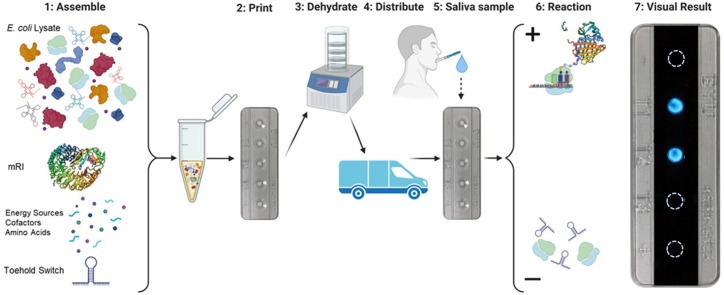
Graphical abstract
Illustration of the assembly, distribution, and point-of-care use of a rapidly-deployable, cell-free COVID-19 biosensor: 1) Assemble: Assembling CFPS reagents by mixing E. coli lysate, murine RNase Inhibitor (mRI), energy sources, cofactors, and toehold switch riboregulator plasmid. 2) Print: aliquoting CFPS reagents onto paper substrates housed in a plastic test cassette. 3) Dehydrate: lyophilizing CFPS reagents on paper substrates. 4) Distribute. 5) Saliva sample: applying saliva samples onto cassette without pretreatments. 6) Reaction: bioluminescent protein expression in presence of target RNA (+), or ribosome detachment in absence of target RNA (-). 7) Visual result: bioluminescent output in the presence of target RNA and NanoLuc luciferase expression.
Abbreviations: CFPS, cell-free protein synthesis; RT-PCR, real-time reverse transcription polymerase chain reaction; mRI, Murine RNase inhibitor; PoC, point of care; LDPE, low density polyethylene; sfGFP, superfolder green fluorescent protein
Keywords: Cell-free protein synthesis, CFPS, TXTL, Toehold switch, Covid-19, Nucleic acid diagnostic
Abstract
The COVID-19 pandemic has illustrated the global demand for rapid, low-cost, widely distributable and point-of-care nucleic acid diagnostic technologies. Such technologies could help disrupt transmission, sustain economies and preserve health and lives during widespread infection. In contrast, conventional nucleic acid diagnostic procedures require trained personnel, complex laboratories, expensive equipment, and protracted processing times. In this work, lyophilized cell-free protein synthesis (CFPS) and toehold switch riboregulators are employed to develop a promising paper-based nucleic acid diagnostic platform activated simply by the addition of saliva. First, to facilitate distribution and deployment, an economical paper support matrix is identified and a mass-producible test cassette designed with integral saliva sample receptacles. Next, CFPS is optimized in the presence of saliva using murine RNase inhibitor. Finally, original toehold switch riboregulators are engineered to express the bioluminescent reporter NanoLuc in response to SARS-CoV-2 RNA sequences present in saliva samples. The biosensor generates a visible signal in as few as seven minutes following administration of 15 μL saliva enriched with high concentrations of SARS-CoV-2 RNA sequences. The estimated cost of this test is less than 0.50 USD, which could make this platform readily accessible to both the developed and developing world. While additional research is needed to decrease the limit of detection, this work represents important progress toward developing a diagnostic technology that is rapid, low-cost, distributable and deployable at the point-of-care by a layperson.
Introduction
The global SARS-CoV-2, or COVID-19, pandemic has emphasized the need for rapid, low-cost, distributable diagnostic testing platforms. Rapid diagnosis, contact tracing and isolation help thwart viral transmission and lower overall morbidity and mortality rates [[1], [2], [3]]; therefore, accurate and widespread laboratory diagnostic testing becomes crucial for an informed and effective public health response [4], especially for viral pathogens like COVID-19 which exhibit asymptomatic and presymptomatic transmission [5]. A low-cost, rapidly deployable, point-of-care (PoC) nucleic acid diagnostic test which could easily be mass produced and distributed would be a life-saving asset during any infectious outbreak [6]. Specifically, such a technology would improve widespread testing capabilities which inform critical decisions to reduce transmission, help sustain the economy, reduce the psychological and emotional burdens of uncertainty and unnecessary quarantine and, most importantly, preserve health and lives.
Most conventional nucleic acid diagnostic procedures, however, are not amenable to large-scale, rapid deployment. For instance, widely used COVID-19 tests employ nucleic acid quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) assays to detect viral RNA during the acute phase of infection [7]. RT-PCR testing requires trained personnel, high-complexity laboratories, expensive laboratory equipment, and supply-limited reagents [4]. Consequently, long processing times, limited testing capacity, high costs and supply shortages often precluded timely diagnoses [7]. As the Covid-19 pandemic progressed, researchers around the globe raced to introduce alternative diagnostic procedures to meet the testing demands [8]. These efforts led to PoC tests; however, supply is still limited, and the cost of these tests make them inaccessible for routine use by most of the world.
To help address the need for affordable and rapidly deployable PoC testing, we propose a low-cost E. coli-based cell-free protein synthesis (CFPS) diagnostic platform for the detection of viral pathogens in saliva. The open transcription-translation reaction environment of CFPS systems provides an ideal platform for engineering and deploying synthetic biochemical systems such as biosensors [[9], [10], [11]]. The advantages afforded to CFPS systems enable versatile biosensing applications in a variety of sample matrices. Notably, biosensors utilizing CFPS technology have been developed for the detection of viral and bacterial pathogens and small-molecules in human clinical samples including sputum, urine, fecal and blood [12]. However, a CFPS-based biosensor had yet to be demonstrated in saliva prior to this work.
Significant work has been done to advance the application of CFPS-based biosensors towards facile deployment outside of laboratory settings. Recent work has extended CFPS biosensor technology to paper-based platforms [[13], [14], [15], [16], [17]]. CFPS systems can be freeze-dried onto paper substrates for stable, long-term storage at ambient conditions and have shown viability for weeks to months [[13], [14], [15],17]. The compatibility of CFPS biosensors with paper supports facilitates inexpensive, distributable PoC use [18]. Pioneering work by the Collins laboratory combined programmable riboregulatory toehold switches with CFPS biosensors demonstrating rapid detection of pathogen markers including Zika and Ebola viral sequences [13,14]. The resulting diagnostics proved to be rapid, robust, and sensitive. Further developments from the Collins, Green and Pardee laboratories led to increased sensitivity and an expanded repertoire of potential targets by adding a step where target nucleic acid sequences are exponentially amplified [15,16]. Inspired by this work, we have applied a similar toehold switch combined with CFPS approach to the viral target SARS-CoV-2. Other laboratories, including the Pardee and Green groups, are also applying this approach toward SARS-CoV-2 detection and have made impressive progress [19]. Towards this objective, here we report the advances of (1) extending paper-based CFPS technology to saliva matrices, (2) demonstrating additional paper support types, and (3) the development of additional SARS-CoV-2-specific toehold switches, (4) developing bioluminescent visual output as a potential way to increase the signal strength and lower the detection limit, and (5) reporting shelf-life of the resulting SARS-CoV-2 sensor.
