Abstract
Fifty methicillin-resistant Staphylococcus aureus (MRSA) initial isolates obtained from patients hospitalized in the orthopedic clinic of the Frankfurt University Hospital and 150 methicillin-sensitive Staphylococcus aureus (MSSA) isolates were investigated in this study to determine whether the Slidex Staph-Kit is capable of differentiating between MRSA and MSSA owing to its unique performance characteristics. The Slidex Staph-Kit is a combined latex hemagglutination test designed to detect clumping factor, protein A, and a specific surface immunogen for S. aureus. Clumping factor-positive strains cause erythrocytes sensitized with fibrinogen to hemagglutinate, thereby resulting in visible red clumps. S. aureus strains deficient in clumping factor agglutinate latex particles sensitized with specific antibodies against surface proteins of S. aureus, thereby resulting in visible white clumps. Our results demonstrate that white clumping has a 99% specificity as well as a 98% positive predictive value for MRSA. Clumping factor-negative MRSA, which have been reported to occur in several countries, are epidemic in the Frankfurt area and account for 80% of all MRSA initial isolates in the orthopedic clinic of the Frankfurt University Hospital. Genotyping of all MRSA isolates by macrorestriction analysis of chromosomal DNA revealed that 83% of clumping factor-negative MRSA are closely related to the “southern-German” epidemic strain. This is the first study demonstrating the Slidex Staph-Kit’s capability for identifying epidemic clumping factor-negative S. aureus strains as methicillin resistant even prior to antimicrobial susceptibility testing.
Methicillin-resistant Staphylococcus aureus (MRSA) strains were initially described in 1961 and emerged in the 1980s as a major epidemiological problem in hospital settings (8, 16). MRSA strains with widely different properties have now become endemic in hospitals and are considered important nosocomial pathogens (14, 24, 26). Methicillin resistance rates differ markedly among countries and range from less than 1% in Scandinavia to 60% in Japan (11, 27).
Accordingly, rapid detection of MRSA is crucial for purposes of initiating hygienic measures and preventing further spread of the pathogen. Commercially produced agglutination kits are available for the rapid identification of S. aureus by clinical laboratories. Nonetheless, virtually all of them have the drawback that additional time-consuming antimicrobial susceptibility testing generally is required in order to detect methicillin resistance in S. aureus.
The aim of this study was to assess the capability and specificity with which the Slidex Staph-Kit differentiates between MRSA and methicillin-susceptible S. aureus (MSSA).
MATERIALS AND METHODS
Bacterial strains.
A total of 50 MRSA isolates and 150 MSSA isolates were investigated in this study. The MRSA initial isolates were cultured from clinical specimens obtained from the orthopedic clinic of the Frankfurt University Hospital between 1993 and 1997. All isolates had been classified as MRSA during routine investigations and were stored in stock cultures prior to the study, as described previously (29). The MSSA isolates were cultured consecutively from clinical specimens obtained from the Frankfurt University Hospital. S. aureus ATCC 25923 was employed as the reference strain.
Agglutination kits and further characterization.
The Slidex Staph-Kit (bioMerieux Vitek, Inc., Hazelwood, Mo.) is a latex and erythrocyte combination agglutination system for the detection of clumping factor, protein A, and other specific immunogens of S. aureus. The Staphylase test (Oxoid, Basingstoke, England) is an erythrocyte agglutination kit for the detection of clumping factor. Both agglutination tests were performed according to the manufacturers’ instructions. A tube coagulase test was performed on each isolate. Plasma coagulation was tested with rabbit plasma (Difco, Detroit, Mich.), and the results were read after 2, 4, 6, and 24 h at 37°C. All isolates were screened for hyaluronidase activity by testing the decapsulation reaction on a cross-inoculated streak of a mucous strain of Streptococcus equi (9, 19).
Detection of methicillin resistance.
