Renal tubules positive for IHC anti-L. interrogans demonstrating leptospiral colonization of proximal tubules (black arrows); insert D with detail of colonized tubule. Note that the serial sections showed in A and D are from the same region of one rat kidney, evidencing the co-localization of tubules concomitantly positive in AB (red arrows) and IHC (black arrows). Magnification, x 200.
The population of rats with renal biofilm was homogeneously comprised of 11 males (46%) and 13 females (54%), not statistically significant; 17 (71%) were collected at valley 4 (p = 4.2e-06); and the majority 15 (63%) were sub-adults (p = 9.6e-05) (Table 1). We observed that the mean of leptospires’ shedding in urine was lower in rats with renal biofilm (n = 24; 2.23e+06 GEq), compared with infected rats with no biofilm (n = 41; 2.2e+07 GEq) (p = 2.2e-10) (Table 1). However, generalized linear model (GLM) analysis did not show statistical association between renal biofilm formation and demographic characteristics and leptospiral urine shedding.
We analyzed the association of renal biofilm formation with renal colonization patterns. We observed an average of 229 colonized tubules (CT) count in rats with renal biofilm, while 113 in rats infected but negative for biofilm (p = 0,25) (Table 1). The majority of rats presenting renal biofilm (15; 63%) presented IHC anti-L. interrogans marking pattern of partial to complete occlusion of CTs (p = 0,037) and 21 rats (88%) presented agglomerated CT distribution (p = 0.73) (Table 1). GLM showed that the intensity of colonized tubules increased the chance of renal biofilm formation (Table 1). Furthermore, the partial to complete pattern of tubule colonization increased in more than three times the chance of having renal biofilm (Table 1).
Biofilm matrix is labeled by ruthenium red in scanning electron microscopy
We stained chronically infected rats’ kidneys with ruthenium red (RR) and analyzed by Scanning Electron Microscopy (SEM). We observed dense leptospiral colonization, forming aggregates inside the renal tubules (Fig 2A and 2B–white arrows), with heavy deposition of amorphous extracellular matrix between leptospires, often covering and embedding spirochete bacteria, compatible with biofilm morphology (Fig 2A and 2B–red arrows). When we analyzed the kidneys of infected rats without RR staining, we observed the presence of isolated and agglomerated leptospires inside the renal tubules, without amorphous extracellular matrix in-between and embedded-in (Fig 2C and 2D–white arrows). Finally, we did not observe biofilm nor Leptospira in negative controls (Fig 2E and 2F).
Fig 2. Scanning electron microscopy (SEM) of leptospiral renal biofilm and its matrix in wild naturally colonized Rattus norvegicus.
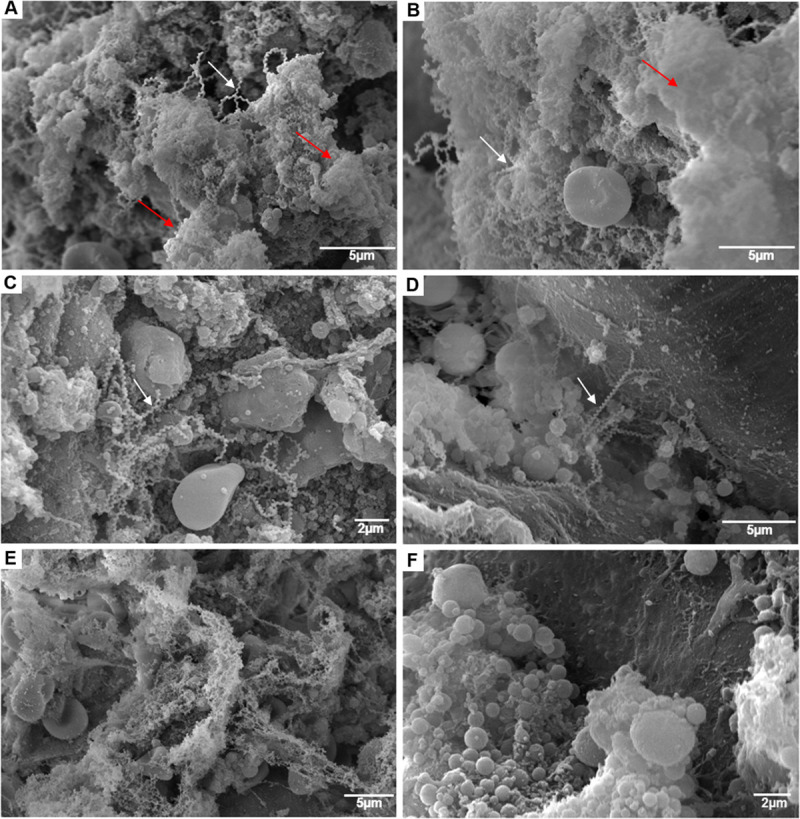
A and B–SEM of colonized kidney with ruthenium red (RR) showed Leptospira agglomerates (white arrows) surrounded by an anionic exopolysaccharidic matrix (red arrows) inside the renal tubules. C and D–SEM of colonized kidney without RR, where leptospires are evidenced agglomerated (C) or isolated (D), without the presence of the matrix. E–SEM using RR of R. norvegicus negative control. F–SEM without RR of R. norvegicus negative control.
