Abstract
Deep-seated trichosporonosis is a lethal opportunistic infection that disseminates rapidly and widely in immunocompromised patients, and early diagnosis is crucial for the treatment of this infection. We developed a novel nested-PCR assay that detects DNA specific for clinically important strains of Trichosporon in serum samples from patients with disseminated trichosporonosis. In this assay, two sets of oligonucleotide primers were derived from the sequence of 26S rRNA genes of Trichosporon asahii. The specific fragment was amplified from T. asahii and T. mucoides, but not from other microorganisms, including some other basidiomycetous fungi (Cryptococcus, Malassezia, Rhodotorula, and Sporobolomyces). Target DNA was detected by the nested PCR with as little as 5 fg of the extracted DNA of T. asahii. In a study using 11 clinical samples, the specific fragment was detected by the nested PCR in 64% (7 of 11) of sera from patients with histologically diagnosed disseminated trichosporonosis, while glucuronoxylomannan antigen was detected in only 54% (6 of 11) of the samples. Our new nested-PCR assay using serum samples can be performed repeatedly throughout the course of the disease. In addition, not only can it be used for early diagnosis of trichosporonosis, but it may also be beneficial for monitoring its progress or response to therapy.
Disseminated trichosporonosis is a lethal opportunistic infection associated with morbidly severe conditions, such as rapidly developing respiratory failure, renal failure, or disseminated intravascular coagulation syndrome, in immunocompromised patients (e.g., those with hematological malignancies) (19). In recent years, the efficacy of different combination therapies for this condition has been reported. These include a variety of antifungal agents and granulocyte-colony stimulating factors (G-CSFs) (3, 14). However, these therapies are only effective if the disease is detected at an early stage, and, therefore, early diagnosis is an important step in the successful management of patients with disseminated trichosporonosis. Unfortunately, an early diagnosis of the disease is very difficult, and in most cases, this disease is only confirmed at autopsy. A firm diagnosis of disseminated trichosporonosis is usually established by histological examination of tissue samples obtained by biopsy as well as by detecting the causative pathogenic fungi in clinical samples. However, invasive examinations, such as biopsy, cannot be repeatedly performed in patients with severe underlying diseases, and even though the detection of causative fungi in the blood is clinically valuable, the results of blood cultures usually become available only after a relatively long period of time.
Various fungal antigen assays have been developed recently for the diagnosis of deep-seated mycosis, and several assays have been applied clinically (6, 13, 18). Even though patients with disseminated trichosporonosis may test positive for the glucuronoxylomannan (GXM) antigen (9, 10) or (1→3)-β-d-glucan (11), patients with other mycoses also test positive in these assays (12), and therefore the specificity of these tests is not satisfactory. PCR has also been used for the diagnosis of deep-seated mycosis, and several attempts have been made to detect fungal DNA from blood samples (2, 5, 7, 23, 24). However, to our knowledge, there are no reports on the use of PCR for direct detection of DNA specific for Trichosporon species in serum samples.
In the present study, we developed a novel PCR assay for the detection of DNA of clinically important strains of Trichosporon species in serum samples from patients with disseminated trichosporonosis and investigated its sensitivity and specificity.
MATERIALS AND METHODS
Organisms and growth conditions.
The fungal and bacterial strains used in this study are listed in Table 1. All of the fungal strains except for Trichosporon asahii OMU239 were supplied by Teikyo University Research Center for Medical Mycology. The fungi were incubated and allowed to proliferate in Sabouraud dextrose broth for 72 h at 30°C, while the bacteria were incubated in Luria-Bertani medium for 24 h at 37°C.
TABLE 1.
