Abstract
Background
Daikenchuto (TU‐100), a Japanese herbal medicine, is widely used for various gastrointestinal diseases. We have previously reported that TU‐100 suppresses CPT‐11‐induced bacterial translocation (BT) by maintaining the diversity of the microbiome. In this study we show that TU‐100 modulates the immune response during BT by inducing PD‐1 expression in Peyer's patches.
Methods
Eighteen male Wistar rats were divided into four groups: a control group; a control + TU‐100 group, given TU‐100 1000 mg/kg orally for 5 d; a BT group, given CPT‐11 250 mg/kg intra‐peritoneal for 2 d; and a TU‐100 group, given TU‐100 1000 mg/kg orally for 5 d with CPT‐11 250 mg/kg intra‐peritoneal on days 4 and 5.
Results
The size of Peyer's patch was significantly bigger in the BT group compared to the control group (9.0 × 104 µm2 vs 29.4 × 104 µm2, P < .05), but improved in the TU‐100 group (15.4 × 104 µm2, P < .005). TU‐100 significantly induced PD‐1 expression in Peyer's patch compared to the control group and the BT group (control vs BT vs TU‐100 = 4.3 ± 4.9 vs 5.1 ± 10.3 vs 17.9 ± 17.8). The CD4+ cells were increased in the BT group (P < .05) compared to the control group but decreased in the TU‐100 group. The Foxp3+ cells were increased in the BT group compared to the control group (P < .05), and further increased in the TU‐100 group compared to the BT group. CPT‐11 significantly increased TLR4, NF‐κβ, TNF‐α mRNA expressions in the BT group. TU‐100 cotreatment significantly reversed these mRNA expressions.
Conclusion
TU‐100 may have a protective effect against BT through PD‐1 expression in Peyer's patch.
Keywords: herbal medicine, inflammatory cytokine, Kampo
Daikenchuto (TU‐100), a Japanese herbal medicine, is widely used for various gastrointestinal diseases. We have previously reported that TU‐100 suppresses CPT‐11‐induced bacterial translocation (BT) by maintaining the diversity of the microbiome. In this study, we show that TU‐100 modulates the immune response during BT by inducing PD‐1 expression in Peyer's patches.
1. INTRODUCTION
Irinotecan hydrochloride (CPT‐11) is a key anticancer drug that is widely used for the treatment of colorectal and other solid tumors.1 Irinotecan therapy is limited by several dose‐limiting toxicities. The most common clinically important side effect is severe diarrhea, which markedly impairs patient quality of life and results in a poor prognosis. We previously established a rat model of bacterial translocation (BT) induced by CPT‐11.2 The intestinal permeability of the large intestine was found to be decreased through disruption of tight junction (TJ) proteins and caused BT.
Daikenchuto (TU‐100) is a Kampo medicine, a traditional Japanese herbal medicine that is made from certain Asian plants. Kampo medicine is very popular and widely used in Japan. In surgical fields, it has been used for a feeling of coldness3 in the abdomen and a postoperative adhesive intestinal ileus.3 Previous reports have shown that TU‐100 improves gastrointestinal motility or transit by modulating cholinergic and serotonergic mechanisms.4 TU‐100 attenuates intestinal inflammation by increasing intestinal blood flow and protecting the luminal surface, producing an antiinflammatory effect.5 We previously reported that TU‐100 protects against by fasting stress6 and CPT‐11‐induced BT.7 One mechanism by which TU‐100 protects against BT is through maintaining the diversity of the microbiome during intestinal inflammation.6 The other mechanism involves inhibiting inflammation and apoptosis by maintaining TJ function.7 However, the mechanism by which TU‐100 protects against BT has not been fully revealed.
A recent study reported that the inhibitory coreceptor programmed cell death‐1 (PD‐1) regulates the gut microbiota through appropriate selection of immunoglobulin A (IgA) plasma cell repertoires.8 In Peyer's patches, IgA plays a key role in maintaining the symbiotic balance between gut bacterial communities and the host immune system.
Hence, this study aimed to elucidate the further mechanism of TU‐100 on preventing BT. We also examined the possible role of TU‐100 through PD‐1 expression in Peyer's patches.
2. METHODS
2.1. Materials
CPT‐11 was purchased from Daiichi Sankyo (Tokyo, Japan) and freshly prepared just prior to the treatment. The rats were given 0.01 mg/kg atropine subcutaneously immediately prior to CPT‐11 treatment to reduce the cholinergic reaction to CPT‐11. TU‐100 was purchased from Tsumura (Tokyo, Japan).
2.2. Experimental design
Male Wistar rats (weighing 230–250 g and aged 6 wk old) were purchased from Charles River (Wilmington, MA). Animals were free of all pathogens and housed under standard conditions (22°C, humidity 50 ± 5%, 12/12 h light/dark cycle). For this study, we used samples from established bacterial translocation model as previously reported.7 The BT was defined as the presence of bacteria in the mesenteric lymph nodes (MLNs).