Here, development of a CFPS technology is described for PoC detection of viral pathogens in saliva. A paper-based Covid-19 biosensor activated by saliva and assayed by visual readout is demonstrated. Novel toehold switch riboregulator RNA strands are reported which respond to RNA sequences from the SARS-CoV-2 genome and generate a rapid bioluminescent signal 7−15 min after initiating the reaction with a saliva sample enriched with significant concentrations of SARS-CoV-2 RNA sequences. To facilitate widespread distribution and deployment of the biosensor, lyophilized CFPS reagents are embedded in a low-cost and widely available paper substrate and housed in a mass-producible LDPE (low density polyethylene) test cassette. The resulting device is shelf stable, can be deployed by a layperson operator at PoC, and is economical with an estimated total cost of less than 0.50 USD. This technology represents important progress towards development of diagnostic technologies for combating viral outbreaks such as the COVID-19 pandemic, although further work to lower the limit-of-detection is needed.
Materials and methods
Materials
Chemical reagents were purchased from Cayman Chemical (Ann Arbor, Michigan) unless otherwise noted. Test cassette was 3D printed using VeroClear PolyJet photopolymer (Stratasys, Minneapolis, Minnesota). Fresh saliva was collected with institutional IRB approval from at least three volunteers and combined before laboratory use.
Cell extract preparation
E. coli extracts were prepared using BL21-Star™ (DE3) (Invitrogen, Carlsbad, CA) and BL21-Star™ (DE3) ΔLac (Invitrogen) strains. Cell extracts were prepared as previously reported [20]. Briefly, cell cultures were grown at 37 °C and 280 rpm with selective antibiotic. Overnight 5 mL cultures were transferred to 100 mL of 2xYT media in 500 mL flasks and grown until an OD600 between 2–4 then added to 900 mL fresh 2xYT media in 2.5 L Tunair baffled shake flasks (IBI scientific, Dubuque, IA). At an OD600 of 0.7, cultures were induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cells were harvested at mid-log phase by centrifugation at 6000 RCF for 30 min. Cell pellets were washed in 10 mL per g wet cells of chilled buffer A (14 mM magnesium acetate, 10 mM tris, 60 mM potassium glutamate, and 1 mM dithiothreitol) and centrifuged again with the same conditions. Cells were resuspended in 1 mL buffer A per g wet cells and lysed by passing 3 times through an Avestin Emulsiflex B-15 cell disrupter (Avestin, Ottawa, Canada) at 21,000 psi. Lysed cells were centrifuged at 12,000 RCF for 30 min and the supernatant was aliquoted in 100 μL volumes, flash-frozen, and stored at −80 °C until single-use.
Cell-free protein synthesis
CFPS reactions were performed using the PANOx-SP system as previously described [21]. LacZα expression reactions were composed of 25 % (v/v) BL21-Star™ (DE3) ΔLac extract, 25 % (v/v) PANOx-SP, 19 mM Mg(Glu)2, 12 or 72 nM LacZα plasmid DNA as template, 2% (v/v) purified in-house T7 RNA polymerase, 0.6 mg/mL LacZα substrate chlorophenol red-β-d-galactopyranoside (CRPG) (Gold Bio, St Louis, MO), trace amounts of pre-synthesized LacZω fragment, and remaining volume distilled deionized water. Expression of the reporter enzyme NanoLuc luciferase (NanoLuc) [22] was conducted similarly except 30 nM linear NanoLuc DNA template and NanoLuc substrate from Nano-Glo® Luciferase Assay System (Promega Corporation, Madison, WI) replaced LacZα plasmid and CRPG, respectively. Liquid CFPS reactions performed in the presence of saliva contained 30 % (v/v) saliva. In early CFPS tests in the presence of saliva, superfolder green fluorescent protein (GFP) was used as the reporter protein with a superfolder sfGFP plasmid as template. Expression yields of sfGFP were determined by fluorescence at 485/510 excitation/emission wavelengths. Murine RNase Inhibitor (mRI) was sourced from New England Biolabs (NEB) or produced in-house [23].
Preparation of paper-based cell-free systems
Paper discs were cut with a standard 8-mm diameter hole punch prior to the addition of CFPS reaction mix. Six different paper types were tested: 100 % cotton cellulose Chromatography Paper (Thick Chromatography Paper Grade 238, VWR 28342−036, VWR International, Radnor, PA, USA), Printer Paper (Double A Everyday Copy Paper 20 lb, Double A, Bangkok, Thailand), Nylon (Corning Filter System 430,767, Corning Inc., Corning, NY, USA), Cellulose Acetate (Corning Filter System 430,767), Cellulose Nitrate (Whatman 7184−002, Whatman, Maidstone, UK), and Ashless Cellulose Filter Paper (Whatman 1444−110, Whatman). CFPS reactions were mixed on ice and pipetted onto paper discs and either immediately air-dried or lyophilized. Air-dried papers were prepared by pipetting CFPS onto paper discs in 3−5 μL increments and promptly air-dried at 37 °C for approximately 5 min between each application for a total reaction volume of 6−15 μL. Paper discs embedded with CFPS were placed in the test cassette receptacles prior to rehydration. After rehydration with water or saliva, receptacles were sealed to prevent evaporation as CFPS reactions proceeded. Rehydration of the paper discs, and accompanying reaction, were performed less than 1 h after air-drying. Lyophilized paper-based CFPS reactions were prepared similarly except 10−15 μL CFPS reagents were applied en masse to paper discs then flash frozen in liquid N2 and dried overnight in a lyophilizer (FreeZone 2.5, Labconco Corporation, Kansas City, MO, USA). Rehydration of discs were performed 1 d after lyophilization. For experiments in which paper discs were blocked with bovine serum albumin (BSA), paper discs were placed on clean parafilm and immersed in 50 μL of a stock solution of 5%, 1% or 0.2 % BSA. After 30 s, all recoverable liquid BSA solution was removed and saturated discs dried on the parafilm. CFPS reaction mix was then applied to discs and air-dried.
Toehold switch biosensor design and construction
Toehold switches were designed using NUPACK software by scanning SARS-CoV-2 protein sense sequences [16] (Supplementary Information S2). Sequences which resulted in ensemble defects of less than 23 % were considered for synthesis (Supplementary Information S3). Overlapping eBlocks (Integrated DNA Technologies, Coralville, IA, USA) were used to clone switches upstream of the NanoLuc gene with Q5 PCR (NEB, Ipswich, MA, USA). EcoRI and XmaI restriction enzymes were then used to clone switch-NanoLuc constructs into vector backbone. Successful cloning was sequence verified using vector primers. Plasmids were purified for use in CFPS reactions using Qiagen Plasmid Maxi Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions.
