Abstract
The activation of the cysteine proteases with aspartate specificity, termed caspases, is of fundamental importance for the execution of programmed cell death. These proteases are highly specific in their action and activate or inhibit a variety of key protein molecules in the cell. Here, we study the effect of apoptosis on the integrity of two proteins that have critical roles in DNA damage signalling, cell cycle checkpoint controls, and genome maintenance—the product of the gene defective in ataxia telangiectasia, ATM, and the related protein ATR. We find that ATM but not ATR is specifically cleaved in cells induced to undergo apoptosis by a variety of stimuli. We establish that ATM cleavage in vivo is dependent on caspases, reveal that ATM is an efficient substrate for caspase 3 but not caspase 6 in vitro, and show that the in vitro caspase 3 cleavage pattern mirrors that in cells undergoing apoptosis. Strikingly, apoptotic cleavage of ATM in vivo abrogates its protein kinase activity against p53 but has no apparent effect on the DNA binding properties of ATM. These data suggest that the cleavage of ATM during apoptosis generates a kinase-inactive protein that acts, through its DNA binding ability, in a trans-dominant-negative fashion to prevent DNA repair and DNA damage signalling.
Apoptosis is of fundamental importance for the homeostasis and development of metazoans (22). This process of cell death occurs in response to a plethora of stimuli (45) and results in distinct cellular features, from the morphological through to the molecular level (1, 11). The regulation of this process is one of high orchestration, and its deregulation has been shown to contribute to a variety of disease states, including cancer and neurodegenerative disorders (43). Key components of the apoptotic machinery have come to light over the last decade or so, with the genetics and cell biology of the nematode Caenorhabditis elegans being a tour de force for their identification (12). One key discovery from studies of this organism was the identification of the Ced-3 gene, whose product was found to be related in sequence to the interleukin 1β-converting enzyme protease (50). This then led to the identification of mammalian interleukin 1β-converting enzyme-like proteases (33, 42). These proteases are now termed caspases (cysteinyl aspartate-specific proteinases), and there are at present 14 mammalian proteases belonging to this family that are believed to be involved in apoptosis (11, 34). Two of these, caspase 3 (CPP32) and caspase 6 (Mch2), have been shown to be the major active caspases of apoptotic cells (14) and have been described as the executioner caspases of apoptotic cell death (31).
To fully understand the function of the caspases, it is of fundamental importance to identify their downstream targets. Despite caspases having been known for several years, targets for the executioner caspases have remained elusive. To date, around 20 targets for caspase 3 and caspase 6 have been identified (11, 34). This lack of substrate identification may be linked to the fact that the caspases are highly specific in their targeting of proteins and appear to cleave only critical components involved in maintaining the integrity of the cell rather than cleaving proteins in a random and inefficient manner. One characteristic of the caspases is that they perform proteolysis at a limited number of sites within their targets and do not totally degrade the protein substrate (36). Two critical proteins involved in DNA repair and DNA damage signalling that have been identified as targets of caspase 3 are poly-(ADP-ribose) polymerase (PARP) (26, 29) and the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) (7, 17, 40). However, it is apparent from cell lines deficient in caspase 3 that other caspases are able to perform these cleavage events in vivo (23, 48). Since nuclear DNA is cleaved during apoptosis by the caspase-activated deoxyribonuclease (CAD) (13), the inhibition of the DNA break-dependent catalytic activities of these two highly abundant enzymes makes not only energetic sense for the dying cell but might inhibit both signalling from and repair processes at the site(s) of damaged DNA.
In light of the facts described above, we have studied the effects of apoptosis on the integrity of two mammalian DNA-PKcs homologues that have been shown to be involved in the maintenance of genomic integrity and in DNA damage detection and its signalling. Thus, we have examined the product of the gene defective in ataxia telangiectasia (A-T), ATM (ataxia telangiectasia mutated) (35, 37), and its relative ATR (ATM related) (9) in cells undergoing apoptosis. A-T is a human autosomal recessive disorder. Characteristics of this disease are the debilitating symptoms of ataxia resulting from cerebellar degeneration, oculocutaneous telangiectasia, immune deficiency, aspects of premature aging, and increased sensitivity to ionizing radiation (IR) (19, 20, 32, 38). A-T cells (both human and those derived from Atm knockout mice) show a high level of chromosomal instability, radioresistant DNA synthesis, and hypersensitivity to IR and radiomimetic agents. A-T cells also display a defective G1/S cell cycle checkpoint after IR-induced DNA damage through, in part, a loss of the ability to signal effectively to p53 (25, 27, 30, 39, 49). Indeed, very recent findings show that ATM is able to mediate the phosphorylation of p53 (2,6). Furthermore, A-T cells have been recently shown to be debilitated in the repair of DNA double-strand breaks (15). Cloning the ATM gene led to the exciting discovery that it encodes a phosphatidylinositol 3-kinase-like protein of approximately 350 kDa (35, 37). Of particular note is that ATM displays high homology across its kinase domain to several other proteins shown or proposed to be involved in maintaining genomic stability (21, 51). This subfamily of phosphatidylinositol 3-kinase-like proteins includes the double-strand DNA break repair protein DNA-PKcs, the Saccharomyces cerevisiae cell cycle checkpoint regulatory protein Mec1p, its Schizosaccharomyces pombe homologue Rad3, and its human homologue ATR (also termed FRP1) (9, 21, 51). Consistent with the phenotypes of yeast defective in Mec1p or Rad3, recent work has shown that ATR functions in DNA damage-induced cell cycle checkpoint processes in mammalian cells (10).
Here, we show that ATM but not ATR is proteolytically cleaved in cells induced to undergo apoptosis by a variety of agents and identify the major caspase 3 cleavage site in ATM. Furthermore, we show that cleavage by caspase 3 does not abrogate ATM’s ability to bind DNA but does affect its ability to phosphorylate p53. Our data not only provide the identification of another protein cleaved during apoptosis but, for the first time, identify an apoptotic cleavage target that has been unequivocally shown to be involved in the signalling of DNA damage to the checkpoint machinery. This strongly supports the argument that the repair and signalling of DNA damage induced during apoptosis are specifically inactivated in apoptotic cells by the action of cell death proteases.
MATERIALS AND METHODS
Reagents.