Simply, they were collected and cultured. Figure 1 shows the experimental design. A total of 21 rats were used and randomly divided into three groups as follows: the control group (n = 6), given TU‐100 1000 mg/kg orally for 5 d; the control + TU‐100 group (n = 3), saline was given intraperitoneally on d 4 and 5; the BT group (n = 6), 250 mg/kg CPT‐11 was given intraperitoneally on d 4 and 5; the TU‐100 group (n = 6), TU‐100 was orally administered for 5 consecutive d from d 1, and CPT‐11 was given intraperitoneally on d 4 and 5. All rats were euthanized on d 6 of the experiment by the administration of an appropriate dose of anesthesia (1.0–1.5% isoflurane), and the small intestine and stool were directly collected from intestine for analysis. The rats in the control + TU‐100 group did not show diarrhea, BT, or body weight loss (there was no difference compared to the control group).
FIGURE 1.
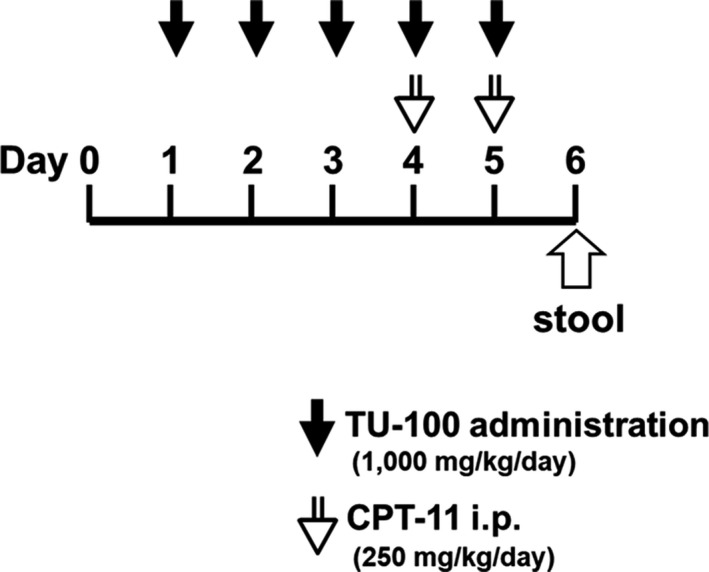
Experimental design
This study was carried out in accordance with NIH guidelines for the care and use of rats. The experiment was approved by the Institute of Animal Committee of Health Bioscience, Tokushima University Graduate School of Medical Science (08093).
2.3. RNA extraction and quantitative real‐time polymerase chain reaction
Total RNA was extracted from collected tissues (small intestine) using a RNeasy mini kit (Qiagen, Chatsworth, CA). Reverse transcription was performed with a high‐capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) Then, quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed using the 7500 real‐time PCR system (Applied Biosystems), TaqMan gene expression assays‐on demand, and the TaqMan universal master mix (Applied Biosystems). The primers used for qRT‐PCR were as follows: TLR4 (Rn00569848_m1), tumor necrosis factor alpha (TNF‐α) (Rn00562055_m1), nuclear factor kappa beta (NF‐κβ) (Rn01399583_m1), interleukin (IL)‐1β (Rn00580432_m1), and IL‐6 (Rn00561420_m1). We used the glyceraldehyde phosphate dehydrogenase (GAPDH) gene (Applied Biosystems, 4352338E) as an endogenous control. The thermal cycler conditions were as follows: 2 min at 50°C, 10 min at 95°C, then 40 cycles of 15 sec at 95°C and 1 min at 60°C. Amplification data were analyzed with an ABI Prism 7500 Sequence Detection System v. 1.3.1 (Applied Biosystems).
2.4. Histopathological analysis
The tissue samples were placed in 10% formalin for 72 h at room temperature, dehydrated by graded ethanol, and embedded in paraffin. The 5‐μm tissue sections were deparaffinized with xylene, stained with hematoxylin and eosin, and observed by optical microscopy. The size of the Peyer's patches was measured with a digital camera photomicroscope (DXM 1200F, Nikon, Tokyo, Japan) in a blinded fashion. The average of all Peyer's patches in each slide was calculated for statistical analyses.
2.5. Immunohistochemical staining
The tissue samples were fixed in 10% formalin and embedded in paraffin. Serial sections (5 µm) were dewaxed, deparaffinized in xylene, and rehydrated with a graded alcohol series. Antigen retrieval in citrate buffer (pH 6.0) was performed with a microwave oven for 20 min. The sections were incubated in 5% goat serum (60 min) to prevent nonspecific antigen binding. Endogenous peroxidase activity was inhibited with 0.3% hydrogen peroxidase for 30 min. Then the primary antibodies were applied and incubated overnight at 4°C. The primary antibodies were diluted as follows: PD‐1 (ab117420, 1:50; Abcam, Cambridge, MA), CD4 (ab237722, 1:4000; Abcam), Foxp3 (ab215206, 1:250; Abcam), and CD8 (bs‐4791R, 1:100; Bioss, Woburn, MA). Then the slides were incubated with a secondary antibody for 60 min. The slides were washed and developed with 3,3‐diaminobenzidine for 10 min. The slides were counterstained with Mayer's hematoxylin and dehydrated with a graded alcohol series.