All of the S. aureus isolates were cultured on Mueller-Hinton agar supplemented with 6 μg of oxacillin/ml and 4% NaCl (Biotest, Heidelberg, Germany) (13, 20). The plates were incubated at 30°C for 48 h and examined for evidence of growth to detect resistance phenotypically. The presence of the mecA gene was proved for all strains by means of PCR. Purification of bacterial DNA was carried out with the QIAmp tissue kit (Qiagen, Hilden, Germany) in accordance with the manufacturers’ instructions. Amplification of the mecA gene was performed with the primers mecA1 (5′-AAA ATC GAT GGT AAA GGT TGG C) and mecA2 (5′-AGT TCT GCA GTA CCG GAT TTG C), yielding a PCR product of 533 bp (15). DNA amplification was carried out for 30 cycles as previously described (25): denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 2 min, with a final extension at 72°C for 5 min. The PCR product was cleaved by the restriction enzyme HhaI (New England BioLabs, Schwalbach, Germany) to confirm the specificity of the mecA primers (25). Electrophoresis of DNA was carried out in 2% agarose gel, which was then stained with ethidium bromide and photographed under UV light.
Genotyping.
All MRSA isolates were analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (29). Briefly, genomic DNA was prepared in low-melting-point agarose plugs and digested with SmaI restriction enzyme (New England BioLabs). Electrophoresis was performed on the CHEF-DR III (Bio-Rad Laboratories, Richmond, Calif.) apparatus. A constant voltage of 6 V/cm was applied, with an increasing pulse time of 5 to 50 s over a period of 22 h in order to separate DNA fragments. Computer-aided analysis of DNA fragment patterns was performed with the DNA fingerprint analysis software Wincam 2.2 (Cybertech, Berlin, Germany).
RESULTS
Strain characterization.
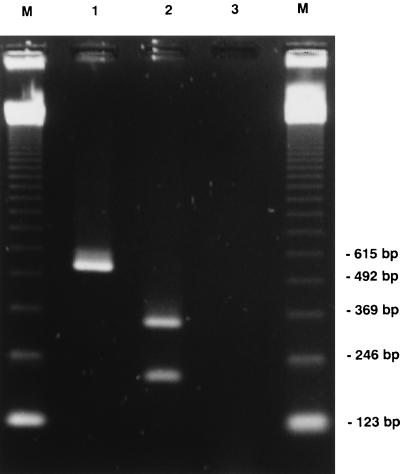
Coagulase production and hyaluronidase activity was demonstrated for all S. aureus isolates in this study. Methicillin resistance was proved genotypically for all isolates by amplification of the mecA gene (Fig. 1) as well as phenotypically by growth on Mueller-Hinton agar supplemented with 6 μg of oxacillin/ml and 4% NaCl.
FIG. 1.
Agarose gel electrophoresis of mecA PCR product. Lanes: M, size markers; 1, mecA-positive S. aureus strain; 2, restriction of amplified DNA with HhaI; 3, mecA-negative S. aureus reference strain ATCC 25923.
Genotyping.
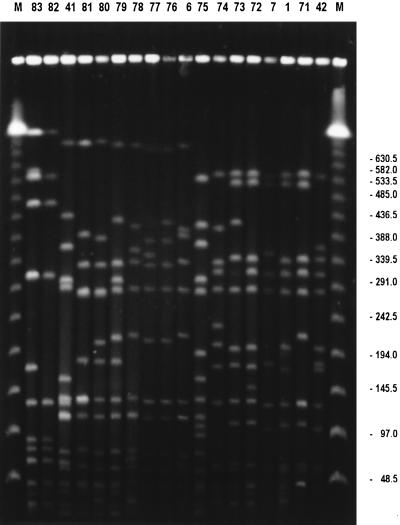
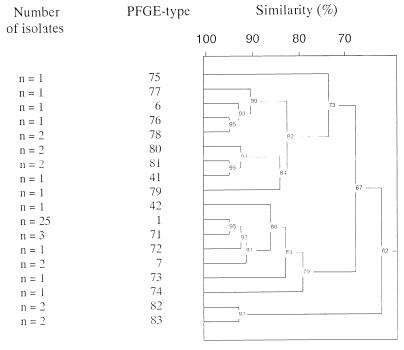
Macrorestriction analysis of 50 MRSA initial isolates obtained from the orthopedic clinic of the Frankfurt University Hospital in a 5-year period revealed 18 different genotypes (Fig. 2). Fifty percent of all isolates belonged to type 1, and strongly corresponding restriction fragment patterns indicating close clonal relatedness between MRSA types 1, 7, 71, and 72 were obvious (Fig. 3).
