Abstract
Background:
The human brain is inherently organized into distinct networks, as reported widely by resting-state functional magnetic resonance imaging (rs-fMRI), which are based on blood-oxygen-level-dependent (BOLD) signal fluctuations. 11C-UCB-J PET maps synaptic density via synaptic vesicle protein 2A, which is a more direct structural measure underlying brain networks than BOLD rs-fMRI.
Methods:
The aim of this study was to identify maximally independent brain source networks, i.e., “spatial patterns with common covariance across subjects”, in 11C-UCB-J data using independent component analysis (ICA), a data-driven analysis method. Using a population of 80 healthy controls, we applied ICA to two 40-sample subsets and compared source network replication across samples. We examined the identified source networks at multiple model orders, as the ideal number of maximally independent components (IC) is unknown. In addition we investigated the relationship between the strength of the loading weights for each source network and age and sex.
Results:
Thirteen source networks replicated across both samples. We determined that a model order of 18 components provided stable, replicable components, whereas estimations above 18 were not stable. Effects of sex were found in two ICs. Nine ICs showed age-related change, with 4 remaining significant after correction for multiple comparison.
Conclusion:
This study provides the first evidence that human brain synaptic density can be characterized into organized covariance patterns. Furthermore, we demonstrated that multiple synaptic density source networks are associated with age, which supports the potential utility of ICA to identify biologically relevant synaptic density source networks.
Keywords: ICA, SV2A, Aging, Positron emission tomography, Synapse, Neuroimaging
1. Introduction
Synaptic vesicle protein 2A (SV2A) is the most monodispersed synaptic vesicle protein and is thus a potentially useful measure of synaptic density (Bajjalieh et al., 1994; Janz and Südhof, 1999; Mutch et al., 2011). 11C-UCB-J is a recently developed PET tracer that binds to SV2A (Finnema et al., 2018, 2016). Previously, we detected changes in synaptic density using 11C-UCB-J PET in several psychiatric and neurodegenerative diseases (Chen et al., 2018; D’Souza et al., 2020; Finnema et al., 2016; Holmes et al., 2019; Matuskey et al., 2020; Mecca et al., 2020). However, no data driven approach to decompose synaptic density PET data has yet been performed.
One such data-driven approach is independent component analysis (ICA), a blind source separation technique. In the resting-state functional magnetic resonance imaging (rs-fMRI) field, it is commonly used to separate statistically independent blood oxygen level dependent (BOLD) components associated both with task-related and spontaneous resting state activity within neuronal networks (Bell and Sejnowski, 1995; Kiviniemi et al., 2003). A major advantage of ICA over traditional hypothesis-driven approaches is that the former is a data-driven approach for differentiating relevant functional brain signals from various sources of noise without a priori assumptions on signal origin (Calhoun et al., 2001; McKeown et al., 2003). ICA separates components from data into a number of spatially or temporally independent components (ICs) based on statistical features of the data (Calhoun et al., 2003; McKeown et al., 2003). Separation occurs by maximizing the non-Gaussianity of the joint density using a projection pursuit methodology. The ICA algorithm initiates projection pursuit with a guess and produces components in random order (Hyvärinen et al., 2001). The number of identified components (i.e., model order) can be selected from 1 to n – 1, where n is the number of imaged brain volumes (spatial ICA), or number of voxels in the volume of interest (temporal ICA). Extracted components consist of voxels which contain related information which we call a ‘source network’. These source networks have been shown to exhibit intersubject covariance and differences between groups.
ICA has more recently been applied to positron emission tomography (PET) data. Spatial components both similar to rs-fMRI-derived resting state networks (RSNs) as well as novel RSNs have been identified by ICA applied to 18F-FDG PET data, a tracer for brain metabolism (Di et al., 2019, 2012; Savio et al., 2017). Recently, we applied ICA to 11C-PHNO PET data (D2/D3 receptors) and found source-based patterns of receptor availability consistent with DA circuitry and predicted receptor occupancy (Smart et al., 2020a; Worhunsky et al., 2017).
In applying ICA, selection of the appropriate number of calculated ICs is an open question. In the previous work with 11C-PHNO PET, a model order of 2,3 was initially estimated and successful as the tracer only binds to D2 and D3 receptors in the dopamine-rich regions of the brain (Worhunsky et al., 2017). In 18F-FDG studies using the whole brain with the aim to identify functional networks, a model order of 18–20 was used, partly due to sample size limitations (Savio et al., 2017). For fMRI data using spatial ICA, there typically are no sample-size related constraints on model order estimates due to the high temporal dimension. The effect of model order on fMRI data has been investigated by Abou-Elseoud and colleagues over a model order range of 10–200. At the lower end, some components comprised multiple distinct sources that separated at higher model orders, and several novel components appeared at model orders > 40. At the extreme end of model order > 100, a decrease in ICA repeatability occurred (Abou-Elseoud et al., 2010). Thus, characterization of model order is a key consideration for initial ICA analyses of a given imaging target.
The aim of this study was to apply a data driven method (ICA) to investigate the utility of 11C-UCB-J PET data in healthy participants to (1) identify spatial patterns, or networks, of coherent synaptic density (SV2A) variability and (2) to examine how these networks may be associated with age and sex. We first set out to assess and characterize 11C-UCB-J components using two independent samples over a range of model orders. We hypothesized that we would find coherent sources, i.e., synaptic density patterns, that would be reliably and reproducibly extracted in the independent samples of healthy individuals. Some of these networks might be expected to be similar to RSNs, based on the biological commonality of synaptic density with neural activity as measured by BOLD-signal (rs-fMRI) and glucose metabolism (18F-FDG PET).
2. Materials and methods
2.1. Participants
Data from 80 cognitively normal, healthy adults were included in the current study. All subjects were screened through a clinical interview, physical examination with medical history, routine blood tests, electrocardiogram and urine toxicology. Individuals were excluded if evaluation revealed a diagnosis of a current and/or lifetime psychiatric disorder, current or past serious medical or neurological illness, metal in body which would result in MRI contraindication, or a history of substance abuse or dependence.
MR images were acquired on all subjects and visually inspected to eliminate subjects with structural brain abnormalities. The study was performed under protocols approved by the Yale University Human Investigation Committee. All subjects signed a written informed consent form. Subjects were pseudo-randomly assigned into two samples of 40 that did not differ in age or gender (Table 1).
Table 1.
Demographic information of study participants, for the total group and the two sample subsets.
| Total | Sample 1 | Sample 2 | |
|---|---|---|---|
|
| |||
| Sample size | 80 | 40 | 40 |
| Sex Male: No. (%) | 47 (58.7) | 23 (57.5) | 24 (60) |
| Female: No. (%) | 33 (41.3) | 17 (42.5) | 16 (40) |
| Age, mean (SD) [range] | 44.7 (17.0) [21.0–82.7] | 44.8 (17.4) [21.0–82.7] | 44.6 (16.8) [21.8–80.6] |
2.2. Image acquisition
11C-UCB-J was synthesized at Yale PET Center as described previously (Nabulsi et al., 2016). Subjects were administered an i.v. injection of 11C-UCB-J (551.00 ± 173.32 MBq; injected mass: 1.57 ± 1.04 μg; range 0.27–5.85 μg) and received one dynamic PET scan (HRRT; Siemens/CTI, Knoxville, TN, USA). In order to obtain an arterial plasma curve, each subject underwent catheterization of the radial artery for blood sampling. Samples were drawn every 10 s for the first 90 s and at 1.75, 2, 2.25, 2.5, 2.75, 3, 4, 5, 6, 8, 10, 15, 20, 25, 30, 45, 60 min after 11C-UCB-J injection. For each sample, plasma was obtained by centrifugation at 4 °C (2930 × g for 5 min). Metabolite analysis and plasma free fraction of 11C-UCB-J were determined for correction of the obtained plasma curve as described previously (Finnema et al., 2018). PET scans (207 slices, 1.2 mm slice separation, reconstructed image resolution ~ 3 mm) were acquired at rest in list mode. Before injection, a transmission scan was obtained for attenuation correction. A T1-weighted structural MR image (repeat time (TR)/echo time (TE) = 2530/3.34, flip angle = 7°, in-plane resolution = 0.98 × 0.98 mm, matrix size = 256 × 256, slice thickness = 1 mm, slices = 176) was also acquired for nonlinear registration into template space.
