Abstract
Steroid receptors are conditional transcription factors that, upon binding to their response elements, regulate the expression of target genes via direct protein interactions with transcriptional coactivators. We have analyzed the functional interactions between the androgen receptor (AR) and 160-kDa nuclear receptor coactivators. Upon overexpression in mammalian cells, these coactivators enhance the transcriptional activity of both the amino-terminal domain (NTD) and the ligand-binding domain (LBD) of the AR. The coactivator activity for the LBD is strictly ligand-controlled and depends on the nature of the DNA-binding domain to which it is fused. We demonstrate that the NTD physically interacts with coactivators and with the LBD and that this interaction, like the functional interaction between the LBD and p160 coactivators, relies on the activation function 2 (AF2) core domain. The mutation of a highly conserved lysine residue in the predicted helix 3 of the LBD (K720A), however, blunts the functional interaction with coactivators but not with the NTD. Moreover, this mutation does not affect the transcriptional activity of the full-size AR. A mutation in the NTD of activation function AF1a (I182A/L183A), which dramatically impairs the activity of the AR, has no effect on the intrinsic transcriptional activity of the NTD but interferes with the cooperation between the NTD and the LBD. Finally, p160 proteins in which the three LXXLL motifs are mutated retain most of their coactivator activity for the full-size AR, although they are no longer functional for the isolated LBD. Together, these data suggest that in the native AR the efficient recruitment of coactivators requires a functional association of the NTD with the LBD and that the binding of coactivators occurs primarily through the NTD.
Steroid receptors, including the androgen receptor (AR), belong to the superfamily of nuclear receptors (NRs) which generally act as ligand-dependent transcription factors. In the absence of ligand, they are maintained in an inactive state complexed to heat shock proteins and/or corepressors. Binding of the cognate ligand results in the dissociation of the receptor from this inactive complex and its subsequent translocation to the nucleus, where it binds, either as a homo- or a heterodimer, to specific response elements in the promoter and/or enhancer regions of responsive genes and up- or downregulates their transcription (37, 49, 62). NRs share a common structural and functional organization and generally harbor two transcription activation functions (AF): a constitutively active AF1, which is located in the highly variable NH2-terminal domain (NTD) of the receptor, and a ligand-dependent AF2, which colocalizes with the more conserved COOH-terminal ligand-binding domain (LBD) (6). Whereas the molecular mechanisms by which AF1 stimulates transcription are still largely unknown, our knowledge of the functioning of AF2 has greatly increased in the past few years, most notably after the discovery of a new class of proteins called NR coactivators and after the crystal structures of apo- and holo-LBDs of several NRs became available (42, 51, 67, 69). The LBD structurally consists of twelve α-helices, the most COOH-terminal of which (helix 12[H12]) is projected away from the hormone-binding pocket in the absence of ligand. After the association of the LBD with an agonist, H12 “flips over” and covers the hydrophobic ligand-binding cavity as a lid (47). Apart from keeping the ligand tightly bound as a “mouse in a trap”, the swinging of H12 also has important structural consequences and induces the formation of a new structural surface which can interact with accessory proteins. A plethora of proteins have been identified which interact, mostly in a ligand-dependent way, with the LBDs of NRs and which can stimulate or repress AF2 activity (reviewed in references 23, 27, and 52). The best-characterized subgroup of receptor-interacting proteins is the family of 160-kDa NR coactivators, also called p160 proteins, comprising (i) steroid receptor coactivator-1 (SRC-1) (29, 30, 45, 57), (ii) the human and rat transcription-intermediary factor 2 (TIF2) (34, 64, 65) and its mouse orthologue glucocorticoid receptor (GR)-interacting protein 1 (GRIP1) (21), and (iii) receptor-associated coactivator 3 (RAC3) (35, 36), also known as ACTR (12), TRAM-1 (58), AIB (3), p/CIP (60), and SRC-3 (56). They interact with the LBD via a centrally located NR-interacting region (NIR) containing three highly conserved α-helical LXXLL motifs, or NR boxes (19, 29, 35, 36, 46, 50, 60, 65). One isoform of SRC-1 (SRC-1a) contains a fourth NR box at its extreme carboxy terminus (29, 30), which is absent in another isoform called SRC-1e. Although this SRC-1a-specific NR box IV can interact with the LBDs of NRs (14, 19, 29, 59), the relevance of this interaction remains unestablished (29, 59). In addition, the carboxy-terminal end of SRC-1a harbors a repressive function that affects its coactivator activity for the estrogen receptor (ER) AF2 function (29).
Mutational analyses of the LBDs of a variety of NRs have revealed that, among others, a conserved lysine residue in H3 and the AF2 core domain located in H12 are essential for the interaction of the receptors with p160 proteins and for transcriptional activity (12, 13, 17, 20, 58, 61). The X-ray structure of a ternary complex of the peroxisome proliferator-activated receptor (PPAR) LBD with rosiglitazone and a fragment of SRC-1 comprising two NR boxes (43) showed that this lysine and a conserved glutamate in H12 form a “change clamp” that forms hydrogen bonds with backbone amides of the LXXLL motifs, whereas the hydrophobic face of the LXXLL helix fits into a hydrophobic cavity. It was reported very recently that the ER AF1 domain may also function via the recruitment of p160 proteins (66), but this interaction does not depend on the LXXLL motifs.
Most of our knowledge of the action mechanism of NRs, and most notably of their AF2s, comes from studies with the retinoic acid and retinoid X receptors (RAR and RXR), the thyroid hormone receptor (TR), and the ER. A striking difference between these receptors and the AR is that for the latter an intrinsic AF2 activity in the LBD has remained elusive (25, 40, 53), despite the relatively high degree of sequence similarity of its LBD with that of other steroid receptors (67, 69). The AF1 function, on the other hand, has received more attention and was characterized by several groups (11, 24, 26). The observation that different activation regions within the NTD are used in a full-length receptor and in a constitutively active mutant, devoid of the LBD, indicates that the activity of the NTD may be controlled by the LBD and raises the possibility of a functional interaction between these domains. Such an interaction could be demonstrated in classical yeast and mammalian two-hybrid experiments (8, 15, 24, 31) and was also observed for the ER (39). Moreover, even in the absence of a heterologous transactivating domain, the human AR (hAR) LBD and NTD can associate to form a transcriptionally active complex (8, 15). It is still unclear whether this interaction is direct or whether a third partner is involved which would act as a bridging factor. Possible candidates for such bridging proteins are the p160 coactivators, since they were shown to enhance the interaction between the NTD and the LBD of the AR and the ER (24, 39).
In this paper, we describe the effects of p160 coactivators on the isolated AF1 and AF2 domains of the hAR. Furthermore, we have analyzed the functional and physical interaction of these AFs with each other and with the coactivators by using point-mutated proteins. Our data suggest that the interaction between the hAR NTD and LBD may be a prerequisite for the efficient recruitment of coactivators to the native receptor and for its transcriptional activity, thus highlighting a novel role for the NTD in the functioning of the AR.
MATERIALS AND METHODS
Materials.
The hAR expression vector pSG5-hAR and the K720A mutant are described elsewhere (2). The other mutants were made by a PCR-based method. The I898A/I899A mutation was generated with the oligonucleotides 5′-GATGGCAGAGGCCGCCTCTGTGCAAGATC-3′ and 5′-GATCTTGCACAGAGGCGGCCTCTGCCATC-3′, in which the underlined sequences represent the mutated codons. To generate the AF1a mutation (I182A/L183A), the following oligonucleotides were used: 5′-CCTTAAAGACGCCGCGAGCGAGGCC-3′ and 5′-GGCCTCGCTCGCGGCGTCTTTAAGG-3′. Expression vectors for the hAR NTD (M1 to R538) and for DNA-binding domain (DBD)-LBD (L539 to Q919) were made by inserting PCR-generated fragments encoding the corresponding regions in the BamHI site of pSG5. Similarly, the DBD-LBD fragment of the rat GR (rGR) (P416 to K777) was PCR amplified from pH6GR (a gift from H. Stunnenberg) and subcloned in the BamHI site of pSG5. The expression vector for the NTD-DBD was a kind gift from A. O. Brinkmann (25). For the expression of receptor domains fused to residues 1 to 147 of the yeast transcription factor GAL4, PCR-generated fragments encoding either the NTD (M1 to R538) or the LBD (A628 to Q919) were inserted in the BamHI restriction site of pAB-Gal4 (4). The reporter plasmid pGal-50hIL6-luc (48), containing two GAL4-binding sites in front of the minimal promoter of the human interleukin 6 (hIL6) gene, was a kind gift of S. Plaisance. For the expression of glutathione S-transferase (GST) fusion proteins, the plasmid pHIL was constructed by inserting two complementary oligonucleotides encoding the vesicular stomatitis virus (VSV)-G epitope tag (NH2-MYTDIEMNRLGKG-COOH) in the BamHI site of pCMV-GST (63). Next, the DNA fragments encoding the DBD-LBD regions of the murine ER (mER) and the hAR were isolated from the corresponding expression plasmids and inserted into pHIL. The resulting vectors drive the expression of fusion proteins consisting of GST, the VSV-G tag, and the entire DBD and LBD of the respective receptors. To generate VP16 fusion proteins for use in mammalian double-hybrid experiments, a modified version of the plasmid pSNATCH was used (10). A cDNA-fragment encoding the residues 454 to 498 of the VP16 activating domain was inserted in pSNATCH, giving rise to pSNATCH-II, which thus directs the expression of the residues 413 to 498 of the VP16 activating region. Finally, the hAR NTD was cloned in frame with the VP16 activating domain in the BglII site of pSNATCH-II. Expression vectors for TIF2 and for SRC-1, and derivatives thereof, were obtained from H. Gronemeyer and M. G. Parker, respectively. All PCRs were performed with the Pwo thermostable DNA polymerase (Boehringer, Mannheim, Germany). Restriction and modifying enzymes were obtained, either from Boehringer, Pharmacia (Uppsala, Sweden), or Life Technologies (Grand Island, N.Y.). The synthetic androgens R1881 (methyltrienolone) and mibolerone were supplied by Dupont-New England Nuclear (Boston, Mass.), and the synthetic glucocorticoid hormone triamcinolone acetonide was from Sigma (St. Louis, Mo.). [3H]mibolerone (82.3 Ci/mmol) was purchased from Amersham (Buckinghamshire, United Kingdom).