FIG. 2.
PFGE patterns of SmaI digests of total DNA from representatives of each pulsotype (indicated by numbers above lanes). Lanes M, size markers. Molecular sizes (in kilobases) are indicated on the right.
FIG. 3.
Computer-aided analysis of different PFGE types, indicating clonal relatedness.
Agglutination performance.
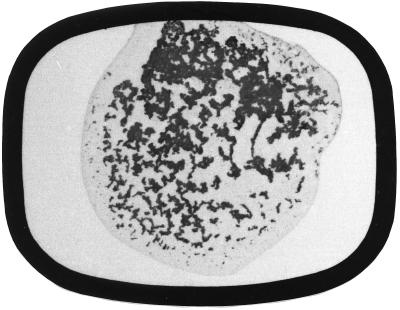
Table 1 compares the performance of the Slidex Staph-Kit with that of the Staphylase agglutination test. It could be demonstrated that all MRSA initial isolates closely related to the epidemic strain, as well as isolates belonging to MRSA types 6, 76, 82, and 83, exhibited white-clumping behavior (Fig. 4). In every instance where the Slidex Staph-Kit exhibited white clumping the Staphylase agglutination test was negative, which indicates that these strains were at least phenotypically clumping factor negative. In contrast, 149 of 150 MSSA isolates classified during routine investigation were determined to be clumping factor positive, i.e., they exhibited red-clumping behavior (Fig. 5).
TABLE 1.
Performance of the Slidex Staph-Kit and Staphylase test for the identification of MRSA and MSSA
| Organism | Type | n | Resultsa
|
|
|---|---|---|---|---|
| Slidex Staph-Kit | Staphylase test | |||
| MRSA | 1 | 25 | W | – |
| 7 | 2 | W | − | |
| 71 | 3 | W | − | |
| 42 | 1 | W | − | |
| 72 | 1 | W | − | |
| 73 | 1 | W | − | |
| 74 | 1 | W | − | |
| 75 | 1 | R | R | |
| 6 | 1 | W | − | |
| 76 | 1 | W | − | |
| 77 | 1 | R | R | |
| 78 | 2 | R | R | |
| 79 | 1 | R | R | |
| 80 | 2 | R | R | |
| 81 | 2 | R | R | |
| 41 | 1 | R | R | |
| 83 | 2 | W | − | |
| 82 | 2 | W | − | |
| MSSA | 150 | R/Wb | ND | |
W, white clumping; R, red clumping; −, no clumping; ND, not determined.
R/W, both R (n = 149) and W (n = 1).
FIG. 4.
Performance characteristics of Slidex Staph-Kit: white clumping. Magnification, ca. ×2.
FIG. 5.
Performance characteristics of Slidex Staph-Kit: red clumping. Magnification, ca. ×2.5.
DISCUSSION
Rapid and accurate identification of methicillin resistance in S. aureus is of ongoing clinical importance for controlling the spread of this pathogen within hospital settings. The plasma coagulase test is generally acknowledged as the “gold standard” for the identification of S. aureus. Nevertheless, the use of commercial agglutination kits for identifying S. aureus is widespread in clinical microbiological laboratories, since these tests are easy to perform and the results are available within minutes. Whereas the first-generation agglutination kits are capable of detecting clumping factor and/or protein A for S. aureus strains while failing to identify certain MRSA strains (3, 22), second-generation agglutination kits, such as the Slidex Staph-Kit, possess the additional feature of being able to detect specific surface antigens for S. aureus. Numerous studies have been published confirming the high sensitivity and specificity of these second-generation kits for the identification of MSSA as well as MRSA (1, 4–6, 10, 17, 23, 30). Definitive detection of methicillin resistance, however, still requires time-consuming antimicrobial susceptibility testing.