2.3. Image processing
Dynamic PET data (frames: 6 × 0.5 min, 3 × 1 min, 2 × 2 min, and 10 × 5 min) were reconstructed using the MOLAR algorithm with corrections for attenuation, normalization, scatter, randoms, deadline, and motion (Carson et al., 2004). Event-by-event head motion correction was included in the reconstruction based on motion detection with a Polaris Vicra® optical tracking system (NDI Systems, Waterloo, Canada) using reflectors mounted on a cap worn by the subject (Jin et al., 2013). Parametric volume of distribution (VT) images using 60 min of dynamic data were generated with a one-compartment model using the metabolite-corrected arterial plasma curve (Finnema et al., 2018). Registration of parametric images to Montreal Neurological Institute (MNI152) template space was performed using SPM12 (Wellcome Trust center for Neuroimaging, London, UK). For each subject, a linear registration of an early sum image (0–10 min post-injection) of motion-corrected PET data to a high-resolution T1-weighted structural image was applied to parametric VT images. Non-linear registration of MR images (Ashburner and Friston, 2005) was then applied to the parametric VT images and lastly smoothed with a 12 mm FWHM Gaussian kernel.
2.4. Independent component analysis
Independent component analysis (ICA) is a data-driven approach that identifies spatial covariation patterns, extracting maximally independent components and associated signal sources. Here, we consider image data from N subjects, each with P pixel values. ICA assumes that the observed data for each subject (x, a P-vector) is comprised of an unknown linear mixture of S independent non-Gaussian sources (s, each a P-vector) with mixing coefficients A. ICA estimates the mixing matrix (Ã) and its corresponding maximally independent sources (y, each a P-vector). Applied to three-dimensional imaging data of a group of N subjects, the output of spatial ICA includes spatial source weights (P-vectors) of M source signals (yi, i = 1,…,M) and subject loading values for component i and subject j (Ãi,j); where M indicates the chosen model order (i.e., the number of components) of the ICA extraction and à is an M-by-N matrix. ICA of biomedical imaging data is typically applied after dimension reduction with principal component analysis (PCA); however, for ease of notation, we define à to represent the final ICA-derived loading parameters. Relative to PET VT images, subject ICA-loadings (Ã) represent the source ‘intensity’ or relative strength of each source for each subject’s VT data, such that an approximation of the original VT maps (ṼT) could be reconstructed as .
A primary goal of this study was to investigate the reliability and reproducibility of ICA-identified components of 11C-UCB-J at different model orders (M). To achieve this, spatial ICA was performed using the source-based morphometry (SBM) module of GroupICA Toolbox (GIFTv4.0b; trendscenter.org/software/gift/) on the two samples of 40 subjects separately. Analyses were constrained to a voxel mask with a mean (across-subject) VT > 10 (Fig. S1) in order to exclude white matter structures (Finnema et al., 2018). The mean within this mask for each scan was subtracted, and VT images of 40 participants were concatenated to form a subject series that was then reduced through PCA prior to ICA using the InfoMax algorithm (Bell and Sejnowski, 1995). ICA was performed at multiple model orders, estimating 8, 12, 18, and 24 components in separate analyses. ICA analyses were performed without normalization (e.g., z-transforming) along either dimension (spatial or subject-wise) before or after extraction to preserve the quantitative VT units and facilitate comparisons with the larger 11C-UCB-J literature.
To assess component stability and repeatability, each extraction was iterated 50 times using ICASSO module implemented in GroupICA Toolbox (Himberg et al., 2004). Given that ICA algorithms are affected by random sampling of the data, ICASSO provides measures of the statistical reliability of estimated components through multiple iterations of the extraction. Consistent with prior investigations of ICA model space (Abou-Elseoud et al., 2010), the repeatability of components was assessed using the cluster-quality index (Iq), which represents a summary metric of the multi-dimensional similarity between the identified mixing matrix solutions (i.e., component loadings, spatial distribution of sources, and relative independences of ICs) across iterated extractions (Himberg et al., 2004). As a measure of between-iteration similarity, Iq ranges between 0 and 1, with greater values representing a higher degree of repeatability and 1 representing identical solutions identified through every random-point initiated iteration. In ICA of fMRI, components with Iq > 0.8 are considered to have been stably and reliably identified (Allen et al., 2011). Final ICA output for each model order represented the average estimate of spatial sources and subject loadings over the 50 iterations.
2.5. Source network replication
The replication of ICA-identified source networks between the two samples of healthy subjects was examined for each model order, and a conjunction of the two constraint masks (i.e., the overlap of the two VT > 10 masks) was generated to limit comparisons to only those voxels present in both analyses. Replication of ICs between samples was determined based on the spatial distribution of source network intensities. Each source network was scaled to estimated ṼT units by multiplying by the average subject loading for that source network. Each of these source networks from sample 1, expressed as P-vectors, were entered as the dependent variable in a multiple regression with the vectors of all source networks from sample 2 as the independent variables. Resulting regression coefficients (betas) for each IC from sample 1 relative to the ICs from sample 2 were considered an index of spatial similarities. The pair of source networks with the overall maximal beta value were visually compared to verify similarity in pattern. Once verified, these two source networks were excluded from subsequent matching, i.e. only one match is reported per source network. The process was repeated until the pair with the maximal remaining beta value did not visually match. At each model order, some ICs did not replicate across samples, based on this procedure.
2.6. Comparison between model orders
First, standard mixed linear models were performed to examine within- and between-sample effects of model order on Iq. Next, the source networks from the independent samples were further examined to assess the persistence of identified components at different model orders. As described above for source network replication, for a given model order, multiple regression analysis was performed comparing each source network (P-vectors) to the source networks derived from the previous model order (i.e., 12→8, 18→12, 24→18) to select similar sources, with matching based on largest beta coefficients followed by visual confirmation. Lastly, matched source networks were excluded from subsequent matches between model orders, such that only the one match is reported.
Exploration of spatial maps and demographic associations of subject loadings
To explore potential physiological implications of the ICA-identified components of 11C-UCB-J VT, ICA procedures were repeated on the full subject dataset (n = 80) at the model order chosen to be optimal based upon the replication and model order analysis. ICs at n = 80 were visually matched to the replicated ICs (Fig. 1) for consistency in numbering. Source networks were region labeled according to the AAL template using xjView toolbox (http://www.alivelearn.net/xjview) in Matlab2015a (Mathworks). Subject component loadings per source network were analyzed in an exploratory fashion using independent sample t-tests to examine sex differences and Pearson correlations to investigate relationships with age, corrected for multiple comparisons with the Bonferroni method. For exploratory reasons, results were also presented without correction for multiple comparisons, and findings were considered significant at P < 0.05.