The evaluation of each slide was performed by a pathologist in a blinded manner. The expression of PD‐1, CD4, and Foxp3 in Peyer's patches was evaluated. The PD‐1‐positive rate (the number of PD‐1‐positive cells / the number of mononuclear cells) was recorded in a ×400 high‐power field.9 The number of CD4‐ and Foxp3‐positive cells at five points in the ×400 high‐power field was recorded.10
2.6. Analysis of the gut microbiome by sequencing 16S rRNA gene clone libraries
To evaluate the influence of TU‐100 on the gut microbiome, we sequenced the 16S rRNA gene (16S rDNA) clone libraries as previously described.6 This analysis was performed for three samples in the control + TU‐100 group, BT group, and TU‐100 group.
DNA was extracted from the rat feces according to the method described by Kataoka et al11 in which lysozyme and achromopeptidase were used as lytic enzymes. Bacterial 16S rDNA was amplified using the universal primers, 27F and 1492R. The amplicons were cloned into pGEM‐T Easy Vectors (Promega, Madison, WI), and libraries were constructed using Escherichia coli DH5α. From each library, 96 colonies were randomly selected, and the inserted DNA fragments were amplified with primers SP6 and T7, which have annealing sites located just outside of the cloning site of the pGEM‐T Easy Vector. The amplicons were purified, and the nucleotide sequences were determined using 27F as a sequencing primer Takara Bio (Shiga, Japan). Of the obtained sequences, low‐quality sequences were removed. The uncertain nucleotides at both ends were also trimmed. The assignment of each 16S rDNA sequence was done by the Classifier program from the Ribosomal Database Project6 with a confidence level of over 80%. Sequences were aligned with the ClustalW2 program and assembled into operational taxonomic units (OTUs) with the parameters of >97% identity and >90% alignment length, and the number of OTUs was used to evaluate the diversity of gut microbiota.
2.7. Statistical analyses
The statistical analyses were performed with JMP 8.0.1 (SAS, Cary, NC). All data are presented as the mean ± standard deviation (SD). The differences between experimental groups were analyzed using a one‐way analysis of variance (ANOVA) or Student's t‐test for unpaired data. Statistical significance was defined as a P < .05.
3. RESULTS
3.1. The Peyer's patch parameters
The number of Peyer's patches was calculated along three regions of small intestine (proximal, middle, and distal). There were few Peyer's patches in proximal small intestine. There was no significant difference in the numbers of Peyer's patches among four groups.
The size of the Peyer's patches was significantly decreased by only TU‐100 administration compared to the control group (10.2 × 104 µm2 vs 19.0 × 104 µm2, P = .03). The size of the Peyer's patches in the BT group was increased compared to that in the control group (19.0 × 104 µm2 vs 29.4 × 104 µm2, P = .04). However, the size increase was reversed by TU‐100 cotreatment (13.1 × 104 µm2, P = .0002, Figure 2A). There was no significant difference in size between the control and TU‐100 groups (P = .07). PD‐1 expression in Peyer's patches in the TU‐100 group (17.9 ± 17.8%) was significantly increased compared to that in the control (4.3 ± 4.9%), P = .001) and BT (5.1 ± 10.3%), P = .007) groups (Figure 2B,C). There was no significant difference in PD‐1 expression between the control and BT groups (P = .73).
FIGURE 2.
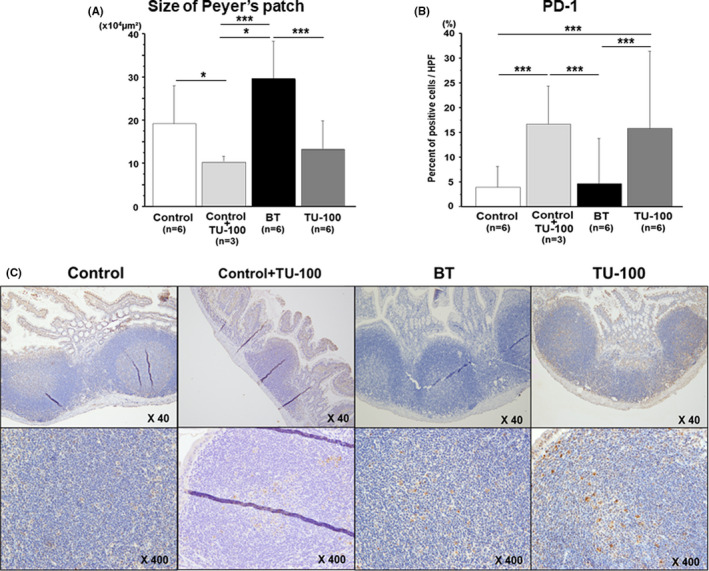
The Peyer's patch size and PD‐1 expression. (A) The Peyer's patch size was significantly greater in the bacterial translocation (BT) group than the control group (P < .05), but the size improved in the TU‐100 group (P < .05). (B) The PD‐1 expression in Peyer's patches in the TU‐100‐treated group was increased compared to that in the control and the BT groups (P < .05). PD‐1 expression was significantly increased by administration of TU‐100 only (P < .05). (C) Representative immunohistochemistry of PD‐1 expression in Peyer's patches. *P < .05, **P< .01, ***P< .001
3.2. Immune cell frequency in the Peyer's patch
The number of CD4+ cells was significantly increased in the BT group than the control group (13.0 ± 7.5 vs 103.8 ± 17.8, P = .0001). However, the frequency was significantly decreased in the TU‐100 group (68.8 ± 16.8, P = .01). There was no difference between the control group and the control + TU‐100 group (P = .15).