Toehold switch biosensor CFPS assay
RNA targets were synthesized from dsDNA templates (Integrated DNA Technologies, Coralville, IA, USA) after amplification with Q5 DNA Polymerase (NEB). T7 HiScribe (NEB) in vitro transcription reactions were digested according to manufacturer’s protocol with DNAseI prior to purification with Monarch RNA cleanup (NEB) and resuspension in RNase-free water. RNA was stored at −80 °C and concentration was determined using SynergyMX Take3 (BioTek, Winooski, VT). CFPS reactions used the PANOx-SP system as described above with 25 % (v/v) standard extract, 25 % (v/v) PANOx-SP, 18 mM Mg(Glu)2, 2 nM toehold switch plasmid templates, and 6 μM RNA target for a total reaction volume of 15 μL. Liquid-based switch CFPS reactions were incubated for 1 h at 37 °C. Following incubation, reactions were diluted and added to Nano-Glo® Luciferase Assay substrate and bioluminescence was measured for 10 min using SynergyMX microplate reader. Paper-based CFPS toehold switch reactions were performed after lyophilization of 10 μL CFPS reagents with 5.4 % (v/v) Nano-Glo® Luciferase Assay substrate onto paper discs as described above. Paper discs were placed in the plastic test cassette, secured with white tape (VWR 89098−058, VWR International, Radnor, PA, USA), rehydrated with 15 μL saliva, sealed with transparent tape (JVCC BOOK-20CC, J.V. Converting Company, Inc., Fairless Fields, PA, USA), and monitored at ∼37 °C for 30 min after the addition of 6 μM target RNA sequences from the SARS-CoV-2 genome (Supplementary Information S2).
Results and discussion
Here the development is reported of a rapidly deployable paper-based SARS-CoV-2 RNA biosensor using CFPS including (1) determining an economical paper support that is inexpensive and not supply-chain limited, (2) creating a simple low density polyethylene (LDPE) cassette to house the biosensor and simplify manufacturing, (3) optimizing CFPS in the presence of saliva, (4) developing new toehold switch riboregulator RNA strands for SARS-CoV-2 RNA sequence sensitivity, and (5) combining the above technologies to create a shelf-stable device which detects SARS-CoV-2 RNA by a bioluminescence signal clear to the naked eye.
Selection of paper substrates for efficient CFPS reactions
During the COVID-19 pandemic many diagnostics suffered material and reagent shortages which severely limited their timely production and deployment [24,25]. These shortages illustrate the need for economic diagnostic platforms which require fewer resources and are more conducive to rapid and widespread deployment. Paper-based diagnostics offer several key advantages over conventional, aqueous-based diagnostics such as RT-PCR assays. Paper-based diagnostics generally require minimal on-site testing equipment and are therefore convenient for PoC use. Furthermore, paper-based platforms can be shipped en masse or individually in envelopes without specialized shipping requirements. Paper substrates are also generally economical and widely available. Finally, paper-based diagnostics can easily be incinerated after use to reduce exposure to the possibly infectious human samples employed in the test [26].
Paper substrates have proven to be a stabilizing support matrix on which CFPS reagents can be dried for convenient and shelf-stable storage. CFPS systems, including biosensors, have been reported on a variety of paper substrates [[13], [14], [15], [16], [17]]. However,certain paper types (such as cellulose acetate which is commonly used for diagnostics) became supply chain limited during the pandemic. This prompted a number of different paper types to be screened for compatibility with CFPS reactions to identify additional paper substrates which maintain high CFPS activity. Screening was concentrated on cellulose-based paper types that are highly accessible (not supply chain limited) and are generally less expensive than previously tested paper matrices.
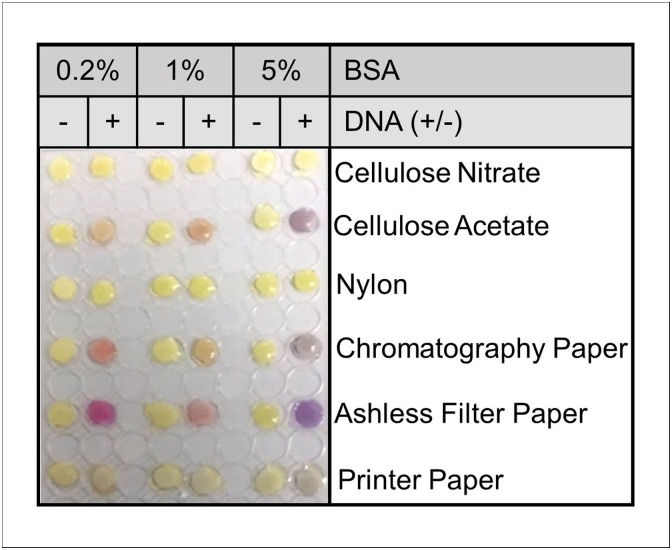
Various paper types were screened for compatibility with CFPS reactions by the expression of the reporter protein LacZα (Fig. 1 ). Reaction performance was evaluated by the colorimetric signal response of CRPG to the expressed β-galactosidase fragment. To reduce possible non-specific interactions between CFPS components and different cellulose-fibers, paper was pretreated with 0.2 %, 1%, or 5% (v/v) of BSA [13]. CFPS reactions on cellulose nitrate and nylon, the only non-cellulose-based matrices tested, did not achieve a detectable response which suggests minimal protein synthesis and poor compatibility with CFPS molecular machinery. In contrast, cellulose acetate, 100 % cotton cellulose chromatography paper, ashless cotton cellulose filter paper and standard 20 lb printer paper supported visible responses and are therefore considered as potential candidates for a low-cost, rapidly deployable paper-based CFPS diagnostic device.
Fig. 1.
Development of CFPS reactions on paper. CFPS of the colorimetric reporter protein LacZα on different paper types pretreated with BSA at % volume. Following BSA pretreatment, 3 μL of CFPS reagents containing CRPG substrate was applied twice to each paper 8-mm diameter disc. Reactions were rehydrated with 6 μL water and photographed for colorimetric response after 24 h.
The strongest visible responses were achieved in cellulose acetate and ashless filter paper. However, cellulose acetate suffered supply chain limitations during the COVID-19 pandemic and is generally more expensive than the other paper types, reducing its potential as a low-cost and widely available test material. Reactions on filter paper, though high performing, proved to be less efficient and thus less conducive to a rapid diagnostic. Reactions on 20 lb printer paper showed only moderate performance with 5 % (v/v) BSA pretreatment; however, printer paper is inexpensive and widely available and therefore could potentially be used in a diagnostic device assembled in low resource settings or during manufacturing delays of other candidate materials.
Of the paper types tested, 100 % cotton cellulose chromatography paper was deemed the most suitable for a low-cost, rapidly deployable paper-based CFPS diagnostic platform. Although chromatography paper did not achieve the strongest visible signal within the initial screening, it demonstrated the fastest signal response. Increasing the volume of embedded CFPS reagents improved reaction performance while still maintaining rapid reaction rates. Furthermore, BSA pretreatment of chromatography paper proved to be inconsequential to reaction performance when CFPS reagents embedded on 8-mm diameter discs exceed 10 μL (Supplementary Information Fig. S1.1). Chromatography paper was therefore considered the optimal matrix as it supports rapid reaction rates, high protein synthesis, and is composed of economical and easily manufactured materials.