This is the first study demonstrating that the Slidex Staph-Kit is a reliable and, above all, rapid method for identifying epidemic clumping factor-negative MRSA even prior to antimicrobial susceptibility testing. Owing to its unique design as a combined latex and hemagglutination test, it reacts, in contrast to all the other latex agglutination kits, in two different ways. Red clumps indicate the presence of clumping factor, with which S. aureus causes erythrocytes sensitized with fibrinogen to agglutinate (Fig. 5). White clumps, on the other hand, signal both the absence of clumping factor and the presence of other immunogens, which trigger the agglutination of the latex particles (Fig. 4). This study shows that white clumping has a 99% specificity and a 98% positive predictive value for the detection of methicillin resistance in S. aureus. Fifty percent of all initial isolates obtained from the orthopedic clinic of the Frankfurt University Hospital and more than 60% of all initial isolates in the Frankfurt metropolitan area, including six community hospitals and the University Hospital (29), belong to one genotype, which has been identified by the Robert Koch Institute, the German national institute of infectious diseases, as the so-called “southern-German” epidemic strain. This epidemic strain is clumping factor negative and exhibits white-clumping behavior. Furthermore, it is not only epidemic in the south but is also widespread in the northern parts of Germany (22, 31). Since bacteria do not respect international boundaries, it came as no surprise when epidemic MRSA strains kindly sent to our institution from Slovakia and Italy exhibited the same southern-German genotype and showed white-clumping behavior (data not shown). As demonstrated by PFGE in this study, it is not only the southern-German epidemic strain which is clumping factor negative; other MRSA genotypes also manifest this typical agglutination characteristic.
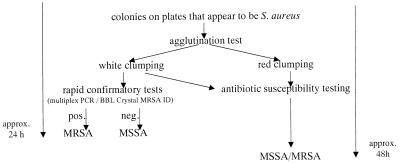
The Slidex Staph-Kit, of course, is neither designed nor licensed to detect resistance, and as such, white clumping can be regarded only as a clue to methicillin resistance. Nonetheless, to the extent that the white clumping serves as a strong indicator for MRSA, rapid tests can be conducted which confirm methicillin resistance and, upon request, species identity within 4 hours. This validation can be achieved genotypically, for example, by multiplex PCR, a method that enables the detection of the mecA gene and a species-specific gene for S. aureus within 5 to 6 h (2, 21). Quicker and less labor intensive is the phenotypical verification of MRSA with the BBL Crystal MRSA ID system from Becton Dickinson, which can be performed within 4 h (12, 18, 28). Finally, the phenotypical test for plasma coagulation can be performed to confirm species specificity, analogous to the genotypic confirmation by multiplex PCR (7) (Fig. 6).
FIG. 6.
Procedure for the rapid identification of clumping factor-negative MRSA with the Slidex Staph-Kit.
The results of this study, as well as the fact that a modern second-generation agglutination test was required owing to the rising incidence of clumping factor-negative MRSA, indicate that all clumping factor-negative MRSA strains are capable of being recognized by the Slidex Staph-Kit with its differentiated agglutination characteristics. In view of these findings, the Slidex Staph-Kit is a valuable tool in clinical laboratories with a frequent occurrence of clumping factor-negative MRSA.
In conclusion, this study confirms the Slidex Staph-Kit’s ability within the context of the clinical microbiological laboratory to pinpoint certain epidemic MRSA strains 24 h prior to the final results provided by antimicrobial susceptibility testing. In turn, the initiation of effective infection control measures may reduce the spread of this pathogen.