The number of Foxp3+ cells was rarely observed in the control group (1.0 ± 2.2, Figure 3B). There was no difference between the control group and the control + TU‐100 group. Compared to the control group, the CPT‐11‐treated group showed a significantly increased number of Foxp3+ cells (1.0 ± 2.2 vs 11.0 ± 2.5, P = .0002). Furthermore, TU‐100 cotreatment further increased the number of Foxp3+ cells (21.0 ± 5.4: vs control group, P < .0001: vs BT group, P = .005).
FIGURE 3.
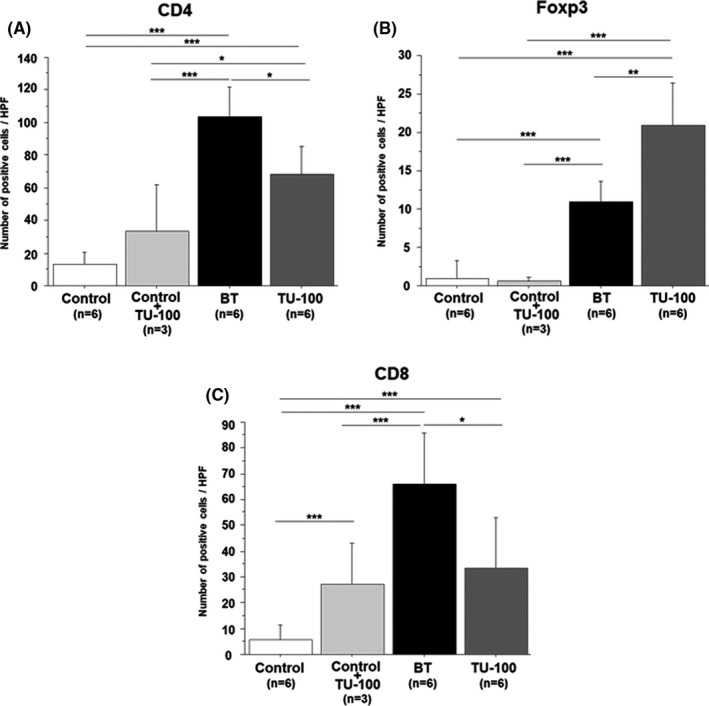
Immune cell populations in the Peyer's patch. (A) The CD4+ cell population was increased in the bacterial translocation (BT) group (P < .05) compared to the control group but decreased in the TU‐100 group (P < .05). (B) The Foxp3+ cell population was increased in the BT group compared to the control group (P < .05) and further increased in the TU‐100 group compared to the BT group. (C) The CD8+ cell population was increased in the BT group (P < .05) compared to the control group but significantly decreased in the TU‐100 group. Only administration of TU‐100 also induced the increase of CD8+ cell population (P < .05). *P < .05, **P < .01, ***P < .001
The number of CD8+ cells was more frequently observed in the control + TU‐100 group than the control group (5.7 ± 5.3 vs 26.1 ± 16.6, P = .004, Figure 3C) in Peyer's patches. CD8+ cells were significantly increased in the BT group than the control group (5.7 ± 5.3 vs 66.3 ± 19.7, P < .0001). The frequency was significantly decreased in the TU‐100 group (33.3 ± 19.6, P = .01).
3.3. Inflammatory cytokine mRNA expression in small intestine
Pattern recognition receptor‐like Toll‐like receptors (TLRs) activate inflammatory signaling pathways. TLR4 mRNA expression was significantly increased by administration of only TU‐100 (0.19 ± 0.11 vs 0.74 ± 0.17, P = .0005, Figure 4A). TLR4 mRNA expression was significantly higher in the BT group than the control group (0.19 ± 0.11 vs 1.43 ± 0.59, P = .0005, Figure 4A). However, TU‐100 cotreatment significantly downregulated CPT‐11‐induced TLR4 mRNA expression (0.67 ± 0.23, P = .005).
FIGURE 4.