Kidneys’ histopathological analyses of R. norvegicus positive for renal biofilm
Histopathological analysis revealed minimal alterations in the kidneys of rats with renal biofilm (Table 2). We observed the occurrence of hyaline-goticular degeneration (n = 12; 50%), mesangial matrix hyperplasia (n = 5; 21%); mesangial hypercellularity (n = 2; 8%), and epithelial tubular regeneration (n = 1; 4%) (Table 2).
Table 2. Histopathological alterations observed in chronically infected Rattus norvegicus with the presence of renal biofilm.
| Alteration | Renal biofilm positive samples No. (%) |
|---|---|
| Total | 24 |
| Mesangial hypercellularity | 2 (8) |
| Mesangial matrix hyperplasia | 5 (21) |
| Hyaline-goticular degeneration | 12 (50) |
| Cylinders | 3 (13) |
| Epithelial tubular regeneration | 1 (4) |
| Minimal glomerular alterations | 3 (13) |
| Focal proliferative glomerulonephritis | 0 |
| Focal mesangial proliferative glomerulonephritis | 0 |
| Focal mesangial segmental proliferative glomerulonephritis | 1 (4) |
| Acute tubular necrosis | 0 |
| Moderate chronic interstitial nephritis | 4 (17) |
| Discrete chronic interstitial nephritis | 7 (29) |
| Kidney within normal parameters | 4 (17) |
Demographic data and leptospiral shedding in rat’s urine are associated with CT count
We observed a homogeneous distribution of infected rats between males (n = 38) and females (n = 40), although uneven among negative rats (8 females and 1 male) (p = 0.03) (Table 3). Infected rats were mainly sub-adult (n = 43, 55%) (p-value = 0.006), collected at valley 4 (n = 44, 56%) (p = 0.24), and presented mean of leptospiral shedding of 1.45e+07 GEq (Table 3). For the characterization of leptospiral colonization patterns, we analyzed 68 rats, which were positive for IHC and/or IF anti-Leptospira (Fig 3). There was a range from 1 to 612 CT count, with an average of 154 CT count (Table 3). Twenty-eight rats (41%) presented IHC anti-L. interrogans marking partial to complete pattern of CT occlusion (Table 3 and Fig 3C), whilst 40 rats (59%) presented marking restricted to the membrane of renal epithelial cells, characterizing partial occlusion of CTs (Table 3 and Fig 3D). Ten rats (15%) presented an isolated distribution pattern (Table 3 and Fig 3E), whereas 58 (85%) were agglomerated (Table 3 and Fig 3F).
Table 3. Demographic characteristics, patterns of renal colonization and Leptospira shedding in wild Rattus norvegicus (Norway rat) naturally infected.