Transfections.
All transfections were performed in COS 7 cells or in CV-1 cells, obtained from the American Type Culture Collection (Rockville, Md.). One day prior to transfection, the cells were seeded in 24-well culture plates in Dulbecco’s modified eagle’s medium (Life Technologies) containing 5% dextran-coated charcoal-stripped fetal bovine serum (DCC) at a density of 7.5 × 104 per well. They were transfected, either by the calcium phosphate coprecipitation method or with the Fugene 6 transfection reagent (Boehringer). After transfection, the cells were incubated for 24 h with DCC-treated medium, either supplemented or not with hormones (1 nM). Finally, they were washed twice with phosphate-buffered saline (PBS) and lysed in 100 μl of passive lysis buffer (Promega Corp., Madison, Wis.). The protein concentration was measured with the Coomassie protein assay reagent from Pierce (Rockford, Ill.). The luciferase and β-galactosidase activities were measured on 10 μl of the extracts with the assay systems from Promega and Clontech (Palo Alto, Calif.), respectively, and corrected for protein content. The luciferase activity was corrected for transfection efficiency by normalizing it against β-galactosidase activity. The total amount of DNA that was transfected depended on the transfection method that was used, but the relative proportions were always 10/1/10 for reporter-receptor-coactivator cotransfections. The amount of pCMV-β-GAL was fixed at 50 ng per well. Where applicable, empty pSG5 vector was added to keep the total amount of transfected DNA constant. The transfection results are always shown as the measured luciferase activity, relative to a standard condition as mentioned in the legends to the respective figures. The values shown are the means of at least three measurements ± the standard error of the mean (SEM).
In vivo ligand binding and immunoblotting.
Androgen binding was measured in transiently transfected COS 7 cells as follows: on day 1 the cells were transfected in 175-cm2 tissue culture flasks with 10 μg of expression vector for either the wild-type hAR or the K720A or I898A/I899A mutants by the calcium phosphate coprecipitation technique. The precipitate was left on the cells for 16 h, after which they received a 15% glycerol shock for 2 min and were incubated in DCC-treated medium. On day 3, they were trypsinized and seeded in 24-well plates at a density of 105 per well and were seeded in 60-mm-diameter dishes at a density of 2.5 × 105 per dish. On day 4, the cells in the 24-well plates were incubated with DCC-treated medium containing increasing concentrations of [3H]mibolerone, (0.1 to 100 nM), either in the absence or in the presence of a 200-fold excess of unlabeled hormone to eliminate nonspecific binding. After 90 min at 37°C, the plates were put on melting ice and the cells were washed twice with ice-cold PBS. They were lysed in 100 μl of 0.1 N NaOH–1% Triton X-100, and the radioactivity was measured in a scintillation counter. Kd values were calculated by the Scatchard method. For Western blotting, the cells in the 60-mm-diameter dishes were lysed by two cycles of freezing in liquid N2 and thawing at room temperature in 20 mM Tris (pH 7.8)–300 mM NaCl–1 mM EDTA–0.1% Nonidet P-40 and soluble extracts were prepared by centrifugation. Ten microliters of each extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in an 8% gel and blotted onto polyvinylidene difluoride membranes. The membranes were probed with a polyclonal antiserum against the NH2 terminus of the hAR (38), and immunoreactive proteins were visualized with the ECL system (Amersham).
GST pull down.
For GST pull-down experiments, COS cells in 9-cm-diameter plates were cotransfected with expression vectors for the respective GST fusion proteins (described above) and FLAG-tagged SRC-1e or the NTD by the calcium phosphate coprecipitation procedure. After transfection, the cells were incubated for 24 h with medium with or without 100 nM of the appropriate hormone. The cells were washed twice with ice-cold PBS, and extracts were prepared by two cycles of freezing in liquid nitrogen and thawing on ice in 50 μl of NENT300-Mo (20 mM TRIS [pH 6.8], 300 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 25% glycerol, 20 mM Na molybdate), followed by centrifugation to pellet insoluble material. The extracts were diluted with 150 μl of NENT0-Mo (like NENT300-Mo but lacking NaCl) and loaded onto glutathione-Sepharose beads (Pharmacia). The GST fusion proteins were then allowed to bind to the matrix for 16 h at 4°C on a rotating wheel. The beads were washed extensively with NENT100-Mo, and bound proteins were eluted by boiling the matrix in SDS-PAGE sample buffer, subjected to SDS-PAGE, and finally blotted onto polyvinylidene difluoride membranes. Hormone (100 nM) was included in all buffers, where applicable. To visualize FLAG-SRC-1e, Western blots were probed with an M2 anti-FLAG antibody (Stratagene, La Jolla, Calif.), and to detect the hAR NTD, a polyclonal antiserum recognizing the first 20 residues of the hAR was used (38). To ensure adequate expression of all GST fusion proteins, aliquots of the COS extracts were immunoblotted with a monoclonal anti-VSV-G antibody (Sigma).
RESULTS
Both the hAR AF1- and AF2-containing domains, when expressed separately, can be stimulated by p160 coactivators.
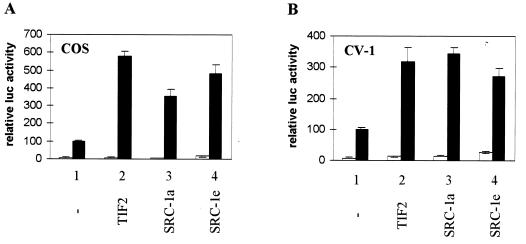
The observation that, contrary to many other NRs, the hAR LBD has no detectable intrinsic AF2 activity in mammalian cells (26, 40, 53) prompted us to analyze the effect of p160 coactivators on the transcriptional activity of this receptor. Cotransfection experiments in COS cells, using the androgen-responsive MMTV-luc vector as a reporter, reveal that p160 proteins stimulate the ligand-dependent transcriptional activity of the hAR (Fig. 1A). In the absence of ectopically expressed coactivator, the luciferase activity is stimulated approximately 10-fold in response to 1 nM synthetic androgen R1881 (Fig. 1, bar 1), whereas cotransfection of TIF2, SRC-1a, or SRC-1e results in a further 4- to 6-fold increase of transcription (bars 2 to 4). To ensure that the presence in COS cells of the large T antigen and the concomitant high expression levels of proteins encoded by pSG5-based plasmids did not influence the observed effects, we also performed the experiment in CV-1 cells. As shown in Fig. 1B, similar results were obtained under these conditions. It should be noted that the absolute levels of luciferase activity measured in CV-1 cells are on average 5 to 10 times lower than those measured in COS cells under the same experimental conditions.
FIG. 1.
p160 coactivators stimulate the transcriptional activity of the full-size hAR. COS 7 cells (A) and CV-1 cells (B) were cotransfected with pMMTV-luc (500 ng) and pSG5-hAR (50 ng), along with 500 ng of either empty pSG5 (bar 1) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (bar 2 to 4). The cells were incubated with DCC-treated medium, either supplemented (solid bars) or not (open bars) with 1 nM R1881 24 h before being harvested. The luciferase activities were corrected for transfection efficiency by using the β-galactosidase activities. The data are presented as relative activities (± SEM). The luciferase activity measured in the presence of ligand but without cotransfected coactivator was arbitrarily set to 100, and test values were calculated accordingly.