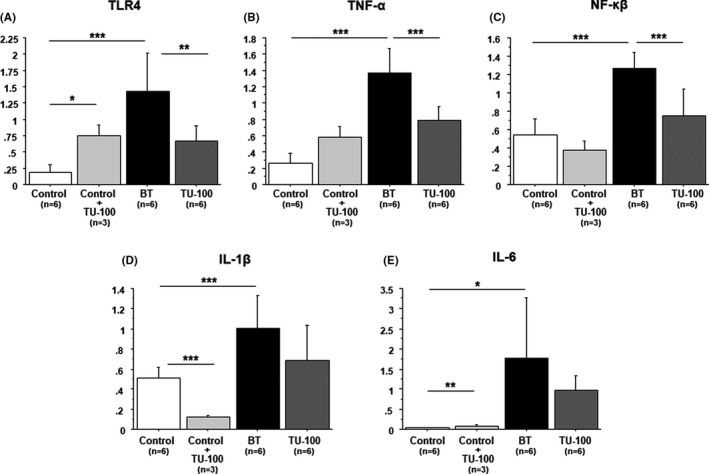
Inflammatory cytokine mRNA expression in small intestine (A) TLR4, (B) TNF‐α, (C) NF‐κβ, (D) IL‐1β and (E) IL‐6. CPT‐11 caused a significant increase in TLR4, TNF‐α, NF‐κβ, IL‐1β, and IL‐6 mRNA expression in the bacterial translocation group. TU‐100 cotreatment significantly reversed increases of TLR4, TNF‐α, and NF‐κβ expression. *P < .05, **P < .01, ***P < .001
The administration of TU‐100 was not affected to TNF‐α and NF‐κβ mRNA expression. Compared with the control group, the CPT‐11‐treated group showed significantly increased mRNA expression of TNF‐α (0.26 ± 0.12 vs 1.37 ± 0.30, P < .0001) and NF‐κβ (0.54 ± 0.17 vs 1.27 ± 0.18, P < .0001). Conversely, compared with the CPT‐11‐treated group, the TU‐100‐cotreatment group showed significantly improved expression of TNF‐α (0.74 ± 0.29, P = .002, Figure 4B) and NF‐κβ (0.79 ± 0.17, P < .0001, Figure 4C).
On the other hand, IL‐1β and IL‐6 mRNA expression was significantly decreased by administration of only TU‐100 (Figure 4D,E). IL‐1β and IL‐6 mRNA expression were significantly higher in the BT group than the control group (IL‐1β: 0.51 ± 0.11 vs 1.00 ± 0.31, P = .004, IL‐6:0.03 ± 0.01 vs 1.76 ± 0.6, P = .01). Although there was no significant difference, the TU‐100‐cotreatment group showed improved expression of IL‐1β (0.69 ± 0.35, P = .10) and IL‐6 (0.96 ± 0.38, P = .16).
3.4. Composition of the gut microbiota
The average of total sequence reads in each group was 22,177 ± 1425 in the control + TU‐100 group, 10,950 ± 1393 in the BT group, and 26,713 ± 18 712 in the TU‐100 group. The diversity of the microbiome was significantly decreased in the BT group compared to the control + TU‐100 group (P = .0006). TU‐100 improved the diversity of the microbiome (P = .21).
Figure 5 shows the major microbiome composition of the BT and TU‐100 groups. In the BT group, the abundance Lactobacillus was remarkably decreased compared to the control + TU‐100 group. The TU‐100 group showed a recovered ratio of Lactobacillus. On the other hand, the abundance of Oscillibacter, Shigella, Eschericha, Enterobacter, Hydrogenoanaerobacterium, Blautia, and Parabacteroides was increased in the BT group compared to the control + TU‐100 group and decreased in the TU‐100 group compared to the BT group. The abundance of Akkermansia, Allobaculum, and Dorea was increased in the BT group compared to the control + TU‐100 group. Moreover, these abundances were increased in the TU‐100 group compared to the BT group.
FIGURE 5.
