Abstract
Segmentation of infections from CT scans is important for accurate diagnosis and follow-up in tackling the COVID-19. Although the convolutional neural network has great potential to automate the segmentation task, most existing deep learning-based infection segmentation methods require fully annotated ground-truth labels for training, which is time-consuming and labor-intensive. This paper proposed a novel weakly supervised segmentation method for COVID-19 infections in CT slices, which only requires scribble supervision and is enhanced with the uncertainty-aware self-ensembling and transformation-consistent techniques. Specifically, to deal with the difficulty caused by the shortage of supervision, an uncertainty-aware mean teacher is incorporated into the scribble-based segmentation method, encouraging the segmentation predictions to be consistent under different perturbations for an input image. This mean teacher model can guide the student model to be trained using information in images without requiring manual annotations. On the other hand, considering the output of the mean teacher contains both correct and unreliable predictions, equally treating each prediction in the teacher model may degrade the performance of the student network. To alleviate this problem, the pixel level uncertainty measure on the predictions of the teacher model is calculated, and then the student model is only guided by reliable predictions from the teacher model. To further regularize the network, a transformation-consistent strategy is also incorporated, which requires the prediction to follow the same transformation if a transform is performed on an input image of the network. The proposed method has been evaluated on two public datasets and one local dataset. The experimental results demonstrate that the proposed method is more effective than other weakly supervised methods and achieves similar performance as those fully supervised.
Keywords: COVID-19, infection segmentation, weakly supervised learning, transformation consistency, uncertainty
1. Introduction
The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global pandemic since the beginning of 2020. It has resulted in over 4,400,284 deaths as of 20 August 2021, and over 209,876,613 cases infected, according to the report from World Health Organization (WHO).
For COVID-19 screening, the reverse-transcription polymerase chain reaction (RT-PCR) is regarded as the gold standard [1]. As an important complement to RT-PCR tests, imaging techniques, such as X-rays, computed tomography (CT), and ultrasound, have also demonstrated their effectiveness in infection detection, follow-up assessment, and evaluation of disease evolution [2]. Among the imaging techniques, CT screening has a three-dimensional view of the lung and high contrast in discriminating lesions from normal tissues [3], [4]. Therefore, CT is an effective disease surveillance imaging tool that can help diagnose COVID-19 in clinical practice quickly.
In the application of CT dealing with COVID-19, segmentation of the infection lesions from CT volumes is essential for quantitative measurement of disease progression [2], [5]. Manual segmentation of the lesions from CT volumes is time-consuming, labor-intensive, and subject to inter- and intra-observer variabilities. It can take up to 7 hours to delineate one CT volume with 250 slices [6]. Automatic segmentation of lesions is highly desirable. However, automatic segmentation of COVID-19 lesions from CT scans is challenging. First, infection lesions are often with different appearances, such as ground-glass opacity (GGO), consolidation (CO), crazy pavement, and others [7]. Second, the positions and sizes of the lesions often vary significantly with the progress of infection and across different patients. Besides, lesions often have irregular shapes, blurry boundaries, and low tissue contrast (especially for GGO). These challenges make it hard to obtain accurate manual annotations for training and introduce obstacles for automatic segmentation of the lesions.
In recent years, deep learning has achieved state-of-the-art performance for many medical image processing tasks [8], [9], [10], [11], [12], [13], [14], [15]. Deep learning has also been proposed to detect patients infected with COVID-19 [16], [17]. A weakly supervised deep learning approach has been proposed for COVID-19 classification and lesion localization by using CT volumes [16]. Specifically, the lung region was first segmented with a pre-trained U-Net, then the segmented lung region was fed into a deep neural network for classification, and finally, the COVID-19 lesions were located by combining activation regions from the classification model and the unsupervised connected component analysis.
Despite numerous existing approaches to assist the diagnosis of COVID-19, only a few works have investigated the segmentation of lesions in CT slices [5], [17], [18]. Fan et al. [17] proposed a network, namely Inf-Net, that used a parallel partial decoder to aggregate the high-level features and generate a global map. Then, implicit reverse attention and explicit edge-attention were used to model the boundaries. The method was further extended to the semi-supervised segmentation scenario to alleviate the label scarcity problem. A noise-robust framework for the segmentation of COVID-19 lesions was proposed in [5] to deal with noise in manual annotations. By combining the mean absolute error loss robust against noisy labels, and Dice loss which is insensitive to foreground-background imbalance, they designed a noise-robust Dice loss function. An adaptive self-ensembling framework was proposed to suppress the effect of noisy labels on training. Laradji et al. [18] proposed a weakly supervised COVID-19 lesion segmentation method, which requires only point annotations, , one pixel for each infected region on a CT slice is used as the ground-truth. This can significantly reduce the labor cost of annotation.
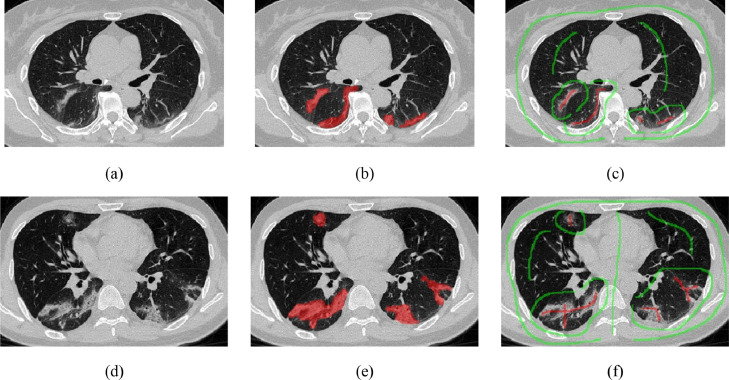
However, we observed several shortcomings in the existing COVID-19 segmentation approaches: 1) For COVID-19 infection segmentation, most existing methods require dense annotations, which are labor-intensive and time-consuming. It is worth noting that the work in [17] was investigated on a small labeled dataset with only 110 slices. While the training annotations in [5] can be noisy, time-consuming dense annotation is still required. Point annotations are used in [18] to reduce the labeling cost. However, it did not perform well on the irregular shape of infections (see Fig. 1 ), and the segmentation performance is limited since the supervision information is scarce. 2) Due to the blurred boundary and low contrast of GGO in CT slices, some pixels are more difficult to segment than other pixels. During the utilization of unlabeled pixels (images), their reliability needs to be considered to avoid meaningless guidance, which is often overlooked by the existing methods. 3) Cross-entropy, Dice loss, or their variants are typically used as the loss functions to train Convolutional Neural Networks (CNNs) [19], [20], while they are not sufficient when dealing with weakly-supervised segmentation, and other guidance is required. It is known that CNNs with pooling layers are less sensitive to some spatial transformations, , the positional variation. This spatial-invariant characteristic is an advantage for classification problems, however, a disadvantage for segmentation problems [21], [22]. If an input image is transformed ( rotated), the expected prediction should be transformed in the same way. However, this favorable characteristic for segmentation is not preserved in CNN generally [21], [23]. To solve the problem, transformation consistent loss [23] is often adopted in semi-supervised tasks [23] or weakly-supervised segmentation tasks [18], [21] to improve the performance.
Fig. 1.
Example of COVID-19 infected CT images and the corresponding annotations. (a) and (d) are original images, (b) and (e) are fully-annotations of infections, (c) and (f) are our scribble-level annotations. Red and green lines denote the infection regions and backgrounds, respectively.
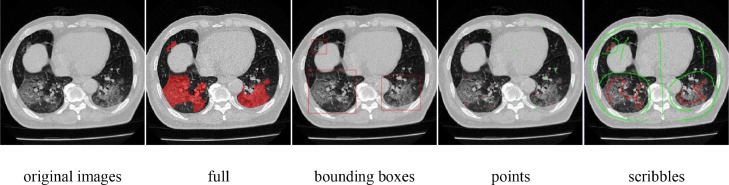
In this work, we propose a novel weakly-supervised method for the segmentation of COVID-19 infections in CT slices with scribble supervision. Specifically, the method is Uncertainty-aware Self-ensembling and Transformation-consistent Mean Teacher Model, namely USTM-Net, with scribble-level annotation of lung infections (see the comparison with the fully-annotation in Fig. 1). According to our statistics, manual scribbles only take about 16 seconds per slice on average in our dataset. This is an order of magnitude faster than dense annotations of the dataset (around 200 seconds per slice on average). The box-level annotations on each image took about 20 seconds (similar to the scribble annotation), while the point-level annotations took only a few seconds. Besides, the cost of scribble labeling is comparable to point labeling [18]. In contrast, the scribbles can better deal with the irregular shape of infections (see Fig. 2 for comparison) and incorporate more supervision information. Inspired by semi-supervised segmentation works [23], [24], we incorporated uncertainty-aware self-ensembling techniques into the weakly supervised segmentation method to make the prediction robust to perturbations. In a typical semi-supervised segmentation scenario, only part of the images have dense annotations, and the perturbation consistency is required on both labeled and unlabeled images [24]. In our method, perturbation consistency is required on the same weakly annotated images. We build a teacher model and a student model. The parameters of the teacher model are the exponential moving average (EMA) of the student's parameters, inspired by the method in [25]. The student model gradually learns from the reliable targets by exploiting the uncertainty information of the teacher model. With a Monte Carlo Dropout [24] based uncertainty measurement, the proposed method can better deal with the low contrast in GGO infections. Image augmentation has been widely used in supervised deep learning to improve the generalization capability and alleviate overfitting [26], [27], usually performed on labeled images. To alleviate the affection of limited supervision, we further incorporated transformation consistency loss into the proposed method, which is also desired by CNNs for segmentation.