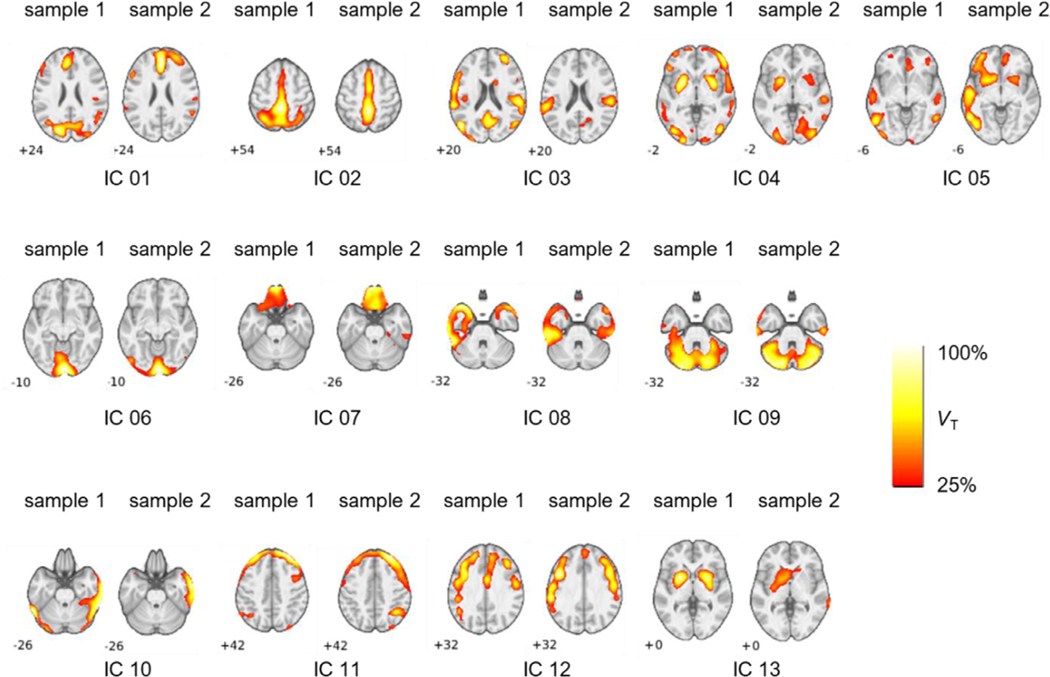
Fig. 1.
Results from source network replication across independent samples from ICA performed at model order 18. The matched components across samples were determined by maximal regression beta (β) from multiple regression analysis. Images are scaled relative to peak ṼT. Depicted slices are chosen to present peak VT coordinates.
3. Results
3.1. Replicated source networks of 11C-UCB-J
In order to assess the replication and persistence of ICA-identified sources networks, ICA was performed in two separate samples of the dataset (Table 1) at a range of model orders (8, 12, 18, and 24), and the identified components within the same model order were compared between samples using multiple regression. Detailed results are presented in Table S1 including Iq values and the peak ṼT in each source network. Replications across independent samples for model order 18 are presented in Fig. 1. At model order 8, 4 ICs replicated between samples (mean ± SD beta: 0.42 ± 0.23). At model order 12, 8 ICs replicated across samples (beta: 0.46 ± 0.14). Thirteen ICs replicated at model order 18 (mean beta: 0.31 ± 0.16). Lastly, at model order 24, 16 ICs (mean beta: 0.30 ± 0.11) replicated across independent samples. Images of replicated source networks at all model orders are shown in Fig. S2.
3.2. Comparison between model orders
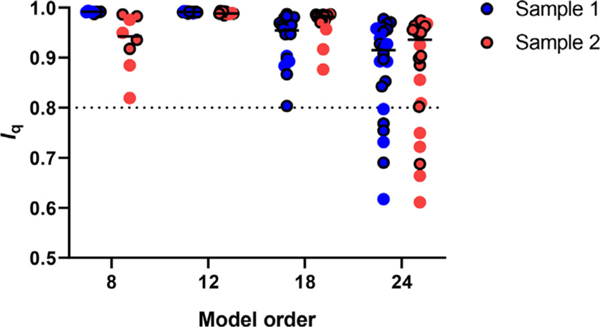
Repeatability of ICA decomposition was evaluated using the mean cluster-quality index (Iq) for all source networks that were found across all model orders (Fig. 2). There was a main effect of model order on Iq (F3,116 = 16.23, p < 0.001), but there was no sample * model-order interaction (F3,116 = 1.59, p = 0.20) or sample group difference (F1,116 = 0.24, p = 0.620) in Iq. Post-hoc testing indicated average Iq was lower for model order 24 relative to the other model orders correcting for multiple comparisons (p < 0.001). Iq values below 0.8 have been considered unreliable, and at model order 8, 12, and 18, there were no Iq values below 0.8 in either sample (sample 1: 0.99 ± 0.002, 0.99 ± 0.002, 0.94 ± 0.050, and 0.88 ± 0.100; sample 2: 0.93 ± 0.057, 0.99 ± 0.003, 0.97 ± 0.029, 0.88 ± 0.113 for model orders 8, 12, 18, and 24, respectively). At model order 24 however, 6 out of 24 components had an Iq below 0.8 for sample 1, and 5 out of 24 components had an Iq below 0.8 for sample 2. When assessing the Iq values of only the replicated ICs between samples, five ICs at model order 24 (IC 05, IC 06, IC 08, IC 10 and IC 12) had Iq below 0.8 in one or both samples, suggesting that these would be considered unreliable (bolded values in Table S1).
Fig. 2.
Cluster quality index (Iq) per sample for all components at each respective model order. Iq decreases as model order size increases indicating that extraction reliability decreases as model order increases. Dashed line at Iq = 0.8 represents cutoff for reliable ICs. Circles outlined in black indicate replicated source networks across samples.
Across investigated model orders, replicated source networks that were identified at one extraction dimension were also identified in higher dimension extractions (Table S2). The replicated ICs from model order 8, IC 01 – IC 04, were also identified at model orders 12, 18, and 24 (Fig. S2). At model order 12, a further four ICs emerged: IC 05–08. At model order 18, an additional five ICs were identified. Lastly, at model order 24, three novel ICs were identified. Representative images for all ICs across model orders identified in samples 1 and 2 are presented in Fig. S2.
Based on the Iq values and the number of replicated source networks, model order 18 appears to be within the optimal range to extract the highest number of robustly replicable ICs (i.e., Iq > 0.8). Model order 24 identified more reproducible components but some ICs, including those reliably identified at lower model orders, had Iq values below 0.8 which would be considered unreliable.