Dimethyl sulfoxide, phenylmethylsulfonyl fluoride (PMSF), 1-1-chloro-3-(4-tosylamido)-7-amino-2-heptone (TLCK), antipain, E-64, leupeptin, etoposide, bleomycin, EGTA, ethanol, and cycloheximide were purchased from Sigma. Tumor necrosis factor alpha (TNF-α) was obtained from Peprotech EC Ltd. (London, England). Staurosporin and Z-Val-Ala-Asp (OMe)-fluoromethylketone (ZVAD-FMK) were purchased from Alexsis Corporation. Antibodies ATM-B and ATM-V and monoclonal antibodies 2C1 and SYR10G31 have been described previously (2, 8, 28). The polyclonal rabbit antiserum ATM-N was raised against a recombinant ATM fragment comprising amino acid residues 1 to 306. The rabbit antiserum ATR-A was raised against a recombinant ATR fragment comprising amino acid residues 2122 to 2644. The monoclonal anti-glutathione S-transferase (GST) antibody (B-14) was purchased from Santa Cruz, and the anti-PARP monoclonal antibody (isotype immunoglobulin G1) was purchased from Serotec, Oxford, United Kingdom. Anti-lamin B antisera was a gift from J. Pines, Cambridge University, Cambridge, United Kingdom.
Cells and extraction and analysis of apoptotic DNA.
HL60 cells, human lymphoblastoid cells (GM02184D), and A-T lymphoblastoid cells (GM00718B) were maintained in RPMI 1640 media supplemented with 15% fetal calf serum (Sigma), 100 IU of penicillin per ml, and 10 IU of streptomycin (Sigma) per ml at 37°C with 7% CO2. HeLa cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Sigma), 100 IU of penicillin per ml, and 10 IU of streptomycin per ml at 37°C with 7% CO2. For the induction of apoptosis, HL60 cells were treated with 68 μM etoposide for various times or with 5 μM STS for 5 h, 100 μg of bleomycin per ml for 6 h, 5% ethanol for 4 h, or 5 mM EGTA for 5 h. Apoptosis in HeLa cells was induced with TNF-α (10 ng/ml) and cycloheximide (20 μg/ml). For inhibitor studies, HL60 cells were grown in the presence of an inhibitor and etoposide for 4 h prior to harvesting. DNA was extracted from cells undergoing apoptosis by using the reagent DNAzol (Gibco-BRL) as described by the manufacturer. Isolated DNA (20 μg for each time point) was electrophoresed on a 1.5% agarose gel in Tris-acetate-EDTA buffer at 40 mA. Fluorescence-activated cell sorting (FACS) analysis was performed with a Becton Dickinson FACSort apparatus to determine sub-G1 chromosomal DNA levels as an indicator of the apoptotic population. Cells were harvested, washed twice in phosphate-buffered saline (PBS), and fixed with ice-cold 70% methanol overnight. Cells were then washed in PBS and suspended in PBS containing 1% Tween 20, 10 μg of RNase A per ml, and 25 μg of propidium iodide per ml, prior to FACS analysis.
Nuclear extract preparation and partial purification of ATM.
Approximately 5 × 107 HL60 cells or ∼1 × 107 HeLa cells, control cells or those induced to undergo apoptosis, were used as starting material to prepare nuclear extracts. An enriched pool of ATM, termed ATM-Q, was purified from 10 ml (200 mg) of HeLa nuclear extract (Computer Cell Culture Centre, Mons, Belgium) by Q-Sepharose (Pharmacia) anion-exchange chromatography. Nuclear extract was diluted twofold in buffer A (20 mM HEPES [pH 7.6], 10% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF) before being applied to a 20- by 1.5-cm Q-Sepharose column previously equilibrated in buffer A containing 50 mM KCl. After the extract was loaded and washed with 5 column volumes of buffer A containing 50 mM KCl, a 10-column volume gradient of 50 to 500 mM KCl in buffer A was applied. ATM fractions, judged by Western blot analysis, were obtained at KCl concentrations between 150 and 200 mM, pooled, dialyzed against buffer A containing 50 mM KCl, and stored at −80°C.
Caspase 3 expression and purification.
Recombinant His-tagged caspase 3 was purified on Ni2+-nitrilotriacetic acid agarose. Caspase 6 was purchased from Pharmingen.
In vitro cleavage of ATM and ATM fragments.
Partially purified ATM (see above), recombinant GST-ATM-caspase 3 site (CS) (see below), or in vitro-translated ATM fragments (see below) were incubated with various amounts of caspase 3 or caspase 6 in a final buffer composition of 20 mM HEPES (pH 7.6), 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 20 mM dithiothreitol, and 5 mM EDTA at 37°C for 30 min. Reactions were terminated by the addition of an equal volume of sodium dodecyl sulfate (SDS)-protein sample buffer to the reaction mixture.
Recombinant ATM domain production and purification.
The regions of ATM containing potential caspase 3 cleavage sites (comprising amino acid residues 727 to 859 or 819 to 939) were amplified from the cDNA with the following primers: for amino acids 727 to 859, 5′GGATCCTAATACGACTCACTATAGGAACAGACCACCATGGGCTGCTACTGTTACATG-3′ and 5′TCAGTTAAATAGATTCATGGA-3′; for amino acids 819 to 939, 5′GGATCCTAATACGAC TCAC TATAGGAACAGACCACCATGTG TAAAAG T T TAGCATCC - 3 ′ and 5′TCAGGTAGGTTCTAGCGTGCT-3′. The amplified cDNAs were placed directly into a rabbit reticulocyte in vitro transcription and translation system (L1170) (Promega) to produce the relevant ATM fragments. For bacterial expression, the region of ATM containing the potential caspase 3 cleavage sites (amino acids 721 to 939) was amplified from the cDNA, cloned into the bacterial expression plasmid pGEX-TK, and expressed and purified as a GST fusion protein. This construct is termed GST-ATM-CS. Primers used to amplify this region were 5′-CATCGGATCCATTACAAATTCAGAAACT-3′ and 5′-GATGTGCTCGAGTCAAACATCTTGGTCACGACG-3′.
Western blotting and immunoprecipitation.
For in vivo analysis of ATM and ATR cleavage during apoptosis, a nuclear extract (50 μg) was subjected to electrophoresis on SDS–7% polyacrylamide gels followed by transfer to nitrocellulose membranes. GST-ATM-CS and in vitro-translated products were resolved on 12 and 18% gels, respectively. Western blots were developed with the ECL reagents (Amersham). Partially purified ATM was cleaved or mock treated with caspase 3 prior to incubation on ice for 1 h with the N-terminal directed antiserum ATM-N or preimmune serum. Protein A-Sepharose (25 μl), equilibrated in Tris-buffered saline (TBS) containing 0.1% Nonidet P-40 (NP-40), was then added, followed by a further 1-h incubation on ice, before the Sepharose beads were washed 10 times with 1 ml of TBS containing 0.1% NP-40. Pellets were resuspended in an equal volume of SDS-protein sample buffer, resolved on an SDS–7% polyacrylamide gel, transferred to nitrocellulose, and probed with the monoclonal antibody 2C1 directed to the kinase domain region of ATM (9).
ATM DNA binding and p53 kinase assays.