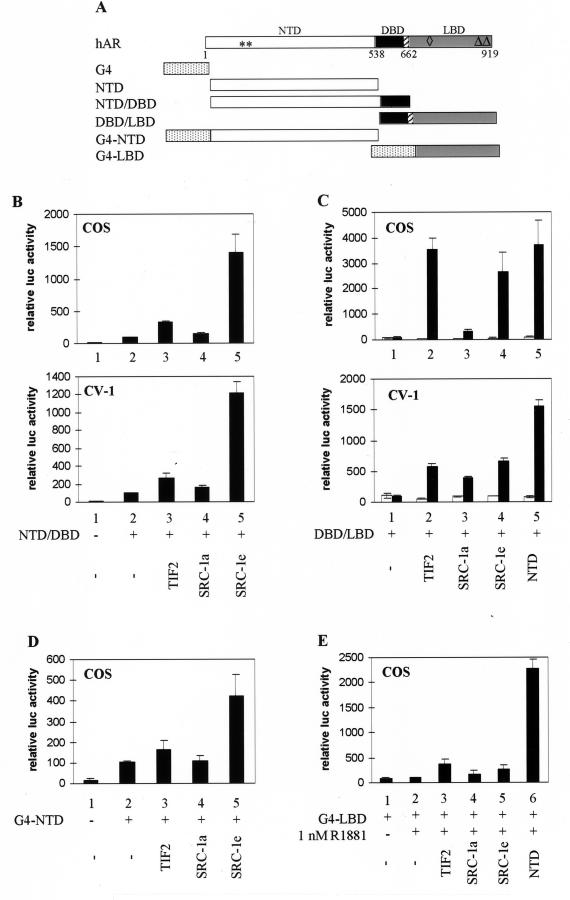
Originally, p160 coactivators were thought to stimulate only the ligand-dependent AF2 transcription activation function of NRs. However, it has been reported that SRC-1 can also interact, in a ligand-independent way, with NRs mutated in the AF2 region (28), and recent data indicate that p160 coactivators may also stimulate AF1 and promote the functional interaction between the two AFs (24, 46, 66). To determine the effect of these coactivators on either AF of the hAR, COS cells or CV-1 cells were cotransfected as above, except that expression vectors encoding either the NTD and the DBD (NTD-DBD) or the DBD and the LBD (DBD-LBD) (Fig. 2A) were used instead of a plasmid encoding the entire receptor. Consistent with previously reported results (26), the NTD has intrinsic transcription activation properties (AF1) (Fig. 2B, compare bars 2 and 1). Coexpression of TIF2 or SRC-1a results in only a slight increase of AF1 activity (Fig. 2B, compare bars 3 and 4 to bars 2), whereas coexpression of SRC-1e results in a 15-fold and a 12-fold induction of transcription (bar 5 versus bar 2) in COS cells and in CV-1 cells, respectively. The effect of the p160 coactivators on the transcriptional activity of the LBD was analyzed in a similar way. The DBD-LBD construct has no intrinsic ligand-dependent transcription activation properties (Fig. 2C, bars 1), but coexpression of either TIF2 or SRC-1e results in a strong induction of transcription (compare bars 2 and 4 to bars 1), as does coexpression of the hAR NTD (bars 5). SRC-1a, on the other hand, is a less potent coactivator for the hAR DBD-LBD construct (bars 3), especially under the conditions of high expression levels in COS cells (Fig. 2C, upper panel). Similar to the effects on the full-size receptor (Fig. 1), the induction of transcriptional activity of the DBD-LBD fragment by overexpression of p160 proteins is strictly ligand dependent. These experiments were also performed in COS cells with constructs encoding the DBD of the yeast transcription factor GAL4 fused to either the hAR NTD or the hAR LBD (Fig. 2A) and a luciferase reporter gene, driven by five copies of a GAL4 DNA-binding site in front of a TATA box [(G4)5-tata-luc]. The results obtained with the G4-NTD construct (Fig. 2D) are very similar to those obtained with the NTD-DBD construct and the MMTV-luc reporter described in Fig. 2B. Whereas TIF2 and SRC-1a influence the activity of G4-NTD only weakly (Fig. 2B, compare bars 3 and 4 with bar 2), SRC-1e clearly coactivates this construct to a much greater extent (bar 5). Although the absolute levels of transcription stimulation of G4-NTD are somewhat lower than those obtained for the NTD-DBD construct (4-fold versus 15-fold for SRC-1e), the effects of either p160 protein are comparable under both conditions. Surprisingly, this is not the case when DBD-LBD and G4-LBD are compared. Whereas TIF2 and SRC-1e strongly stimulate the activity of DBD-LBD (Fig. 2C), they have only minor effects on G4-LBD (Fig. 2E; compare bars 3 to 5 with bar 2). hAR-NTD, on the other hand, does strongly stimulate the transcriptional activity of G4-LBD (to 23-fold [bar 6]).
FIG. 2.
The hAR AF1 and AF2 functions are both stimulated by p160 coactivators. (A) Schematic representation of the hAR and the constructs used in this study. Depicted are the NTD, the centrally located DBD, the hinge region (hatched box), and the C-terminal LBD of the hAR, and the DBD of the yeast transcription factor GAL4 (G4). The numbers refer to amino acid positions. ∗∗, position of the residues mutated in the AF1a mutant (I182A/L183A); ◊, position of the H3 mutant (K720A); ▵▵, position of the AF2 mutant (I898A/I899A). (B) COS cells (upper graph) and CV-1 cells (lower graph) were transfected with pMMTV-luc (250 ng) without (bars 1) or with (bars 2 to 5) an expression vector encoding NTD-DBD (125 ng) and with either empty pSG5 (bars 1 and 2) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (900 ng). After 24 h, the cells were harvested and luciferase and β-galactosidase activities were measured as described in Materials and Methods. The activity of NTD-DBD in the absence of exogenously added coactivator was given the value 100. All other activities were calculated accordingly. (C) COS cells (upper graph) and CV-1 cells (lower graph) were cotransfected with pMMTV-luc (250 ng), pSG5-DBD/LBD (25 ng), and either empty pSG5 (bar 1) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (250 ng) (bars 2 to 5) and incubated for 24 h in DCC-treated medium, either supplemented (solid bars) or not (open bars) with 1 nM R1881. They were harvested and processed as described in Materials and Methods. (D) COS 7 cells were transfected with the p(G4)5-tata-luc reporter (250 ng) without (bar 1) or with (bars 2 to 5) an expression vector for G4-NTD (25 ng) and with either empty pSG5 (bars 1 and 2) or pSG5-TIF2, pSG5-SRC-1a, or pSG5-SRC-1e (bars 3 to 5; 250 ng) and treated as described for panel B. (E) COS 7 cells were transfected as for Fig. 2C, except that the p(G4)5-tata-luc reporter vector was used and pSG5-DBD/LBD was replaced by an expression vector encoding G4-LBD. The cells were treated with 1 nM R1881 as indicated. The luciferase activity measured in the presence of R1881 but without ectopically expressed coactivator (bar 2) was set to 100, and test values were calculated accordingly. The error bars indicate the SEM.
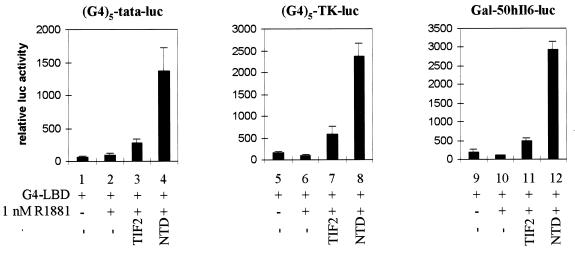
A recent report suggested that the coactivator activity of TIF2 for the hAR AF2 function might be promoter dependent (8). To verify that the differences we observed between DBD-LBD and MMTV-luc on the one hand (Fig. 2C) and G4-LBD and (G4)5-tata-luc on the other hand (Fig. 2E) are not merely the result of using a complex MMTV-luc reporter versus a minimal (G4)5-tata-luc reporter, we extended the experiments with G4-LBD to two other GAL4-driven reporter constructs with more complex promoters: (G4)5-TK-luc and Gal-50hIL6-luc containing 5 and 2 GAL4 DNA-binding sites, respectively, in front of the thymidine kinase (TK) promoter or the minimal promoter of the human interleukin 6 gene (48). As shown in Fig. 3, irrespective of the reporter used, coexpression of the NTD always results in a potent activation of the transcriptional activity of G4-LBD, whereas coexpression of TIF2 results in only a very poor stimulation of transcription.
FIG. 3.
hAR NTD efficiently stimulates the transcriptional activity of G4-LBD, independent of the promoter context. COS 7 cells were cotransfected with either p(G4)5-tata-luc (left graph), p(G4)5-TK-luc (center graph), or pGal-50hI16-luc (right graph), along with pG4-LBD and either empty pSG5 (bars 1, 2, 5, 6, 9, and 10), pSG5-TIF2 (bars 3, 7, and 11), or pSG5-NTD (bars 4, 8, and 12), as for Fig. 2E. The cells were incubated with DCC-treated medium, either supplemented (+) or not (−) with 1 nM R1881 for 24 h and subsequently processed as described in Materials and Methods. In each case, the luciferase activity measured in the presence of hormone but in the absence of exogenously added coactivator was arbitrarily set to 100, and the values for the other conditions were calculated accordingly. The error bars indicate the SEM.
The hAR NTD physically interacts with SRC-1e and with the hAR LBD in COS cells.