3.3. Physiological and functional associations
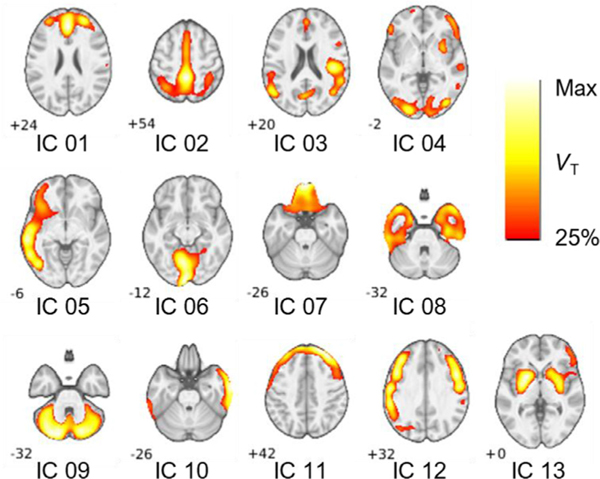
Source networks of the identified ICs from the ICA performed with the full dataset (n = 80) at model order 18 are shown in Fig. 3. For our purposes, we selected to further analyze ICs from model order 18 as they represent a balance of being highly repeatable (i.e. high Iq, none below 0.8), robustly sourced (i.e. high peak VT with loading weights significantly different from zero), and a regionally distinct and diverse variety of potentially functionally relevant source networks of 11C-UCB-J binding. Following anatomical labeling using the AAL template, the ICs are described in Table 2 along with cluster size and MNI coordinates of peak ṼT. The ICs have a peak ṼT value of 9.9 ± 2.1 mL/cm3. Typically, gray matter VT values for 11C-UCB-J are in the range of 20 mL/cm3.
Fig. 3.
Selected slices display the main regional source network distribution/characteristics of ICs extracted at model order 18 in the n = 80 dataset. Identified ICs were as follows: an anterior prefrontal (IC 01), a posterior cortical (IC 02), a superior/middle temporal (IC 03), a corticostriatal (IC 04), a left middle/inferior temporal (IC 05), a calcarine (medial occipital, IC 06), an orbitofrontal (IC 07), a frontotemporal pole (IC 08), a cerebellar (IC 09), right temporal (IC 10), frontoparietal (IC 11), middle frontal (IC 12), and striatal (IC 13). Images in ṼT units (25–100% of per IC peak ṼT). Slice values were chosen to display peak ṼT coordinates.
Table 2.
Peak ICA-estimated volume of distribution contribution (ṼT, mL/cm3) of identified networks at model order 18 in the n = 80 dataset. Total cluster size in voxels (k), x, y, z coordinates (in mm) of peak ṼT in MNI space.
| Identified network | k | x | y | z | Peak ṼT | |
|---|---|---|---|---|---|---|
|
| ||||||
| IC 01 | Anterior prefrontal | 16,714 | − 2 | 26 | 38 | 9.1 |
| IC 02 | Posterior cortical | 12,288 | 0 | − 42 | 56 | 14.36 |
| IC 03 | Superior/mid temporal | 25,852 | 50 | 12 | − 4 | 7.1 |
| IC 04 | Corticostriatal | 24,406 | 36 | − 60 | 40 | 7.4 |
| IC 05 | Left middle/inferior temporal | 18,470 | − 60 | − 28 | − 10 | 9.3 |
| IC 06 | Calcarine (medial occipital) | 15,463 | − 8 | − 64 | − 8 | 9.5 |
| IC 07 | Orbitofrontal | 6581 | − 2 | 52 | − 28 | 13.05 |
| IC 08 | Left frontotemporal | 6016 | − 24 | 6 | − 44 | 12.3 |
| Right frontotemporal | 7162 | 30 | 14 | − 44 | 11.1 | |
| IC 09 | Cerebellar | 19,798 | − 18 | − 80 | − 30 | 8.4 |
| IC 10 | Right temporal | 14,720 | 66 | − 36 | − 16 | 9.6 |
| IC 11 | Frontoparietal | 12,881 | 22 | 52 | 38 | 10.3 |
| IC 12 | Left middle frontal | 11,518 | − 50 | − 40 | 40 | 8.3 |
| Right middle frontal | 7403 | 44 | 4 | 46 | 8 | |
| IC 13 | Striatal | 11,984 | − 22 | 6 | − 4 | 10.2 |
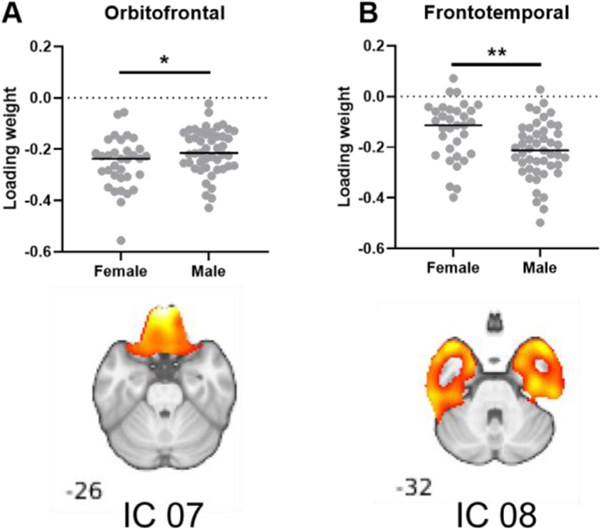
The effects of sex and age on subject ṼT mixtures (i.e., subject loading weights) for identified ICs were assessed (Figs. 4, 5). Significant sex differences were found in two ICs: an orbitofrontal source network (Fig. 5A, p < 0.05, IC 07) and a frontotemporal source network (Fig. 5B, p < 0.01, IC 08). However, these two results did not survive the Bonferroni correction for multiple comparisons.
Fig. 4.
Components from ICA of the total dataset (n = 80) with significantly different loading weights between men and women. Loading weights were significantly different in (A) IC 07, an orbitofrontal network and (B) in IC 08, a frontotemporal network. * p < 0.05, ** p < 0.01. Statistical analysis performed using unpaired t-tests. Results were not significant after Bonferroni correction.
Fig. 5.
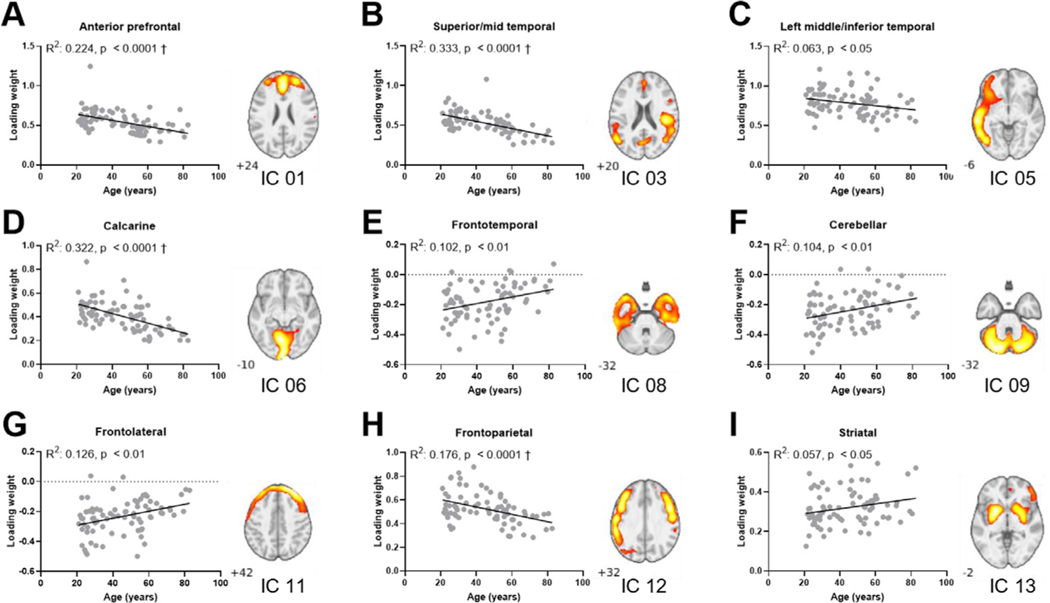
Correlation plots between age and subject loading weights from ICA of the total dataset (n = 80). Correlation plot for the anterior prefrontal network displays a significant change in loading weights with age (A, IC 01, R2: 0.224, p < 0.0001). Further changes in loading weight with age were found in the superior/mid temporal network (B, IC 03, R2: 0.333, p < 0.0001), left middle/temporal spatial (C, IC 03, R2: 0.063, p < 0.05), calcarine (D, IC 06, R2: 0.322, p < 0.0001), frontotemporal (E, IC 08, R2: 0.102, p < 0.01), cerebellar (F, IC 09, R2: 0.104, p < 0.01), frontoparietal (G, IC 11, R2: 0.126, p < 0.01), middle frontal (H, IC 12, R2: 0.176, p < 0.0001), and the striatal network (I, IC 13, R2: 0.057, p < 0.05). † denotes correlations that remain significant following Bonferroni correction.