Fig. 2.
Example of different types of weak annotations on COVID-19 infected CT images.
The main contributions of this work are summarized as follows:
-
1)
We propose a weakly supervised COVID-19 infection segmentation method with scribble supervision. To the best of our knowledge, it is the first work adopting scribble-level supervision in COVID-19 segmentation.
-
2)
An uncertainty-aware mean teacher framework is incorporated into the proposed method to guide the model training, encouraging the segmentation predictions to be consistent under different perturbations for an input image. With the pixel level uncertainty measure on the predictions of the teacher model, the student model is guided with reliable supervision. We further regularize the model with a transformation-consistent strategy, which is beneficial for the segmentation task and makes our approach easier to deal with the segmentation of irregular lesion areas.
-
3)
We evaluated the proposed method on three datasets and compared it with other advanced approaches. The results demonstrated the superiority of the proposed method.
The remaining of the paper is organized as follows. Section II reviews the related work. Section III introduces the proposed approach in detail. Section IV presents experimental results. And Section V concludes the paper.
2. Related Work
This section reviews some related works, including 1) annotation-efficient medical image segmentation, 2) COVID-19 infection segmentation in CT images, and 3) the transformation consistency techniques.
2.1. Annotation-efficient Deep Learning in Medical Image Segmentation
Various methods have been proposed to lessen the cost of annotation in medical image segmentation. Some unsupervised anomaly detection methods can identify anomaly regions, but they cannot identify whether the anomaly is related to a specific disease [28]. For the COVID-19, Yao et al. [29] proposed a label-free COVID-19 lesion segmentation method for CT images. Based on the patterns of tracheae and vessels in CT scans, lesions were synthesized with several operations and superimposed on the lung regions of healthy images. The segmentation model is trained on the synthesized dataset. However, many specially designed operations are handcrafted, including threshold selection, morphological processing, elastic deformation, etc. The performance gap between unsupervised methods and supervised methods is large due to the limited supervision information.
Recently, semi-supervised learning (SSL) has been widely investigated in image classification and segmentation [24], [30], [31]. The main goal of SSL is to improve the model performance by using a large amount of unlabeled data, besides the small number of labeled data [30]. These methods usually optimize a supervised loss on the labeled data along with an unsupervised loss imposed on the unlabeled data [31] or both the labeled and unlabeled data [24]. For medical image segmentation, the lack of pixel-level labeled data is common due to the requirement of expertise and time. In contrast, a large number of unlabeled images may be available [32]. Cui et al. [33] adopted a mean teacher framework [25] for the stroke lesion segmentation in MR images. The mean teacher framework was originally proposed for image classification based on the assumption that CNN models should favor functions that produce consistent outputs for similar inputs. The ensemble of predictions of a network in different training steps can be more accurate than the single latest model [33]. In this method, two models with the same structure are constructed: one is the student model, and the other is the teacher model. The parameters in the teacher model are updated by the student model with an exponential moving average strategy. For unlabeled data, the predictions from the student model and the teacher model are required to be consistent. Uncertainty is incorporated into the mean teacher framework in [23] for semi-supervised left atrium segmentation. Besides the mean teacher framework, generative adversarial network (GAN) is also popular in semi-supervised segmentation of medical images [34], [35], [36]. An adversarial learning framework for unsupervised domain adaptation was proposed in [36], which aimed to accurately segment chest organs for unlabeled data by learning domain invariant feature representations from public datasets. The model can also be trained in a semi-supervised manner and had performed well. Similarly, in order to reduce the workload of dense annotations, it is common for training data to be partially labeled. For example, in the partially-labeled multi-organ segmentation dataset, only a few organs are fully labeled. Hence, some work has been done in partially-supervised learning [37], [38]. Zhou et al. [37] proposed the Prior-aware Neural Network, which used domain-specific knowledge to guide the training process by combining prior anatomical knowledge about abdominal organ size.
Recently self-supervised learning methods have also been proposed to avoid the cost of manual annotations [39], [40]. Grill et al. [39] introduced a new self-supervised image representation learning method called Bootstrap Your Own Latent, which consisted of two networks named the online network and the target network. They trained the online network to predict the target network's representation from an augmented view, while the parameters of the target network are updated from the slow-moving average of the online network. Chen et al. [40] demonstrated that simple Siamese networks could learn meaningful representations even without negative sample pairs, large batches, or momentum encoders. In the overall framework, both our method and these methods use two branches for training, and only one branch is backpropagated, however, the strategy used in our method is different. Other methods took two different image augmentations on the original data before feeding them separately into the encoder (aiming to better learn the representations of the images by maximizing the similarity under different conditions), whereas our method enforces the transformation consistency strategy, in which one model transforms the image before it is input, and the other model transforms the output result. Our method aims to alleviate the affection of limited supervision and alleviate the influence of the undesired characteristic of a typical CNN in segmentation; that is, a CNN is not transformation equivariant generally. Besides, other methods were proposed for unsupervised image classification problems, while our method is proposed for weakly supervised image segmentation.
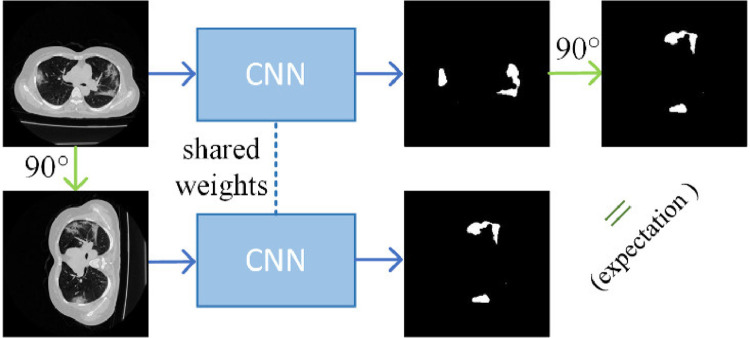
To combat the high cost associated with dense annotations, approaches that require only weak annotations have also been explored recently. Typical weak annotations include image-level labels [41], bounding-boxes [42], points [18], [43], and scribbles [44]. Wu et al. [41] proposed a weakly supervised 3D brain lesion segmentation with image-level labels. A dimensional independent attention mechanism was applied on the top of the class activation maps [45] to improve lesion localization. The estimated lesion regions and normal tissues were then used to train the segmentation network. Rajchl et al. [42] combined a CNN with fully connected conditional random field (CRF) for brain and lung segmentation with bounding box supervision. Matuszewski et al. [43] performed virus segmentation in microscopy images given point annotations. Based on the statistical size of virus, foreground and background masks were obtained by dilating the manual annotations. A scribble-level annotation-based weakly supervised cell segmentation framework was proposed in [44]. They combined pseudo-labeling and label filtering to generate reliable labels under weak supervision. The pseudo labels were improved by leveraging the consistency of the predictions during iterations. Fig. 2 shows that the scribble is the most suitable description for COVID-19 segmentation among different types of weak annotations, which balances the cost of labeling and plenty of supervision, and also can deal with the irregular shapes of infections. Our proposed USTM -Net is thus based on scribble-level annotations.
2.2. COVID-19 Infection Segmentation in CT Images
Although infection lesion segmentation is important for quantitative assessment of the disease, due to the short time of the emergence and the shortage of annotated image datasets, automatic segmentation of COVID-19 infections has not been widely investigated [6], [7]. Shan et al. [46] proposed to automatically segment and quantify the infection regions and the entire lungs from CT scans. VB-Net was employed to accelerate manual delineation of CT scans, and a human-involved-model-iterations strategy was adopted to assist radiologists in refining the automatic annotations. Fan et al. [17] proposed Inf-Net for lung infection segmentation from CT slices. Features from high-level layers were aggregated with a parallel partial decoder, and a global map was generated as the initial guidance areas from the combined features. A noise-robust framework for the segmentation of COVID-19 lesions was proposed in [5] to deal with noises in manual annotations. By combining the mean absolute error loss and Dice loss, they designed a noise-robust Dice loss function. An adaptive self-ensembling framework was proposed to suppress the effect of noisy labels on training. Laradji et al. [18]proposed a weakly supervised COVID-19 infection segmentation method, which requires only point annotations. The method can greatly reduce the labor cost of annotations. Tilborghs et al. [47] compared twelve deep learning algorithms for lung and lung lesion segmentation in CT scans, including Inf-Net. They demonstrated that deep learning-based lesion segmentation methods achieved an average volume error better than human raters, suggesting those methods may be mature for clinical practice.