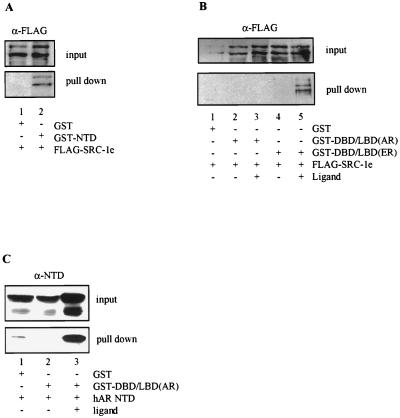
To examine whether the stimulatory effects we observed upon cotransfection of p160 proteins with either the isolated NTD or LBD can be correlated with direct protein-protein interactions, we performed GST pull-down experiments from transfected-cell extracts. Therefore, vectors were constructed directing the expression of the hAR NTD or of the DBD-LBD of the hAR and the mER, respectively, fused in frame to GST and the VSV-G tag, which were then cotransfected in COS cells with a FLAG-tagged SRC-1e expression vector. Protein complexes associated with the GST fusion proteins were precipitated from whole-cell extracts with a glutathione-Sepharose affinity matrix and subsequently immunoblotted with anti-FLAG antibody. As shown in Fig. 4A, SRC-1e can be precipitated specifically by GST-NTD, indicating that the hAR NTD physically interacts with SRC-1e in COS cells. Using this technique, however, we were unable to demonstrate a strong physical interaction between the hAR DBD-LBD and SRC-1e, either in the absence or in the presence of ligand (Fig. 4B, lanes 2 and 3). Nevertheless, under the same experimental conditions we saw a good ligand-dependent interaction of SRC-1e with the GST–DBD-LBD construct of the mER (lanes 4 and 5). Aliquots of the extracts were immunoblotted and probed with anti-VSV-G antibody to ensure adequate expression of the mER and hAR GST–DBD-LBD fusion proteins (data not shown). This result indicates that the hAR LBD has very low intrinsic affinity for p160 proteins. However, in yeast and mammalian double-hybrid experiments we saw a ligand-dependent interaction between the AR LBD and TIF2.5, a fragment of TIF2 harboring the three NR box motifs (65). Compared to the interaction observed with the LBD of a panel of NRs with a strong intrinsic AF2 activity, however, the AR LBD-TIF2.5 interaction is much weaker (our unpublished results).
FIG. 4.
hAR NTD physically interacts with SRC-1e and with the hAR LBD in COS cells. (A) COS cells were transfected with expression vectors for either GST alone or a fusion protein of GST with the hAR NTD, along with an expression vector for FLAG-tagged SRC-1e. After 24 h of incubation at 37°C, extracts were prepared and protein complexes bound to GST or GST-NTD were precipitated with glutathione Sepharose beads, immunoblotted, and stained with monoclonal anti-FLAG antibody. (B) COS cells were transfected and treated as described for panel A, except that GST fusion proteins of the DBD-LBD regions of the hAR and the mER were used. The cells were treated without hormone (lanes 1, 2, and 4) or with 100 nM of either R1881 (lane 3) or estradiol (lane 5) 24 h prior to the preparation of protein extracts and GST pull down. (C) COS cells were transfected and treated as for panel B, except that an expression vector for the hAR NTD was used instead of FLAG-tagged SRC-1e. The immunoblots were probed with a polyclonal anti-AR antiserum. +, present; −, absent.
The GST pull-down experiment was also performed with an expression vector for the hAR NTD instead of FLAG-tagged SRC-1e, and the precipitated complexes were immunoblotted with a polyclonal antiserum against the hAR NTD (38). The hAR NTD can be precipitated specifically and in a ligand-dependent way with the GST–DBD-LBD fusion protein (Fig. 4C, lanes 2 and 3), pointing to a physical interaction between the NTD and the LBD of the hAR.
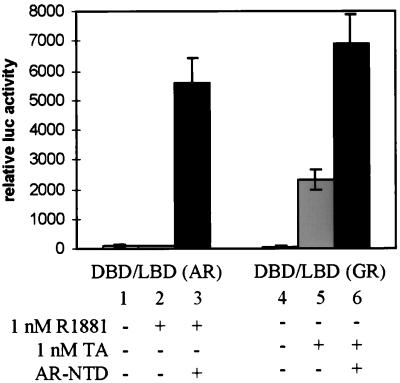
The hAR NTD stimulates the transcription activation function of the hAR LBD more efficiently than that of the rGR LBD.
The observation that the hAR NTD efficiently stimulates the hAR AF2 function raises the question whether this is a receptor-specific effect or whether the AR NTD can also activate the AF2 function of another steroid receptor, such as the rGR. To test this, a construct was made encoding the entire DBD and LBD of the rGR [here called DBD-LBD(GR)], similar to the DBD-LBD construct of the hAR [here called DBD-LBD(AR)]. Both constructs were transfected in COS cells, along with the MMTV-luc reporter plasmid and with or without an expression vector for the hAR NTD. As shown in Fig. 5 and consistent with the results described above, the hAR AF2 activity is efficiently activated upon coexpression of the hAR NTD (Fig. 5, compare bar 3 to bar 2). Contrary to DBD-LBD(AR), however, DBD-LBD(GR) has a good intrinsic ligand-dependent AF2 function (Fig. 5, compare bars 5 and 4), and the addition of the synthetic glucocorticoid hormone triamcinolone acetonide results in a 35-fold increase of reporter gene expression. Cotransfection of the hAR NTD, however, stimulates the DBD-LBD(AR) 59-fold (Fig. 5, bar 3 versus bar 2), whereas it has less pronounced effects (3-fold induction) on the AF2 activity of DBD-LBD(GR). Alternatively, the rGR NTD does not stimulate the transcriptional activity of the hAR LBD, nor does it strongly affect the rGR AF2 activity (data not shown). Note that the total transcriptional activities of either DBD-LBD construct in the presence of the hAR NTD are comparable for the hAR and the rGR (Fig. 5, compare bars 3 and 6).
FIG. 5.
Coexpression of the hAR NTD strongly stimulates the transcriptional activity of the hAR LBD but has only weak effects on the rGR AF2 function. COS 7 cells were transfected with 250 ng of pMMTV-luc, along with 25 ng of either pSG5-DBD/LBD(AR) or pSG5-DBD/LBD(GR) and 250 ng of either empty pSG5 (bar 1, 2, 4, and 5) or pSG5-NTD (bar 3 and 6), and treated for 24 h without hormone (bar 1 and 4), with 1 nM R1881 (bars 2 and 3), or with 1 nM triamcinolone acetonide (TA) (bars 5 and 6). Luciferase and β-galactosidase activities were measured as described in Materials and Methods. The luciferase activity measured with DBD-LBD(AR) in the presence of R1881 and in the absence of NTD (bar 2) was arbitrarily set to 100. The activities of all other conditions were calculated relative to this standard. The error bars indicate the SEM.
Effect of mutations in H3 and H12 of the LBD.
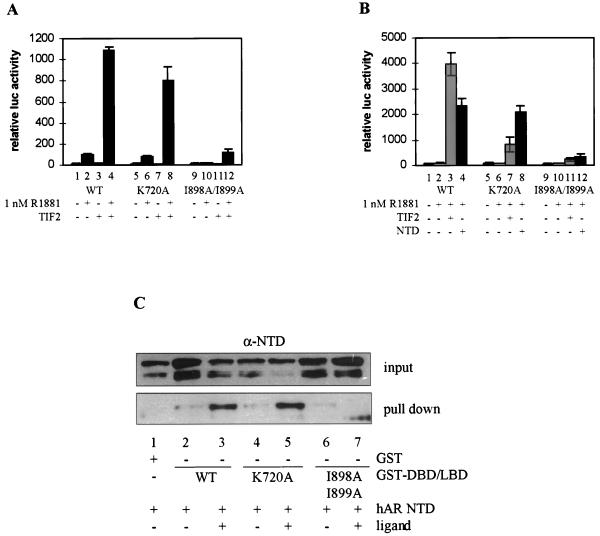
The interaction between the p160 coactivators and nuclear receptors has been reported to occur primarily through residues in H3, -4, -5, and -12 of the LBD (17, 20, 42). We have exchanged a highly conserved lysine residue in H3 (K720) and two bulky hydrophobic residues in H12 (I898 and I899) of the hAR LBD for alanines, both of which dramatically impair the transcription activation function of the mER (13, 20). The mutants are efficiently expressed, as could be judged from Western blots of extracts of transfected COS cells (data not shown). Their ability to bind ligand was analyzed in a whole-cell ligand-binding assay. The following Kd values were calculated for the synthetic hormone mibolerone (mean ± SEM): 2.53 nM (±0.15) for the native receptor and 4.08 nM (±0.31) and 3.13 nM (±0.38) for the K720A and the I898A/I899A mutants, respectively. Thus, these mutations do not substantially influence ligand binding and probably do not alter the overall structure of the receptor.
Next, the effects of these amino acid changes on the transcriptional activity of the hAR were examined. In the absence of exogenous TIF2, the mutant K720A can be induced as well as the wild-type receptor by 1 nM R1881 (Fig. 6A, compare bars 5 and 6 to bars 1 and 2), whereas the mutant I898A/I899A is transcriptionally inactive (bars 9 and 10). All three receptors, however (the wild-type protein as well as both mutants), can be stimulated by coexpressing TIF2. Compared to control conditions (without ectopic expression of TIF2), the unmutated receptor and the K720A mutant are stimulated 10- and 8-fold, respectively, upon cotransfection of TIF2 (Fig. 6A, compare bar 4 to bar 2 and bar 8 to bar 6). Although the absolute level of luciferase activity measured with the I898A/I899A mutant in the presence of TIF2 and R1881 (Fig. 6A, bar 12) is much lower, coexpression of TIF2 still enhances the activity of the I898A/I899A mutant (compare bar 12 to bar 10), possibly reflecting a coactivator activity of TIF2 on the NTD. This hypothesis is further supported by the observation that SRC-1e, which is a better coactivator for the NTD, stimulates the I898A/I899A mutant to a greater extent than TIF2 (data not shown).
FIG. 6.