| Leptospira infection | CT count¥ | CT distribution (n = 68)¥ | CT occlusion pattern (n = 68) ¥ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Rats No. | Positive No. (%⁂) | OR (CI) | Mean | OR (CI) | A. No. (%) | I. No. (%) | OR (CI) | P. No. (%) | P/C No. (%) | OR (CI) |
| Total | 87 | 78 (90)* | 154* | 58 (85) | 10 (15) | 40 (59) | 28 (41) | ||||
| Sex categories | |||||||||||
| Intercept | 5.0 (2.3–10.7) | 146.2 (142.3–150.1) | 0.12 (0.04–0.34) | 0.54 (0.28–1.06) | |||||||
| Female | 48 | 40 (83)* | Ref. | 146 | Ref. | 33 (57) | 4 (40) | Ref. | 24 (60) | 13 (46) | Ref. |
| Male | 39 | 38 (97) | 7.6 (0.9–63.6) | 164 | 1.12** (1.08–1.16) | 25 (43) | 6 (60) | 1.98 (0.50–7.77) | 16 (40) | 15 (54) | 1.66 (0.65–4.59) |
| Weight categories | |||||||||||
| Intercept | 1,2E+8 (0–Inf) | 174.4 (167.8–181.2) | 0.07 (0.01–0.54) | 0.87 (0.32–2.41) | |||||||
| Adult | 15 | 15 (100)* | Ref. | 174 | Ref. | 14 (24) | 1 (10) | Ref. | 8 (20) | 7 (25) | Ref. |
| Juvenile | 27 | 20 (74) | 0.00 (0–Inf) | 207 | 1.18** (1.12–1.25) | 10 (17) | 3 (30) | 4.20 (0.38–46.49) | 8 (20) | 5 (18) | 0.71 (0.16–3.23) |
| Sub-adult | 45 | 43 (95) | 0.00 (0–Inf) | 130 | 0.74** (0.71–0.78) | 34 (59) | 6 (60) | 2.47 (0.27–22.44) | 24 (60) | 16 (57) | 0.76 (0.23–2.52) |
| Site of collection | |||||||||||
| Intercept | 1,2E+8 (0–Inf) | 175.1 (167.4–183.1) | 0.10 (0.01–0.78) | 0.83 (0.25–2.73) | |||||||
| Valley 1 | 11 | 11 (100) | Ref. | 175 | Ref. | 10 (17) | 1 (10) | Ref. | 6 (15) | 5 (18) | Ref. |
| Valley 2 | 8 | 6 (75) | 0.00 (0–Inf) | 162 | 0.93 (0.86–1.00) | 5 (9) | 1 (10) | 2.0 (0.1–39.1) | 4 (10) | 2 (7) | 0.60 (0.08–4.76) |
| Valley 4 | 50 | 44 (88) | 0.00 (0–Inf) | 146 | 0.83** (0.79–0.88) | 31 (53) | 5 (50) | 1.61 (0.17–15.5) | 21 (52) | 15 (54) | 0.86 (0.2–3.3) |
| Leptospiral shedding in urine | |||||||||||
| Intercept | 81.5 (75.4–88.1) | 0.70 (0.1–4.0) | 0.26 (0.07–1.0) | ||||||||
| Mean qPCR urine | 7.16 | 1.09** (1.08–1.10) | 5.2 | 4.21 | 0.77 (0.6–1.1) | 3.8 | 5.4 | 1.1 (0.94–1.28) | |||
*Results statistically different (p-value < 0.05);
** Bold items indicating significant OR in GLM analysis (P<0.05);
⁂ Percentage relative to the number rats;
¥Data referring to histopathological analysis of 68 positives rats for IHQ and/or IF anti-Leptospira. Ref.: reference category; refers to the default chosen category against which other categories are compared to in regression models when we use categorical dependent variables. Intercept: regression constant; predicts a linear value when all predictor variables are set to zero.
Abbreviations: OR, Odds Ratios; CI, Confidence intervals; A., Agglomerated; I., Isolated; P., Partial; P/C, Partial to complete.
Fig 3. Patterns of kidney colonization marking of naturally infected Rattus norvegicus.
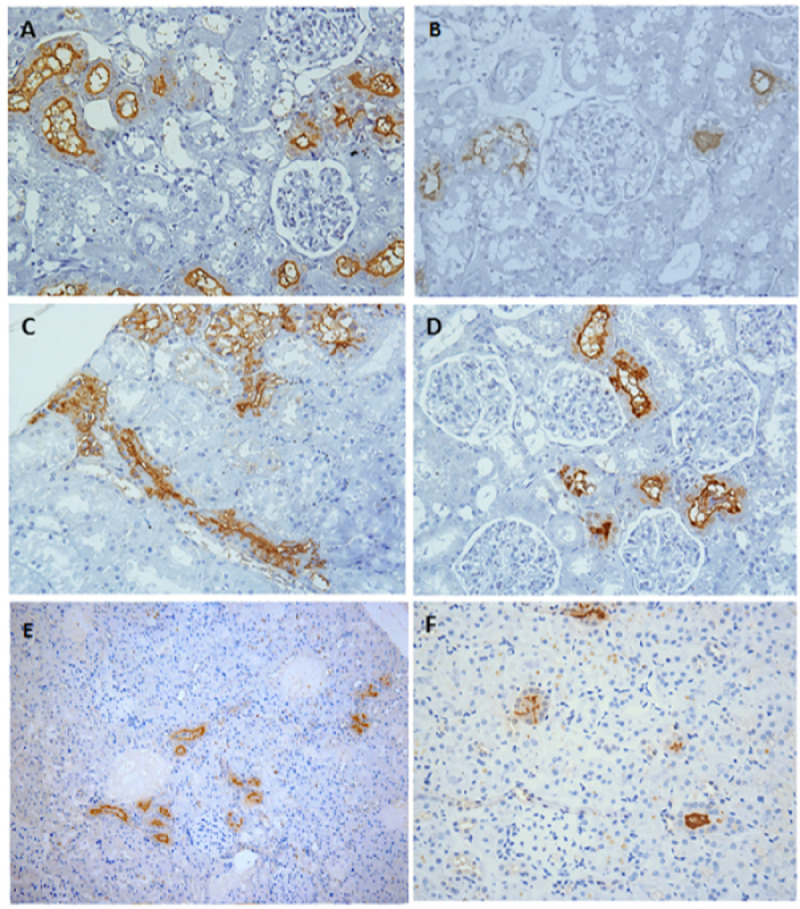
Immunohistochemical representative images of kidney with (A) high intensity of colonized tubules (CTs); (B) low intensity of CTs; (C) partial to complete pattern of CT lumen occlusion; (D) marking restricted to the membrane of renal epithelium (partial occlusion); (E) agglomerated CTs distributed in the renal cortex; (F) isolated CTs distributed in the cortex. A, B, C and D: magnification, x 400. E and F: magnification, x 200.