Housing paper-based CFPS diagnostic biosensor in a deployable test cassette
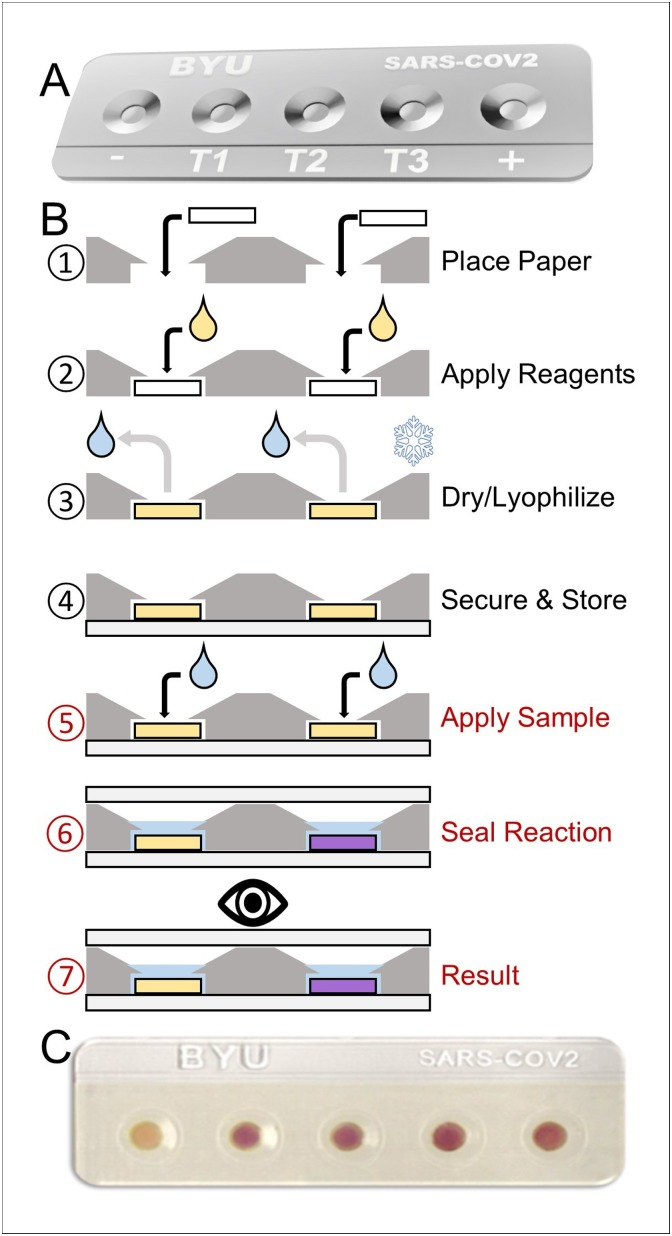
It was determined that 100 % cotton cellulose chromatography paper was an optimal support matrix for a low-cost yet efficient paper-based CFPS diagnostic platform. To facilitate practical deployment by lay-person operator, a portable, low-cost LDPE test cassette (Fig. 2 A) to house paper-based CFPS biosensors was developed. This cassette promotes ease of use and protects paper substrates and paper-based reactions while still maintaining the desirable attributes of paper-based platforms: low-cost materials, cost-effective manufacturing and convenience in transportability.
Fig. 2.
Test cassette for housing paper-based CFPS biosensor. (A) CAD rendering of test cassette. Cassette is designed with 5 clearly labeled receptacles for positive (+) and negative (-) controls and multiple sensor tests (T1, T2, T3). (B) Schematic representing the preparation and deployment of the paper-based CFPS diagnostic test. Steps 1-4, shown in black, can be completed prior to distribution. Steps 5-7, shown in red, can be implemented at point-of-care by operator. (C) Paper-based CFPS of the colorimetric reporter protein LacZα housed in cassette. Paper discs were embedded with 15 μL CFPS reagents and air-dried. Discs were placed in the receptacles and secured with white tape. Reactions were rehydrated with 15 μL saliva. Receptacles were sealed with transparent tape to prevent evaporation while reaction proceeded for 1 h. Receptacles from left to right: 1- negative LacZα template (yellow) and 2-5 - positive LacZα template (purple).
The test cassette is small and lightweight to facilitate widespread distribution such as shipment in small envelopes (See Supplementary Information Fig. S1.2). Additionally, the minimalistic plastic design has the potential to be manufactured in bulk at low cost by injection molding at millions per day. The LDPE cassette is also recyclable. The cassette holds receptacles for five discrete paper-based CFPS reactions. This accommodates positive and negative controls, replicate tests, or the detection of multiple pathogen nucleic acid sequences within the same test. Receptacles act as small chemical bioreactors upon rehydration with saliva samples and are uniquely tapered to optimally sequester 15 μL of saliva to the reaction site. The preparation and deployment of a diagnostic test within the cassette is simple and straightforward, as illustrated in Fig. 2B. A single 8-mm paper disc is placed within each receptacle, embedded with CFPS reagents, and secured with white tape. Once secured, the test is ready to be deployed or stored for later use; the end user simply fills receptacles with saliva, ∼15 μL per well, and seals the cassette with transparent tape to prevent evaporation during the ensuing reactions. A prototype of the test cassette was 3D printed and evaluated following these steps. The cassette successfully housed paper-based CFPS reactions expressing LacZα after rehydration with 15 μL saliva. Reactions achieved clearly visible LacZα mediated color changes with discernible difference between LacZα negative (yellow) or LacZα positive (purple) reactions (Fig. 2C). Results were distinct for each reaction with no observed reagent seepage or spreading outside individual receptacles.
Engineering CFPS in the presence of saliva
Saliva is an attractive clinical medium with expanding diagnostic applications. Clinical saliva samples are simple to obtain through non-invasive means, yet still maintain an extensive profile of potential biomarkers [27]. For example, saliva-based diagnostics have been able to characterize salivary biomarkers for viral, cancer, and autoimmune diseases [28]. Therefore, engineering CFPS systems to maintain protein synthesis capabilities in saliva samples could greatly expand the potential applications of CFPS biosensor technology. CFPS has previously been performed in a variety of other biological samples including sputum, urine, and blood [12]. The addition of these biological samples to a native CFPS environment inhibits protein synthesis; however, protein yields have been at least partially preserved by the addition of RNase inhibitors. This suggests that endogenous RNases present in biological samples have a significant inhibitory effect on CFPS systems [[29], [30], [31]]. Like other biological samples, saliva is rich in various RNases and shows extensive RNase activity. Indeed, exogenous RNA in saliva has an estimated half-life of 0.4 min [[32], [33], [34], [35]]. Because it has been shown that mRI sustains CFPS in the presence of other biological samples [[29], [30], [31]], it was hypothesized that the addition of RNase inhibitor to CFPS systems would be effective, and even essential, for the successful implementation of CFPS in saliva samples.