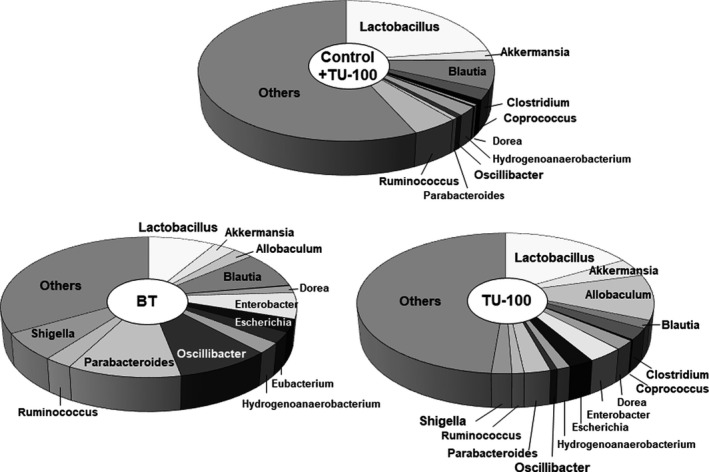
Composition of the gut microbiota. The abundance of Oscillibacter, Shigella, Eschericha, Enterobacter, Hydrogenoanaerobacterium, Blautia, and Parabacteroides was increased in the bacterial translocation (BT) group compared to the control + TU‐100 group and decreased in the TU‐100 group compared to the BT group
4. DISCUSSION
CPT‐11 has been used as an extremely effective anticancer drug for a variety of solid tumors. However, the safety of the drug is limited, particularly in debilitated patients, due to its toxicity‐associated side effects, including severe diarrhea. Therefore, another treatment combination should be investigated to block the cytotoxic effect of CPT‐11 in normal cells but not in tumor cells. TU‐100 has negligible side effects, which have been investigated in previous in vivo, in vitro, and clinical trial studies. However, TU‐100 has multiple functions, including the downregulation of inflammatory mediators.13, 14 Our previous findings revealed that TU‐100 cotreatment prevents CPT‐11‐induced BT through the inhibition of inflammation and apoptosis. Furthermore, TU‐100 treatment improved CPT‐11‐related TJ dysfunction.7 However, the influence of TU‐100 on the immune system has not been investigated thus far.
The recent emergence of immunotherapy provides opportunities for herbal medicines to reduce the side effects of chemotherapy because of their safety. Herbal medicines play a role in enhancing immunity induced with cancer vaccines and immune responses of cancer immunotherapy.14 Recently, the immunomodulatory ability of herbal medicines has attracted much attention. Several reports have demonstrated that TJ‐41 (Hochu‐ekki‐to) enhances natural killer (NK) cell activity, activates anticancer immunity, and protects against infections in animal models.15 NJT (Ninjinto) also increases NK cell activity in peripheral blood mononuclear cells and effectively controls functional gastrointestinal disorders associated with chronic intestinal failure.16 A recent study showed that TJ‐35 (Shigyakusan) prolongs the survival of fully allogeneic cardiac allografts and may induce regulatory CD4+CD25+Foxp3+ cell production.17 These reports suggest the possible use of herbal medicines as modulators of the immune response. Our present and previous data showed that TU‐100 treatment clearly suppressed BT‐induced inflammation (TNF‐α and NF‐κβ), possibly through the TLR pathway.7 The frequency of both CD4+ and Foxp3+ cells was increased by BT, reflecting inflammation. In contrast, the frequency of CD4+ cells was decreased, but Foxp3+ cell numbers were further increased by treatment with TU‐100. The transcription factor Foxp3 is a well‐established marker of regulatory T cells (Tregs) and is required for Treg cell differentiation and function.18 Foxp3+ Tregs play a crucial role in maintaining immune homeostasis, preventing autoimmunity, and regulating excessive immune responses. The number of Foxp3+ cells in the BT group might be increased to suppress excessive inflammation. Moreover, the Foxp3+ expression was increased by TU‐100 cotreatment. An additional possible mechanism is immune system modulation via PD‐1 expression in Peyer's patches. PD‐1 (CD28 family member) is expressed on many immune cells, such as myeloid cells, B cells, and activated T cells.19 PD‐1 downregulates inflammatory responses through the inhibition of cytokine production. PD‐1 activates the transcription factor Foxp3, which is also known as a strong regulator of T cells.20 Our data showed that PD‐1 expression in Peyer's patches was strongly induced by TU‐100 treatment. The induction of PD‐1 by TU‐100 might activate Foxp3 and regulate excess inflammation through inducing Treg production.
A recent study showed that PD‐1 regulates the gut microbiota through appropriate selection of IgA, which maintains homeostasis at mucosal surfaces.8 The study showed that PD‐1 deficiency perturbs the balance of bacterial communities in the gut. We previously showed that TU‐100 prevents the reduction in microbiome diversity in a rat fasting stress model.18 Although the diversity was not significantly changed by TU‐100 treatment in that study, the OTU was increased in the TU‐100 group. However, The TU‐100 changed the composition of gut microbiota dramatically. In the TU‐100 group, the abundance of Lactobacillus was remarkably increased. In line with our result, the recent study showed Lactobacillus ameliorates TNF‐α‐induced BT in Caco‐2 cells through regulation of TLR4.21 On the other hand, in the abundance of pathogenic bacteria causing the colonic inflammation such as Oscillibacter, Shigella 22, Eubacterium 23, Enterobacter 24, Escherichia, and Hydrogenoanaerobacterium 25 were increased in the BT group and decreased in the TU‐100 group. Oscillibacter mediates HFD‐induced gut dysfunction and correlates with gut permeability.26 A recent report showed that IgA could suppress the cell growth of E. coli and improve the intestinal environment.27 The role of Akkermancia is still controversial for colitis but the ratio of Akkermancia was further increased in the TU‐100 group compared to the control + TU‐100 group and the BT group.29, 30
Only two beneficial bacteria, Parabacteroides 31 and Blautia 32, which had a protective effect for colitis, were decreased in the TU‐100 group. Our data suggest TU‐100 exerts a protective effect through inducing Lactobacillus and reducing several pathogenic bacteria. The correlations among these changes in gut microbiota and PD‐1 expression needs to be confirmed by further investigation.