The effect of the K720A and I898A/I899A mutations on the transcriptional activities of the native hAR and on the DBD-LBD construct. (A) COS 7 cells were cotransfected with 250 ng of pMMTV-luc and 25 ng of expression vector for either the full-size wild-type hAR, the K720A mutant, or the I898A/I899A mutant, along with 250 ng of either empty pSG5 (bars 1, 2, 5, 6, 9, and 10) or pSG5-TIF2 (bars 3, 4, 7, 8, 11, and 12) and incubated for 24 h with DCC-treated medium without (open bars) or with (solid bars) 1 nM R1881. The measured luciferase activities were corrected for protein content and β-galactosidase activity and are expressed as relative values, with the activity of the native receptor in the presence of R1881 taken as 100. (B) COS 7 cells were transfected as described for panel A, but instead of expression vectors for full-size receptors, 25 ng of the plasmids encoding the DBD-LBD fragments of either the wild-type receptor, the K720A or the I898A/I899A mutants were used, along with 250 ng of either empty pSG5 (bars 1, 2, 5, 6, 9, and 10), pSG5-TIF2 (bars 3, 7, and 11) or pSG5-NTD (bars 4, 8, and 12). The cells were either left untreated (bars 1, 5, and 6) or treated with 1 nM R1881 (bars 2 to 4, 6 to 8, and 10 to 12) for 24 h before being harvested. The luciferase activity measured for the wild-type DBD-LBD construct in the presence of hormone was taken as 100, and test values were calculated. (C) GST pull-down experiments were performed as described in Materials and Methods and in the legend to Fig. 4. The immunoblots were probed with an anti-AR antiserum. +, present; −, absent. The error bars indicate the SEM.
To eliminate such effects and to analyze the consequences of the above-mentioned mutations solely on the AF2 function, DBD-LBD constructs were made carrying the K720A and the I898A/I899A mutations. The K720A mutant can be activated by cotransfection of TIF2 (Fig. 6B, compare bar 7 to bar 6), although clearly less efficiently than the unmutated DBD-LBD (bar 7 versus bar 3). Mutation of the two isoleucines in H12 (I898A/I899A) has even more pronounced effects, and cotransfection of TIF2 results in a maximal twofold stimulation of transcription (Fig. 6B, bar 11 versus bar 10). The effects of these mutations on the stimulation of AF2 by the hAR NTD were also studied. As shown in Fig. 6B, the K720A mutation does not affect stimulation by the NTD (Fig. 6B, compare bar 8 to bar 4), whereas I898A/I899A does (compare bar 12 to bars 4 and 8). In keeping with this are the results of GST pull-down experiments (Fig. 6C). The hAR NTD can be precipitated from COS cell extracts, in a ligand-dependent way, by GST fusion proteins of the wild-type and the K720A mutated DBD-LBD (Fig. 6C, lanes 1 to 5) but not by the I898A/I899A mutant (lanes 6 and 7).
Mutation of the LXXLL motifs in p160 proteins affects their coactivator properties for hAR LBD but not for hAR NTD.
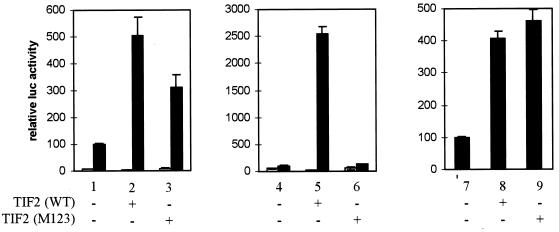
In vitro and in vivo binding studies with several p160 family members have identified a centrally located NIR containing three highly conserved LXXLL motifs, which are sufficient for interaction with the NR AF2 domain (19, 29, 46, 60, 65). Our observation that the hAR AF1 domain, but also a full-size receptor in which the AF2 function is mutated, can be stimulated by 160-kDa coactivators prompted us to look at the effect of the mutation of the LXXLL motifs on the coactivator properties for the full-size hAR, as well as for the isolated AF1 and AF2 functions.
Both native TIF2 and a molecule in which the three LXXLL motifs are mutated (M123) act as efficient coactivators for the full-length hAR with MMTV-luc as a reporter (Fig. 7, left graph), although the activity of the M123 mutant is somewhat (∼30%) lower than that of wild-type TIF2 (compare bars 2 and 3 to bar 1). The same experiment was performed with the DBD-LBD construct (Fig. 7, center graph). As expected, mutation of the three LXXLL motifs results in a complete loss of coactivator activity of TIF2 for the isolated LBD (compare bars 5 and 6 to bar 4). AF1, on the other hand, can be stimulated by both native and mutated TIF2 (Fig. 7, right graph). No significant differences were detected between the wild-type and the M123 forms with respect to their ability to stimulate the transcriptional activity of the NTD-DBD construct (compare bars 8 and 9 to bar 7).
FIG. 7.
The effect of mutation of the three LXXLL motifs in TIF2 on the transcriptional activity of the hAR. COS-7 cells were transfected with 250 ng of pMMTV-luc reporter and 25 ng of pSG5-hAR (left graph), 25 ng of pSG5-DBD/LBD (center graph), or 125 ng of NTD-DBD expression vector (right graph), along with either 250 ng (bars 1 and 4) or 900 ng (bar 7) of empty pSG5, 250 ng (bars 2 and 5) or 900 ng (bar 8) of pSG5-TIF2(WT), or 250 ng (bars 3 and 6) or 900 ng (bar 9) of pSG5-TIF2(M123). The cells transfected with NTD-DBD (right graph) were incubated for 24 h in DCC-treated medium, and those transfected with the native hAR or DBD-LBD (left and center graphs) received DCC-treated medium supplemented with 1 nM R1881 (solid bars) or vehicle (ethanol) alone (open bars). The luciferase activities were corrected for transfection efficiency by using the β-galactosidase activities. +, present; −, absent. The error bars indicate the SEM.
Since SRC-1e has more pronounced effects on the activity of the hAR NTD (Fig. 2B), we also analyzed the effects of comparable mutations in SRC-1e and found that they are identical to the effects described here for TIF2 and TIF2(M123) (data not shown).
Mutation of AF1a blunts the functional interaction between the hAR NTD and LBD.
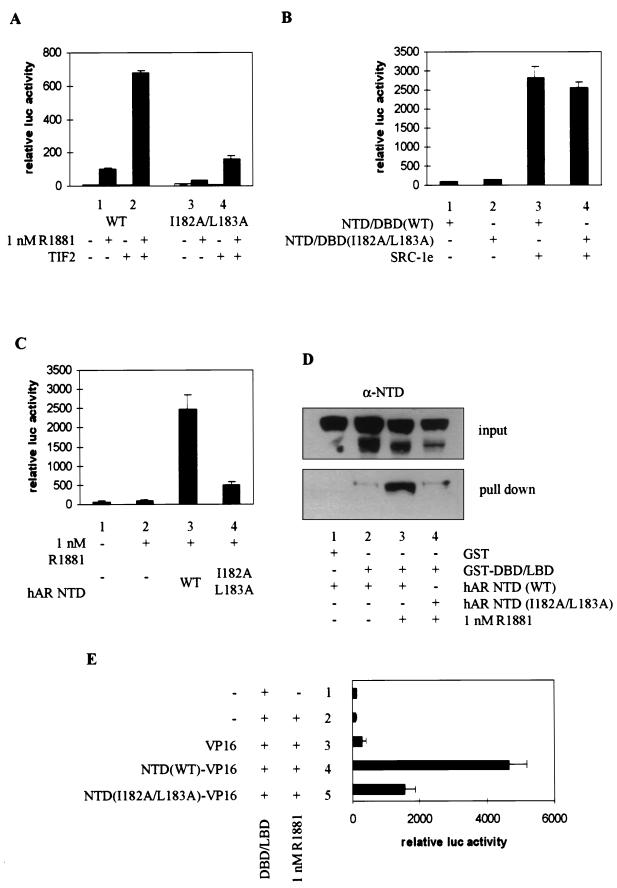
Chamberlain et al. (11) reported that the mutation of an isoleucine and a leucine in the NTD of the rAR, located in a region they termed AF1a, dramatically impairs the transcriptional activity of the receptor.
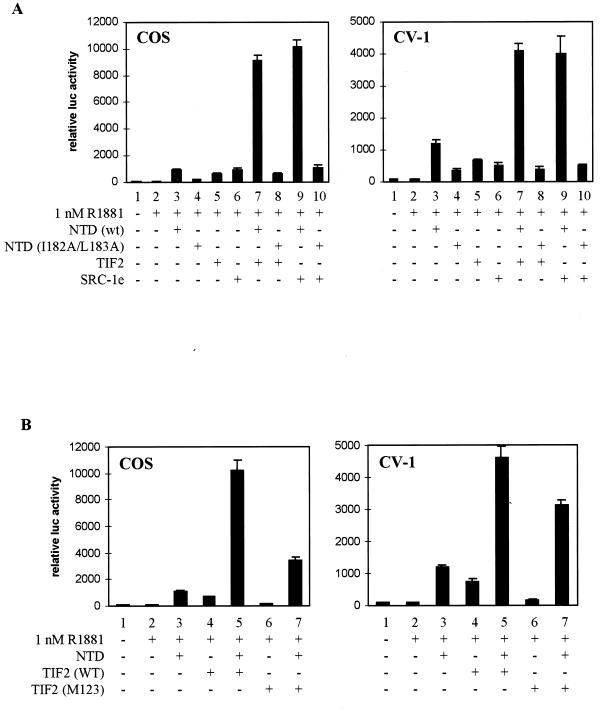
We mutated these residues to alanines (I182A/L183A) and looked at the effect of this point mutation on the transcriptional activity of the native receptor, both in the absence and in the presence of cotransfected TIF2. Whereas transcription is induced 10-fold by the addition of 1 nM R1881 to the wild-type receptor (Fig. 8A, bar 1), the I182A/L183A mutant can only be induced 2-fold (bar 3). Coexpression of TIF2, however, results in an efficient (∼7-fold) stimulation of the ligand-dependent transcriptional activity of both receptors (bars 2 and 4 versus bars 1 and 3), possibly reflecting the stimulatory effect of p160 coactivators on the intact AF1 and/or AF2 function.