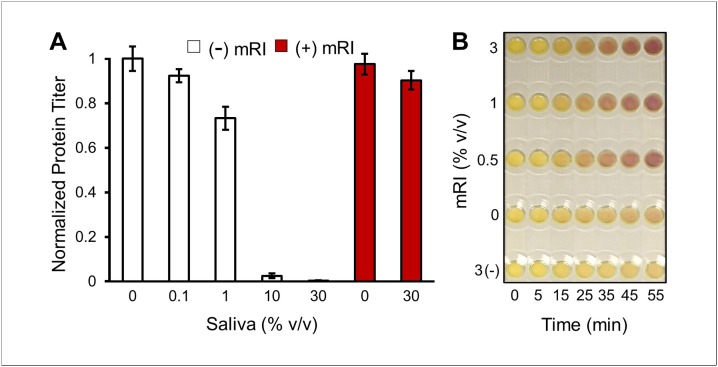
To test the hypothesis that mRI enables CFPS in saliva, the effect of saliva on CFPS was assessed first in standard liquid-based CFPS reactions expressing superfolder green fluorescent protein (sfGFP). As shown in Fig. 3 A, protein expression of a native CFPS reaction is completely inhibited with the addition of >10 % saliva v/v. However, as anticipated, protein expression was restored by adding murine RNase inhibitor (mRI) to the reaction mixture. As demonstrated, CFPS performance of reactions with 30 % saliva is almost completely preserved by the addition of 1% (v/v) mRI. The utility of mRI addition was further evaluated for use in paper-based CFPS reactions by expressing the colorimetric reporter protein LacZα on a paper substrate (Fig. 3B). It was determined that 1% (v/v) mRI sustains CFPS sufficiently when reactions lyophilized on paper are rehydrated with pure saliva. Thus, the addition of mRI in the CFPS system enables a “just-add-saliva” paper-based CFPS platform.
Fig. 3.
CFPS reactions in saliva samples. (A) Liquid-based CFPS of superfolder green fluorescent protein (sfGFP) in saliva at the volume percent indicated with or without 1% (v/v) murine RNase Inhibitor (mRI). 1% (v/v) mRI corresponds to 0.4 U/μL mRI where 1 U is defined as the amount of enzyme required for 50 % reduction of 5 ng RNase A. Results were normalized to sfGFP expression without the presence of saliva which achieved 1.07 μg/μL. Error bars represent one standard deviation for n = 3. (B) Paper-based CFPS of the colorimetric reporter protein LacZα in the presence of varying mRI concentrations. “(-)” indicates negative DNA template control. 15 μL CFPS reactions were air-dried on chromatography paper discs. Reactions were placed in plastic cassettes and rehydrated with 15 μL pure saliva and photographed for colorimetric response over the course of 1 h. Without the presence of mRI, CFPS reaction did not achieve appreciable protein yields as compared to the negative LacZα template control. With the addition of mRI, protein yields improved as evidenced by the LacZα mediated color change. Photograph brightness was holistically enhanced to resemble more closely in-person observations.
Paper-based CFPS of bioluminescent reporter protein NanoLuc
To date, all paper-based CFPS biosensors have utilized either fluorescent or colorimetric reporter technology. However, fluorescent reporter proteins such as GFP or mCherry have limited application in low-resource settings as they require light excitation and electronic optical readers to decipher biosensor output signals. To overcome this challenge, low-cost fluorometric devices have been developed in tandem with paper-based CFPS biosensors [13,14,17] but inevitably, additional testing equipment complicates simple user analysis and increases overall costs and resources. Colorimetric assays offer signal outputs more compatible to analysis with the naked eye; however, at the lower limits of detection, they too may require optical readers to decipher subtle color changes.
Alternatively, bioluminescent reporters offer simple assay analysis since signal output can be visible to the naked eye when viewed in the dark and does not require light excitation. Bioluminescence assays can also deliver 10- to 1000- fold higher sensitivity than fluorescent assays, providing much lower limit of detection (LoD) [36]. In particular, bioluminescence assays which utilize the enzyme luciferase report high sensitivities, low signal backgrounds, and wide linear ranges [12]. The bioluminescent reporter NanoLuc luciferase (NanoLuc) was therefore chosen for the diagnostic platform. NanoLuc is a commercially available luciferase engineered to be even more responsive and versatile than conventional luciferase. NanoLuc is a small stable protein which folds rapidly and is highly responsive to the substrate furimazine [22]. These characteristics make it an ideal reporter candidate for a highly sensitive and rapid diagnostic device.
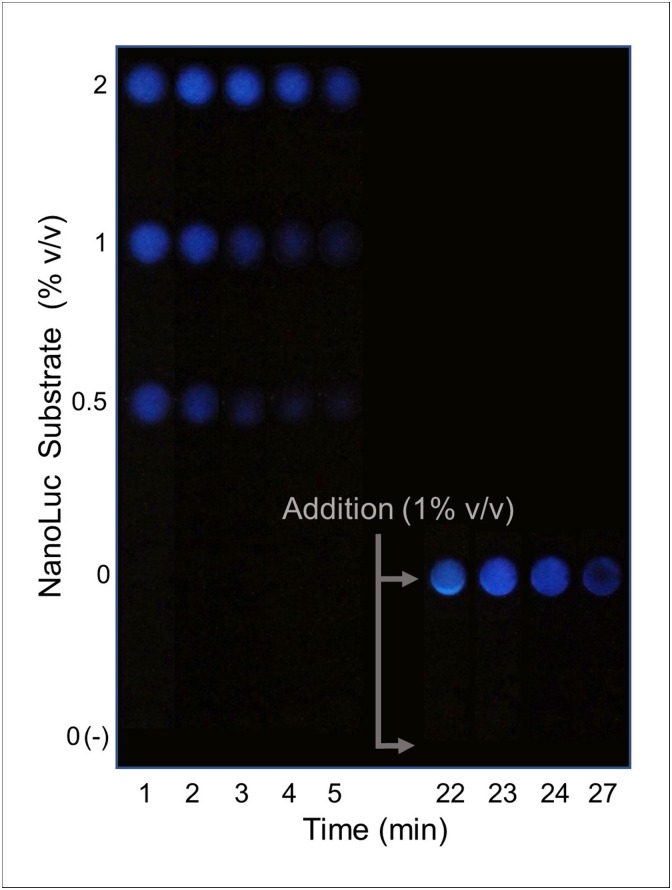
Bioluminescent reporters have not previously been expressed in CFPS on paper mediums. It was therefore necessary to test the expression and bioluminescent activity of NanoLuc on paper. Additionally, NanoLuc substrate is generally added to the reaction after completion so it was also requisite to determine if addition of substrate prior to NanoLuc synthesis would impede bioluminescent response. Single-pot CFPS reactions, including NanoLuc substrate at different volume percentages, were performed on paper as shown in Fig. 4 . All test reactions exhibited visible bioluminescent signals with up to 2% (v/v) substrate without evidence of interference to protein synthesis.