2.3. Transformation Consistency
Image augmentation is widely used in deep learning-based classification and segmentation. For the fully supervised semantic segmentation, if a transformation is performed on an input image, its pixel-level prediction should follow the same transformation. This is named transformation consistency property. Transformation consistency has been investigated recently in image label based weak segmentation [21], semi-supervised segmentation [23], and self-supervision technique [18]. Wang et al. [21] proposed a self-supervised equivariant attention mechanism (SEAM) to discover additional supervision when dealing with image label based weakly supervised segmentation. Class activation map (CAM) [45] was widely used in weakly supervised segmentation.The CAM usually only cover the most discriminative part of the objects and highlight unrelated background regions. To deal with the lack of dense annotations, they required the CAM to be transformation consistent. Li et al. [23] proposed a semi-supervised medical image segmentation method. The network was optimized by a weighted combination of supervised loss on labeled inputs and a regularization loss on both the labeled and unlabeled data. The regularization loss is based on transformation consistency property, encouraging consistent predictions of the network for the same inputs under different perturbations. The self-ensembling model was integrated with the transformation-consistent strategy. Laradji et al [18] integrated a transformation-consistent strategy into a point based weakly supervised segmentation for COVID-19 lesions. This paper integrates a transformation consistency strategy with scribble-based supervision for COVID-19 infection segmentation in CT images.
3. The Proposed Method
In this section, the proposed USTM-Net for COVID-19 infection segmentation in CT images is described in detail. In the first part, we present the scribble-supervised segmentation framework. In the second part, we present several core network components. In the third part, we introduce an extended framework for multi-class segmentation to segment different types of lung infections.
3.1. Framework of the Proposed Method
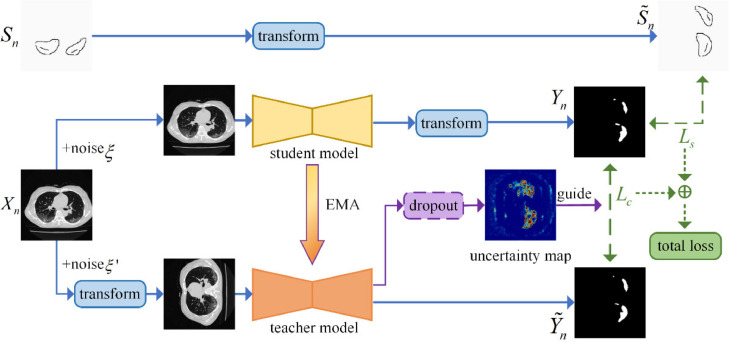
Fig. 3 presents the overall framework of the proposed method for weakly supervised COVID-19 infection segmentation in CT images. We treat the segmentation task as a binary classification problem, which classifies each pixel to the infection or the background regions. In this problem, the training set consists of input images in total. We leverage a data set for training, where is an input image and is the annotation corresponding to . includes three categories of labels: 0 for the scribbled background pixels, 1 for the scribbled pixels in infections, and 2 for unannotated pixels. Teacher and student models with the same network structure are employed as two branches to obtain two segmentation outputs in the framework. The proposed method consists of a training phase and a testing phase, which are described as follows.
Fig. 3.
The overall framework of our uncertainty-aware transformation-consistent model for scribble-supervised medical image segmentation. The teacher and student models share the same architecture. The student model is trained by the supervised loss and consistency loss with the guide of uncertainty.
The parameters of the teacher and student models are randomly initialized in training. And the input sources are the images and the corresponding scribbles. We randomly sample data from the training dataset under different noises into the teacher model and the student model, respectively. The images are transformed before the images are fed into the teacher model. The transformation includes rotation, flip, and scale. The same transformation is also performed on the given scribbles to obtain and on the outputs of the student model to obtain . To train the student model, is compared with the scribble-level label using the scribbled pixel loss and with the output of the teacher model using the uncertainty-aware consistency loss . The loss is used to guide the training of the student model with estimated uncertainty in the teacher model, in which only reliable segmentation regions will be used. The loss and are only applied to backpropagate the parameters of the student model. The teacher model is updated by the exponential moving average (EMA) algorithm, with a step of learning iterations of the student model. The goal is to train an effective segmentation model (, the student model) to segment the infection regions in the CT image.
In the testing phase, only the trained student network is employed to predict the infections of the images. Note that perturbations and transformation operations are not used during the testing phase.
3.2. Network Architecture
3.2.1. Scribble-supervised Segmentation
We tackle the task of scribble-supervised segmentation using a small set of manual scribbles on both infection and background regions to lessen the burden of manual labeling. Here, a standard cross-entropy loss function is applied on scribbled pixels to train the network.
A recent study [33] shows that self-ensembling exploits the intermediate information during the training process. The ensembled prediction can be closer to the correct result than the single latest model. The teacher and student framework is a typical self-ensembling model. Considering that the learning information provided by the scribble-level labels is limited, our method is expected to produce consistent outputs in the teacher and student models. Therefore, we employ the mean teacher based framework [25] to improve the quality of the predictions. To train the framework, we update the teacher model's parameters as the EMA of the student's weights at training step through:
| (1) |
where and are the parameters of the student model and teacher model, respectively. is the smoothing coefficient hyperparameter that controls the updating rate. and specify the contributions of and , respectively. According to the previous work in [25], the performance is the best with, thus, we set to 0.99 in our experiments.
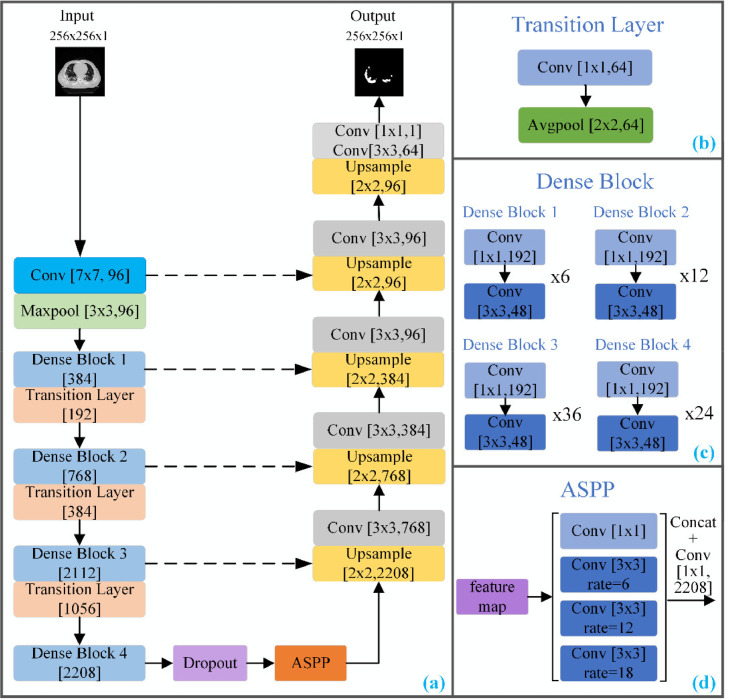
For both student and teacher models, we use a modified 2D DenseUNet architecture [48], with the detailed network structure shown in Fig. 4 . Compared with the standard U-Net [49], the convolution layers in the encoder path are replaced with the dense block layers. Moreover, different from DenseUNet [48], to better deal with infections at different scales, we add an Atrous Spatial Pyramid Pool (ASPP) module [50] at the intersection between the encoder and the decoder (see Fig. 4). The ASPP module consists of four layers of parallel convolutions with different expansion rates. The outputs of ASPP by four parallel convolutions are concatenated, and then used as the input of the decoder path. We also add a dropout layer after the last layer of the encoder, which will be explained in the later section.
Fig. 4.
The proposed segmentation network was improved by DenseUNet, where Conv[3 × 3, 96] represents a 3 × 3 convolution, and the output is 96 channels.
3.2.2. Uncertainty-Aware Mean Teacher Framework
The parameters of the teacher model are updated by EMA, making them more stable than the student model. Therefore, the student model is trained by the teacher model by a consistency loss [24], [25]. However, for the pixels without scribble annotations, the teacher model's predicted results may also be unreliable. Therefore, we design an uncertainty-aware scheme to enable the student model to gradually learn from only reliable predictions. For each training batch, besides the segmentation predictions, the teacher model also estimates the uncertainty of the predictions at pixel-level to guide the calculation of consistency loss, letting the calculation of the consistency loss only focus on reliable regions. In this way, the unlabeled infection regions are robustly learned by the teacher model, thus improving its segmentation performance.
We employ Monte Carlo Dropout [24] to estimate the uncertainty of segmentation predictions from the teacher model. In this case, we encourage consistent predictions of the network under different perturbations, , Gaussian noise, network dropout, and randomized data transformation. Specifically, under different input Gaussian noises and random dropouts, each input image is stochastically passed forward times through the teacher model. For each pixel in an input image, we obtain a set of softmax probability vector: . These probability vectors are averaged to get the final prediction probability score of the teacher model, which can be written as:
| (2) |
where is the number of classes (in our case). is the probability of the -th class in the -th time prediction, and is the average of the probabilities of the -th class. In our experiment, we set to balance the quality and training efficiency. We also choose predictive entropy as the measure of uncertainty, and the uncertainty can be computed by [24]:
| (3) |
where represent the uncertainty score in pixel level, and the uncertainty map of the whole image is . The uncertainty score reflects different levels of difficulties for the teacher model in segmenting different regions on a CT slice. If the predictions for a pixel in different forward passes are scattered, then the uncertainty value is high and indicates the pixel's prediction is uncertain. Under the guidance of the estimated uncertainty, the unreliable predictions are filtered, and only reliable predictions are used to guide the student model (see Eq. (6) below).