Partially purified ATM (200 μg) was mock treated or treated with caspase 3 prior to DNA binding by using double-stranded DNA (50 bp) coupled via a 5′ biotin group to streptavidin-coated iron-oxide particles as described elsewhere (39a). Protein was visualized by Western blotting with antiserum ATM-B or ATM-N. The ATM kinase assay was performed by using nuclear extracts prepared from HeLa, HL60, or etoposide (68 μM)-treated HL60 cells (2). Baculovirus-expressed and purified human p53 (100 ng) was used as the substrate. Phosphorimage analysis and quantification was performed with a Fuji BAS-2500 phosphorimager.
Microsequencing.
Recombinant GST-ATM-CS (10 μg) was incubated with 500 ng of caspase 3 for 30 min at 37°C. Samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 12% gel prior to being transferred onto a polyvinylidine difluoride membrane. The membrane was stained with 0.1% Coomassie blue R250 in 50% methanol and 1% acetic acid. After destaining with 50% methanol, a band of approximately 10 kDa, not present in the uncut control or samples with caspase 3 alone, was excised and sequenced by Edman degradation by using an Applied Biosystems Procise sequencer according to the manufacturer’s protocols.
RESULTS
ATM cleavage in HL60 cells treated with etoposide.
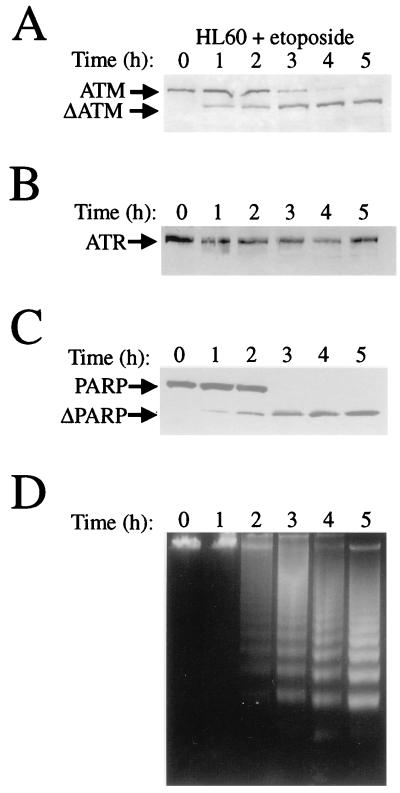
To study whether ATM and/or ATR is proteolytically cleaved during apoptosis, we induced HL60 cells to undergo apoptosis with the topoisomerase II inhibitor etoposide. The HL60 apoptotic system has been well characterized previously (16, 17). ATM and ATR protein integrity was followed by Western blot analysis of nuclear extracts prepared from HL60 cells that had been treated with drug over a time course of 5 h. Polyclonal antibodies raised against ATM (ATM-B [28]) or ATR (see Materials and Methods) were used. The former antibody was raised against a C-terminal portion of ATM (amino acid residues 1980 to 2337), and the ATR antibody was raised against the kinase region of ATR (amino acid residues 2122 to 2644). As shown in Fig. 1A, the ATM protein becomes shifted to a faster migrating product, ΔATM, in cells undergoing apoptosis. This mobility shift appears to start before the onset of the DNA laddering, a classic characteristic of the apoptotic response (Fig. 1D). FACS analysis was used to quantify the population of cells that had chromosomal DNA sub-G1 as an indicator of the population of cells undergoing apoptosis. This revealed that, after 1 h of etoposide treatment, 4% of the cells were undergoing apoptosis, after 2 h 14% of the cells were apoptotic, after 3 h 61% of the cells were apoptotic, after 4 h 76% of the cells were apoptotic, and after 5 h 79% of the cells were apoptotic. Similar to what occurs with PARP cleavage (Fig. 1C), ATM appears to be degraded as early as 1 h after the induction of apoptosis with etoposide. In stark contrast, the ATR protein is not altered in its mobility, as determined by Western blotting (Fig. 1B). Therefore, ATM but not ATR exhibits an altered electrophoretic mobility upon the induction of apoptosis by etoposide in the HL60 system.
FIG. 1.
ATM, but not ATR, is cleaved during apoptosis in HL60 cells treated with etoposide. HL60 cells were grown in the presence of 68 μM etoposide for the times indicated. Cells were harvested, nuclear extracts were prepared, and protein (50 μg for each lane) was subjected to Western blot analysis with polyclonal rabbit anti-ATM antiserum ATM-B (A), polyclonal rabbit anti-ATR antiserum ATR-A (B), or monoclonal anti-PARP antibody PARP (C). (D) Time course of genomic DNA fragmentation in HL60 cells treated with 68 μM etoposide and visualized by ethidium bromide staining after agarose gel electrophoresis (1.5%).
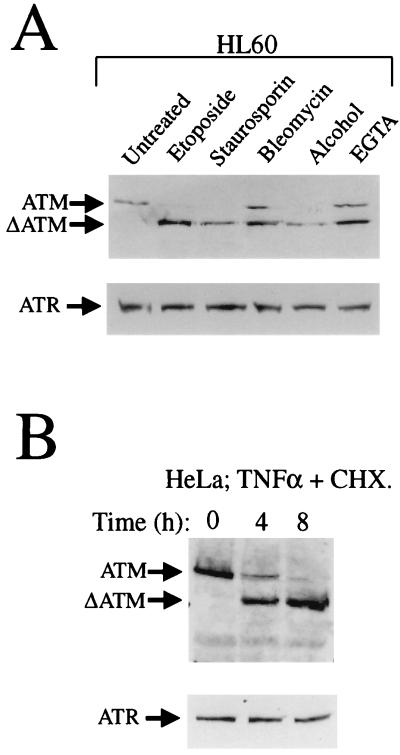
ATM cleavage occurs in HL60 and HeLa cells treated with any of a variety of apoptotic inducers.
The studies described above utilized etoposide, which results in the generation of DNA double-strand breaks. To investigate whether ATM is cleaved during apoptotic responses that are initiated by other agents, we treated HL60 cells with a variety of insults that have been reported to act as apoptotic inducers in this cell line. The results of these studies revealed that ATM is cleaved to a breakdown product similar to that induced by etoposide (ΔATM) when staurosporin, bleomycin, alcohol, or EGTA is employed (Fig. 2A). FACS analysis was used to quantify the population of cells that had chromosomal DNA sub-G1, as an indicator of the population of cells undergoing apoptosis by these different inducing agents. This revealed that for staurosporin treatment 61% of the cells were apoptotic, for bleomycin treatment 37% of the cells were apoptotic, for alcohol treatment 69% of the cells were apoptotic, and for EGTA treatment 22% of the cells were apoptotic. The cleavage of ATM is not unique to HL60 cells, since HeLa cells treated with TNF-α and cycloheximide also exhibit cleavage of the ATM protein to its faster migrating form, ΔATM (Fig. 2B). For the HeLa cell treatment, FACS analysis revealed that for the 4- and 8-h time points the population was 12 and 51% apoptotic, respectively, judged by the chromosomal DNA that was sub-G1. In contrast, ATR is not cleaved detectably under any of the apoptosis-inducing conditions that we have tested (Fig. 2A and B, lower panels).