There are two major limitations to this study that could be addressed in future research. First, the sample size was relatively small. Second, the proteins of interest were assessed by only one experiment (mRNA level or protein level). Moreover, the detailed mechanism of PD‐1 induction in Peyer's patches was not fully investigated in this study, and further investigation is needed.
5. CONCLUSION
In conclusion, TU‐100 may protect against BT via PD‐1 expression regulation in Peyer's patches. Our data suggest a new immunomodulatory effect of TU‐100. Our finding will be a new aspect of TU‐100 for the treatment of BT.
DISCLOSURE
Funding: This study was partly supported by a Cancer Research Project Cooperated by TAIHO Pharmaceutical Co., Ltd. and the University of Tokushima and by Japan Society for the promotion of Science (Grant‐in‐Aid for Scientific Research B: No. 23791536). Mitsuo Shimada received a research grant from Taiho Pharmaceutical Co. Ltd.
AUTHOR CONTRIBUTION
C.T. and K.Y. designed the experiment. C.T., H.K and T.Y performed the experiments and analyzed the data. C.T wrote the manuscript in discussions with M.N and T.T. H.O analyzed IHC experiment with C.T.. K.M performed the revised experiments with C.T.. S.M. supervised the whole project and Y.M. directed the revised project.
ACKNOWLEDGMENT
The authors thank Dr. Toru Kono (Center for Clinical and Biomedical Research, Sapporo Hisgashi Tokushukai Hospital, Hokkaido, Japan) for helpful discussions and valuable advice on this study.
Takasu C, Mitazaki K, Yoshikawa K, et al. Effect of TU‐100 on Peyer's patches in a bacterial translocation rat model. Ann Gastroenterol Surg. 2021;5:683–691. 10.1002/ags3.12460
REFERENCES
- 1.Beppu N, Yoshie H, Kimura F, Aihara T, Doi H, Kamikonya N, et al. Clinicopathological outcomes of preoperative chemoradiotherapy using S‐1 plus irinotecan for T4 lower rectal cancer. Surg Tod. 2016;46(7):852–9. [DOI] [PubMed] [Google Scholar]
- 2.Nakao T, Kurita N, Komatsu M, Yoshikawa K, Iwata T, Utusnomiya T, et al. Irinotecan injures tight junction and causes bacterial translocation in rat. J Surg Res. 2012;173(2):341–7. [DOI] [PubMed] [Google Scholar]
- 3.Sato Y, Inoue S, Katagiri F, Itoh H, Takeyama M. Effects of Pirenzepine on Dai‐kenchu‐to‐Induced Elevation of the Plasma Neuropeptide Levels in Humans. Biological and Pharmaceutical Bulletin. 2006;29(1):166–71. 10.1248/bpb.29.166 [DOI] [PubMed] [Google Scholar]
- 4.Itoh T, Yamakawa J, Mai M, Yamaguchi N, Kanda T. The effect of the herbal medicine dai‐kenchu‐to on post‐operative ileus. J Int Med Res. 2002;30(4):428–32. [DOI] [PubMed] [Google Scholar]
- 5.Manabe N, Camilleri M, Rao A, Wong BS, Burton D, Busciglio I, et al. Effect of daikenchuto (TU‐100) on gastrointestinal and colonic transit in humans. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G970–G975. [DOI] [PubMed] [Google Scholar]
- 6.Kono T, Omiya Y, Hira Y, Kaneko A, Chiba S, Suzuki T, et al. Daikenchuto (TU‐100) ameliorates colon microvascular dysfunction via endogenous adrenomedullin in crohn's disease rat model. J Gastroenterol. 2011;46(10):1187–96. [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa K, Shimada M, Kuwahara T, Hirakawa H, Kurita N, Sato H, et al. Effect of Kampo medicine "Dai‐kenchu‐to" on microbiome in the intestine of the rats with fast stress. J Med Invest. 2013;60(3–4):221–7. [DOI] [PubMed] [Google Scholar]
- 8.Takasu C, Yismaw WG, Kurita N, Yoshikawa K, Kashihara H, Kono T, et al. TU‐100 exerts a protective effect against bacterial translocation by maintaining the tight junction. Surg Tod. 2017;47(10):1287–94. [DOI] [PubMed] [Google Scholar]
- 9.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, et al. The inhibitory receptor PD‐1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336(6080):485–9. [DOI] [PubMed] [Google Scholar]
- 10.Eto S, Yoshikawa K, Nishi M, Higashijima J, Tokunaga T, Nakao T, et al. Programmed cell death protein 1 expression is an independent prognostic factor in gastric cancer after curative resection. Gastric Cancer. 2016;19(2):466–71. [DOI] [PubMed] [Google Scholar]
- 11.Flammiger A, Weisbach L, Huland H, Tennstedt P, Simon R, Minner S, et al. High tissue density of FOXP3+ T cells is associated with clinical outcome in prostate cancer. Eur J Cancer. 2013;49(6):1273–9. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka K, Kibe R, Kuwahara T, Hagiwara M, Arimochi H, Iwasaki T, et al. Modifying effects of fermented brown rice on fecal microbiota in rats. Anaerobe. 2007;13(5–6):220–7. [DOI] [PubMed] [Google Scholar]