The relationship between the subject loading weights and age were more pronounced (Fig. 5). In an exploratory analysis, significant correlations of loading weight with age were found in 9 source networks: an anterior prefrontal network (Fig. 5A, IC 01, R2: 0.224, p < 0.0001), superior/mid temporal network (5B, IC 03, R2: 0.333, p < 0.0001), left middle/temporal (5C, IC 05, R2: 0.063, p < 0.05), calcarine (5D, IC 06, R2: 0.322, p < 0.0001), frontotemporal (5E, IC 08, R2: 0.102, p < 0.01), cerebellar (5F, IC 09, R2: 0.104, p < 0.01), frontoparietal (5G, IC 11, R2: 0.126, p < 0.01), middle frontal (5H, IC 12, R2: 0.176, p < 0.0001), and a striatal network (5I, IC 13, R2: 0.057, p < 0.05). IC 01, IC 03, IC 05, IC 06, and IC 12 had positive subject loadings and the correlation with age is negative (Fig. 5A–D, H), which reflects a decrease in ṼT with age. IC 08, IC 09, and IC 11 had negative subject loadings and the correlation with age was positive (Fig. 5E–G), i.e., the magnitude of ṼT variability from these source networks reduce with age. Lastly, the subject loadings and slope of the relationship between IC 13 (Fig. 5I) and age were positive, which would represent an increase in ṼT from this source network with age.
When corrected for multiple comparisons (Bonferroni), four of the 9 IC relationships with age remained: the anterior prefrontal (IC 01), superior/mid temporal (IC 03), calcarine (IC 06), and the middle frontal network (IC 12). These four ICs all had positive subject loadings and negative slopes with age.
4. Discussion
In this study, we present results from a data-driven method for identifying source networks of 11C-UCB-J synaptic density (SV2A) PET data. We first validated the robustness and persistence of these source networks by performing ICA in two separate age- and sex-matched samples of 40 healthy subjects each. Next, as the ‘true’ number of source networks in 11C-UCB-J data is unknown, we assessed the effect of model order selection using the two samples. For the purposes of our study, we determined that estimating 18 components appeared optimal based on Iq and the observation that at model 18, 5 new ICs not found at the lower model orders were identified. At the highest investigated model order 24, 3 additional ICs were identified, but a greater fraction of the estimated components had Iq < 0.8 and thus the extraction was considered less reliable. When estimating 18 components, we were able to identify 13 source networks that robustly replicated across both samples. Using this model order, ICA of the entire 80-subject dataset reliably extracted the same source networks found previously in the two separate samples. The relationships between the subject loadings for these source maps and sex and age were assessed, and we found that the subject loadings of several source networks significantly correlated with age.
All of the ICs first extracted at lower model orders (8, 12) continued to be extracted at higher model orders (18, 24). Based on the high within-cohort correlations of the spatial regression coefficients across model orders, and confirmed visually, the source networks differed little across model orders (Table S2). This contrasts with reports of model order effects on RSNs in rs-fMRI data; as model order increases, spatial features including volume (number of contributing voxels), mean z-score, and repeatability decreased significantly with model order (Abou-Elseoud et al., 2010). Furthermore, as the neuroanatomical precision increased (i.e., the specificity and differentiation of spatial loading on different anatomical structures increased), the repeatability of the ICA decomposition was reduced (Abou-Elseoud et al., 2010). Similarly, the identified 11C-UCB-J source networks tended to display reduced coverage as model order increased (IC 01 – IC 04, Fig. S2). Here we conclude that within the tested range of model orders (i.e., 8–24), ICs could reliably be extracted from 11C-UCB-J PET data. However, at model order 24, a significant fraction of extracted components had an Iq below 0.8 (sample 1: 6/24, sample 2: 5/24), which was not the case for model order 18 and lower. Based on this, a model order in the range of 18 appears to be a suitable number of estimated components for this dataset.
When comparing ICs between model orders, we did not account for multiple matches nor did we investigate any “splitting” of ICs (i.e., branching of one IC at lower model order to multiple ICs at higher model order) as these were not the focus of our replication assessment. Additionally, ICA identified several ICs that did not replicate across samples at different model orders. In limited sample sizes and real-world medical imaging data, it is likely that some unstructured and/or truly random variance will be present, and thus those portions of the ICA solution, which aims to model all data variance (i.e. unlike PCA which aims to model only primary vectors of variance and does not account for residual error) will be sample-dependent to a degree. Thus, while we validated 13 ICs within the constraints of our dataset, future analyses using larger samples may identify additional replicable ICs of synaptic density. The reliable source networks that we identified are as follows: an anterior prefrontal source network (Fig. 1; IC 01), posterior cortical (IC 02), superior/mid temporal (IC 03), corticostriatal (IC 04), left middle/inferior temporal (IC 05), calcarine (i.e. medial occipital, IC 06), orbitofrontal (IC 07), frontotemporal (IC 08), cerebellar (IC 09), right temporal (IC 10), frontoparietal (IC 11), middle frontal (IC 12), and striatal (IC 13).
A number of 11C-UCB-J ICA source networks (Fig. 3) appear spatially consistent with RSNs derived from ICA on 18F-FDG PET data. Among them, the anterior prefrontal (IC 01), the superior/mid temporal (IC 03), the calcarine (medial occipital, IC 06), the cerebellar (IC 09), and the striatal network (IC 13) were reported both by Di et al. (2012) and Savio et al. (2017). Another three 11C-UCB-J ICs were also found by Di and colleagues, namely the posterior cortical (IC 02), the orbitofrontal (IC 07), the right temporal (IC 10), and the middle frontal (IC 12).
Synaptic density might also be associated to structural volumes, and 11C-UB-J ICA source networks were examined in relation to sources of covariance reported in structural MRI (Di and Biswal, 2016; Xu et al., 2009). The 11C-UCB-J superior/middle temporal (Fig. 3, IC 03) and striatal (Fig. 3, IC 13) source networks visually resemble previously reported structural sources by Xu and colleagues. Furthermore, the following 7 11C-UCB-J source networks strongly resemble gray matter sources as reported by Di and Biswal (2016) : anterior prefrontal (Fig. 3, IC 01), posterior cortical (IC 02), superior/middle temporal (IC 03), corticostriatal (IC 04), frontotemporal (IC 08), cerebellar (IC 09), and the striatal source network (IC 13). These qualitative comparisons are further consistent with evidence of shared associations between loss in gray matter volume and synaptic density in neurodegenerative disease (Mecca et al., 2020) and support exploration in 11C-UCB-J ICs in clinical populations.