FIG. 8.
Effect of mutation of the AF1a function on the functional NTD-LBD interaction. (A) COS 7 cells were cotransfected with 250 ng of pMMTV-luc and 25 ng of expression vector for the wild-type hAR or the I182A/L183A mutant, along with 250 ng of either empty pSG5 (bars 1 and 3) or pSG5-TIF2 (bars 2 and 4) and incubated with DCC-treated medium, supplemented with vehicle alone (ethanol) (open bars) or 1 nM R1881 (solid bars). The measured luciferase activities were corrected for β-galactosidase activity and are expressed as relative values, with the activity of the wild-type receptor in the presence of hormone but without coexpression of TIF2 set to 100. (B) COS cells were transfected with pMMTV-luc (250 ng), expression vectors for either the wild-type (bars 1 and 3) or the I182A/L183A mutated (bars 2 and 4) NTD-DBD constructs and either empty pSG5 (bars 1 and 2) or pSG5-SRC-1e (bars 3 and 4). Extracts were prepared, and luciferase activities and β-galactosidase activities were measured as described in Materials and Methods. (C) COS 7 cells were transfected with 250 ng of pMMTV-luc and 25 ng of expression plasmid for the wild-type DBD-LBD fragment, along with 250 ng of either empty pSG5 (bars 1 and 2) or expression vector for the wild-type hAR NTD (bar 3) or the NTD carrying the I182A/L183A mutation (bar 4). The cells were treated for 24 h without (bar 1) or with (bars 2 to 4) 1 nM R1881. The luciferase activities are presented relative to the activity measured in the presence of hormone but without cotransfected NTD. (D) GST pull-down experiments were carried out as for Fig. 4C with either GST alone or a GST–DBD-LBD fusion protein and the wild-type (lanes 1 to 3) or I182A/L183A mutated (lane 4) hAR NTD. (E) COS 7 cells were transfected as for panel C, except that plasmids directing the expression of the NTD fused to the VP16 activating domain were used. The luciferase activity measured in the presence of ligand but in the absence of VP16 fusion protein was taken as 100. Test values were calculated accordingly. +, present; −, absent. The error bars indicate the SEM.
To investigate whether the effect of the AF1a mutation might be due to an impaired intrinsic transcription activation function of the hAR NTD or to a loss of stimulation of AF1 by p160 proteins, transfections were performed with NTD-DBD constructs, either carrying or not carrying the I182A/L183A mutation, with or without cotransfection of SRC-1e. The mutated NTD-DBD construct has intrinsic transcription activation properties comparable to those of a wild-type construct (Fig. 8B, compare bars 2 and 1). Moreover, both constructs are stimulated equally well by coexpression of SRC-1e, indicating that these hydrophobic residues are probably not involved in the interaction of the hAR NTD with p160 coactivators.
Finally, the effect of the AF1a mutation on the functional interaction of the hAR NTD with the LBD was studied (Fig. 8C). Whereas the wild-type NTD efficiently stimulates the hAR AF2 function (compare bar 3 to bar 2), this effect is markedly reduced by the mutation of AF1a (bar 4). The ability of the mutated NTD to physically interact with the LBD was analyzed in the GST pull-down assay in COS extracts (Fig. 8D). In the presence of androgen, the wild-type hAR NTD interacts with the GST–DBD-LBD fusion protein (lanes 1 to 3) whereas the NTD carrying the I182A/L183A mutations does not (lane 4). Similar conclusions can be drawn from mammalian double-hybrid experiments (Fig. 8E). Whereas the hAR LBD efficiently interacts with a fusion protein of the unmutated hAR NTD with the VP16 acidic transactivating domain (bar 4), this interaction is severely impaired by the mutation of AF1a (bar 5 versus bar 4).
The hAR NTD enhances the coactivator properties of TIF2 for the hAR LBD.
Finally, we analyzed the effect on the transcriptional activity of the LBD of the simultaneous expression of the NTD and p160 proteins. As shown in Fig. 9A, both in COS cells (left graph) and in CV-1 cells (right graph) the NTD and TIF2 or SRC-1e cooperatively stimulate the transcriptional activity of the hAR LBD. This effect is most pronounced in COS cells. Whereas, under these conditions, cotransfection of the DBD-LBD construct with either the NTD or p160 protein alone results in a ∼10-fold induction of transcription (compare bars 3, 5, and 6 with bar 2), the simultaneous expression of the NTD and 160-kDa coactivators results in an almost-100-fold induction of transcription (compare bars 7 and 9 with bar 2). This effect relies on an intact AF1a region, since cotransfection of the p160 coactivators with the I182A/L183A mutated NTD does not result in cooperativity (bars 8 and 10).
FIG. 9.
hAR NTD and TIF2 cooperatively stimulate the transcriptional activity of the hAR LBD. (A) COS-7 cells (left graph) or CV-1 cells (right graph) were cotransfected with 250 ng of pMMTV-luc, 25 ng of expression vector for the DBD-LBD construct, and 250 ng of expression vector for either the wild-type or I182A/L183A mutated NTD, for TIF2, or for SRC-1e as indicated. Under the conditions where only one cofactor was transfected (bars 1 to 6), empty pSG5 plasmid was added to keep the total amount of transfected DNA constant. The cells were incubated for 24 h without (bars 1) or with (bars 2 to 10) 1 nM R1881 before harvesting and measurement of luciferase and β-galactosidase activities. The luciferase activity measured in the presence of hormone, but without cotransfected NTD or coactivator (bars 2), was set to 100, and test values were calculated accordingly. (B) COS-7 cells (left graph) or CV-1 cells (right graph) were cotransfected with 250 ng of pMMTV-luc, 25 ng of expression vector for the DBD-LBD construct, and 250 ng of expression vector for the hAR NTD, TIF2(WT), or TIF2(M123), as indicated. For bars 1 to 4 and bars 6, empty pSG5 was added to keep the total amount of transfected DNA constant. The cells were further processed as described for panel A. +, present; −, absent. The error bars indicate the SEM.
To analyze the importance of the three NR boxes in TIF2 for this cooperative stimulation of the transcriptional activity of the LBD, the same type of experiment was performed with the wild-type NTD and with the wild-type or mutated TIF2. As shown in Fig. 9B, both in COS cells (left graph) and in CV-1 cells (right graph), cotransfection of the NTD with TIF2(M123) results in a marked increase of the transcriptional activity of the LBD, compared to conditions where either of these two components is transfected alone (compare bars 7 to bars 3 and 6). Although the absolute reporter activity measured with the combination of the NTD and TIF2(M123) is lower than that measured with the combination of the NTD and wild-type TIF2 (Fig. 9B, compare bars 7 to bars 5), cotransfection of the NTD still reveals coactivator properties in the mutated TIF2. Note that the results of the cotransfection of the NTD and the LBD with either wild-type or mutated TIF2 (bars 5 and 7) are comparable to the effects seen after cotransfection of the full-size hAR with TIF2 or TIF2(M123) (Fig. 7).
DISCUSSION
Upon cotransfection in COS or in CV-1 cells, 160-kDa NR coactivators stimulate the ligand-dependent transcriptional activity of the hAR. The currently accepted model to explain these effects proposes that ligand binding induces conformational changes in the LBD, resulting in a novel molecular surface that interacts with the coactivators (42, 61). These coactivators, like the receptors themselves, interact with basal transcription factors and serve to bring the transcription preinitiation complex and RNA polymerase II to the promoter (7, 57). Furthermore, the coactivators have intrinsic histone-acetylase (HAT) activity (12, 55), and interact with yet other HATs, such as CREB-binding protein (CBP) (5, 44) and p300/CBP-associated factor (p/CAF) (71), which in turn also bind to the receptors (1, 9, 18). Following the acetylation of lysine residues in the tails of core histones, the tight chromatin structure is relaxed and the transcription apparatus can gain access to the DNA (27, 52).
The hAR AF1 and AF2 functions can both be stimulated by p160 proteins.
Our observation and those of others (26, 40, 53) that the LBD of the hAR has no intrinsic AF2 transcription activation function does not entirely fit into this model and prompted us to study the role of p160 proteins in the functioning of this receptor in more detail. Cotransfection experiments in COS cells and in CV-1 cells with the isolated NTD and LBD revealed that the AF1 function can be stimulated by 160-kDa NR coactivators and that, only under the conditions of p160 overexpression, the LBD displays a transcription activation function that has all the characteristics of a classical AF2. The fact that the LBD has no measurable AF2 activity in the absence of ectopically expressed coactivator is consistent with the observation that it has low affinity for SRC-1e in GST pull-down experiments. Under the same conditions, the mER LBD binds well to SRC-1e, which explains the high intrinsic AF2 activity of this receptor (13). Such differences in the affinities of distinct NR LBDs for coactivators have also bee reported by others (14, 50, 72). Note that in most cases the LBDs of receptors that have a strong intrinsic AF2 activity (such as ER, TR, RAR, and RXR) also display the highest affinity for coactivators.