FIG. 2.
ATM, but not ATR, is cleaved in HL60 cells induced to undergo apoptosis by any of a variety of agents and in apoptotic HeLa cells. (A) Cleavage of ATM during HL60 cell apoptosis induced by etoposide (68 μM for 4 h), staurosporin (5 μM for 4 h), bleomycin (100 μg/ml for 6 h), alcohol (5% for 4 h), or EGTA (5 mM for 5 h), as indicated. Nuclear extracts were prepared and 50 μg of protein was loaded per lane. (B) Cleavage of ATM but not ATR in HeLa cells treated with TNF-α (10 ng/ml) and cycloheximide (CHX; 20 μM) for the times shown. Nuclear extracts were prepared, and 50 μg of protein was loaded per lane.
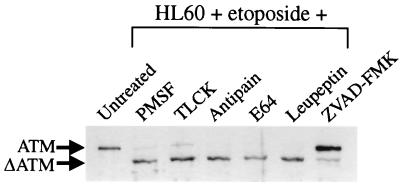
Inhibition of ATM proteolysis in vivo.
In order to make an initial identification of the class of protease involved in ATM cleavage, we grew etoposide-treated HL60 cells in the presence of a range of protease inhibitors (Fig. 3). Most notably, we found that the cell-permeable caspase inhibitor ZVAD-FMK was effective in inhibiting ATM breakdown. In contrast, inhibitors of other classes of protease had little or no effect. Treatment of the cells with etoposide and ZVAD-FMK resulted in only 8% of the population being apoptotic, while the population was over 70% apoptotic for all other treatments. Although these studies do not indicate exactly what protease(s) is involved in the cleavage of ATM, they suggest strongly that the protease is an apoptotic cysteine protease and serve as the basis for the more detailed studies described below.
FIG. 3.
ATM cleavage is inhibited in vivo by the caspase inhibitor ZVAD-FMK. HL60 cells were treated with 68 μM etoposide for 4 h in the presence of the following protease inhibitors: 200 μM PMSF, 200 μM TLCK, 100 μM antipain, 200 μM E64, 200 μM leupeptin, and 20 μM ZVAD-FMK. Nuclear extracts were prepared, and 50 μg of protein was loaded per lane.
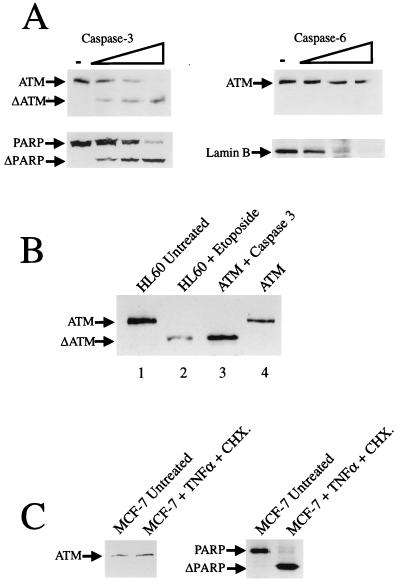
Caspase 3 but not caspase 6 cleaves ATM efficiently in vitro.
It has been demonstrated previously that caspase 3 and caspase 6 are the two most prevalent proteases activated during apoptosis that cleave critical cellular proteins (14). These caspases have therefore been described as the executioners of apoptosis. However, it must be borne in mind from studies utilizing cells defective in caspase 3 that other caspases are carrying out cleavage events during apoptosis (23, 48). Taking this information together with our data on ATM proteolysis in vivo, we asked whether partially purified ATM can be cleaved by recombinant caspase 3 or caspase 6 in vitro and, if so, whether the pattern of degradation seen in vitro mirrors that observed in vivo? As shown in Fig. 4A, partially purified ATM is cleaved efficiently by caspase 3. The efficiency of cleavage by caspase 3 of ATM is similar to that observed when PARP is cleaved by this enzyme in vitro (Fig. 4A, lower panel). However, unlike lamin B, caspase 6 does not cleave ATM under our assay conditions (Fig. 4A). Notably, additional analyses reveal that, as is the case for ATM cleavage in vitro by caspase 3, the cleavage of ATM in HL60 cells undergoing apoptosis in vivo also yields the major product, ΔATM (Fig. 4B). These data therefore strongly suggest that caspase 3 or a caspase 3-like enzyme is the active ATM protease in cells undergoing programmed cell death. To try to support this suggestion we looked at ATM cleavage in the caspase 3-deficient MCF-7 cell line. As shown in Fig. 4C, ATM is not cleaved in MCF-7 cells treated with TNF-α and cycloheximide to undergo apoptosis, while as has been reported previously, PARP is cleaved (23).
FIG. 4.
(A) Caspase 3, but not caspase 6, cleaves partially purified ATM in vitro. Partially purified ATM pool (ATM-Q; 50 μg of total protein) was incubated with no caspase (−) or increasing amounts (33, 100, or 300 ng) of caspase 3 or caspase 6 for 30 min at 37°C. The cleavage of PARP by caspase 3 is shown in the left, lower panel, and the cleavage of lamin B by caspase 6 is shown in the right, lower panel. (B) Caspase 3-mediated cleavage of ATM in vitro yields the same proteolytic degradation pattern as that seen in HL60 cells undergoing apoptosis. Lane 1, 50 μg of nuclear extract from untreated HL60 cells; lane 2, 25 μg of nuclear extract from HL60 cells treated with 68 μM etoposide for 5 h; lane 3, 20 μg of partially purified ATM (ATM-Q) incubated with 300 ng of caspase 3; lane 4, 20 μg of untreated partially purified ATM (ATM-Q). Western blot analysis was performed with anti-ATM antiserum ATM-B. (C) The caspase 3-deficient cell line MCF-7 does not cleave ATM after treatment with TNF-α (30 ng/ml) and cycloheximide (CHX) (10 μg/ml) for 20 h. Cleavage of PARP in the apoptotic MCF-7 cells is shown in the right-hand panel.
Antibody mapping of ATM fragments generated by caspase 3.