Lastly, nine of the 11C-UCB-J ICA-derived networks visually resemble RSNs found with rs-fMRI ICA (Allen et al., 2011; Di et al., 2012; Laird et al., 2011; Savio et al., 2017; Smith et al., 2009). Based on visual comparison, the anterior prefrontal (IC 01) source network resembles the anterior part of the default-mode network (DMN), as shown in Fig. S3. Further, the superior/mid temporal (IC 03) resembles the auditory functional network, the calcarine (medial occipital, IC 06) resembles a visual network, and the cerebellar (IC 09) and striatal (IC 13) networks are also similar to rs-fMRI networks. While the aforementioned networks were identified in all the above fMRI studies, a further two networks that visually matched our findings were reported only by Laird et al.: the orbitofrontal (IC 07) as well as the frontotemporal network (IC 08). BOLD images can have significant signal dropout and spatial distortion in the orbitofrontal and anterior temporal cortices (Yeo et al., 2011), which may have limited detection of these two networks other fMRI studies.
In general, 11C-UCB-J PET ICs demonstrate left-right symmetry, i.e., most of the ICs are symmetric between hemispheres (11 out of 13 ICs, Fig. 3). The two notable ICs that are present only in one hemisphere each are the left middle/inferior temporal network (IC 05) and the right temporal network (IC 10). Thus, while most of the ICs represent covariance that spans both hemispheres, the two asymmetric ICs may represent a network that is unique to each of their respective hemisphere. Although these two asymmetric 11C-UCB-J ICs do not resemble any of those reported in 18F-FDG PET or rs-fMRI literature, other asymmetric ICs have been found in 18F-FDG PET and rs-fMRI studies (Di et al., 2012; Smith et al., 2009).
One interesting discrepancy between 11C-UCB-J networks and rs-fMRI RSNs lies with networks consisting of anterior-posterior connectivity. We did not find a spatial source resembling the posterior default-mode network in our study at any model order. Though we did identify an anterior prefrontal network (IC 01) resembling the anterior part of the DMN, we did not detect either the posterior DMN or the DMN in its entirety. Similarly, the posterior DMN was not identified in one 18F-FDG ICA study (Savio et al., 2017). Taken together, these intra-modality similarities and differences suggest that 11C-UCB-J networks likely contain both complementary and unique information relative to 18F-FDG and rs-fMRI networks.
Using the full 80-subject cohort, we assessed the effects of sex and age on component loadings. In our initial exploratory analysis, a significant difference in loading weights between sex was found in the or- bitofrontal network (IC 07, Fig. 4A) and in the frontotemporal network (IC 08, Fig. 4B), though these did not survive a correction for multiple comparisons. In 9 11C-UCB-J ICs, there was a significant correlation between IC loading weights and age. Of these, 4 correlations survived the correction for multiple comparisons: the anterior prefrontal (IC 01), calcarine (IC 06), superior/mid temporal (IC 03), and middle frontal (IC 12) IC. These 4 ICs all had positive subject loadings as well as a negative slope with age. Interestingly, the striatal network (IC 13) had positive subject loadings and a positive correlation with age, however, this correlation did not survive the correction for multiple comparisons. The correlations that did not survive Bonferroni correction (Fig. 5C, E, F, G, I) had R2 values below 0.1. Thus, these correlations are at best weak, and not clearly associated with age.
The brain is affected by the aging process, with many cognitive functions declining with age, such as attentional resources, working and episodic memory, processing speed, and inhibitory capacities (Craik and Byrd, 1982; Salthouse, 2000). Brain structure also changes with age both in white and gray matter (Damoiseaux et al., 2009; Driscoll et al., 2009), and links between structural changes and cognition have been established (Hedden et al., 2016). Furthermore, aging has been associated with decreased functional connectivity in DMN and dorsal attention networks (Tomasi and Volkow, 2012). One interpretation of the age-related correlations with the IC loading weights is that the age-related decrease in subject loadings represents a decrease in synaptic density in the network, potentially reflecting a weaker independent source of variance. Age-effects on ICA-identified components of gray-matter structural images have similarly reported a negative relationship between age and volume of the network (Xu et al., 2009). However, it stands to note that prior to ICA, the global per-subject mean is removed and thus the remaining variance detected is separate from any global brain effects and can therefore be positive or negative relative to the mean. As such, these aging effects should be interpreted with caution, given that positive aging effects may reflect stability relative to global effects, and significant age-associated decreases may be missed because they are similar to the global effects. Interestingly, some of the 11C-UCB-J ICs exhibited positive relationships between age and ṼT contributions. From the perspective of ICs representing mixture of variance sources within regions, this counterintuitive observation may reflect a condensing of variance (i.e., a loss of source diversity) within particular regions or networks with age. Such a process would also be consistent with models of generally reducing connective diversity with age. However, it is known that age-related volume loss is not uniform across brain regions, and thus some of our findings may be in part due to volume loss. Additionally, brain size differences (as assessed with total intracranial volume, TIV) may potentially affect 11C-source networks. Two ICs (IC 07 and IC 09) exhibited an association (p-vals < 0.05) with TIV across all subjects. While these relationships did not replicate in the subsample analyses, the potential influence of registration algorithms on sources of variance in 11C-UCB-J ICs, particularly those in orbitofrontal and cerebellar regions requires attention in future research.
A major methodological difference between ICA on fMRI data and PET data is that voxels in fMRI are assigned to a component based on similarity in temporal fluctuations within-subject as well as across-subject, while such assignment occurs across subjects for PET data. At rest, the DMN is one of the most highly active networks (Raichle, 2015). Such activation might be captured with BOLD signal in the temporal dimension by the continuous fluctuations “as the mind wanders”, but when investigated in a static manner (i.e., PET ICA), such temporal dynamics might be lost. Second, fMRI, 18F-FDG PET, and 11C-UCB-J PET measure different aspects of neural activity. The BOLD signal from fMRI reflects cerebral blood flow as an indirect measure of metabolic oxygen rate, and 18F-FDG PET measures uptake of FDG which mirrors energy metabolism; these are both dynamic processes. This is unlike 11C-UCB-J PET, which quantifies the vesicular SV2A protein, and reflects synaptic density which is considered a structural measure and unlikely to fluctuate with increase or decreases in neural activity. Furthermore, a recent study found that during visual-activation tasks, 11C-UCB-J VT remain unchanged, while blood flow increased as reflected in increased K1 values (Smart et al., 2020b). Thus, differences in ICA-derived networks between these modalities are not unexpected. The PET VT data have physiological meaning, being linearly related to SV2A density, thus, ICA analyses were performed in a manner to preserve the quantitative VT units in our data. Alternatively, normalization or other pre- and post-processing measures may be performed before or after ICA extraction and may provide a different representation of the distribution and variability of source weights across voxels or component loadings across subjects. Lastly, our study reports the ICs using a quantitative measurement (ṼT) which may have great interpretive value.