Fig. 4.
Bioluminescent response of NanoLuc protein expressed in CFPS reactions on paper substrate. 12 μL CFPS reagents were air-dried on blotting paper with the indicated amount of NanoLuc substrate. “(-)” indicates negative DNA template control. Reactions were rehydrated with 12 μL water and response was monitored over the course of 30 min. Bioluminescent response was photographed at regular intervals in a dark room with a camera shutter speed of 2 s and high ISO, which resulted in signal detection sensitivity only slightly higher than that of the naked eye of two observers. Reaction in which no substrate was added prior to rehydration had substrate added 20 min after reaction was initiated.
In all cases, bioluminescent response was rapid but ultimately short-lived. The rapid signal response observed in the test reactions attests to the robust nature of CFPS reactions and the short activation of the NanoLuc protein. The short-lived signal response is due to substrate depletion as evidenced by the high dependency of the signal response on the concentration of substrate added. As the concentration of substrate increased, so too did the intensity and duration of the bioluminescent signal. Furthermore, adding substrate to a reaction 20 min after initiation resulted in a more intense and abrupt signalcompared to the same substrate concentration added prior to the reaction, due to the higher number of enzymes acting on the substrate.
Development of toehold switch biosensors to detect SARS-CoV-2 RNA
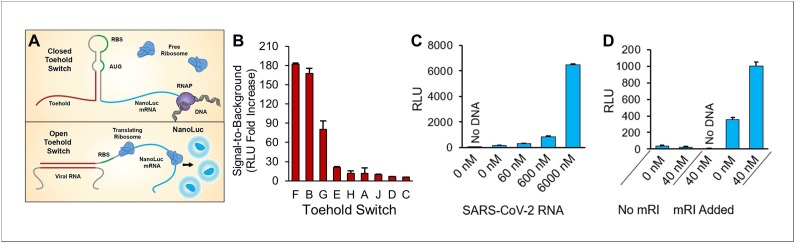
Toehold switch biosensors are programmable riboregulators which use de novo designed RNA sequences to provide versatile and highly programmable gene regulation [37,38]. Toehold switches have successfully been designed to detect viral, bacterial and host biomarker nucleic acid sequences and have been demonstrated in CFPS systems (Fig. 5 A) [[13], [14], [15], [16],39]. NUPACK software [40] was used to scan SARS-CoV-2 protein sense sequences for switch designs likely to respond to SARS-CoV-2 RNA. Two of these represent sense sequences from the membrane glycoprotein mRNA (Toehold E, J), and the remaining 7 represent sense sequences from the capsid protein mRNA. Toehold switch designs which employed both 11 bp and 13 bp stems were considered, and one construct contained a 27 bp toehold region. In total, 9 toehold switches were constructed, each placed upstream of the NanoLuc reporter protein, in a plasmid backbone under T7 RNAP control. Switch constructs were then expressed in CFPS reactions with or without ∼6 μM RNA sequences from the SARS-CoV-2 genome. Synthesized RNA molecules with identical sequences to ∼100 bp of the SARS-CoV-2 genome, which included the complementary toehold recognition sequence, were used for experimental validation to simulate more closely detection of clinical SARS-CoV-2 RNA. All toehold switches responded to the SARS-CoV-2 sequence, and two responded with signal-to-background >150-fold (Fig. 5B), both of which targeted sense RNA sequences from the SARS-CoV-2 capsid protein. Factors which could affect the ability of each switch mRNA to respond to the intended construct sequence include long-range intermolecular interactions and the secondary structure of the target RNA molecule itself. Indeed, it was recently reported that one potential predictor of a given mRNA sequence window is a lack of inherent secondary structure in that portion of the target sequence [41]. Sequences and details of NUPACK structure prediction and analysis for each constructed switch and experimental target are included in Supplementary Information Supplementary S2. These results indicate that NUPACK is a useful tool for RNA secondary structure prediction and switch design, and that experimental validation identifies the optimal performers.
Fig. 5.
SARS-CoV-2 Toehold switch performance. (A) Illustration of a CFPS-based biosensor utilizing toehold switch riboregulators and NanoLuc reporter. Top: Depiction of the toehold switch sequence components and secondary structure without the presence of the target viral RNA. When unbound, the hairpin loop remains “closed” and sequesters the ribosome binding site (RBS) repressing translation. Within the CFPS reaction, toehold switch RNA is actively transcribed from DNA template. Bottom: Binding of the target viral RNA sequence to the toehold domain “opens” the switch releasing the RBS for gene translation resulting in the synthesis of the bioluminescent reporter NanoLuc in the CFPS system. (B) Performance of toehold switches designed, constructed and tested in this work as measured by the ratio of relative bioluminescence of CFPS toehold switch reaction in the presence of 6 μM SARS-CoV-2 target RNA sequence to CFPS toehold switch reaction without target RNA. Error bars represent one standard deviation for n = 2 liquid-based CFPS reactions. (C) CFPS performance of Toehold Switch B in the presence of the indicated concentration of SARS-CoV-2 target RNA. “No DNA” indicates bioluminescence of negative control to which no switch plasmid was added. Error bars represent one standard deviation for n = 2 liquid-based CFPS reactions. (D) Detection of 40 nM SARS-CoV-2 target RNA in human saliva at 30 % (v/v) in CFPS enabled by the addition of murine RNase Inhibitor (mRI). “No DNA” indicates bioluminescence of negative control to which no switch plasmid was added. Error bars represent one standard deviation for n = 2 liquid-based CFPS reactions.
Toehold B was selected for further biosensor development as it demonstrated a high signal-to-background ratio and a more efficient plasmid preparation over the similarly performing Toehold F. The performance of Toehold B was tested for bioluminescent response at lower target RNA concentrations (Fig. 5C). An observable signal was obtained at 60 nM SARS-CoV-2 synthetic target RNA. Toehold B was then evaluated for its ability to detect SARS-CoV-2 RNA sequence in human saliva. Reactions without mRI exhibited no signal in the presence of 40 nM SARS-CoV-2 target RNA, while reactions with mRI exhibited strong signals at 40 nM target RNA (Fig. 5D). While the LoD exhibited by these switches may be insufficient for direct detection in patient samples, recent developments in RNA amplification technology [42] preserve the clinical potential of these novel riboregulators.