3.2.3. Transformation-Consistent Scheme
In weakly supervised learning, one prominent difference between classification and segmentation problems is that the former tend to be transformation (, translation, rotation, and flipping) invariant while the latter is transformation equivariant. The main reason is that, when using convolutional neural networks (CNNs) to tackle the segmentation tasks, the CNNs with pooling layers are not transformation equivariant generally [21]. It is easy to find that transformation has profound effects on the result of the segmentation problems in Fig. 5 . For example, the feature maps do not necessarily rotate in a meaningful manner if the inputs rotate before they are fed to the convolutions [51]. Formally, we denote the semantic segmentation function as, and suppose there is a transformation function for each image sample . The segmentation function is expected to be equivariant,. However, the two results are not equal in most cases. In addition, in the segmentation of COVID19 on CT images, the shapes of lesion regions vary, which makes the segmentation difficult. To enhance the accuracy of the segmentation, it is worth incorporating the transformation consistency strategy.
Fig. 5.
Segmentation is desired to be rotation equivariant. If the rotation is performed on an input image, the ground truth mask should be rotated in the same manner. After the CNNs, the generated output from the rotated image should be the same as the rotation of the original output, which is generally different in fact.
To keep the consistency of the teacher and student model, we randomly employed several types of transformations, including random flipping, scaling as well as rotation operations. Flipping operation flips an image horizontally or vertically. Our scaling operations use scaling ratios between 0.8 and 1.2, and four rotation operations use angles of , where During each training pass, one type of these transformations is randomly chosen and applied to the network. It is noted that our approach is different from traditional data augmentation. Specifically, our method utilized the data without requiring its annotation by minimizing the output difference in teacher and student models under different transformations.
3.3. Loss Functions
3.3.1. Total Loss
The weakly-supervised segmentation network is trained by minimizing a weighted combination of the scribble-level supervised loss and consistency (, uncertainty-aware transformation consistency regularized unsupervised) loss. The combined objective function can be written as follows:
| (4) |
where is the number of the scribble annotated images in the training set. and are the supervised loss and consistency loss, respectively. A time-dependent Gaussian warming up function is used to control the trade-off between the two losses. Following [25], we use to control the balance of the two losses, where denotes the current training step and is the maximum training step. At the beginning of the training phase, is small and the supervised loss is dominant. This design can ensure the network learns accurate information from scribble annotations, which avoids getting stuck in a degenerate solution [52]. The first term of the loss function is represented by a standard cross-entropy loss on scribbled pixels. The second term of the combined function is represented by the weighted difference between the predictions of the student model and the teacher model.
3.3.2. Supervised Loss
Ignoring unscribbled pixels (pixels with label 2), we use the standard cross-entropy to define the supervised loss, which is defined as follows:
| (5) |
where is the set of the scribbled pixels for the image , and denote the coordinates of a pixel on the image . is the transformation of the scribbled label at pixel, and denote the prediction of the student model at pixel after transformation.
3.3.3. Consistency Loss
The mean teacher framework originally proposed for semi-supervised learning [25] typically divides data into labeled and unlabeled data in the training process. The labeled data with full annotations is used to calculate the loss between the segmentation result of the student model and the ground truth, and the consistency between the segmentation results generated by the two models. And the unlabeled data is only used to compute the consistency. All images are used to calculate the consistency in our method since pixel-wise ground truth of COVID19 CT images is not provided. When calculating the consistency loss , instead of using all the pixels equally, the uncertainty estimated by the teacher model is used to exclude the unreliable pixels. Based on the above analysis, loss is defined as the mean square error (MSE) of the prediction discrepancy between the teacher model and the student model as follows:
| (6) |
where is the indicator function and is the estimated uncertainty at the -th pixel (see Eq. (3)). The function is equal to 1 if is less than, and 0 otherwise. is a threshold to select the most certain targets. and are the prediction of the teacher model and the transformed prediction of the student model, respectively. We use the Gaussian ramp-up paradigm same to [24], to ramp up the uncertainty threshold from to , where is the maximum uncertainty value (, ln2 in our case).
The procedure of the proposed method USTM -Net is listed in Algorithm IX . In the training phase, the transformation consistency scheme is incorporated into our framework to regularize it. We use the same transformation operation randomly three times. The first operation is applied to transform the scribble annotations to get , the second operation is applied to transform the original image before it is input to the teacher model, and the third operation is applied to transform the predicted output from the student model to get . The supervised information for the model comes from the scribble annotations. is compared with using the supervised loss. At the same time, the teacher model guides the prediction results of the student model by calculating the pixel level uncertainty. The consistency loss is used to encourage the student model and teacher model predictions to become consistent without requiring their annotations.
Algorithm IX.
The proposed algorithm
| Input:; //: Original COVID-19 CT train set; : Scribbled labels corresponding to ; : The number of times that the teacher model is passed forward. |
| Output: with parameters ; with parameters ; //: The student model; : The teacher model. |
| 1: fort in [1, epochs]do |
| 2: foreach batchdo |
| 3: get image and scribble annotationfrom; |
| 4: randomly choose transformation operation; |
| 5: ; // transform the scribble labels |
| 6: add different Gaussian noises to the imageas the input of the student modeland the teacher model; |
| 7: fordo |
| 8: ; // transform the input of the teacher model |
| 9: ; // get the prediction from the teacher model |
| 10: end for; |
| 11: ; // obtain relatively reliable predictions and average them |
| 12: ; // get the prediction from the student model |
| 13: ; // transform the output of the student model |
| 14: ; // denotes the scribble-level supervised loss, see equation (5) |
| 15: ; // is the uncertainty-aware transformation consistency regularized unsupervised loss, see equation (6) |
| 16: ; // update the student model parameters, see equation (4) |
| 17: ; // update the teacher model parameters, see equation (1) |
| 18: end for; |
| 19: end for; |
3.4. Extension to Multi-Class Infection Labeling
Our USTM-Net can effectively identify and provide information about overall lung infections to doctors for diagnosis. However, it is necessary to evaluate different kinds of lung infections ( ground-glass opacity, and consolidation) in some clinical applications. The proposed USTM-Net can deal with multi-class segmentation to provide richer information for further diagnosis and treatment of COVID-19. To further separate ground-glass opacity from consolidation, the corresponding scribble annotation for the original image is changed to . The difference from the scribbles of the two classes is that both 1 and 2 are for scribbled infection pixels, which represent ground-glass opacity and consolidation, respectively, and 3 denotes unknown pixels.
4. Experiments and Results
4.1. Datasets
To evaluate our method, we conducted experiments on three COVID-19 CT datasets, including a local dataset (, uAI 3D) and two public datasets (, IS-COVID dataset [17] and Lesion Segmentation dataset [53]).
4.1.1. uAI 3D dataset
The uAI 3D CT dataset consists of 30 volumetric COVID-19 chest CT images from 30 patients and was obtained from Shanghai United Imaging Intelligence Inc.. The size of data varied from to , and the voxel resolution varied from to . All images of the dataset had been labeled with pixel-level ground truth by two experts with consensus. We extracted 4,000 2D CT axial slices to annotate scribbled labels. We removed the images which contained non-lung regions and also removed the slices without infections. For image pre-processing, we truncated the image intensity values of all slices to the HU range of [-800, 100] [29] to remove the irrelevant details. To improve the efficiency of the network training, the selected slices were resized to using bicubic interpolation and normalization. Our experiments randomly divided the dataset into two subsets, one of which was used for training and validation (3,200 slices from 24 patients), and the remaining one (800 slices from 6 patients) was used for testing. Only testing data requires full segmentation labels for comparison. On the whole, the scribbled pixels accounted for about 2% of the full labeled pixels, which indicated that scribble annotations were efficient and time-saving.
4.1.2. IS-COVID dataset [17]
IS-COVID dataset is a dataset with 110 axial lung CT slices from more than 40 patients with COVID-19 that were converted from openly accessible JPG images. All the CT slices were collected by the Italian Society of Medical and Interventional Radiology. Each slice was annotated for infection regions by a radiologist. The slice size ranged from to . The scribbles were generated in the same manner as the CC-COVID dataset. To balance the segmentation performance and computational cost, we first uniformly resized all the inputs to a fixed dimension of using bicubic interpolation before training. Ninety slices randomly selected from the 110 slices were used to build the training and validation set, and the remaining 20 were used for testing.
4.1.3. Lesion Segmentation (CC-COVID) dataset [53]
The CT images (slices) and metadata in this dataset were derived from the China Consortium of Chest CT Image Investigation (CC-CCII) [53]. All the CT images were classified into novel coronavirus pneumonia (NCP) caused by SARS-CoV-2 virus, common pneumonia, and normal controls. The dataset used in our experiments included 750 CT slices with the size of from 150 COVID-19 patients. Each slice was manually segmented into background, lung field, ground-glass opacity (GGO), and consolidation (CO). The original slices were resized to as the input to our network. Scribble annotations on CC-COVID were also manually drawn by referencing the fully annotated ground-truth labels provided in the dataset. We randomly selected 600 CT slices to create the training and validation set and the remaining 150 slices to create the test set.