REFERENCES
- 1.Adams J, Van Enk R. Use of commercial particle agglutination systems for the rapid identification of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1994;13:86–89. doi: 10.1007/BF02026132. [DOI] [PubMed] [Google Scholar]
- 2.Brakstad O G, Tveten Y, Nato F, Fournier J M. Comparison of various methods and reagents for species identification of Staphylococcus aureus positive or negative for the mecA gene. APMIS. 1993;101:651–654. doi: 10.1111/j.1699-0463.1993.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 3.Croize J, Gialanella P, Monnet D, Okada J, Orsi A, Voss A, Merlin S. Improved identification of Staphylococcus aureus using a new agglutination test. Results of an international study. APMIS. 1993;101:487–491. [PubMed] [Google Scholar]
- 4.Davies S. Detection of methicillin-resistant Staphylococcus aureus: the evaluation of rapid agglutination methods. Br J Biomed Sci. 1997;54:13–15. [PubMed] [Google Scholar]
- 5.Felten A, Lepage E, Lagrange P. Critical analysis of tests for rapid detection of Staphylococcus aureus, Pastorex Staph plus, Slidex Staph-kit and Staph aureus in clinical isolates. Pathol Biol. 1995;43:471–476. [PubMed] [Google Scholar]
- 6.Fournier J M, Bouvet A, Mathieu D, Nato F, Boutonnier A, Gerbal R, Brunengo P, Saulnier C, Sagot N, Slizewicz B, Mazie J C. New latex reagent using monoclonal antibodies to capsular polysaccharide for reliable identification of both oxacillin-susceptible and oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:1342–1344. doi: 10.1128/jcm.31.5.1342-1344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda W M, Ristow K, Novak D. Evaluation of RapiDEC Staph for identification of Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus saprophyticus. J Clin Microbiol. 1994;32:2056–2059. doi: 10.1128/jcm.32.9.2056-2059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jevons M P. “Celbenin”-resistant staphylococci. Br Med J. 1961;1:124–125. [Google Scholar]
- 9.Kaffka A. Erfahrungen mit einfachen Untersuchungsmethoden zum Nachweis von Hyaluronidase, Fibrinolysin und Phosphatase bei Staphylokokken. Zentbl Bakteriol. 1957;168:381–385. [PubMed] [Google Scholar]
- 10.Kampf G, Weist K, Swidsinski S, Kegel M, Rüden H. Comparison of screening methods to identify methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1997;16:301–307. doi: 10.1007/BF01695635. [DOI] [PubMed] [Google Scholar]
- 11.Kimura A, Igarashi H, Ushioda H, Okuzuki K, Kobayashi H, Otsuka T. Epidemiological study of Staphylococcus aureus isolated from Japanese national university and medical college hospitals. Kansenshogaku Zasshi. 1992;66:1543–1549. doi: 10.11150/kansenshogakuzasshi1970.66.1543. [DOI] [PubMed] [Google Scholar]
- 12.Knapp C C, Ludwig M D, Washington J A. Evaluation of BBL Crystal MRSA ID system. J Clin Microbiol. 1994;32:2588–2589. doi: 10.1128/jcm.32.10.2588-2589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie A M R, Richardson H, Lannigan R, Wood D. Evidence that the National Committee for Clinical Laboratory Standards disk test is less sensitive than the screen plate for detection of low-expression-class methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1995;33:1909–1911. doi: 10.1128/jcm.33.7.1909-1911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michel M, Gutmann L. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: therapeutic realities and possibilities. Lancet. 1997;349:1901–1906. doi: 10.1016/s0140-6736(96)11192-2. [DOI] [PubMed] [Google Scholar]