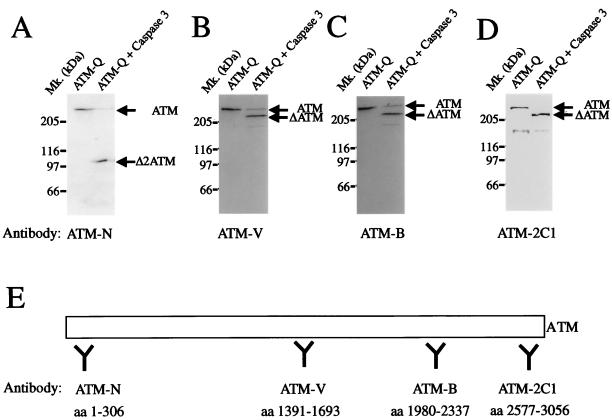
In an initial attempt to identify the major site of ATM cleavage by caspase 3, we treated partially purified ATM with caspase 3 and subjected the products to Western blot analysis with antibodies to various regions of the protein. By using antiserum ATM-N, which was raised against an N-terminal region of ATM (amino acid residues 1 to 306), a major product of 100 kDa is observed (Δ2ATM; Fig. 5A). In contrast, antisera ATM-V and ATM-B recognize a major fragment of 240 kDa (ΔATM) and a very minor ∼150-kDa fragment (Fig. 5B and C), while an antibody that recognizes the kinase domain (ATM-2C1) also recognizes a 240-kDa fragment (Fig. 5D). Since the former antibody was raised against an N-terminal fragment and the latter antibodies were raised against central and C-terminal portions of the ATM protein, it can be concluded that a major site of ATM cleavage lies approximately 100 kDa from the amino terminus of the protein.
FIG. 5.
Antibody mapping of ATM fragments generated by caspase 3. Partially purified ATM (ATM-Q; 50 μg of total protein) was incubated with 300 ng of recombinant caspase 3 for 30 min at 37°C. Western blot analysis was performed with polyclonal rabbit antiserum raised against the N-terminal portion of ATM (ATM-N) (A), the central region of ATM (ATM-V) (B), the C-terminal region of ATM (ATM-B) (C), or the kinase domain of ATM (ATM-2C1) (D). (E) Schematic representation of ATM and the regions against which the antibodies were raised. Mk., molecular size markers; aa, amino acids.
Identification of caspase 3 cleavage sites in ATM.
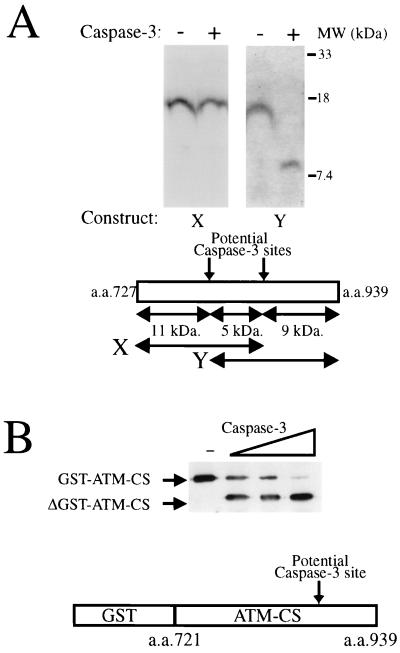
Previous studies have established that caspase 3 cleaves carboxy-terminally to sequences conforming to the optimal consensus sequence DEXD (41, 44). Notably, around 100 kDa from the N terminus of the ATM protein are two potential caspase 3 cleavage sites: the first comprising amino acid residues 814 to 818 with the sequence DIAD/I (the slash indicates the cleavage site in the amino acid sequence) and the second corresponding to amino acid residues 860 to 864 with the sequence DYPD/S. To identify which one of these (or both) is targeted by caspase 3, we generated by in vitro translation overlapping regions of the ATM protein that span these two potential cleavage sites (constructs X and Y; Fig. 6A). The in vitro-translated products were then incubated with caspase 3 and resolved by SDS-PAGE. The pattern of products generated revealed that no detectable proteolysis takes place at the more N-terminal of the two sites (Fig. 6A), whereas efficient caspase 3-mediated cleavage occurs in the more C-terminal site, corresponding to amino acid residues 860 to 864 (DYPD/S; Fig. 6A); note that the smaller part of the cleaved construct Y cannot be seen clearly due to it containing only two radiolabelled methionine residues. Although this site appears to be a poor match for the caspase 3 recognition site, the sterol regulatory element binding protein 2 has been shown to be cleaved by caspase 3 at a similar site (DEPD/S) (46). To unequivocally identify the caspase 3 cleavage site in ATM, we overexpressed the region of ATM corresponding to amino acids 721 to 939 of the protein as a GST fusion protein. Cleavage of this fusion protein by caspase 3, as judged by Western blotting with a monoclonal antibody to GST, is shown in Fig. 6B. We next sought to identify the sequence of the cleaved C-terminal fragment derived from caspase 3-cleaved GST-ATM-CS. After transferring the cleavage reaction to polyvinylidine difluoride membranes, N-terminal sequencing by Edman degradation of an ∼10-kDa fragment derived from cleavage of the GST-ATM-CS protein was performed. This analysis resulted in the amino acid sequence SSVSDA being derived. This sequence confirms that in ATM, the caspase 3 cleavage site is between Asp863 and Ser864 in the sequence DYPDSSVSDA (amino acid residues 860 to 869). While searching the ATM protein for other potential caspase 3 cleavage sites, we identified the motif DIVD/G corresponding to amino acid residues 2913 to 2917. However, this site is only weakly cleaved by caspase 3 in vitro, as judged by the cleavage of a GST-ATM kinase domain fusion protein (data not shown). Moreover, we have been unable to see cleavage at this site in vivo (data not shown).
FIG. 6.
Identification of caspase 3 cleavage sites in ATM. (A) The ATM region containing the motif DYPD/S, but not the region containing the motif DIAD/I, is cleaved by caspase 3 in vitro. As shown in the top panel, the region of ATM comprising amino acid (a.a.) residues 727 to 859 (region X) or the region comprising residues 819 to 939 (region Y) was transcribed and translated in vitro in the presence of 35S-radiolabelled methionine before being incubated for 30 min at 37°C in the absence or presence of 300 ng of caspase 3, as indicated. Reaction products were detected by SDS-PAGE (18%) followed by autoradiography. The bottom panel is a schematic representation of the regions of ATM utilized in the above-mentioned studies together with potential caspase 3 cleavage sites. (B) Cleavage of the ATM caspase 3 cleavage site as a GST fusion. Bacterially expressed and purified GST-ATM-CS fusion protein (2 μg) was incubated with no caspase 3 (−) or with increasing amounts (33, 100, or 300 ng) of caspase 3 for 30 min at 37°C. Products were analyzed by SDS-PAGE (12% gel) followed by Western immunoblotting with an anti-GST monoclonal antibody.
ATM cleavage products retain an ability to bind to one another and to DNA.