4.2. Comparison Methods and Metrics
To investigate the infection segmentation performance of the proposed USTM-Net, we compared it with other five state-of-the-art methods, including Scribble2Label (S2L) model [44], weakly-supervised salient object detection (WSOD) method [54], partial U-Net (p-UNet) [55], weakly-supervised consistency-based learning (WSCL) method [18], and U-Net [49]. The first three methods are weakly supervised learning methods using scribbled annotations, and the fourth method is a weakly supervised method using point-level annotations. Although S2L is also based on scribble annotations, they focus on cell segmentation, which is different from our problem. The core idea of S2L is to combine pseudo-labeling with label filtering to generate reliable labels from weak supervision, while our method filters out the unreliable predictions using an uncertainty-aware scheme. WSOD introduces an auxiliary edge detection network and a gated structure-aware loss to place constraints on the scope of structure to be recovered. It also uses a scribble boosting scheme to iteratively integrate scribble annotations. WSCL proposes using a consistency-based loss function and two branches with shared weights in the network to encourage the consistency between the output predictions. The first branch takes the original images as the inputs whereas the second branch takes the transformed images as the inputs. One of the main differences between our framework and WSCL is that we use a teacher-student model instead of two that shared-weighted convolutional networks. Although it takes less time to annotate points than scribbles, scribble-level learning can incorporate more supervision information. For fair comparison, we re-implemented these methods on the abovementioned three datasets with the same network backbone as our methods. Partial U-Net uses scribble-level labels and it only calculates the cross-entropy loss with scribbled foreground and background pixels. In addition, we used full pixel-wise annotations to train a U-Net.
Our method used the improved 2D DenseUNet architecture [48] for both the teacher and student models. For the uAI 3D dataset, the network was trained with a batch size of 4 for a total of 20000 iterations with the Adam optimizer [56], and the initial learning rate was set to 0.0001. Data augmentation including rotation, flipping, and scaling are utilized on the student model with respect to scribble supervision. The same augmentation procedure is also applied in other compared methods. Our model was implemented using PyTorch, and was accelerated by an NVIDIA GeForce 1080 Ti GPU. The training time was around 18 hours on the uAI dataset and the testing time was around 40 milliseconds per slice.
Based on previous papers related to COVID-19 segmentation [5], [17], we utilized five widely adopted metrics to quantitatively evaluate the method, including dice coefficient (DI), Jaccard index (JA), sensitivity (SE), specificity (SP), and Mean Absolute Error (MAE). DI is a set similarity metric commonly used to measure the similarity between the predictions and the ground truth. DI is defined as:
| (7) |
where and refer to the number of true positives, true negatives, false positives, and false negative pixels of all the images in the test set, respectively. The equations for the other three metrics are expressed as follows:
| (8) |
MAE is also introduced to measure the pixel-wise errors between the prediction map and the ground-truth, which can be formulated as follows:
| (9) |
where and are the final prediction maps and the object-level segmentation ground-truth, respectively. and denote the image width and height.
4.3. Results on uAI Dataset
This section presents the results of qualitative and quantitative comparisons between the proposed USTM-Net and several comparison methods on uAI Dataset.
4.3.1. Qualitative Results
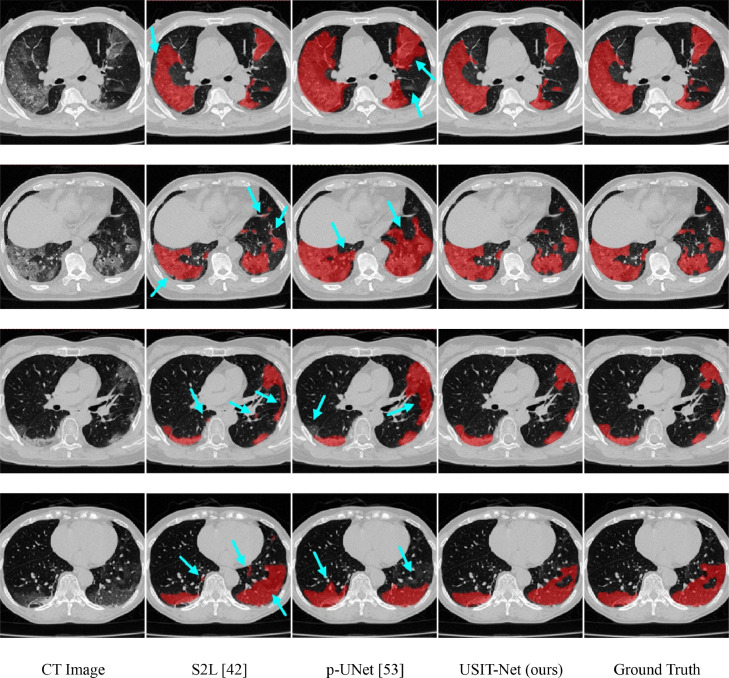
Fig. 6. shows the segmentation results of our scribble-supervised method and other two methods (S2L and p-UNet) on the uAI 3D CT dataset. In the experiment, we only segmented the lesion and the background areas without distinguishing the lesion types. By visually checking the segmentation results in Fig. 6., we can find that, for the large and clear infections (as shown in the first row), all the segmentation methods can get relatively good results. However, for cases of small lesion areas (as shown in the second and third rows) and lesions of ambiguous boundaries (as shown in the fourth row), our method achieved closer results to the ground truth than the other two weakly supervised methods. Although S2L (see in the second column) adopted a technique to combine pseudo-labels with scribbled labels, which still created unsatisfactory results and could not capture the boundary accurately. S2L also made several false positive results (, label normal tissues as infections) and some false negative results. Because p-Unet learned using only scribble annotations, p-Unet (in the third column) failed in yielding accurate segmentation results and produced more false positive results. These observations showed the advantage of the proposed uncertainty-aware mean teacher framework, which made our segmentation results (see in the fourth column) have a higher overlap ratio with the ground truth. In addition, the success of USTM-Net was due to the strategy of transformation consistency, which made our approach easier to deal with the segmentation of the irregular lesion areas. Overall, the results in Fig. 6 can reflect the effectiveness of the strategies in our method.
Fig. 6.
Examples of lung infection segmentation results of three methods on UAI dataset, where the red denotes the result of the segmented lesion and the blue arrows highlight some mis-segmentations.
4.3.2. Quantitative Results
We employed five metrics to perform quantitative comparisons. Table I lists the specific quantitative results obtained from the experiments on the uAI 3D dataset. As shown in Table I, our method achieved the highest performance on all the evaluation metrics. Compared with p-UNet, WSOD achieved competitive performance in most evaluation metrics. WSOD outperformed p-UNet with 2.3% improvement on DI, from 68.3% to 70.6%, and the performance on JA was also improved by 2.7%. Because it was learned only by scribbles, while WSOD explicitly used edge structure auxiliary information, p-UNet failed in predicting the results as accurately as WSOD. Under the same supervision, S2L performed better than other scribble-level weakly supervised methods. Our method achieved the best performance among all the weakly supervised methods, with DI of 76.2% and JA of 62.8%. Compared with S2L[42], the improvements were 1.7% on DI and 2.1% on SE. Compared with WSCL [18], our method improved by at least 4.3% on four metrics, including DI, JA, SE and SP. A smaller MAD value also indicated that the predictions of our method were closer to the ground truth. The results also confirmed that using uncertainty guidance and scribbles was effective in generating accurate boundaries, comparable to the point-level segmentation. Furthermore, it was observed that the performance obtained by our method was comparable to the performance obtained by full supervision. In terms of specificity, the proposed method outperformed the fully supervised U-Net model by 0.6%. This observation demonstrated that transformation consistency played an important role in our scribble label based weak segmentation and made our model produce better predictions.
Table I.
The quantitative results of infection areas on our uAI dataset compared to the corresponding ground-truth. The best results are highlighted in bold.
| Label | Methods | DI | JA | SE | SP | MAE |
|---|---|---|---|---|---|---|
| Scribble | p-UNet [55] | 0.683 | 0.531 | 0.837 | 0.836 | 0.127 |
| WSOD [54] | 0.706 | 0.558 | 0.871 | 0.885 | 0.093 | |
| S2L [44] | 0.745 | 0.607 | 0.843 | 0.956 | 0.087 | |
| USTM -Net (proposed) | 0.762 | 0.628 | 0.882 | 0.984 | 0.076 | |
| Point | WSCL [18] | 0.719 | 0.573 | 0.826 | 0.929 | 0.114 |
| Full | U-Net [49] | 0.788 | 0.663 | 0.885 | 0.978 | 0.071 |
4.4. Ablation Study
We introduced the uncertainty-aware self-ensembling and transformation-consistent techniques in the network to improve the segmentation predictions in the proposed method. This section will first describe the ablation experiments to evaluate the two techniques’ impacts on our framework. Then, we describe the experiments only using one type of transformation operation. We also provide the experiments with the U-Net [49] architecture and VGG16-FCN8 [57] network as the backbone networks. Finally, we explore the effects of weighting parameters on the loss function. All the experiments were performed on the uAI dataset.
4.4.1. Effectiveness of Uncertainty-aware and Transformation-consistent Strategies
To investigate the effectiveness of the proposed uncertainty-aware scheme and transformation-consistent strategy, we conducted ablation experiments on the uAI dataset, and the results are shown in Table II . Table II. “baseline” refers to scribble-supervised DenseUNet with ASPP, without transformation consistency or uncertainty guidance. In other words, we used the original teacher and student model with scribble annotations in the training process. “USTM-T” refers to the baseline with the transformation consistency strategy. “USTM-U” refers to the baseline with an uncertainty guidance scheme. Our proposed “USTM” refers to the baseline with both the transformation consistency strategy and uncertainty guidance scheme. As shown in Table II, both strategies independently contributed to the performance of the weakly supervised learning. By introducing the transformation consistency strategy into the baseline, the performance was boosted from 71.4% to 74.3% in terms of DI, and from 56.9% to 60.4% in terms of JA. The DI of “USTM-U” is 73.6%, which was a 2.2% increment compared with the baseline framework. From these two comparisons, we can find that the improvement of transformation-consistent strategy was very competitive, compared with the uncertainty guidance scheme. It is worth mentioning that the performance can be further enhanced when they were employed together, with 76.2% in terms of DI and 62.8% in terms of JA. This significantly outperformed the baseline framework by 4.8% in terms of DI and 5.9% in terms of JA.