- 13.Chikakiyo M, Shimada M, Nakao T, Higashijima J, Yoshikawa K, Nishioka M, et al. Kampo medicine "Dai‐kenchu‐to" prevents CPT‐11‐induced small‐intestinal injury in rats. Surg Tod. 2012;42(1):60–7. [DOI] [PubMed] [Google Scholar]
- 14.Ueno N, Hasebe T, Kaneko A, Yamamoto M, Fujiya M, Kohgo Y, et al. TU‐100 (Daikenchuto) and ginger ameliorate anti‐CD3 antibody induced T cell‐mediated murine enteritis: microbe‐independent effects involving Akt and NF‐kappaB suppression. PLoS One. 2014;9(5):e97456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodeker G. Integrative oncology meets immunotherapy: new prospects for combination therapy grounded in Eastern medical knowledge. Chin J Integr Med. 2012;18(9):652–62. [DOI] [PubMed] [Google Scholar]
- 16.Kimura M, Sasada T, Kanai M, Kawai Y, Yoshida Y, Hayashi E, et al. Preventive effect of a traditional herbal medicine, Hochu‐ekki‐to, on immunosuppression induced by surgical stress. Surg Tod. 2008;38(4):316–22. [DOI] [PubMed] [Google Scholar]
- 17.Uehara S, Ogawa K, Arimitsu J, Okuyama H. "Ninjinto" (Ginseng Decoction), a traditional Japanese herbal medicine, improves gastrointestinal symptoms and immune competence in patients with chronic intestinal failure. Evid Based Complement Alternat Med. 2015;2015:462586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin E, Uchiyama M, Niimi M. Induction of regulatory CD4(+) Cells and prolongation of fully major histocompatibility complex mismatched murine cardiac allograft by Shigyakusan. Transplant Proc. 2018;50(1):274–82. [DOI] [PubMed] [Google Scholar]
- 19.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–61. [DOI] [PubMed] [Google Scholar]
- 20.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD‐1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11(11):3887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahearne MJ, Bhuller K, Hew R, Ibrahim H, Naresh K, Wagner SD. Expression of PD‐1 (CD279) and FoxP3 in diffuse large B‐cell lymphoma. Virchows Arch. 2014;465(3):351–8. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Li J, Wang S, Hao Y, Zhao X, Chen J. Lactobacillusplantarum ameliorates tumour necrosis factor‐induced bacterial translocation in Caco‐2 cells by regulation of TLR4 expression. J Med Microbiol. 2018;67(7):982–91. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, Nakanishi K, Tsutsui H, Iwai H, Akira S, Inohara N, et al. A novel caspase‐1/toll‐like receptor 4‐independent pathway of cell death induced by cytosolic Shigella in infected macrophages. J Biol Chem. 2005;280(14):14042–50. [DOI] [PubMed] [Google Scholar]
- 24.Kanauchi O, Fukuda M, Matsumoto Y, Ishii S, Ozawa T, Shimizu M, et al. Eubacteriumlimosum ameliorates experimental colitis and metabolite of microbe attenuates colonic inflammatory action with increase of mucosal integrity. World J Gastroenterol. 2006;12(7):1071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith JW, Dong Q, Sorbara MT, Becattini S, Sia JK, Gjonbalaj M, et al. Impact of antibiotic‐resistant bacteria on immune activation and clostridioides difficile infection in the mouse intestine. Infect Immun. 2020;88(4):e00362‐19. 10.1128/iai.00362-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinoso Webb C, den Bakker H, Koboziev I, Jones‐Hall Y, Rao Kottapalli K, Ostanin D, et al. Differential susceptibility to T cell‐induced colitis in mice: role of the intestinal microbiota. Inflamm Bowel Dis. 2018;24(2):361–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet‐induced obese mice. PLoS One. 2012;7(3):e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okai S, Usui F, Yokota S, Hori‐I Y, Hasegawa M, Nakamura T, et al. High‐affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol. 2016;1(9):16103. [DOI] [PubMed] [Google Scholar]
- 29.Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al. Administration of akkermansia muciniphila ameliorates dextran sulfate sodium‐induced ulcerative colitis in mice. Front Microbiol. 2019;10:2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14(10):609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp‐regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One. 2016;11(1):e0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515(7527):423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