Assembly of paper-based CFPS biosensor device
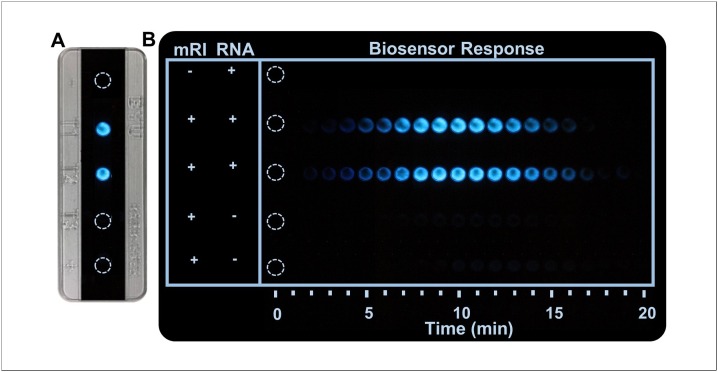
As a demonstration of a rapidly deployable, PoC CFPS-based diagnostic platform, a paper-based CFPS toehold switch biosensor was assembled and deployed. The biosensor assay was performed with lyophilized CFPS reagents and Toehold B template on paper discs housed in the test cassette. 15 μL saliva samples were applied to the paper discs to activate the CFPS reaction. As proof-of-concept, saliva samples were enriched with ∼6 μM synthetic SARS-CoV-2 RNA sequences to simulate clinical samples. The paper-based CFPS toehold switch biosensor achieved a visible, switch dependent response to SARS-CoV-2 RNA sequences in saliva (Fig. 6 A). Bioluminescence was only visible when reactions proceeded in the presence of SARS-CoV-2 RNA sequences and maximum signal response occurred between 7−12 min at which time bioluminescence was clearly visible with the naked eye in a dark room as shown in Fig. 6 B (for time-lapse video see Supplementary Video 1). The reaction volume as well as the short NanoLuc reporter gene may have helped to promote the rapid response achieved here. In principle, larger reactions allow greater accumulation of reporter proteins and thus stronger signal output. Housing the biosensor reactions in the plastic test cassette allows for larger reactions than those previously demonstrated on paper substrates. It has also been suggested that the size of a reporter gene may directly affect assay response times. Shorter genes with rapid activations promote faster reactions and increased detection speed [15]. Therefore, the small size of NanoLuc (510 bp) may aid in rapid expression and thus rapid biosensor output response. Finally, an initial shelf-life assessment suggests the biosensor reagents maintain functional activity up to 7 weeks at room temperature (Supplementary S4).
Fig. 6.
Performance of assembled SARS-CoV-2 biosensor using CFPS reagents, Toehold B switch template, and NanoLuc substrate lyophilized on chromatography paper. (A) Response of assembled biosensor after 10 min reaction. Duplicate reactions were rehydrated with saliva including mRI in excess and 6 μM of SARS-CoV-2 target RNA as indicated. Bioluminescent response was only achieved in reactions which contained target RNA. (B) Time course of biosensor response. Reactions were photographed for bioluminescent response over the course of 20 min at regular intervals in a dark room with a camera shutter speed of 2 s and high ISO, which resulted in signal detection sensitivity only slightly higher than that of the naked eye as described by two observers. For a time-lapse video of the biosensor response see Supplementary Video 1.
Conclusion
This work represents progress toward a diagnostic platform which is rapid, low-cost, distributable, and deployable at PoC by laypersons. A rapidly deployable, paper-based CFPS biosensor was engineered which responds when high concentrations of SARS-CoV-2 RNA sequences are added to human saliva. First, cost-effective paper substrates were identified and a portable plastic cassette was fabricated with strategic saliva receptacles for five CFPS biosensor reactions. Next, CFPS reactions were engineered with mRI to preserve protein synthesis capability even in the presence of 100 % human saliva. This feature enabled a shelf-stable “just-add-saliva” paper-based CFPS platform which is the biochemical foundation of the biosensor. Finally, toehold switch riboregulators were engineered to respond to the SARS-CoV-2 RNA sequence by expressing the bioluminescent reporter protein NanoLuc. These riboregulators respond with dynamic range approaching 200 and limit of detection near 10 nM RNA. Because of the flexibility of the toehold switch design process, this nucleic acid diagnostic platform could be applied to other RNA targets of interest for detecting a myriad of pathogens. The resulting portable biosensor device can be manufactured for an estimated cost of less than 0.50 USD (Supplementary S5). Because of its operational simplicity and economy, this platform could potentially lead to a viable diagnostic test which could be used by laypersons in both the developed and the developing worlds as a global tool for combating pandemics [6].
Author contributions
J. Porter Hunt: Conceptualization, Methodology, Investigation, Validation, Formal Analysis, Visualization, Supervision, Funding acquisition, Writing – Original Draft, Writing – Review and Editing. Emily Long Zhao: Methodology, Investigation, Validation, Formal Analysis, Visualization, Resources, Writing – Original Draft, Writing – Review and Editing. Tyler J. Free: Methodology, Investigation, Validation, Visualization, Writing – Review and Editing. Mehran Soltani: Methodology, Investigation, Validation, Visualization, Writing – Original Draft, Writing – Review and Editing. Chandler A. Warr: Methodology, Investigation. Alex B. Benedict: Methodology, Investigation. Melissa K. Takahashi: Methodology. Joel S. Griffitts: Conceptualization, Methodology. William G. Pitt: Project Administration, Supervision, Methodology, Funding Acquisition. Bradley C. Bundy: Project Administration, Supervision, Conceptualization, Methodology, Validation, Funding Acquisition, Writing – Original Draft, Writing – Review and Editing.
Declaration of Competing Interest
Authors have filed a patent application regarding work reported herein.