Table II.
Ablation of our USTM-Net method on uAI dataset. “T” denotes transformation. “U” denotes uncertainty guidance. “TU” denotes both of these operations. The best results are highlighted in bold.
| Setting | DI | JA | SE | SP | MAE |
|---|---|---|---|---|---|
| baseline | 0.714 | 0.569 | 0.822 | 0.924 | 0.094 |
| USTM-T | 0.743 | 0.604 | 0.852 | 0.957 | 0.082 |
| USTM-U | 0.736 | 0.592 | 0.834 | 0.940 | 0.085 |
| USTM (proposed) | 0.762 | 0.628 | 0.882 | 0.984 | 0.076 |
4.4.2. Ablation Experiments for Different Types of Transformations
In this section, we analyze the impact of different types of transformation operations. Three different types of transformation operations (, flip, scale, rotation) were investigated in the experiments. The results are listed in Table III . From Table III, we can find that random rotation improved the results noticeably, and other transformations also positively affected the results. Compared with the baseline framework, the random rotations improved the performance by 1.8% in DI and 2.5% in JA. For the flipping operations, there was also a slight increase in the performance (from 71.4% to 72.1% in DI compared with the baseline framework). In addition, compared with “USTM-T” in Table II, it is observed that the generalized form of transformation-consistent strategy improved the weak-supervised learning more effectively.
Table III.
Ablation experiments for different types of transformations on uAI dataset. The best results are highlighted in bold.
| Transformation operations | DI | JA | SE | SP | MAE |
|---|---|---|---|---|---|
| baseline | 0.714 | 0.569 | 0.822 | 0.924 | 0.094 |
| Flip | 0.721 | 0.575 | 0.833 | 0.937 | 0.090 |
| Scale | 0.725 | 0.581 | 0.836 | 0.951 | 0.089 |
| Rotate | 0.732 | 0.594 | 0.841 | 0.942 | 0.086 |
4.4.3. Effectiveness of the Backbone Segmentation Network
As abovementioned, our framework can use a variety of segmentation networks. We conducted two experiments with a U-Net architecture [49] and VGG16-FCN8 network [57] as the backbone network, respectively. As shown in Table IV , using U-Net [49] architecture as the backbone network, our method performed better on four quality metrics than other comparison methods. For example, compared with WSCL [18], the proposed method improved the performance from 70.2% to 75.1% in DI. Using VGG16 FCN8 [57] as the backbone network, our method was also superior to other comparative methods on all metrics. Overall, when using one of these two networks as the backbone network, the results were inferior to the one using the improved DenseUNet network on uAI 3D dataset, which proved the effectiveness of our segmentation network.
Table IV.
The quantitative results of infection areas on our uAI30 dataset compared to the corresponding ground-truth using U-Net architecture [49] and VGG16 FCN8 network [57]. The best results are highlight in bold.
4.4.4. Effects of Weighting Parameters on Loss Function
The segmentation network was optimized by the joint loss function defined in Eq. (4). The joint function consisted of two components, which influenced the training of the network through different weight parameters. A time-dependent Gaussian warming up function [24] was used to weight the two losses. For comparison, we also set the weighting parameter be 0.05, 0.1. 0.5 and 1 to train the network and used the trained network to test. The experimental results are shown in Table V . From the results in Table V, we can find that our setup achieved the best performance.
Table V.
Average result of metrics on our proposed method with different weights for segmentation network function
| λ | DI | JA | SE | SP |
|---|---|---|---|---|
| λ=0.05 | 0.758 | 0.621 | 0.880 | 0.962 |
| λ=0.1 | 0.754 | 0.619 | 0.879 | 0.966 |
| λ=0.5 | 0.749 | 0.613 | 0.873 | 0.954 |
| λ=1 | 0.743 | 0.605 | 0.864 | 0.951 |
| 0.762 | 0.628 | 0.882 | 0.984 |
4.4.5. Sensitivities to Scribble Quality
Since the scribbles are manually annotated, the quality of the scribbles depends on the habit and experience of the annotators. In this case, we investigated the influence of the annotation quality on the performance of our method. Since labeling the dataset with different scribbles requires additional time and labor, we used existing scribble annotations to generate new scribbles, similar to [58]. Specifically, we used the existing annotations as a baseline to scale down the length of each scribble. We randomly selected a point from the original scribble as one of the end-points of the shortened scribble, and the length of the reduction determined the other end-point.
Table VI shows the results of our USTM-Net method with different lengths of scribbles for supervision. When the length ratio dropped from 1 to 0.7, the performance of the network decreased slowly, the performance of DI and JA was only reduced by 0.016 and 0.025, respectively. The stable performance indicated that our method was quite robust for the scribble quality. When the length ratio was reduced from 0.7 to 0.5, the performance of the network decreased faster. It deserves to note that it is not difficult to obtain reasonable quality scribbles in practice.
Table VI.
Sensitivities to scribble length evaluated on the uAI dataset. The length ratio is the ratio by which a scribble is shortened. The best results are highlighted in bold.
| length ratio | DI | JA | SE | SP |
|---|---|---|---|---|
| 1(proposed) | 0.762 | 0.628 | 0.882 | 0.984 |
| 0.9 | 0.759 | 0.623 | 0.879 | 0.978 |
| 0.8 | 0.752 | 0.616 | 0.874 | 0.971 |
| 0.7 | 0.746 | 0.603 | 0.867 | 0.964 |
| 0.6 | 0.735 | 0.591 | 0.855 | 0.952 |
| 0.5 | 0.723 | 0.572 | 0.841 | 0.938 |
4.5. Results on IS-COVID Dataset
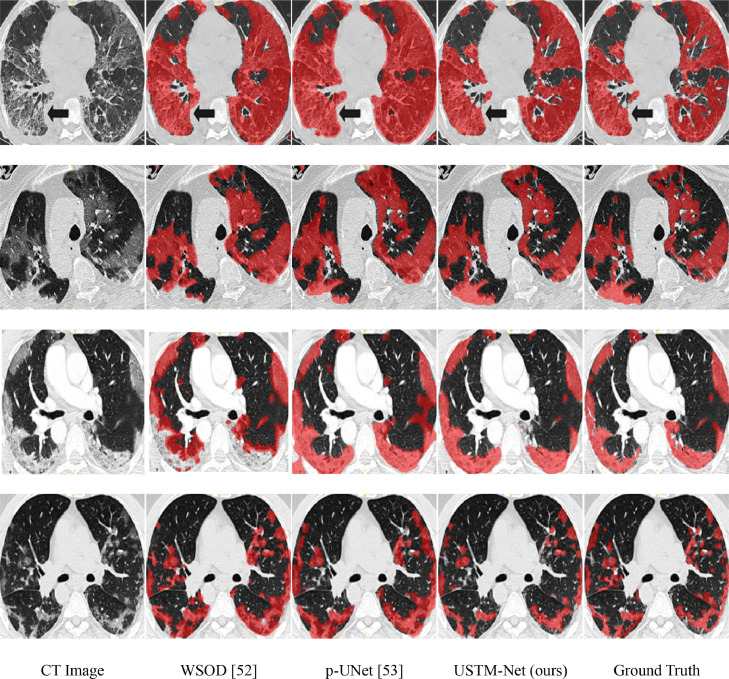
IS-COVID [17] is a smaller dataset compared with uAI 3D dataset and CC-COVID dataset. In this dataset, we also segmented the infections besides the background without distinguishing the infection types. Fig. 7 shows the segmentation results on IS-COVID dataset by two other weakly supervised methods. The experiments showed that WSOD may face difficulty in correctly identifying some tissue areas and lesions, resulting in missing lesion segments and ambiguous boundaries. The segmentation of p-UNet was not very accurate. Compared with other methods, the predictions of the proposed method had the highest match with the ground truth. The quantitative results for each comparison method are listed in Table VII . S2L performed slightly better than WSOD on the IS-COVID dataset. The results showed that the proposed USTM-Net achieved the best performance among all compared weakly-supervised methods.
Fig. 7.
Examples of lung infection segmentation results of three methods on IS-COVID dataset [17]. Red denotes the results of the segmented lesions.
Table VII.
The quantitative results of infection areas on IS-COVID dataset [17]. The best results are highlighted in bold.