We used generalized linear models to analyze leptospiral renal colonization patterns associations with demographic data and intensity of leptospiral shedding in the rats’ urine. The intensity of colonized tubules was associated with demographic data and leptospiral shedding in urine. Male and juvenile rats had a higher chance of having more CT count, while rats captured at valley 4 were more likely to have fewer (Table 3). Finally, the greater the intensity of leptospiral shedding in the rats’ urine, the greater is the chance of having more CT count (Table 3).
Discussion
We identified and characterized leptospiral biofilm formation inside the renal tubules of R. norvegicus naturally infected with pathogenic Leptospira. By analyzing demographic data and histological patterns of renal colonization by leptospires, we identified the risk factors associated with biofilm formation. Additionally, we investigated histopathological alterations in the rats’ kidneys positives for biofilm. Finally, we characterized R. norvegicus infection by pathogenic Leptospira, analyzing demographic data, leptospiral shedding intensity in urine, and histological patterns of colonization.
Reservoir hosts infected with Leptospira and experimentally infected rats (chronic model of disease) present dense renal tubules colonization [9,13]. In the present study, from the 65 infected Norway rats analyzed, more than a third (37%) had pathogenic Leptospira forming biofilms inside renal tubules. Biofilm formation by pathogenic bacteria has been described in other hosts. Borrelia burgdorferi, the causative agent of Lyme disease, are spirochetes capable of forming biofilms in vitro, in the midguts of infected ticks, and in the skin tissue of borrelial lymphocytoma patients [28,29,40]. Yersinia pestis, the causative agent of bubonic plague, form biofilm inside the flea gut [41]. In both cases, biofilm formation is described as having a role in the transmission of the pathogen to the accidental host. Additionally, bacterial biofilms are considered colonization factors since they may contribute to bacterial evasion from the immune system [42–44]. Finding Leptospira biofilms in rats’ renal tubules may have implications in Leptospira survival and transmission.
We noticed a positive correlation between the intensity of CT count and biofilm formation, that is, the greater the number of colonized tubules, greater was the chance of renal biofilm formation by pathogenic Leptospira. If we consider the number of CT as a measure of Leptospira population inside the organ, this finding may indicate that biofilm formation in vivo by Leptospira is dependent on leptospiral numbers during renal colonization, suggesting the presence of quorum sensing mechanisms [45]. Moreover, our data showed that Norway rats with renal biofilm excreted ten times less leptospires in the urine than rats positive for renal infection but negative for biofilms, what we believe is a consequence of leptospires maintenance in the renal biofilm. The majority of animals presenting biofilms had a marking pattern of IHC anti-L. interrogans of partial/complete occlusion of CTs, as previously observed for naturally infected rats with no information about biofilm occurrence [11,12]. This complete pattern of renal colonization increased in more than three times the chance of renal biofilm development. Furthermore, 21 Norway rats naturally infected and with renal biofilm had agglomerated pattern of CT. Thus, it is possible to hypothesize that single leptospires infect the kidneys diffusely, coming from the circulatory system, migrate through tissue, multiply and colonize into the proximal renal tubules, where they form cell aggregates and biofilms [13].