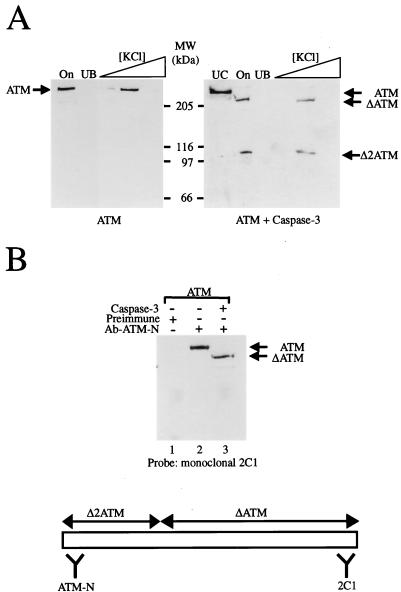
We have shown recently that, like DNA-PKcs, ATM has the ability to bind to DNA and has preference for binding to DNA ends (39a). To study whether caspase 3 has any effect on the DNA binding function of ATM, we subjected untreated ATM and caspase 3-treated ATM to a DNA pull-down assay in which ATM is tested for binding to iron-oxide particles containing a double-stranded DNA oligonucleotide. As seen in Fig. 7A, both the N-terminal 100-kDa fragment (Δ2ATM) and the C-terminal 240-kDa fragment (ΔATM) of cleaved ATM retain the ability to bind DNA in this assay (as expected from our previous analyses of DNA binding by ATM, no binding of either fragment was observed if the iron-oxide particles lacked DNA [data not shown]). This therefore indicated either that both protease-generated ATM fragments can bind DNA independently or that one binds to DNA and retains the other via protein-protein interactions. To probe for possible interactions between the two ATM fragments, we conducted immunoprecipitation studies. Thus, we immunoprecipitated caspase 3-treated ATM with the ATM-N antibody, which recognizes the N-terminal ATM fragment, and then probed the resulting immunoprecipitated material by Western blotting with the monoclonal antibody 2C1 (8), which recognizes the C-terminal fragment (Fig. 7B). The results of these studies show clearly that the larger ΔATM fragment coimmunoprecipitates with the N-terminal fragment Δ2ATM (Fig. 7B, lane 3). These results therefore indicate that the caspase 3-generated ATM fragments are still able to interact with one another.
FIG. 7.
Proteolytically cleaved ATM fragments retain an ability to bind DNA and bind to each other. (A) Untreated (left-hand panel) or caspase 3-treated (right-hand panel) ATM was bound to DNA-coated iron-oxide particles, and then these were washed in 100 mM KCl to remove unbound material before elution of ATM protein with 150, 250, and then 500 mM concentrations of KCl. On, input material; UB, unbound material; UC, uncleaved ATM control; MW, molecular size markers. Samples were subjected to Western blot analysis with a mixture of anti-ATM antisera ATM-B and ATM-N. (B) Untreated or caspase 3-treated ATM pool was immunoprecipitated with either preimmune or ATM-N antiserum, as indicated. Following capture on protein A-Sepharose and extensive washing with TBS containing 0.1% NP-40, samples were processed by Western blotting with monoclonal antibody 2C1 that recognizes the C terminus of the ATM protein. Lane 1, ATM plus preimmune serum; lane 2, ATM plus antiserum ATM-N; lane 3, ATM plus caspase 3 plus antiserum ATM-N.
Apoptotic cleavage of ATM abrogates its p53 kinase function.
It has recently been shown that ATM protein immunoprecipitated from wild-type cells can effectively phosphorylate p53 at serine-15 (2, 6). Furthermore, this kinase potential has been shown to be dependent on an intact ATM protein. Thus, the truncation of as few as 10 amino acid residues from the C terminus of the ATM kinase domain destroys its ability to phosphorylate p53 (2). To study whether apoptotic cleavage of ATM leads to a loss of its p53 kinase function, we immunoprecipitated ATM from HeLa nuclear extract or HeLa nuclear extract treated with caspase 3 in vitro (Fig. 8B, left-hand panel) (control immunoprecipitations are shown in Fig. 8A). Importantly, Western immunoblot analysis reveals that immunoprecipitation from these two sources yielded similar amounts of ATM and ΔATM and that ATM had become efficiently cleaved during the caspase 3 treatment to generate ΔATM (Fig. 8B, left-hand panel). Samples of the immunoprecipitated materials were tested in parallel for kinase activity by incubating them with p53, and radiolabelled ATP, and then detecting radiolabelled p53 by autoradiography. Strikingly, these studies revealed that ATM immunoprecipitated from caspase 3-treated nuclear extract displays significantly reduced p53 kinase activity (13% of control levels, determined by phosphorimage analysis) compared to that of full-length ATM immunoprecipitated from the mock treated extract (Fig. 8B, right-hand panel).
FIG. 8.
Caspase 3-mediated cleavage of ATM disrupts its p53 kinase activity. (A) Control immunoprecipitations to show the specificity of the monoclonal antibody raised against amino acid residues 819 to 844 of ATM (SYR 10G31). Full-length ATM was immunoprecipitated from HeLa nuclear extract (300 μg) or nuclear extracts (300 μg) prepared from control (wt) or A-T lymphoblastoid cells with monoclonal antibody SYR 10G31. A control immunoprecipitation with an isotype-matched antibody (anti-PARP) and HeLa nuclear extract (300 μg) was also carried out. ATM was visualized by probing a Western blot of the immunoprecipitated materials with the polyclonal antiserum ATM-B. Samples of the immunoprecipitates were also incubated with recombinant p53 and [γ-32P]ATP in an in vitro kinase assay, and radiolabelled p53 was detected by autoradiography (lower panel). (B) Full-length ATM or caspase 3-cleaved ATM was immunoprecipitated from HeLa nuclear extract (N.E.) (300 μg) incubated at 37°C for 15 min in the absence or presence of recombinant caspase 3 (3 μg). ATM and ΔATM were visualized as described for panel A. Immunoprecipitated ATM samples were analyzed for p53 kinase function as described for panel A. (C) ATM and ΔATM were immunoprecipitated, as described for panel A, from nuclear extracts prepared from untreated HL60 cells or HL60 cells treated with 68 μM etoposide for 4 h (left-hand panel). Immunoprecipitated ATM samples were analyzed for p53 kinase function as described for panel A.
Furthermore, ATM kinase function was examined after immunoprecipitation of ATM or ΔATM from extracts of untreated HL60 cells or from extracts of HL60 cells that had been treated with etoposide for 4 h. Western immunoblot analysis reveals that immunoprecipitation from these two sources yielded similar amounts of ATM and that ATM had become efficiently cleaved during the etoposide treatment to generate ΔATM (Fig. 8C, left-hand panel). Analyses, as described above, confirmed that the etoposide-induced cleavage of ATM in HL60 cells diminishes its p53 kinase function to 30% of control levels (Fig. 8C). In line with previous studies (2, 6), analysis with antisera capable of detecting p53 only after it has become phosphorylated on serine-15 confirmed that p53 phosphorylation by ATM-containing immunoprecipitates occurs at this site (data not shown). Using these two complementary approaches to study the effect of ATM cleavage, we can conclude that the p53 kinase function of ATM is impaired by caspase 3 or a caspase 3-like protease in cells undergoing apoptosis (Fig. 8C).