4.6. Results on CC-COVID Dataset
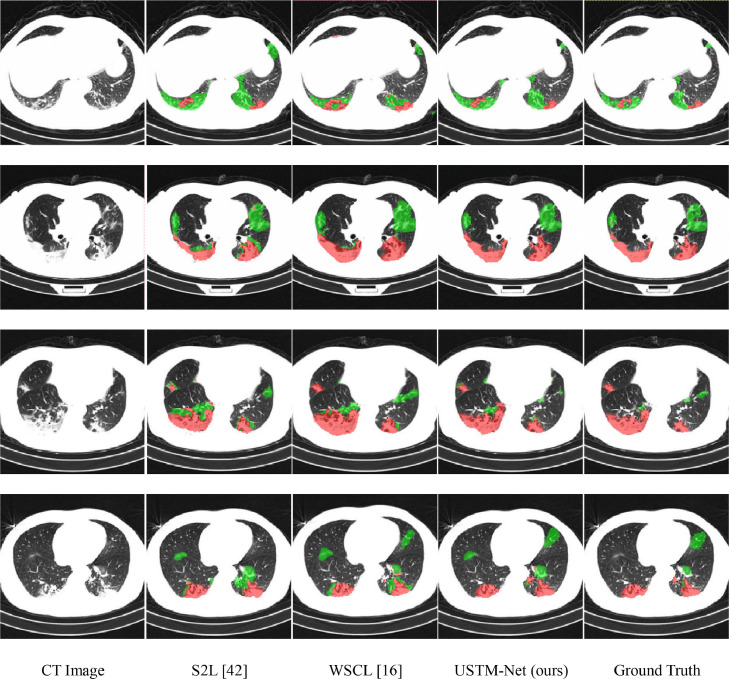
In clinical diagnosis, distinguishing different types of infections can provide more quantitative information about the infected areas. Therefore, we extended our method to multi-class (, GGO and CO) labeling. Then, we evaluated our multi-class weakly supervised learning model on CC-COVID dataset [53]. The challenge of the multi-class segmentation problem is that the two lesions (GGO and CO) are difficult to distinguish. Fig. 8 shows typical multi-class infection segmentation results on CC-COVID dataset [53]. From Fig. 8, we can find that both the segmentation results of GGO and CO obtained by the proposed USTM-Net had higher overlap with the ground truth than other methods, which further indicated the effectiveness of our model. In contrast, other weakly supervised methods obtained unsatisfactory results, where neither GGO nor CO can be accurately segmented. For example, WSCL mistakenly segmented part of the background as infections in some images. Besides, both WSCL and S2L confused these two types of infections, leading to inaccurate segmentation. It was obvious that the proposed USTM-Net, obtained the best performance among all the weakly-supervised methods.
Fig. 8.
Visual comparison of multi-class lung infection segmentation results on CC-COVID dataset [53], where the green and red labels indicate the GGO and CO, respectively.
The quantitative comparison is listed in Table VIII . As can be seen from the table, the proposed USTM-Net outperformed other weakly supervised methods in terms of DI, SE, and SP to the segmentation of the infections. For GGO, our method also achieved the best results among all the weakly supervised methods. For instance, our weakly supervised method improved the performance by 6.6% in terms of DI, compared with p-UNet on GGO. Compared with S2L, our method improved from 71.1% to 72.3% in terms of DI on GGO and CO. In addition, our proposed method achieved similar results with the fully supervised U-Net [49] on average. Both the quantitative and qualitative comparisons demonstrated that the proposed scribble supervision was able to outperform other weakly supervised methods and performed competitively compared to the fully supervised methods.
Table VIII.
Quantitative results of GGO and CO on CC-COVID dataset [53]. Bold fonts show the best results.
| Label | Methods | Consolidation |
Ground-Glass Opacity |
Average |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DI | SE | SP | DI | SE | SP | DI | SE | SP | ||
| Scribble | p-UNet [55] | 0.672 | 0.806 | 0.908 | 0.643 | 0.789 | 0.894 | 0.658 | 0.798 | 0.901 |
| WSOD [54] | 0.695 | 0.833 | 0.917 | 0.674 | 0.801 | 0.902 | 0.685 | 0.817 | 0.910 | |
| S2L [44] | 0.724 | 0.857 | 0.934 | 0.698 | 0.840 | 0.928 | 0.711 | 0.849 | 0.931 | |
| USTM-Net (proposed) | 0.736 | 0.862 | 0.958 | 0.709 | 0.829 | 0.947 | 0.723 | 0.846 | 0.953 | |
| Point | WSCL [18] | 0.705 | 0.827 | 0.920 | 0.681 | 0.803 | 0.916 | 0.693 | 0.815 | 0.918 |
| Full | U-Net [49] | 0.748 | 0.874 | 0.966 | 0.713 | 0.825 | 0.952 | 0.731 | 0.850 | 0.959 |
5. Conclusion
This paper proposed a novel and effective weakly supervised method for the segmentation of COVID-19 infections in CT slices with scribble supervision. The whole framework was constructed with a mean teacher framework and optimized by a weighted combination of the supervised and unsupervised losses. Specifically, we introduced a transformation-consistent technique to enhance the accuracy of the segmentation. We also explored the uncertainty-aware self-ensembling strategy to improve the quality of segmentation. The comparisons with other weakly supervised methods demonstrated the effectiveness of the proposed method. The experiment results on a local dataset and two public datasets demonstrated the performance of the proposed method, which outperformed four state-of-the-art weakly supervised methods, and achieved similar performance even to the fully supervised methods. In the future work, we will further explore more transformations (such as deformable transformation) in our segmentation framework and investigate scribble-based COVID-19 infection segmentation on 3D CT images directly instead of slices. Besides, we need to avoid the occurrence of false annotations since annotation errors in weak supervision may degrade the performance of the proposed method. Although some methods have been proposed to deal with noise annotations in segmentation with dense annotations, few of them can deal with weak supervision-based segmentation [5], [59], [60]. In prospective work, we will try to research an effective method to overcome the impact of noise annotations on the network in the case of a large number of inaccurate labels.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by the National Natural Science Foundation of China under Grant 62176190.
Biographies
Xiaoming Liu received the Ph.D degree from Zhejiang University, China, in 2007. From 2014 to 2015, he was a Visiting Scholar in the University of North Carolina at Chapel Hill, NC USA. Currently, he is a Professor with the School of Computer Science and Technology, Wuhan University of Science and Technology, Wuhan, China. His research interests include medical image processing, pattern recognition, and machine learning.
Quan Yuan is currently a graduate student in the School of Computer Science and Technology, Wuhan University of Science and Technology, Wuhan, China. Her research interests are medical image processing and machine learning.
Yaozong Gao received the B.S. degree in software engineering and the M.S. degree in medical image analysis from Zhejiang University, in 2008 and 2011, respectively, and the Ph.D. degree from the Department of Computer Science, University of North Carolina at Chapel Hill. He was a Computer Vision Researcher with Apple Inc. He is currently directing the Deep Learning Group, United Imaging Intelligence, China. He has published over 90 papers in the international journals and conferences, such as MICCAI, TMI, and MIA. His research interests include machine learning, computer vision, and medical image analysis.
Kelei He received the Ph.D. degree in computer science and technology from Nanjing University, China. He is currently the assistant dean of National Institute of Healthcare Data Science at Nanjing University. He is also an assistant researcher of Medical School at Nanjing University, China. His research interests include medical image analysis, computer vision and deep learning.
Shuo Wang is currently a graduate student in the School of Computer Science and Technology, Wuhan University of Science and Technology, Wuhan, China. Her research interests are medical image processing and machine learning.
Xiao Tang is with Department of Medical Imaging, Tianyou Hospital Affiliated to Wuhan University of Science and Technology, he has over 20 years’ experience in radiology.
Jinshan Tang is currently a professor at George Mason University. He received his Ph.D. from Beijing University of Posts and Telecommunications, and got post-doctoral training in Harvard Medical School and National Institute of Health. He has published more than 100 papers. His research interests include biomedical image processing, biomedical imaging. He is a leading guest editor of several journals on medical image processing and computer aided cancer detection.
Dinggang Shen Professor, IEEE Fellow, AIMBE Fellow, IAPR Fellow. His research interests include medical image analysis, computer vision, and pattern recognition. He has published more than 1000 papers in the international journals and conference proceedings, with h-index of 105. He serves as an editorial board member for eight international journals, and was General Chair for MICCAI 2019.
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.F. Shan, Y. Gao, J. Wang, W. Shi, N. Shi, M. Han, Z. Xue and Y. Shi, Lung infection quantification of covid-19 in ct images with deep learning, arXiv preprint arXiv:2003.04655, 2020.
- 3.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan H., Qi F., Xue Z., Chen L., Shen D. Multimodality image registration by maximization of quantitative–qualitative measure of mutual information. Pattern Recognition. 2008;41(1):285–298. [Google Scholar]
- 5.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Transactions on Medical Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. Ma, Y. Wang, X. An, C. Ge, Z. Yu, J. Chen, Q. Zhu, G. Dong, J. He and Z. He, Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation, arXiv preprint arXiv:2004.12537, 2020.