In previous in vitro work developed by RISTOW et al. (2008), Leptospira biofilms were analysed by scanning and transmission electron microscopy [15]. The authors observed biofilms formed by a network of leptospires embedded in the extracellular matrix, although they did not use extracellular matrix specific stains. Thibeaux and collaborators (2020) analysed leptospiral charge in urine and kidneys of experimentally infected rodents, by qPCR, but did not explore structural aspects of biofilms formed in vivo [20]. Finally, Yamaguchi and collaborators (2018) used TEM to analyze the renal tubules of infected mice and suggested the presence of biofilm-like structures during tubular colonization by pathogenic leptospires; although they did not use specific dyes for biofilms [13].
We observed that R. norvegicus renal biofilms stained by AB in light turquoise blue but did not stain by MM. SEM using RR revealed the ultrastructure of the tubular biofilms with agglomerates of Leptospira involved in an acidic exopolysaccharidic matrix. Alcian blue, Mayer mucicarmine and ruthenium red are staining methods used to visualize the exopolymeric matrix. AB pH 2.5 stains alginate, acid mucopolysaccharides and sialomucins; MM stains sulfated and carboxylated mucins; and RR stains acid polysaccharides [46–48]. Those stains are commonly used to characterize extracellular matrix of bacterial biofilms, both in in vitro studies [28] and in in vivo studies [27,29,48–50]. A recent in vitro study showed that alginate lyase treatment did not digest the biofilm of Leptospira, suggesting that alginate is not a component of the exopolymeric matrix [20]. Besides, in the transcriptome study of Leptospira biflexa saprophytic species in mature biofilm (48 h) there was downregulation of alginate-related genes [19]. Altogether, those data indicate that the exopolysaccharidic matrix of Leptospira biofilm inside the renal tubules of chronically infected R. norvegicus is composed of acid mucopolysaccharides, but not alginate.
The histopathology of infected rats is well known in the literature [9,51]. Hence, in the present work, we focused on the novelty of the histopathology of R. norvegicus naturally infected with Leptospira and positive for renal biofilm. Histopathological analysis revealed minimal alterations. The most frequent alterations were hyaline-goticular degeneration and chronic inflammation, both occurring in 50% of the rats. Another frequent alteration was the interstitial nephritis (discrete and moderate) that together occurred in 46% of the rats. Those results are in agreement with previous studies of naturally infected R. norvegicus [9,10,52]. In the biofilm phenotype, although bacteria are more sessile and encased in a exopolymeric matrix, it can result in some tissue alterations [53], as we observed in this study. However, since synanthropic R. norvegicus are exposed to many environmental factors, including other pathogens, it was not possible to determine if those kidney alterations were due to the presence of leptospires or their biofilms, or a consequence of the multiple factors the rats are exposed in the environment.
The prevalence of Leptospira in naturally infected R. norvegicus was 90%, in agreement with previous literature for Brazil and other countries (BOEY; SHIOKAWA; RAJEEV, 2019; COSTA et al., 2015b). Leptospiral shedding in urine was 1.45e+07 GEq, in accordance with the previous literature, where a range of 1.0e+05 to 1e+07 leptospires were count per mL of rat urine, by dark field microscopy [54,55]. 97% of the rat males captured and all the adult rats were positive for Leptospira infection. The intensity of colonization varied greatly among infected rats and was associated with demographic data, with the greatest amount of CT in males and juveniles. Male and adult Rattus norvegicus have a social behavior of huddling and fighting, aside from active search for food. Juveniles, on the other side, have a social play behavior in which they learn adult behavior [56]. Those juvenile rats’ behavior could augment the direct contact with contaminated environment, leading to the observed prevalence of infection.
One limitation of our study was that we were not able to directly quantify live leptospires in the rats’ kidneys and urine, but rather estimated Leptospira quantity using qPCR. Another limitation was the small sample of Rattus norvegicus with renal biofilm formation (n = 24). Thus, the prevalence here described should be considered as suggestive, instead of typical.
To date, most of the studies of in vivo biofilms were developed with experimentally infected hosts. Here, we demonstrated that Leptospira interrogans produce renal biofilm during infection in naturally infected Rattus norvegicus captured from an endemic site. This is an important finding on the biology of this host-adapted pathogen. Leptospira biofilm formation in rats’ renal tubules may have implications in bacterial survival and transmission. The biofilm phenotype in animal host reservoirs may probably impact the disease transmission cycle and should be further investigated.