- 15.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panlilio A L, Culver D H, Gaynes R P, Banerjee S, Henderson T S, Tolson J S, Martone W J. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Infect Control Hosp Epidemiol. 1992;13:582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- 17.Personne P, Bes M, Lina G, Vandenesch F, Brun Y, Etienne J. Comparative performances of six agglutination kits assessed by using typical and atypical strains of Staphylococcus aureus. J Clin Microbiol. 1997;35:1138–1140. doi: 10.1128/jcm.35.5.1138-1140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qadri S M H, Ueno Y, Imambaccus H, Almodovar E. Rapid detection of methicillin-resistant Staphylococcus aureus by Crystal MRSA ID system. J Clin Microbiol. 1994;32:1830–1832. doi: 10.1128/jcm.32.7.1830-1832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raus J, Lovee D N. Characterization of coagulase-positive Staphylococcus intermedius and Staphylococcus aureus isolated from veterinary clinical specimens. J Clin Microbiol. 1983;18:789–792. doi: 10.1128/jcm.18.4.789-792.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmalreck A F, Friedrich W, Kottmann I, Reiser A, Ruffer U, Schlenk R, Tränkle P, Vanca E, Wildfeuer A. Resistenzmuster von Staphylokokken und Nachweis des mecA-Gens. Chemother J. 1996;4:195–202. [Google Scholar]
- 21.Schmitz F J, Hofmann B, Finken-Eigen M, Idel H, Hadding U, Heinz H P, Köhrer K. Etablierung einer Multiplex-PCR mit Aussagen zur Taxonomie, Pathogenität und Methicillin-Resistenz bei Staphylokokken. J Lab Med. 1996;20:543–550. [Google Scholar]
- 22.Schwarzkopf A, Karch H, Schmidt H, Lenz W, Heesemann J. Phenotypical and genotypical characterization of epidemic clumping factor-negative, oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:2281–2285. doi: 10.1128/jcm.31.9.2281-2285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smole S C, Aronson E, Durbin A, Brecher S M, Arbeit R D. Sensitivity and specificity of an improved rapid latex agglutination test for identification of methicillin-sensitive and -resistant Staphylococcus aureus isolates. J Clin Microbiol. 1998;36:1109–1112. doi: 10.1128/jcm.36.4.1109-1112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speller D C E, Johnson A P, James D, Marples R R, Charlett A, George R C. Resistance to methicillin and other antibiotics in isolates of Staphylococcus aureus from blood and cerebrospinal fluid, England and Wales, 1989–95. Lancet. 1997;350:323–325. doi: 10.1016/s0140-6736(97)12148-1. [DOI] [PubMed] [Google Scholar]
- 25.Ungeheuer J, Wilms S, Sietzen W. mecA-Gen bei Staphylokokken—vereinfachter Nachweis mit der PCR. Chemother J. 1994;1:31–35. [Google Scholar]
- 26.Vincent J L, Bihari D J, Suter P M, Bruining H A, White J, Nicolas-Chanoin M H, Wolff M, Spencer R C, Hemmer M. The prevalence of nosocomial infection in intensive care units in Europe. JAMA. 1995;274:639–644. [PubMed] [Google Scholar]
- 27.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]
- 28.Wallet F, Roussel-Delvallez M, Courcol R J. Choice of a routine method for detecting methicillin-resistance in staphylococci. J Antimicrob Chemother. 1996;37:901–909. doi: 10.1093/jac/37.5.901. [DOI] [PubMed] [Google Scholar]
- 29.Wichelhaus T A, Schulze J, Hunfeld K P, Schäfer V, Brade V. Clonal heterogeneity, distribution, and pathogenicity of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1997;16:893–897. doi: 10.1007/BF01700555. [DOI] [PubMed] [Google Scholar]
- 30.Wilkerson M, McAllister S, Miller J M, Heiter B J, Bourbeau P P. Comparison of five agglutination tests for identification of Staphylococcus aureus. J Clin Microbiol. 1997;35:148–151. doi: 10.1128/jcm.35.1.148-151.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witte W, Cuny C, Braulke C, Hueck D. Clonal dissemination of two MRSA strains in Germany. Epidemiol Infect. 1994;113:67–73. doi: 10.1017/s0950268800051475. [DOI] [PMC free article] [PubMed] [Google Scholar]