Using the yeast two-hybrid assay, Ding et al. (14) demonstrated that the hAR LBD fails to interact with the central NIR of SRC-1. Yet, in their experiments an interaction was seen with the carboxy-terminal NR box IV that is specific to the SRC-1a isoform. We have not analyzed the interaction of the AR with SRC-1a or with a fragment containing the NR box IV in vitro, since our functional data clearly demonstrate that SRC-1a is a less potent coactivator for the hAR AF2 function than SRC-1e. In addition, Kalkhoven et al. (29) reported that the mutation of NR box IV does not affect the ability of SRC-1a to stimulate the transcriptional activity of the mER and Takeshita et al. (59) showed that, although the TR can interact with NR box IV in GST pull-down experiments, this interaction is undetectable with DNA-bound TR. Together, these data raise the question of the physiological relevance of the LBD-box IV interaction. Recently, Hong et al. (22) identified an auxiliary region, specific to the GRIP1-TIF2 subgroup of coactivators, that is required for the efficient interaction of the GRIP1 NIR with the AR LBD in yeast, thus providing yet another possible explanation of why we were unable to detect a strong LBD–SRC-1e interaction in GST pull down. However, our functional data demonstrate that both TIF2 and SRC-1e coactivate the hAR LBD, as well as the full-size AR, to similar extents and thus suggest that both coactivators interact with the receptor in similar ways.
Contrary to the LBD, we saw a good physical interaction between the hAR NTD and SRC-1e. Likewise, weak but specific interactions of SRC-1 with the NTD of the TRβ (28) and of SRC-1 and TIF2 with the NTD of the ER have been described (33, 66), and the progesterone receptor (PR) LBD and NTD were both shown to interact in vitro with TIF2 and SRC-1 (46). Moreover, cotransfection of SRC-1 stimulates the transcriptional activity of an ER molecule in which AF2 is mutated (54) and of the isolated AF1 functions of the PR, GR, and ER (46, 66). We have not characterized the interaction of the hAR NTD with p160 proteins in more detail, but as for the ER NTD (66), the three LXXLL motifs that interact with the LBD are dispensable.
A novel role for the NTD in the functioning of the native AR.
Functional interactions between the hAR LBD and the NTD have been described in yeast and in mammalian cells (8, 15, 31). We demonstrate here that these domains physically interact in GST pull-down experiments. Since the NTD also interacts with coactivators, this suggests that a ternary complex of the NTD, the LBD, and coactivators might exist, as could also be concluded from experiments by others (24, 39, 46). From our GST pull-down data it remains unclear whether the NTD and the LBD are engaged in a direct interaction or whether the pull down results from an indirect interaction mediated by a third partner present in the cell extract, which would act as a bridging factor. Although p160 proteins have been suggested to build a “molecular bridge” between the NTD and the LBD (24, 39), two observations argue against such a model. Firstly, the NTD-LBD interaction can be demonstrated in the yeast double-hybrid system (8, 15), although yeast cells lack functional homologues of p160 coactivators. Secondly, the AF1a mutation has no effect on the functional interaction between the NTD and p160 proteins. Thus, if p160 coactivators acted as bridging factors, one would expect the NTD-LBD interaction to be unaffected by this mutation. Our GST pull-down and mammalian two-hybrid experiments, however, demonstrate the opposite. Still, the involvement of other (yet unidentified) bridging factors in our cell lysates cannot be excluded a priori.
The model that we propose is that the interaction of the hAR NTD with the LBD generates a novel platform for the recruitment of coactivators to the receptor, which compensates for the low intrinsic affinity of the isolated hAR LBD for coactivators. The interaction of coactivators with the native hAR would thus occur through both strong contacts with the NTD and weak contacts with the LBD, and the apparent coactivator properties of the NTD for the LBD result from the recruitment of endogenous coactivators to the NTD-LBD complex. Alternatively, the ligand-dependent binding of the NTD to the LBD could relieve some inhibitory function located in the LBD. Although we do not have clear indications of an inhibitory region in the AR LBD, such function has been assigned to the carboxy terminus of the PR LBD (70).
The nature of the DBD influences the coactivator properties of TIF2 for the hAR LBD.
Our observation that a DBD-LBD construct is activated efficiently by both p160 proteins and the NTD whereas G4-LBD is activated efficiently only by the NTD suggests that the mode of interaction of coactivators with the LBD, in the absence of NTD, differs from the binding of the NTD to the LBD. Possibly, DNA binding through the homologous DBD induces subtle conformational changes in the LBD that are essential for efficient binding of coactivators. Alternatively, dimers of functional AF2 domains are essential for the interaction of SCR-1e with the LBD (29), and the conformation of a DBD-LBD homodimer could differ from that of a G4-LBD homodimer. Although the hAR contains a leucine-rich motif comparable to the one that is essential for dimerization in the ER (16), its main dimerization interface resides within the second zinc finger of the DBD (68). Thus, in the GAL-LBD homodimer, dimerization can be assumed to occur through the GAL4 DBD, resulting in a suboptimal alignment of the LBDs, whereas the DBD-LBD construct dimerizes on DNA through the hAR DBD, resulting in a dimer that efficiently binds coactivators. Yet another possibility is that the hAR DBD also binds accessory proteins, such as p/CAF (9) or small nuclear RING finger protein (41), which could cooperate with the p160 proteins to stimulate transcription.
Mutation in the hAR LBD and NTD and their effects on transcriptional activity.
We generated point mutations in the hAR to validate the above-described model. A conserved lysine residue at the distal end of H3 of the LBD was shown to be essential for interaction with coactivators and for high intrinsic AF2 activity in many NRs (17, 20, 42, 43). When this mutation is introduced in a DBD-LBD construct, it dramatically reduces the coactivator properties of TIF2 for the hAR AF2, similar to the effects described for the ER and the TR. However, the interaction with the NTD remains intact. Thus, the K720A mutation discriminates between the interaction of the LBD with either p160 proteins or the NTD. Since this mutation has no severe effects on the transcriptional activity of the full-size hAR, this result indicates that in the native receptor the reduced interaction of the coactivators with the LBD can be compensated for by additional interactions, for instance, with the NTD. The observation that the full-size ER carrying the corresponding mutation is transcriptionally inactive (20) and that the activity of the TR is reduced to ∼40% (17) indicates that for these receptors the contacts between the LBD and the coactivators are crucial, whereas for the hAR their contribution is of minor importance, relative to the NTD-coactivator interactions.
Contrary to the K720A exchange, the mutation of two bulky hydrophobic residues in H12 (I898A/I899A) (13) completely destroys all transcriptional activity of the full-size hAR and impairs the interaction of the isolated LBD with both p160 proteins and the hAR NTD. Similar effects are observed when an adjacent conserved glutamate is mutated to a glutamine (8) or when the LBD is occupied by the antagonist hydroxyflutamide (our unpublished results). Yet, the mutated receptor is activated by overexpression of p160 proteins, similar to the effects observed with an ER molecule in which AF2 is mutated (33). These effects could be due to the binding of the coactivator to the intact NTD.
The mutation of isoleucine 182 and leucine 183 to alanines dramatically impairs the activity of the full-size receptor (11). This effect can be explained by the observation that the functional and physical interactions between the NTD and the LBD are impaired, thus not allowing the efficient recruitment of coactivators to the mutated full-size receptor. In addition, the NTD and p160 coactivators cooperatively stimulate the transcriptional activity of the hAR LBD, when expressed simultaneously, as was also demonstrated for the PR (46). The importance of the AF1a region is further underlined by the observation that this cooperativity fully relies on an intact AF1a domain. Two other mutations in the AR LBD were described that cause androgen insensitivity and that affect the NTD-LBD interaction (32) (V889M and R752Q). At physiological hormone concentrations, the transcriptional activity of a receptor molecule carrying one of these mutations is considerably lower than that of the native receptor, and efficient transactivation occurs only at 103- to 104-fold-higher androgen concentrations. According to our model, the disruption of the NTD-LBD interaction by these mutations would not allow the efficient binding of coactivators, resulting in a deficient receptor and androgen insensitivity.
Deletion-mapping experiments were performed previously (8, 31) to define the region of the hAR NTD that binds to the LBD. These results indicated that the extreme NH2 terminus (amino acids 15 to 30) is essential for this interaction. Earlier reported results, however, showed that a receptor devoid of the first 142 residues still retains 70% of the activity of the native protein (26). Our observation that the mutation of AF1a also affects the binding of the hAR NTD to the LBD demonstrates that the extreme NH2 terminus alone is not sufficient for this interaction and suggests that an extended region of the NTD is required, as was also suggested by Ikonen et al. (24). Moreover, in addition to a correctly folded LBD, the interaction most likely also requires the correct folding of the NTD. The introduction of point mutations, as we did in this study, probably has less severe effects on the three-dimensional structure of the different domains of the receptor than the deletion of large parts of the protein described in other studies (8, 31) and would therefore better conserve the original conformation of the protein.
Mutation of the three LXXLL motifs in p160 proteins and the effect on their coactivator activity for the hAR.