As part of ICA, the relative contribution of each source component is determined for each subject. These individual loading weights can be examined in clinically relevant analyses, as we have done here with the aging correlations (Fig. 5). This approach may be of interest in studies of neurodegenerative and psychiatric disorders. Previously, we and others have reported differences in synaptic density as measured with 11C-UCB-J PET in epilepsy, Alzheimer’s disease, PTSD, Parkinson’s disease and psychiatric disorders (Chen et al., 2018; D’Souza et al., 2020; Finnema et al., 2020, 2016; Holmes et al., 2019; Matuskey et al., 2020; Onwordi et al., 2020). As well, multi-modality approaches utilizing ICA have already been applied to find altered network integrity in dementing disorders (Ripp et al., 2020). While an individual-subject ICA could be performed across multiple time points, the structures of within-subject temporal changes/covariance in UCBJ distribution cannot be assumed to be the same as those identified across subjects. This certainly represents an area of future interest, particularly in clinical populations with marked neurodegeneration, and warrants additional research using longitudinal data. Thus, the application of ICA to 11C-UCB-J PET, which allows examination of coherent and distinct source networks of synaptic density variability, may hold significant diagnostic applicability to these and to other neurodegenerative and psychiatric disorders related to synaptic loss and/or pathology.
5. Conclusion
ICA identified source networks of 11C-UCB-J PET data that were sample-independent and extracted consistently across a range of model orders. From this dataset, we identified 13 synaptic density source networks, several of which were consistent with sources derived from structural MR gray matter and 18F-FDG PET, and to a lesser degree also showed some spatial similarity with rs-fMRI RSNs. The relative contribution of individual subject variations was related to age for 4 ICs. Synaptic density networks can be reliably and reproducibly identified in healthy adults, show specificity in associations with physiological processes like aging, and should be applied in psychiatric and neurodegenerative populations as possible imaging biomarkers of disease.
Supplementary Material
Acknowledgments
The authors would like to thank the Yale PET Center staff for their expertise and assistance. This publication was also made possible by CTSA grant UL1 TR000142 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Funding: This work was supported by National Institutes of Health grants R01NS094253, R21DA043831-02, and R01AG052560, and funding from the National Center for PTSD, and the Nancy Taylor Foundation.
Footnotes
Data statement
In vivo data from this study are available on request from the corresponding author pending the institute Ethics approval.
Declarations of Competing Interest
The authors declare that they have no conflicts of interest.
Credit authorship contribution statement
Xiaotian T. Fang: Conceptualization, Methodology, Formal analysis, Writing-original draft, Visualization. Takuya Toyonaga: Software, Resources, Data curation, Writing-review & editing. Ansel T. Hillmer: Conceptualization, Methodology, Formal analysis, Writing-original draft. David Matuskey: Investigation, Writing-review & editing. Sophie E. Holmes: Investigation, Resources, Writing-review & editing. Rajiv Radhakrishnan: Investigation, Resources, Writing-review & editing. Adam P. Mecca: Investigation, Resources, Writing-review & editing. Christopher H. van Dyck: Investigation, Writing-review & editing. Deepak Cyril D’Souza: Investigation, Writing-review & editing, Funding acquisition. Irina Esterlis: Investigation, Writing-review & editing, Funding acquisition. Patrick D. Worhunsky: Conceptualization, Methodology, Formal analysis, Writing-original draft. Richard E. Carson: Conceptualization, Methodology, Formal analysis, Writing-original draft, Supervision, Funding acquisition.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2021.118167.
References
- Abou-Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V, 2010. The effect of model order selection in group PICA. Hum. Brain Mapp. 31, 1207–1216. doi: 10.1002/hbm.20929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD, 2011. A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 5, 1–23. doi: 10.3389/fn-sys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ, 2005. Unified segmentation. Neuroimage 26, 839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bajjalieh SM, Frantz GD, Weimann JM, McConnell SK, Scheller RH, 1994. Differential expression of synaptic vesicle protein 2 (SV2) isoforms. J. Neurosci. doi: 10.1523/jneurosci.14-09-05223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ, 1995. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 7, 1129–1159. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Hansen LK, 2003. ICA of functional MRI data: an overview. Proc. …. 10.1.1.3.7473 [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ, 2001. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum. Brain Mapp. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson RE, Barker WC, Liow Jeih-San, Johnson CA, 2004. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. In: Proceedings of the 2003 IEEE Nuclear Science Symposium. Conference Record (IEEE Cat. No.03CH37515). IEEE, pp. 3281–3285. doi: 10.1109/nss-mic.2003.1352597. [DOI] [Google Scholar]
- Chen MK, Mecca AP, Naganawa M, Finnema SJ, Toyonaga T, Lin S, Najafzadeh S, Ropchan J, Lu Y, McDonald JW, Michalak HR, Nabulsi NB, Arnsten AFT, Huang Y, Carson RE, van Dyck CH, 2018. Assessing synaptic density in alzheimer disease with synaptic vesicle glycoprotein 2A positron emission tomographic imaging. JAMA Neurol. doi: 10.1001/jamaneurol.2018.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik FIM, Byrd M, 1982. Aging and cognitive deficits. In: Aging and Cognitive Processes. Springer US, Boston, MA, pp. 191–211. doi: 10.1007/978-1-4684-4178-9_11. [DOI] [Google Scholar]
- D’Souza DC, Radhakrishnan R, Naganawa M, Ganesh S, Nabulsi N, Najafzadeh S, Ropchan J, Ranganathan M, Cortes-Briones J, Huang Y, Carson RE, Skosnik P, 2020. Preliminary in vivo evidence of lower hippocampal synaptic density in cannabis use disorder. Mol. Psychiatry doi: 10.1038/s41380-020-00891-4. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Smith SM, Witter MP, Sanz-Arigita EJ, Barkhof F, Scheltens P, Stam CJ, Zarei M, Rombouts SARB, 2009. White matter tract integrity in aging and Alzheimer’s disease. Hum. Brain Mapp 30, 1051–1059. doi: 10.1002/hbm.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Biswal BB Alzheimer’s Disease Neuroimaging Initiative, 2012. Metabolic Brain Covariant Networks as Revealed by FDG-PET with Reference to Resting-State fMRI Networks. Brain Connect 2, 275–283. doi: 10.1089/brain.2012.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Biswal BB, 2016. Similarly expanded bilateral temporal lobe volumes in female and male children with autism spectrum disorder. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. doi: 10.1016/j.bpsc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Wölfer M, Amend M, Wehrl H, Ionescu TM, Pichler BJ, Biswal BB, 2019. Interregional causal influences of brain metabolic activity reveal the spread of aging effects during normal aging. Hum. Brain Mapp. doi: 10.1002/hbm.24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll I, Davatzikos C, An Y, Wu X, Shen D, Kraut M, Resnick SM, 2009. Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology doi: 10.1212/WNL.0b013e3181a82634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnema SJ, Nabulsi NB, Eid T, Detyniecki K, Lin SF, Chen MK, Dhaher R, Matuskey D, Baum E, Holden D, Spencer DD, Mercier J, Hannestad J, Huang Y, Carson RE, 2016. Imaging synaptic density in the living human brain. Sci. Transl. Med. 8. doi: 10.1126/scitranslmed.aaf6667, 348ra96. [DOI] [PubMed] [Google Scholar]