Supporting information
(TIF)
(XLSX)
(PDF)
Acknowledgments
We thank Prof. Jamie Childs, Prof. Albert Ko, and Fleur Porter for contributing to the study design; the staff of the Zoonosis Control Center of Salvador and the Service of Histotechnology of IGM/FIOCRUZ for technical assistance; and Larissa Vasconcelos for the critical reading of the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
AA and PSR were fellows of National Council for Scientific and Technological Development (CNPq - https://www.gov.br/cnpq/pt-br). This study was supported by Oswaldo Cruz Foundation (FIOCRUZ - https://www.bahia.fiocruz.br/), Federal University of Bahia (UFBA - www.ufba.br), the Secretary of Health Surveillance of the State of Bahia (http://www.saude.ba.gov.br/suvisa), the National Institutes of Health (NIH - https://www.nih.gov) (R01 AI052473, R01 TW009504, R25 TW009338), the Wellcome Trust (https://wellcome.org) (102330/Z/13/Z), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol [Internet]. 2009;7(10):736–47. Available from: doi: 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis. 2015;9(9):0–1. doi: 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casanovas-Massana A, Costa F, Riediger IN, Cunha M, de Oliveira D, Mota DC, et al. Spatial and temporal dynamics of pathogenic Leptospira in surface waters from the urban slum environment. Water Res [Internet]. 2018;130:176–84. Available from: doi: 10.1016/j.watres.2017.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reis RB, Ribeiro GS, Felzemburgh RDM, Santana FS, Mohr S, Melendez AXTO, et al. Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl Trop Dis. 2008;2(4):11–8. doi: 10.1371/journal.pntd.0000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casanovas-Massana A, Hamond C, Santos LA, de Oliveira D, Hacker KP, Balassiano I, et al. Leptospira yasudae sp. Nov. and Leptospira stimsonii sp. nov., two new species of the pathogenic group isolated from environmental sources. Int J Syst Evol Microbiol. 2020;70(3):1450–6. doi: 10.1099/ijsem.0.003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019;13(5):25. doi: 10.1371/journal.pntd.0007270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levett PN. Systematics of Leptospiraceae. 2015; [DOI] [PubMed] [Google Scholar]
- 8.Costa F, Elsio A. Wunder J, Oliveira D De, Bisht V, Rodrigues G, Reis MG, et al. Patterns in Leptospira Shedding in Norway Rats (Rattus norvegicus) from Brazilian Slum Communities at High Risk of Disease Transmission. PLoS Negl Trop Dis [Internet]. 2015;9(6):e0003819. Available from: doi: 10.1371/journal.pntd.0003819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucunduva de Faria M, Athanazio D a., Gonçalves Ramos EA, Silva EF, Reis MG, Ko AI. Morphological Alterations in the Kidney of Rats with Natural and Experimental Leptospira Infection. J Comp Pathol. 2007;137(4):231–8. doi: 10.1016/j.jcpa.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Agudelo-flórez P, Murillo VE, Londoño AF, Rodas JD. Histopathological kidney alterations in rats naturally infected with Leptospira. 2013;33:82–8. [PubMed] [Google Scholar]
- 11.Athanazio D a., Silva EF, Santos CS, Rocha GM, Vannier-Santos MA, McBride AJA a, et al. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Trop. 2008;105(2):176–80. doi: 10.1016/j.actatropica.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 12.Santos AAN, Figueira CP, Reis MG dos, Costa F, Ristow P. Heterogenic colonization patterns by Leptospira interrogans in Rattus norvegicus from urban slums. Brazilian J Microbiol. 2015;46(4):1161–4. doi: 10.1590/S1517-838246420140873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi T, Higa N, Okura N, Matsumoto A, Hermawan I, Yamashiro T, et al. Characterizing interactions of Leptospira interrogans with proximal renal tubule epithelial cells. BMC Microbiol. 2018;18(1):1–11. doi: 10.1186/s12866-017-1144-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratet G, Veyrier FJ, Fanton d’Andon M, Kammerscheit X, Nicola M-A, Picardeau M, et al. Live Imaging of Bioluminescent Leptospira interrogans in Mice Reveals Renal Colonization as a Stealth Escape from the Blood Defenses and Antibiotics. PLoS Negl Trop Dis [Internet]. 2014;8(12):e3359. Available from: doi: 10.1371/journal.pntd.0003359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristow P, Bourhy P, Kerneis S, Schmitt C, Prevost MC, Lilenbaum W, et al. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology. 2008;154(5):1309–17. doi: 10.1099/mic.0.2007/014746-0 [DOI] [PubMed] [Google Scholar]