We also analyzed the effects of mutations in the three conserved LXXLL motifs in p160 coactivators and found that these mutated proteins can still stimulate the transcriptional activity of the native hAR, although to a slightly lesser extent than the wild-type coactivators. Nevertheless, their coactivator properties for the isolated LBD are dramatically impaired, so the residual effect that is seen with the full-size receptor results from their coactivator activity on the NTD and strengthens the hypothesis that the interaction of p160 proteins with the NTD-LBD complex in the full-size receptor can occur through residues outside of the NIR. In addition, the cooperativity between the NTD and TIF2 for the stimulation of the transcriptional activity of the hAR LBD is conserved with a TIF2 molecule in which the three NR boxes are mutated. The observation that similar mutations have more severe effects on the coactivator properties for the ER, PPAR, RXR, and GR (29, 46) is yet another indication that for these receptors the contribution of the LBD to the interaction with coactivators is most important. For the hAR, however, the AF2 function is much less prominent, while the AF1 function (and thus the NTD) is the major contributor to the total transcriptional activity.
ACKNOWLEDGMENTS
We thank A. O. Brinkmann, H. Gronemeyer, M. G. Parker, S. Plaisance, and H. Stunnenberg for the generous gifts of plasmids; R. Bollen and H. De Bruyn for excellent technical assistance; and V. Feytons for the expert synthesis of oligonucleotides.
This work was supported by a grant from Geconcerteerde Onderzoeksactie van de Vlaamse Gemeenschap, by grants from the Belgian Fonds voor Geneeskundig Wetenschappelijk Onderzoek, and by a grant from the Interuniversity Poles of Attraction Programme, Belgian State, Prime Minister’s Office, Federal Office for Scientific, Technical and Cultural Affairs. F.C. is a Senior Research Assistant of the Fund for Scientific Research Flanders (Belgium).
REFERENCES
- 1.Aarnisalo P, Palvimo J J, Jänne O A. CREB-binding protein in androgen receptor-mediated signaling. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alen P, Claessens F, Schoenmakers E, Swinnen J V, Verhoeven G, Rombauts W, Peeters B. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1α with multiple steroid receptors and identification of an internally deleted ELE1β isoform. Mol Endocrinol. 1999;13:117–128. doi: 10.1210/mend.13.1.0214. [DOI] [PubMed] [Google Scholar]
- 3.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad A, Kohne A C, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Beato M, Chavez S, Truss M. Transcriptional regulation by steroid hormones. Steroids. 1996;61:240–251. doi: 10.1016/0039-128x(96)00030-x. [DOI] [PubMed] [Google Scholar]
- 7.Beato M, Sanchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocrine Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- 8.Berrevoets C A, Doesburg P, Steketee K, Trapman J, Brinkmann A O. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor 2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 9.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchert M, Schneider S, Adams M T, Hefti H P, Moelling K, Hovens C M. Useful vectors for the two-hybrid system in mammalian cells. BioTechniques. 1997;23:396–402. doi: 10.2144/97233bm10. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain N L, Whitacre D C, Miesfeld R L. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem. 1996;271:26772–26778. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interaction protein 1(GRIP1) and steroid receptor coactivator 1 (SRC1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 15.Doesburg P, Kuil C W, Berrevoets C A, Steketee K, Faber P W, Mulder E, Brinkmann A O, Trapman J. Functional in vivo interaction between the amino-terminal and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 16.Fawell S E, Lees J A, White R, Parker M G. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60:953–962. doi: 10.1016/0092-8674(90)90343-d. [DOI] [PubMed] [Google Scholar]
- 17.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 18.Fronsdal K, Engedal N, Slagsvold T, Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- 19.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 20.Henttu P M A, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 24.Ikonen T, Palvimo J J, Jänne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 25.Jenster G, van der Korput H A G M, van Groonhoven C C J, Van der Kwast T H, Trapman J, Brinkmann A O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 26.Jenster G, van der Korput H A G M, Trapman J, Brinkmann A O. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 27.Jenster G. Coactivators and corepressors as mediators of nuclear receptor function: an update. Mol Cell Endocrinol. 1998;143:1–7. doi: 10.1016/s0303-7207(98)00145-2. [DOI] [PubMed] [Google Scholar]
- 28.Jeyakumar M, Tanen M R, Bagchi M K. Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor. Mol Endocrinol. 1997;11:755–767. doi: 10.1210/mend.11.6.0003. [DOI] [PubMed] [Google Scholar]
- 29.Kalkhoven E, Valentin J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 31.Langley E, Zhou Z X, Wilson E M. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 32.Langley E, Kemppainen J A, Wilson E M. Intermolecular NH2-carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 33.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T-M, Schiff R, Del-Rio A L, Ricolte M, Ngo S, Gemsch J, Hilsenbeck S G, Osborne C K, Glass C K, Rosenfeld M G, Rose D W. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leers J, Treuter E, Gustaffson J A. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol Cell Biol. 1998;18:6001–6013. doi: 10.1128/mcb.18.10.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Gomes P J, Chen D J. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1996;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Chen D. The receptor-associated coactivator 3 activates transcription through CREB-binding protein recruitment and autoregulation. J Biol Chem. 1998;273:5948–5954. doi: 10.1074/jbc.273.10.5948. [DOI] [PubMed] [Google Scholar]
- 37.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marivoet S, Hertogen M, Verhoeven G, Heyns W. Antibodies against synthetic peptide recognise the human and rat androgen receptor. J Steroid Biochem Mol Biol. 1990;37:39–45. doi: 10.1016/0960-0760(90)90370-z. [DOI] [PubMed] [Google Scholar]
- 39.McInerney E M, Tsai M J, O’Malley B W, Katzenellenbogen B S. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moilanen A, Rouleau N, Ikonen T, Palvimo J J, Jänne O A. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1997;412:355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- 41.Moilanen A, Poukka H, Karvonen U, Häkli M, Jänne O A, Palvimo J J. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol Cell Biol. 1998;18:5128–5139. doi: 10.1128/mcb.18.9.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 43.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M, Kurokawa R, Rosenfeld M, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 44.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 45.Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 46.Onate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interaction domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 47.Parker M G, White R. Nuclear receptors spring into action. Nat Struct Biol. 1996;3:113–115. doi: 10.1038/nsb0296-113. [DOI] [PubMed] [Google Scholar]
- 48.Plaisance S, VandenBerghe W, Boone E, Fiers W, Haegeman G. Recombination signal sequence binding protein J kappa is constitutively bound to the NF-kappa B site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quigley C A, De Bellis A, Marschke K B, El-Awady M K, Wilson E M, French F S. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocrine Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt S, Baniahmad A, Eggert M, Schneider S, Renkawitz R. Multiple receptor interaction domains of GRIP1 function in synergy. Nucleic Acids Res. 1998;26:1191–1197. doi: 10.1093/nar/26.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1999;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 52.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Role of coactivators and corepressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–165. [PubMed] [Google Scholar]
- 53.Simental J A, Sar M, Wilson E M. Domain functions of the androgen receptor. J Steroid Biochem Mol Biol. 1992;43:37–41. doi: 10.1016/0960-0760(92)90185-l. [DOI] [PubMed] [Google Scholar]
- 54.Smith C L, Nawaz Z, O’Malley B W. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 55.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 56.Suen C S, Berrodin T J, Mastroeni R, Cheskis B J, Lyttle C R, Frail D E. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem. 1998;273:27645–27653. doi: 10.1074/jbc.273.42.27645. [DOI] [PubMed] [Google Scholar]
- 57.Takeshita A, Yen P M, Misti S, Cardona G R, Liu Y, Chin W W. Molecular cloning and properties of a full-length putative thyroid hormone receptor coactivator. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 58.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 59.Takeshita A, Yen P M, Ikeda M, Cardona G R, Liu Y, Koibuchi N, Norwitz E R, Chin W W. Thyroid hormone response elements differentially modulate the interactions of thyroid hormone receptors with two receptor binding domains in the steroid receptor coactivator-1. J Biol Chem. 1998;273:21554–21562. doi: 10.1074/jbc.273.34.21554. [DOI] [PubMed] [Google Scholar]
- 60.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 61.Torchia J, Glass C, Rosenfeld M. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 62.Tsai M J, O’Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 63.Tsai R Y L, Reed R R. Using a eukaryotic GST fusion vector for proteins difficult to express in E. coli. BioTechniques. 1997;23:794–800. doi: 10.2144/97235bm06. [DOI] [PubMed] [Google Scholar]
- 64.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 65.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S M, Subramanian S, McKinerney E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 67.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 68.Wong C I, Zhou Z X, Sar M, Wilson E M. Steroid requirements for androgen receptor dimerization and DNA binding. J Biol Chem. 1993;268:19004–19012. [PubMed] [Google Scholar]
- 69.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 70.Xu J, Nawaz Z, Tsai S Y, Tsai M J, O’Malley B W. The extreme C-terminus of the progesterone receptor contains a transcriptional repressor domain that functions through a putative corepressor. Proc Natl Acad Sci USA. 1996;93:12195–12199. doi: 10.1073/pnas.93.22.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang X J, Ogryzko V V, Nishikawa J I, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;283:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 72.Zhou G, Cummings R, Li Y, Mitra S, Wilkinson H A, Elbrecht A, Hermes J D, Schaeffer J M, Smith R G, Moller D E. Nuclear receptors have distinct affinities for coactivators: characterization by fluorescence resonance energy transfer. Mol Endocrinol. 1998;12:1594–1604. doi: 10.1210/mend.12.10.0176. [DOI] [PubMed] [Google Scholar]