DISCUSSION
The identification of substrates for the cell death-activated caspases is of fundamental importance for the study of downstream events that take place during apoptosis. It is becoming increasingly apparent that the caspases are more specific in their action than was first presumed (11, 36). Caspases can be divided into two classes: those that are involved in the activation of apoptosis (e.g., caspase 8 and caspase-10) and those that are involved in the execution of apoptosis (e.g., caspase 3, caspase 6, and caspase 7) (11). Caspases have the ability to both activate and inactivate their protein substrates. The specificity of the caspases is highlighted by the fact that relatively few substrates have been identified to date. In this paper, we have shown that ATM, the gene product defective in the human autosomal recessive disorder A-T, is cleaved specifically in cells undergoing apoptosis. The cleavage of ATM not only occurs after apoptotic induction by DNA-damaging agents but appears to be a common event in cells undergoing apoptosis induced by a variety of different stimuli.
Previous work has established that ATM is a predominantly nuclear protein (5, 8, 28, 47) that is involved in the signalling of DNA damage to the cell cycle checkpoint machinery (20, 35, 51). ATM has also been implicated in having a direct role in DNA double-strand break repair (15). In regard to these points, it is noteworthy that apoptotic cleavage of ATM is reminiscent of that seen for DNA-PKcs (7, 17, 40) and PARP (26, 29), two other proteins involved in controlling DNA repair and genomic stability (21, 24). Indeed, the time course for apoptotic cleavage of ATM is very similar to that seen for the classic caspase substrate PARP. In contrast to ATM, DNA-PKcs, and PARP, however, the ATM-related protein ATR, which is also involved in DNA damage signalling and maintaining genomic stability, is not cleaved detectably in the situations that we have studied.
Through using a range of protease inhibitors, we established that a caspase was the most likely candidate for mediating ATM cleavage in apoptotic cells. Recent evidence has indicated that caspase 3 and caspase 6 are the two most prevalent proteases activated during apoptosis that are involved in the cleavage of downstream targets (14), although importantly not the only ones (23, 48). We find that ATM is cleaved efficiently in vitro by caspase 3, although caspase 6 does not cleave ATM under the assay conditions we have employed. Notably, the cleavage pattern of ATM in vivo mirrors that produced when partially purified ATM is treated in vitro by caspase 3. Furthermore, by performing kinetic studies, we have found that ATM appears to be as good a substrate for caspase 3 as PARP. It was also found that ATM was not cleaved in the caspase 3-deficient MCF-7 cell line. Taken together, our data strongly implicate caspase 3 or a caspase 3-like enzyme in the cleavage of ATM during programmed cell death.
Mapping the regions on ATM that are targeted by caspase 3 revealed one major site ∼100 kDa from the N terminus. Although no specific function has yet been ascribed to the N-terminal region of ATM, it is clear that even relatively minor alterations within the ATM protein lead to the loss of ATM function and the generation of the human A-T phenotype. Indeed, we have demonstrated that caspase 3-mediated cleavage diminishes the ability of ATM to phosphorylate p53, a known downstream effector of ATM-dependent DNA damage signalling. We have shown recently that ATM binds to DNA and has specificity towards DNA ends. Perhaps surprisingly, we have found that DNA binding by ATM is not abrogated upon caspase 3-mediated proteolysis. Indeed, both regions of the cleaved ATM protein were still found to be retained on iron-oxide particles bearing oligonucleotide DNA. Interestingly, we have found by coimmunoprecipitation studies that, after caspase 3 cleavage, the 100-kDa N-terminal region and the 240-kDa C-terminal region of ATM remain complexed with one another, presumably via intramolecular interactions. It will be of great interest to further define these interactions and to investigate whether it is the N-terminal region, the C-terminal region, or both that mediate DNA binding.
Although it is possible that ATM plays a key role in the triggering or execution of apoptosis under normal physiological circumstances, the fact that A-T patients and Atm knockout mice develop normally suggests that this is not the case (3, 49). Another possibility is that ATM functions in the triggering of apoptosis in response to certain DNA-damaging agents. Indeed, whereas mice treated with IR show a normal thymic apoptotic response (4), certain regions of the mouse brain do not undergo effective apoptosis after IR exposure in the absence of Atm function (18). Hence, signalling by ATM to the apoptotic machinery might be a tissue-dependent phenomenon. Nevertheless, our data indicate that a major involvement of ATM in apoptosis is as a ubiquitous downstream target for caspase-mediated proteolysis. In this regard, it is noteworthy that the cleavage of ATM appears to occur before the onset of a characteristic feature of apoptosis, DNA laddering, which is brought about by CAD (13). Since we have shown that proteolysis diminishes the catalytic function of ATM, it is tempting to speculate that the degradation of this protein prevents it from signalling this CAD-induced DNA damage to the cell cycle checkpoint machinery and/or prevents attempts to repair the damaged DNA. Finally, it is notable that cleaved ATM, while catalytically inactive, retains its ability to bind to DNA. An attractive model, therefore, is that this results in a trans-dominant-negative protein which can bind to DNA but is inactive, a model similar to that which has been proposed for the cleavage of PARP. Given the demonstrated preference of ATM to bind to DNA ends in vitro, this raises the possibility that cleaved ATM binds to genomic DNA double-strand breaks when they are generated during the apoptotic process, preventing their recognition by other components. In regard to the above issues, it will clearly be of great interest to generate mutated derivatives of ATM that cannot be cleaved during apoptosis and establish the effects that such mutations have on the highly orchestrated process of apoptotic progression.
ACKNOWLEDGMENTS
This work was funded by grants SP2143/0103 and SP2143/0501 from the Cancer Research Campaign and by grants from the Kay Kendall Leukaemia Fund and the A-T Childrens’ Project.
We thank members of the SPJ laboratory for their advice and support. We thank Y. Shiloh for generously providing us with the anti-ATM monoclonal antibody used for the ATM kinase assay and Eva Lee for provision of the anti-ATM monoclonal antibody 2C1. The caspase-3 expression construct was a kind gift from G. Cohen at the University of Leicester and the p53 protein was provided by Byron Hann, UCSF. Thanks also to Mike Waldon at the Protein Nucleic Acid Chemistry facility, Biochemistry Department, University of Cambridge, for technical assistance with N-terminal sequence analysis.