- 7.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Reviews in Biomedical Engineering. 2020;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 8.Cao X., Yang J., Zhang J., Nie D., Kim M., Wang Q., Shen D. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2017. Deformable image registration based on similarity-steered CNN regression; pp. 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z., Tang J., Wang Z., Zhang K., Zhang L., Sun Q. Deep learning for image-based cancer detection and diagnosis− a survey. Pattern Recognition. 2018;83:134–149. [Google Scholar]
- 10.Shen D., Wu G., Suk H.-I. Deep learning in medical image analysis. Annual review of biomedical engineering. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Yu A., Wei X., Pan Z., Tang J. Multimodal MR Image Synthesis Using Gradient Prior and Adversarial Learning. IEEE Journal of Selected Topics in Signal Processing. 2020;14(6):1176–1188. [Google Scholar]
- 12.Liu X., Zhu T., Zhai L., Liu J. Mass classification of benign and malignant with a new twin support vector machine joint l2,1-norm. International Journal of Machine Learning and Cybernetics. 2019;10(1):155–171. [Google Scholar]
- 13.Zhang J., Liu M., Shen D. Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Transactions on Image Processing. 2017;26(10):4753–4764. doi: 10.1109/TIP.2017.2721106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J., Cao X., Yap P.-T., Shen D. BIRNet: Brain image registration using dual-supervised fully convolutional networks. Medical image analysis. 2019;54:193–206. doi: 10.1016/j.media.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Shi F., Lin W., Gilmore JH., Shen D. Automatic segmentation of neonatal images using convex optimization and coupled level sets. NeuroImage. 2011;48(3):805–817. doi: 10.1016/j.neuroimage.2011.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE transactions on medical imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 17.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Transactions on Medical Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 18.Laradji I., Rodriguez P., Manas O., Lensink K., Law M., Kurzman L., Parker W., Vazquez D., Nowrouzezahrai D. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 2021. A weakly supervised consistency-based learning method for covid-19 segmentation in ct images; pp. 2453–2462. [Google Scholar]
- 19.Goceri E. Deep learning based classification of facial dermatological disorders. Computers in Biology and Medicine. 2021;128 doi: 10.1016/j.compbiomed.2020.104118. [DOI] [PubMed] [Google Scholar]
- 20.Goceri E. Diagnosis of skin diseases in the era of deep learning and mobile technology. Computers in Biology and Medicine. 2021;134 doi: 10.1016/j.compbiomed.2021.104458. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Zhang J., Kan M., Shan S., Chen X. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Self-supervised equivariant attention mechanism for weakly supervised semantic segmentation; pp. 12275–12284. [Google Scholar]
- 22.Goceri E. CapsNet topology to classify tumours from brain images and comparative evaluation. IET Image Processing. 2020;14(5):882–889. [Google Scholar]
- 23.Li X., Yu L., Chen H., Fu C.-W., Xing L., Heng P.-A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Transactions on Neural Networks and Learning Systems. 2020;32(2):523–534. doi: 10.1109/TNNLS.2020.2995319. [DOI] [PubMed] [Google Scholar]
- 24.Yu L., Wang S., Li X., Fu C.-W., Heng P.-A. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation; pp. 605–613. [Google Scholar]
- 25.Tarvainen A., Valpola H. Advances in neural information processing systems. 2017. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results; pp. 1195–1204. [Google Scholar]
- 26.Goceri E. 2020 IEEE 4th International Conference on Image Processing, Applications and Systems (IPAS) IEEE; 2020. Image Augmentation for Deep Learning Based Lesion Classification from Skin Images; pp. 144–148. [Google Scholar]
- 27.Goceri E. 2019 ninth international conference on image processing theory, tools and applications (IPTA) IEEE; 2019. Challenges and recent solutions for image segmentation in the era of deep learning; pp. 1–6. [Google Scholar]
- 28.Schlegl T., Seeböck P., Waldstein S.M., Schmidt-Erfurth U., Langs G. International conference on information processing in medical imaging. Springer; 2017. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery; pp. 146–157. [Google Scholar]
- 29.Q. Yao, L. Xiao, P. Liu and S. K. Zhou, Label-Free Segmentation of COVID-19 Lesions in Lung CT, arXiv preprint arXiv:2009.06456, 2020. [DOI] [PMC free article] [PubMed]
- 30.Van Engelen J.E., Hoos H.H. A survey on semi-supervised learning. Machine Learning. 2020;109(2):373–440. [Google Scholar]
- 31.Lee D.-H. Workshop on challenges in representation learning, ICML. Vol. 3. 2013. Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. [Google Scholar]
- 32.Tajbakhsh N., Jeyaseelan L., Li Q., Chiang J.N., Wu Z., Ding X. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Medical Image Analysis. 2020 doi: 10.1016/j.media.2020.101693. [DOI] [PubMed] [Google Scholar]
- 33.Cui W., Liu Y., Li Y., Guo M., Li Y., Li X., Wang T., Zeng X., Ye C. International Conference on Information Processing in Medical Imaging. Springer; 2019. Semi-supervised brain lesion segmentation with an adapted mean teacher model; pp. 554–565. [Google Scholar]
- 34.Liu X., Cao J., Fu T., Pan Z., Hu W., Zhang K., Liu J. Semi-supervised automatic segmentation of layer and fluid region in retinal optical coherence tomography images using adversarial learning. IEEE Access. 2018;7:3046–3061. [Google Scholar]
- 35.Nie D., Gao Y., Wang L., Shen D. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2018. ASDNet: Attention based semi-supervised deep networks for medical image segmentation; pp. 370–378. [Google Scholar]
- 36.Dong N., Kampffmeyer M., Liang X., Wang Z., Dai W., Xing E. International conference on medical image computing and computer-assisted intervention. Springer; 2018. Unsupervised domain adaptation for automatic estimation of cardiothoracic ratio; pp. 544–552. [Google Scholar]
- 37.Zhou Y., Li Z., Bai S., Wang C., Chen X., Han M., Fishman E., Yuille A.L. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. Prior-aware neural network for partially-supervised multi-organ segmentation; pp. 10672–10681. [Google Scholar]
- 38.Shi G., Xiao L., Chen Y., Zhou S.K. Marginal loss and exclusion loss for partially supervised multi-organ segmentation. Medical Image Analysis. 2021;70 doi: 10.1016/j.media.2021.101979. [DOI] [PubMed] [Google Scholar]
- 39.Grill J.-B., Strub F., Altché F., Tallec C., Richemond P.H., Buchatskaya E., Doersch C., Pires B.A., Guo Z.D., Azar M.G. Conference on Neural Information Processing Systems. 2020. Bootstrap your own latent: A new approach to self-supervised learning. [Google Scholar]
- 40.Chen X., He K. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021. Exploring simple siamese representation learning; pp. 15750–15758. [Google Scholar]
- 41.Wu K., Du B., Luo M., Wen H., Shen Y., Feng J. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. Weakly supervised brain lesion segmentation via attentional representation learning; pp. 211–219. [Google Scholar]
- 42.Rajchl M., Lee M.C., Oktay O., Kamnitsas K., Passerat-Palmbach J., Bai W., Damodaram M., Rutherford M.A., Hajnal J.V., Kainz B. Deepcut: Object segmentation from bounding box annotations using convolutional neural networks. IEEE transactions on medical imaging. 2016;36(2):674–683. doi: 10.1109/TMI.2016.2621185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matuszewski D.J., Sintorn I.-M. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) IEEE; 2018. Minimal annotation training for segmentation of microscopy images; pp. 387–390. [Google Scholar]
- 44.Lee H., Jeong W.-K. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2020. Scribble2Label: Scribble-Supervised Cell Segmentation via Self-generating Pseudo-Labels with Consistency; pp. 14–23. [Google Scholar]
- 45.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
- 46.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. Abnormal Lung Quantification in Chest CT Images of COVID-19 Patients with Deep Learning and its Application to Severity Prediction. Medical physics. 2020 doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. Tilborghs, I. Dirks, L. Fidon, S. Willems, T. Eelbode, J. Bertels, B. Ilsen, A. Brys, A. Dubbeldam and N. Buls, Comparative study of deep learning methods for the automatic segmentation of lung, lesion and lesion type in CT scans of COVID-19 patients, arXiv preprint arXiv:2007.15546, 2020.
- 48.Li X., Chen H., Qi X., Dou Q., Fu C.-W., Heng P.-A. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE transactions on medical imaging. 2018;37(12):2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 49.Ronneberger O., Fischer P., Brox T. International Conference on Medical image computing and computer-assisted intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 50.Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. Proceedings of the European conference on computer vision (ECCV) 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 801–818. [Google Scholar]
- 51.Worrall D.E., Garbin S.J., Turmukhambetov D., Brostow G.J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Harmonic networks: Deep translation and rotation equivariance; pp. 5028–5037. [Google Scholar]
- 52.S. Laine and T. Aila, Temporal ensembling for semi-supervised learning, arXiv preprint arXiv:1610.02242, 2016.
- 53.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J., Yu X., Li A., Song P., Liu B., Dai Y. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2020. Weakly-supervised salient object detection via scribble annotations; pp. 12546–12555. [Google Scholar]
- 55.Tang M., Djelouah A., Perazzi F., Boykov Y., Schroers C. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018. Normalized cut loss for weakly-supervised cnn segmentation; pp. 1818–1827. [Google Scholar]
- 56.D. P. Kingma and J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980, 2014.
- 57.Long J., Shelhamer E., Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2015;39(4):640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 58.Lin D., Dai J., Jia J., He K., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Scribblesup: Scribble-supervised convolutional networks for semantic segmentation; pp. 3159–3167. [Google Scholar]
- 59.Zhang T., Yu L., Hu N., Lv S., Gu S. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2020. Robust medical image segmentation from non-expert annotations with tri-network; pp. 249–258. [Google Scholar]
- 60.Shu Y., Wu X., Li W. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2019. Lvc-net: Medical image segmentation with noisy label based on local visual cues; pp. 558–566. [Google Scholar]