- 16.Kumar KV, Lall C, Raj RV, Vedhagiri K, Vijayachari P. Coexistence and survival of pathogenic leptospires by formation of biofilm with Azospirillum. FEMS Microbiol Ecol [Internet]. 2015;91(6):1–27. Available from: 10.1093/femsec/fiv051 [DOI] [PubMed] [Google Scholar]
- 17.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial Biofilms. Annu Rev Microbiol. 1995;49(June 2017):711–45. doi: 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- 18.Flemming H, Wingender J. The biofilm matrix. Nat Rev Microbiol [Internet]. 2010;8(9):623–33. Available from: 10.1038/nrmicro2415\nhttp://www.ncbi.nlm.nih.gov/pubmed/20676145 [DOI] [PubMed] [Google Scholar]
- 19.Iraola G, Spangenberg L, Lopes Bastos B, Graña M, Vasconcelos L, Almeida Á, et al. Transcriptome Sequencing Reveals Wide Expression Reprogramming of Basal and Unknown Genes in Leptospira biflexa Biofilms. mSphere [Internet]. 2016;1(2):1–16. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27303713%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4863578 doi: 10.1128/mSphere.00042-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibeaux R, Soupé-Gilbert ME, Kainiu M, Girault D, Bierque E, Fernandes J, et al. The zoonotic pathogen Leptospira interrogans mitigates environmental stress through cyclic-di-GMP-controlled biofilm production. npj Biofilms Microbiomes [Internet]. 2020;6(1):1–11. Available from: doi: 10.1038/s41522-020-0134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol [Internet]. 2016;14(9):563–75. Available from: http://www.nature.com/doifinder/10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- 22.Parsek MR, Singh PK. Bacterial Biofilms: An Emerging Link to Disease Pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- 23.Costerton JW. Bacterial bofilms: a common cause of persistent infections. Science (80-). 1999;284(1999):1318–22. [DOI] [PubMed] [Google Scholar]
- 24.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11(7):1034–43. doi: 10.1111/j.1462-5822.2009.01323.x [DOI] [PubMed] [Google Scholar]
- 25.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;(136):1–51. doi: 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- 26.Del Pozo JL. Biofilm-related disease. Expert Rev Anti Infect Ther [Internet]. 2018;16(1):51–65. Available from: doi: 10.1080/14787210.2018.1417036 [DOI] [PubMed] [Google Scholar]
- 27.Schaber JA, Triffo WJ, Sang JS, Oliver JW, Hastert MC, Griswold JA, et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect Immun. 2007;75(8):3715–21. doi: 10.1128/IAI.00586-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapi E, Bastian SL, Mpoy CM, Scott S, Rattelle A, Pabbati N, et al. Characterization of Biofilm Formation by Borrelia burgdorferi In Vitro. PLoS One. 2012;7(10):1–11. doi: 10.1371/journal.pone.0048277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sapi E, Balasubramanian K, Poruri A, Maghsoudlou JS, Socarras KM, Timmaraju A V., et al. Evidence of in vivo existence of Borrelia biofilm in borrelial lymphocytomas. Eur J Microbiol Immunol. 2016;6(1):9–24. doi: 10.1556/1886.2015.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008Jan;322:229–48. doi: 10.1007/978-3-540-75418-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes-Solecki M, Santecchia I, Werts C. Animal models of leptospirosis: Of mice and hamsters. Front Immunol. 2017;8(FEB). doi: 10.3389/fimmu.2017.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himsworth CG, Bidulka J, Parsons KL, Feng AYT, Tang P, Jardine CM, et al. Ecology of Leptospira interrogans in Norway Rats (Rattus norvegicus) in an Inner-City Neighborhood of Vancouver, Canada. PLoS Negl Trop Dis. 2013;7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felzemburgh RDM, Ribeiro GS, Costa F, Reis RB, Hagan JE, Melendez AXTO, et al. Prospective Study of Leptospirosis Transmission in an Urban Slum Community: Role of Poor Environment in Repeated Exposures to the Leptospira Agent. PLoS Negl Trop Dis. 2014;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maciel EAP, de Carvalho ALF, Nascimento SF, de Matos RB, Gouveia EL, Reis MG, et al. Household transmission of Leptospira infection in urban slum communities. PLoS Negl Trop Dis. 2008;2(1). doi: 10.1371/journal.pntd.0000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa F, Ribeiro GS, Felzemburgh RDM, Santos N, Reis RB, Santos AC, et al. Influence of Household Rat Infestation on Leptospira Transmission in the Urban Slum Environment. PLoS Negl Trop Dis [Internet]. 