- Finnema SJ, Nabulsi NB, Mercier J, Lin SF, Chen MK, Matuskey D, Gallezot JD, Henry S, Hannestad J, Huang Y, Carson RE, 2018. Kinetic evaluation and test–retest reproducibility of [11C]UCB-J, a novel radioligand for positron emission tomography imaging of synaptic vesicle glycoprotein 2A in humans. J. Cereb. Blood Flow Metab. 38, 2041–2052. doi: 10.1177/0271678X17724947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnema SJ, Toyonaga T, Detyniecki K, Chen MK, Dias M, Wang Q, Lin SF, Naganawa M, Gallezot JD, Lu Y, Nabulsi NB, Huang Y, Spencer DD, Carson RE, 2020. Reduced synaptic vesicle protein 2A binding in temporal lobe epilepsy: a [11C]UCB-J positron emission tomography study. Epilepsia doi: 10.1111/epi.16653. [DOI] [PubMed] [Google Scholar]
- Hedden T, Schultz AP, Rieckmann A, Mormino EC, Johnson KA, Sperling RA, Buckner RL, 2016. Multiple brain markers are linked to age-related variation in cognition. Cereb. Cortex 26, 1388–1400. doi: 10.1093/cercor/bhu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himberg J, Hyvärinen A, Esposito F, 2004. Validating the independent components of neuroimaging time series via clustering and visualization. Neuroimage 22, 1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Scheinost D, Finnema SJ, Naganawa M, Davis MT, DellaGioia N, Nabulsi N, Matuskey D, Angarita GA, Pietrzak RH, Duman RS, Sanacora G, Krystal JH, Carson RE, Esterlis I, 2019. Lower synaptic density is associated with depression severity and network alterations. Nat. Commun. 10, 1529. doi: 10.1038/s41467-019-09562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen A, Karhunen J, Oja E, 2001. What is Independent Component Analysis?, in: Independent Component Analysis. John Wiley & Sons, Inc., pp. 145–164. doi: 10.1002/0471221317.ch7. [DOI] [Google Scholar]
- Janz R, Südhof TC, 1999. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience doi: 10.1016/S0306-4522(99)00370-X. [DOI] [PubMed] [Google Scholar]
- Jin X, Mulnix T, Gallezot JD, Carson RE, 2013. Evaluation of motion correction methods in human brain PET imaging-a simulation study based on human motion data. Med. Phys. 40, 102503. doi: 10.1118/1.4819820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Kantola JH, Jauhiainen J, Hyvärinen A, Tervonen O, 2003. Independent component analysis of nondeterministic fMRI signal sources. Neuroimage doi: 10.1016/S1053-8119(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, Mckay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT, 2011. Behavioral interpretations of intrinsic connectivity networks. J. Cogn. Neurosci. 23, 4022–4037. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuskey D, Tinaz S, Wilcox KC, Naganawa M, Toyonaga T, Dias M, Henry S, Pittman B, Ropchan J, Nabulsi N, Suridjan I, Comley RA, Huang Y, Finnema SJ, Carson RE, 2020. Synaptic changes in parkinson disease assessed with in vivo imaging. Ann. Neurol. 87, 329–338. doi: 10.1002/ana.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Hansen LK, Sejnowsk TJ, 2003. Independent component analysis of functional MRI: what is signal and what is noise? Curr. Opin. Neurobiol. doi: 10.1016/j.conb.2003.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecca AP, Chen MK, O’Dell RS, Naganawa M, Toyonaga T, Godek TA, Harris JE, Bartlett HH, Zhao W, Nabulsi NB, Wyk BCV, Varma P, Arnsten AFT, Huang Y, Carson RE, van Dyck CH, 2020. In vivo measurement of widespread synaptic loss in Alzheimer’s disease with SV2A PET. Alzheimers Dement. 16, 974–982. doi: 10.1002/alz.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch SA, Kensel-Hammes P, Gadd JC, Fujimoto BS, Allen RW, Schiro PG, Lorenz RM, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT, 2011. Protein quantification at the single vesicle level reveals that a subset of synaptic vesicle proteins are trafficked with high precision. J. Neurosci. doi: 10.1523/JNEUROSCI.3805-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabulsi NB, Mercier J, Holden D, Carre S, Najafzadeh S, Vandergeten MC, Lin SF, Deo A, Price N, Wood M, Lara-Jaime T, Montel F, Laruelle M, Carson RE, Hannestad J, Huang Y, 2016. Synthesis and preclinical evaluation of 11C-UCB-J as a PET tracer for imaging the synaptic vesicle glycoprotein 2A in the brain. J. Nucl. Med. 57, 777–784. doi: 10.2967/jnumed.115.168179. [DOI] [PubMed] [Google Scholar]
- Onwordi EC, Halff EF, Whitehurst T, Mansur A, Cotel MC, Wells L, Creeney H, Bonsall D, Rogdaki M, Shatalina E, Reis Marques T, Rabiner EA, Gunn RN, Natesan S, Vernon AC, Howes OD, 2020. Synaptic density marker SV2A is reduced in schizophrenia patients and unaffected by antipsychotics in rats. Nat. Commun. doi: 10.1038/s41467-019-14122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, 2015. The brain’s default mode network. Annu. Rev. Neurosci. 38, 433–447. doi: 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- Ripp I, Stadhouders T, Savio A, Goldhardt O, Cabello J, Calhoun V, Riedl V, Hedderich D, Diehl-Schmid J, Grimmer T, Yakushev I, 2020. Integrity of neurocognitive networks in dementing disorders as measured with simultaneous PET/functional MRI. J. Nucl. Med. 61, 1341–1347. doi: 10.2967/jnumed.119.234930. [DOI] [PubMed] [Google Scholar]
- Salthouse TA, 2000. Aging and measures of processing speed. Biol. Psychol. 54, 35–54. doi: 10.1016/S0301-0511(00)00052-1. [DOI] [PubMed] [Google Scholar]
- Savio A, Fünger S, Tahmasian M, Rachakonda S, Manoliu A, Sorg C, Grimmer T, Calhoun V, Drzezga A, Riedl V, Yakushev I, 2017. Resting-state networks as simultaneously measured with functional MRI and PET. J. Nucl. Med. 58, 1314–1317. doi: 10.2967/jnumed.116.185835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart K, Gallezot JD, Nabulsi N, Labaree D, Zheng MQ, Huang Y, Carson RE, Hillmer AT, Worhunsky PD, 2020a. Separating dopamine D2 and D3 receptor sources of [11C]-(+)-PHNO binding potential: independent component analysis of competitive binding. Neuroimage 214. doi: 10.1016/j.neuroimage.2020.116762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart K, Liu H, Matuskey D, Chen MK, Torres K, Nabulsi N, Labaree D, Ropchan J, Hillmer AT, Huang Y, Carson RE, 2020b. Binding of the synaptic vesicle radiotracer [11C]UCB-J is unchanged during functional brain activation using a visual stimulation task. J. Cereb. Blood Flow Metab. doi: 10.1177/0271678X20946198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF, 2009. Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. 106, 13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND, 2012. Aging and functional brain networks. Mol. Psychiatry 17, 549–558. doi: 10.1038/mp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worhunsky PD, Matuskey D, Gallezot JD, Gaiser EC, Nabulsi N, Angarita GA, Calhoun VD, Malison RT, Potenza MN, Carson RE, 2017. Regional and source-based patterns of [11C]-(+)-PHNO binding potential reveal concurrent alterations in dopamine D2and D3receptor availability in cocaine-use disorder. Neuroimage 148, 343–351. doi: 10.1016/j.neuroimage.2017.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD, 2009. Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum. Brain Mapp. 30, 711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fisch B, Liu H, Buckner RL, 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.