Acknowledgements
This work was supported by the National Institute of Biomedical Imaging and Bioengineering (Award #: 3U54HL143541-02S1) and by the Simmons Center for Cancer Research at Brigham Young University. We would also like to thank Dr. Niles A. Pierce (California Institute of Technology, Pasadena) for consulting on RNA toehold switch design. We also thank Hope Callister Anderson, Heather Mills Beutler, Jordan Barnett Nelson, Landon Ebbert, and Andrew Nelson for their supporting work preparing reagents and assisting in the laboratory. We acknowledge the use of BioRendor.com to create the abstract figure.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.nbt.2021.09.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bell B.P., Damon I.K., Jernigan D.B., Kenyon T.A., Nichol S.T., O’Connor J.P., et al. Overview, control strategies, and lessons learned in the CDC response to the 2014-2016 Ebola epidemic. MMWR Suppl. 2016;65:4–11. doi: 10.15585/mmwr.su6503a2. [DOI] [PubMed] [Google Scholar]
- 2.Kretzschmar M.E., Rozhnova G., Bootsma M.C.J., van Boven M., van de Wijgert J.H.H.M., Bonten M.J.M. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 2020;5 doi: 10.1016/S2468-2667(20)30157-2. e452-e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salathé M., Althaus C.L., Neher R., Stringhini S., Hodcroft E., Fellay J., et al. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med Wkly. 2020;150 doi: 10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- 4.Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., et al. Infectious Diseases Society of America guidelines on the diagnosis of Coronavirus disease 2019. Clin Infec Dis. 2020:760. doi: 10.1093/cid/ciaa760. ciaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi M., Yokoe D.S., Havlir D.V. Asymptomatic transmission, the Achilles’ heel of current strategies to control Covid-19. N Engl J Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly-Cirino C.D., Nkengasong J., Kettler H., Tongio I., Gay-Andrieu F., Escadafal C., et al. Importance of diagnostics in epidemic and pandemic preparedness. BMJ Glob Health. 2019;4 doi: 10.1136/bmjgh-2018-001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC . 2020. Overview of testing for SARS-CoV-2 (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html [Google Scholar]
- 9.Shin J., Noireaux V. An E. Coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 10.Slomovic S., Pardee K., Collins J.J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc Natl Acad Sci U S A. 2015;112:14429–14435. doi: 10.1073/pnas.1508521112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soltani M., Davis B.R., Ford H., Nelson J.A.D., Bundy B.C. Reengineering cell-free protein synthesis as a biosensor: biosensing with transcription, translation, and protein-folding. Biochem Eng J. 2018;138:165–171. doi: 10.1016/j.bej.2018.06.014. [DOI] [Google Scholar]
- 12.Zhang L., Guo W., Lu Y. Advances in cell-free biosensors: principle, mechanism, and applications. Biotechnol J. 2020;15 doi: 10.1002/biot.202000187. [DOI] [PubMed] [Google Scholar]
- 13.Pardee K., Green A.A., Ferrante T., Cameron D.E., DaleyKeyser A., Yin P., et al. Paper-based synthetic gene networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 15.Ma D., Shen L., Wu K., Diehnelt C.W., Green A.A. Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Biomed Pept Proteins Nucleic Acids. 2018 doi: 10.1093/synbio/ysy018. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi M.K., Tan X., Dy A.J., Braff D., Akana R.T., Furuta Y., et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun. 2018;9:3347. doi: 10.1038/s41467-018-05864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gräwe A., Dreyer A., Vornholt T., Barteczko U., Buchholz L., Drews G., et al. A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D., et al. Portable, on-demand biomolecular manufacturing. Cell. 2016;167:248–259. doi: 10.1016/j.cell.2016.09.013. e12. [DOI] [PubMed] [Google Scholar]
- 19.Amalfitano E., Karlikow M., Norouzi M., Jaenes K., Cicek S., Masum F., et al. A glucose meter interface for point-of-care gene circuit-based diagnostics. Nat Commun. 2021;12:724. doi: 10.1038/s41467-020-20639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salehi A.S.M., Smith M.T., Bennett A.M., Williams J.B., Pitt W.G., Bundy B.C. Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system. Biotechnol J. 2016;11:274–281. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- 21.Jewett M.C., Swartz J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 22.England C.G., Ehlerding E.B., Cai W. NanoLuc: A small luciferase is brightening up the field of bioluminescence. Bioconjug Chem. 2016;27:1175–1187. doi: 10.1021/acs.bioconjchem.6b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltani M., Porter Hunt J., Bundy B.C. Rapid RNase inhibitor production to enable low-cost, on-demand cell-free protein synthesis biosensor use in human body fluids. Biotechnol Bioeng. 2021 doi: 10.1002/bit.27874. n/a. [DOI] [PubMed] [Google Scholar]
- 24.Government Accountability Office . 2020. COVID-19: urgent actions needed to better ensure an effective federal response. Report to Congressional Committees.https://www.gao.gov/products/gao-21-191 GAO-21-191. [Google Scholar]
- 25.McMahon D.E., Peters G.A., Ivers L.C., Freeman E.E. Global resource shortages during COVID-19: bad news for low-income countries. PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ Sci Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Duan Y., Saliva A. Potential media for disease diagnostics and monitoring. Oral Oncol. 2012;48:569–577. doi: 10.1016/j.oraloncology.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 28.Wren M.E., Shirtcliff E.A., Drury S.S. Not all biofluids are created equal: chewing over salivary diagnostics and the epigenome. Clin Ther. 2015;37:529–539. doi: 10.1016/j.clinthera.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt J.P., Barnett R.J., Robinson H., Soltani M., Nelson J.A.D., Bundy B.C. Rapid sensing of clinically relevant glutamine concentrations in human serum with metabolically engineered E. Coli-based cell-free protein synthesis. J Biotech. 2021;325:389–394. doi: 10.1016/j.jbiotec.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Salehi A.S.M., Shakalli Tang M.J., Smith M.T., Hunt J.M., Law R.A., Wood D.W., et al. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal Chem. 2017;89:3395–3401. doi: 10.1021/acs.analchem.6b04034. [DOI] [PubMed] [Google Scholar]
- 31.Salehi A.S.M., Yang S.O., Earl C.C., Shakalli Tang M.J., Porter Hunt J., Smith M.T., et al. Biosensing estrogenic endocrine disruptors in human blood and urine: a RAPID cell-free protein synthesis approach. Toxicol Appl Pharmacol. 2018;345:19–25. doi: 10.1016/j.taap.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Eichel H.J., Conger N., Chernick W.S. Acid and alkaline ribonucleases of human parotid, submaxillary, and whole saliva. Arch Biochem Biophys. 1964;107:197–208. doi: 10.1016/0003-9861(64)90322-4. [DOI] [PubMed] [Google Scholar]
- 33.Park N.J., Zhou X., Yu T., Brinkman B.M.N., Zimmermann B.G., Palanisamy V., et al. Characterization of salivary RNA by cDNA library analysis. Arch Oral Biol. 2007;52:30–35. doi: 10.1016/j.archoralbio.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130 doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park N.J., Li Y., Yu T., Brinkman B.M.N., Wong D.T. Characterization of RNA in saliva. Clin Chem. 2006;52:988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon T.M., Kopish A., Kopish K., Co Promega. Luciferase reporter assays: powerful, adapatable tools for cell biology research. Cell Notes. 2008;21:23–26. [Google Scholar]
- 37.Green A.A., Silver P.A., Collins J.J., Yin P. Toehold switches: de-Novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Zhou Y., Carlson P.D., Teichmann M., Chaudhary S., Simmel F.C., et al. De novo-designed translation-repressing riboregulators for multi-input cellular logic. Nat Chem Biol. 2019;15:1173–1182. doi: 10.1038/s41589-019-0388-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albayrak C., Swartz J.R. Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation. Nucleic Acids Res. 2013;41:5949–5963. doi: 10.1093/nar/gkt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., et al. NUPACK: analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 41.Hochrein L.M., Ge T.J., Schwarzkopf M., Pierce N.A. Signal transduction in human cell lysate via dynamic RNA nanotechnology. ACS Synth Biol. 2018;7:2796–2802. doi: 10.1021/acssynbio.8b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo C.H., Jang S., Shin G., Jung G.Y., Lee J.W. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat Biomed Eng. 2020;4:1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.