REFERENCES
- 1.Arends M J, Wyllie A H. Apoptosis—mechanisms and roles in pathology. Int Rev Exp Pathol. 1991;32:223–254. doi: 10.1016/b978-0-12-364932-4.50010-1. [DOI] [PubMed] [Google Scholar]
- 2.Banin S, Moyal L, Shieh S Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 3.Barlow C, Hirotsune S, Paylor R, Liyange M, Eckgaus M, Collins F, Shiloh Y, Crawley J N, Reid T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 4.Barlow C, Brown K D, Deng C X, Tagle D A, Wynshaw-Boris A. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- 5.Brown K D, Ziv Y, Sadanandan S N, Chessa L, Collins F S, Shiloh Y, Tagle D. The ataxia-telangiectasia gene product, a constitutively expressed nuclear protein that is not up-regulated following genome damage. Proc Natl Acad Sci USA. 1997;94:1840–1845. doi: 10.1073/pnas.94.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 7.Casciola-Rosen L A, Anhlat G J, Rosen A. DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J Exp Med. 1995;182:1625–1634. doi: 10.1084/jem.182.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, Lee E Y-H P. The product of the ATM gene is a 370-kDa nuclear phosphoprotein. J Biol Chem. 1996;52:33693–33697. doi: 10.1074/jbc.271.52.33693. [DOI] [PubMed] [Google Scholar]
- 9.Cimprich K A, Shin T B, Keith C T, Schreiber S L. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 12.Ellis H M, Yuan J, Horvitz H R. Mechanisms and functions of cell death. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 13.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 14.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foray N, Priestley A, Arlett C F, Malaise E P. Hypersensitivity of ataxia telangiectasia fibroblasts to ionising radiation is associated with a repair deficiency of DNA double-strand breaks. Int J Radiat Biol. 1997;72:271–283. doi: 10.1080/095530097143266. [DOI] [PubMed] [Google Scholar]
- 16.Han Z, Chatterjee D, He D M, Early J, Pantazis P, Wyche J H, Hendrickson E A. Evidence for a G2 checkpoint in p53-independent apoptosis induction by X-irradiation. Mol Cell Biol. 1995;15:5849–5857. doi: 10.1128/mcb.15.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Z, Malik N, Carter T, Reeves W H, Wyche J H, Hendrickson E A. DNA-dependent protein kinase is a target for a CPP32-like apoptotic protease. J Biol Chem. 1996;271:25035–25040. doi: 10.1074/jbc.271.40.25035. [DOI] [PubMed] [Google Scholar]
- 18.Herzog K H, Chong M J, Kapsetaki M, Morgan J I, McKinnon P J. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 19.Hoekstra M F. Responses to DNA damage and regulation of cell-cycle checkpoints by the ATM protein kinase family. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- 20.Jackson S P. Cancer predisposition; ataxia-telangiectasia at the crossroads. Curr Biol. 1995;5:1210–1212. doi: 10.1016/s0960-9822(95)00238-7. [DOI] [PubMed] [Google Scholar]
- 21.Jackson S P. DNA damage detection by DNA dependent protein kinase and related enzymes. Cancer Surv. 1996;28:261–279. [PubMed] [Google Scholar]
- 22.Jacobson M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 23.Janicke R U, Ng P, Sprengart M L, Porter A G. Caspase-3 is required for a-fodrin cleavage but dispensable for cleavage of other death substrates in apoptosis. J Biol Chem. 1998;273:15540–15545. doi: 10.1074/jbc.273.25.15540. [DOI] [PubMed] [Google Scholar]
- 24.Jeggo P A. DNA repair: PARP—another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 25.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase—an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 27.Khanna K, Lavin M F. Ionizing radiation and UV induction of p53 protein by different pathways in ataxia-telangiectasia. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 28.Lakin N D, Weber P, Stankovic T, Rottinghaus S T, Taylor A M, Jackson S P. Analysis of the ATM protein in wild-type and ataxia telangiectasia cells. Oncogene. 1996;13:2707–2716. [PubMed] [Google Scholar]
- 29.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 30.Lu X, Lane D P. Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 31.Martin S J, Green D R. Protease activation during apoptosis: death by a thousand cuts? Cell. 1995;82:349–352. doi: 10.1016/0092-8674(95)90422-0. [DOI] [PubMed] [Google Scholar]
- 32.Meyn M S. Ataxia-telangiectasia and cellular responses to DNA damage. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- 33.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffen P R, Labelle M, Lazebnick Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V L, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 34.Porter A G, Ng P, Janicke R U. Death proteases come alive. Bioessays. 1997;19:501–507. doi: 10.1002/bies.950190609. [DOI] [PubMed] [Google Scholar]
- 35.Rotman G, Shiloh Y. ATM: from gene to function. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 36.Salvesen G S, Dixit V M. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 37.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashjenazi M, Pecker I, Frydman M, Harnik R, Patanjali S R, Simmons A, Clines G A, Sateil A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G I, Taylor A M R, Arlett C F, Miki T, Weissmann S M, Lovett M, Collins F S, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI 3-kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 38.Shiloh Y. Ataxia-telangiectasia: closer to unraveling the mystery. Eur J Hum Genet. 1995;3:116–138. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- 39.Siliciano J D, Canman C E, Taya Y, Sakaguchi J, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Smith, G. C. M., R. B. Cary, N. D. Lakin, S.-H. Teo, D. J. Chen, and S. P. Jackson. Submitted for publication.
- 40.Song Q, Lees-Miller S P, Kumar S, Zhang Z, Chan D W, Smith G C M, Jackson S P, Alnemari E S, Litwack G, Khanna K K, Lavin M F. DNA-dependent protein kinase catalytic subunit: a target for an ICE-like protease in apoptosis. EMBO J. 1996;15:3238–3246. [PMC free article] [PubMed] [Google Scholar]
- 41.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 42.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 43.Thompson C. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 44.Thornberry N A, Ranon T A, Pieterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 45.Vaux D L. Towards an understanding of the molecular mechanisms of physiological cell death. Proc Natl Acad Sci USA. 1993;90:786–789. doi: 10.1073/pnas.90.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Pai J, Wiedenfield E A, Medina J C, Slaughter C A, Goldstein J L, Brown M S. Purification of an interleukin-1β converting enzyme-related cysteine protease that cleaves sterol regulatory element-binding proteins between the leucine zipper and transmembrane domains. J Biol Chem. 1995;270:18044–18050. doi: 10.1074/jbc.270.30.18044. [DOI] [PubMed] [Google Scholar]
- 47.Watters D, Khanna K K, Beamish H, Birrel G, Spring K, Kedar P, Gatei M, Stenzel D, Hobson K, Koslov S, Zhang N, Farrel A, Ramsay J, Gatti R, Lavin M F. Cellular localisation of the ataxia-telangiectasia (ATM) gene product and discrimination between mutated and normal forms. Oncogene. 1997;14:1911–1921. doi: 10.1038/sj.onc.1201037. [DOI] [PubMed] [Google Scholar]
- 48.Woo M, Hakem R, Soengas M S, Duncan G S, Shahinian A, Kagi D, Hakem A, McCurrach M, Khoo W, Kaufman S A, Senaldi G, Howard T, Lowe S W, Mak T W. Essential contribution of caspase 3 CPP32 to apoptosis and its associated nuclear changes. Genes Dev. 1998;12:805–819. doi: 10.1101/gad.12.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 50.Yuan J, Shaman S, Ledoux S, Ellis H M, Horvitz H R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 51.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]