2014;8(12):e3338. Available from: doi: 10.1371/journal.pntd.0003338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoddard R a., Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis [Internet]. 2009;64(3):247–55. Available from: 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 37.Chagas-Junior AD, da Silva CLR, Soares LM, Santos CS, Silva CDC, Athanazio D a., et al. Detection and quantification of leptospira interrogans in hamster and rat kidney samples: Immunofluorescent imprints versus real-time PCR. PLoS One. 2012;7(2):1–5. doi: 10.1371/journal.pone.0032712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croda J, Figueira CP, Wunder E a., Santos CS, Reis MG, Ko AI, et al. Targeted mutagenesis in pathogenic Leptospira species: Disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun. 2008;76:5826–33. doi: 10.1128/IAI.00989-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris R. An Introduction to R. Quant Geogr Basics. 2018;3:250–86. [Google Scholar]
- 40.Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, et al. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest. 2009;119(12):3652–65. doi: 10.1172/JCI39401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190(4):783–92. doi: 10.1086/422695 [DOI] [PubMed] [Google Scholar]
- 42.Watters C, Fleming D, Bishop D, Rumbaugh KP. Host Responses to Biofilm [Internet]. Vol. 142, Progress in Molecular Biology and Translational Science. Elsevier Inc.; 2016. 193–239 p. Available from: 10.1016/bs.pmbts.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 43.Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. How Biofilms Evade Host Defenses. Microbiol Spectr. 2015;3(3):1–10. doi: 10.1128/microbiolspec.MDNA3-0047-2014 [DOI] [PubMed] [Google Scholar]
- 44.Salyers A a., Wilson B a., Whitt DD, Winkler M. Bacterial Pathogenesis; A Molecular Approach. 2010. 150–151 p. [Google Scholar]
- 45.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73(2):310–47. doi: 10.1128/MMBR.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carson FL, Hladik C. Histotechnology A Self-Instructional Text. 3rd ed.American Society for Clinical Pathology; 2009. [Google Scholar]
- 47.Fassel TA, Edmiston CE. Bacterial biofilms: Strategies for preparing glycocalyx for electron microscopy. Methods Enzymol. 1999;310(Dic):194–203. doi: 10.1016/s0076-6879(99)10017-x [DOI] [PubMed] [Google Scholar]
- 48.Hoffmann N, Rasmussen TBT, Jensen PP, Stub C, Hentzer M, Molin S, et al. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect Immun [Internet]. 2005;73(4):2504–14. Available from: http://iai.asm.org/content/73/4/2504.abstract%5Cnhttp://iai.asm.org/cgi/content/abstract/73/4/2504 doi: 10.1128/IAI.73.4.2504-2514.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44(6):547–58. doi: 10.1002/ppul.21011 [DOI] [PubMed] [Google Scholar]
- 50.Fulcher TP, Dart JKG, McLaughlin-Borlace L, Howes R, Matheson M, Cree I. Demonstration of biofilm in infectious crystalline keratopathy using ruthenium red and electron microscopy. Ophthalmology. 2001;108(6):1088–92. doi: 10.1016/s0161-6420(01)00561-9 [DOI] [PubMed] [Google Scholar]
- 51.Faria MT de, Calderwood MS, Athanazio DA, McBride AJA, Hartskeerl RA, Pereira MM, et al. Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta Trop. 2008;108(1):1–5. doi: 10.1016/j.actatropica.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chagas-Junior AD, McBride AJ a, Athanazio D a., Figueira CP, Medeiros M a., Reis MG, et al. An imprint method for detecting leptospires in the hamster model of vaccine-mediated immunity for leptospirosis. J Med Microbiol. 2009;58(12):1632–7. doi: 10.1099/jmm.0.014050-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9(2). doi: 10.3390/antibiotics9020059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monahan AM, Callanan JJ, Nally JE. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect Immun. 2008;76(11):4952–8. doi: 10.1128/IAI.00511-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nally JE, Chow E, Fishbein MC, Blanco DR, Lovett M a. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect Immun. 2005;73(6):3251–60. doi: 10.1128/IAI.73.6.3251-3260.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schweinfurth MK. The social life of norway rats (Rattus norvegicus). Elife. 2020;9:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