Specificity of PCR for the fungal and bacterial strains used in this study
| Organism | Strain | Result bya:
|
|||
|---|---|---|---|---|---|
| PCR
|
Southern blot hybridization
|
||||
| Single | Nested | TA-p | TM-p | ||
| Fungi | |||||
| Trichosporon asahii var. asahii | TIMM 1318 | + | + | + | − |
| TIMM 1574 | + | + | + | − | |
| TIMM 1705 | + | + | + | − | |
| TIMM 1706 | + | + | + | − | |
| TIMM 2862 | + | + | + | − | |
| OMU 239 | + | + | + | − | |
| Trichosporon mucoides | TIMM 1573 | + | + | − | + |
| Trichosporon montevideense | TIMM 1979 | + | − | − | − |
| Aspergillus fumigatus | TIMM 0069 | − | − | − | − |
| Aspergillus flavus | TIMM 0058 | − | − | − | − |
| Aspergillus nidulans | TIMM 0111 | − | − | − | − |
| Aspergillus niger | TIMM 0113 | − | − | − | − |
| Aspergillus terreus | TIMM 0119 | − | − | − | − |
| Candida albicans | TIMM 1623 | − | − | − | − |
| Candida glabrata | TIMM 1064 | − | − | − | − |
| Candida parapsilosis | TIMM 0292 | − | − | − | − |
| Candida tropicalis | TIMM 0313 | − | − | − | − |
| Penicillium crustosum | TIMM 1332 | − | − | − | − |
| Cryptococcus neoformans | TIMM 0354 | + | − | − | − |
| Malassezia furfur | TIMM 1852 | + | − | − | − |
| TIMM 2782 | + | − | − | − | |
| Malassezia pachydermatis | TIMM 2783 | + | − | − | − |
| Sporobolomyces roseus | TIMM 1517 | + | − | − | − |
| Rhodotorula acheniorum | TIMM 1855 | + | − | − | − |
| Rhodotorula minuta | TIMM 1859 | + | − | − | − |
| Bacteria | |||||
| Staphylococcus aureus | MB 2786 | − | − | − | − |
| Staphylococcus epidermidis | Clinical isolate | − | − | − | − |
| Streptococcus pneumoniae | Clinical isolate | − | − | − | − |
| Klebsiella pneumoniae | ATCC 10031 | − | − | − | − |
| Pseudomonas aeruginosa | PAO 2001 | − | − | − | − |
| Escherichia coli | ATCC 39188 | − | − | − | − |
| Serratia marcescens | ATCC 8100 | − | − | − | − |
| Haemophilus influenzae | Clinical isolate | − | − | − | − |
| Proteus mirabilis | Clinical isolate | − | − | − | − |
| Enterobacter cloacae | Clinical isolate | − | − | − | − |
+, positive; −, negative.
Extraction of DNA from cultured strains.
Extraction of DNA from fungi was performed according to the method described by Yamakami et al. (23). Briefly, 50 ml of Sabouraud glucose broth in a flask was inoculated with conidia and incubated for 72 h at 30°C. The mycelial mat was transferred into a 50-ml polypropylene screw-cap tube containing six glass beads (4 mm in diameter). The tube was then immersed in liquid nitrogen for 10 s and vortexed vigorously for 20 s. In the next step, 2 ml of DNA extraction buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.15 M NaCl, 2% sodium dodecyl sulfate, and 0.5 mg of proteinase K per ml [Sigma Chemical Co., St. Louis, Mo.]) was added, and the mixture was allowed to thaw. The mixture was then incubated for 2 h at 55°C, followed by inactivation of proteinase K by heating it to 95°C for 10 min. A volume of 0.7 ml of the mixture was transferred to a 1.5-ml microcentrifuge tube, and an equal volume of phenol-chloroform (1:1) was added, followed by centrifugation at 10,000 × g for 5 min at 4°C (MRX-150; Tomy Seiko Co., Tokyo, Japan). The supernatant was transferred to a fresh tube, and the same procedure was repeated with chloroform-isoamyl alcohol (24:1). The DNA was precipitated with 2 volumes of ethanol at −20°C and centrifuged at 12,000 × g for 20 min at 4°C, and the pellet was allowed to dry. After being rinsed with 70% ethanol at 4°C, the pellet remained intact, and the sample was air dried. The extracted DNA was then suspended in 50 μl of distilled water, and 5 μl of the suspension was used for PCR. Extraction of DNA from bacteria was performed according to the method described by Sambrook et al. (16), using lysis by alkali and sodium dodecyl sulfate.
Clinical samples and DNA extraction.
The protocol was approved by the Humans Ethics Review Committee of our University, and signed consent was obtained from each subject. Clinical samples were obtained from 20 healthy subjects and 11 patients with disseminated trichosporonosis diagnosed by an immunohistochemical technique described by Tashiro et al. (19). All histological diagnoses were established by autopsy. To isolate Trichosporon species, the blood asamples were cultured on Sabouraud glucose broth at 30 and 37°C for 7 days. Trichosporon species were identified by their morphological and biochemical characteristics (19).
The GXM antigen assay and PCR were performed in a retrospective fashion. Blood samples for these assays were centrifuged at 2,500 × g for 10 min, and the sera were stored at −20°C until use. Extraction of DNA from the serum was performed according to the method described by Tokimatsu et al. (20). In the first step, a serum sample of 100 μl was combined with the same volume of a lysis buffer containing 100 mM KCl, 20 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 0.2 mg of gelatin per ml, and 0.9% polysorbate 20 (Tween 20) solution. Proteinase K was added to a final concentration of 60 μg/ml. The mixture was incubated for 60 min at 55°C. Proteinase K was then inactivated by heating of the mixture to 95°C for 10 min. The supernatant was used for PCR amplification following centrifugation at 12,000 × g for 10 min at 4°C.
Oligonucleotide primers and PCR.
The design of oligonucleotides used in the present study was based on a comparison of the sequences of 26S rRNA genes (rDNA) of Trichosporon species and other fungi available in the GenBank database. Nested PCR was performed with two sets of primers. The outer primer set consisted of TB26-1 (5′AAAGATGAAAAGCACTTTGG3′) and TB26-2 (5′AAGCCATTATGTCAACATCC3′), amplifying a 287-bp sequence. The inner primer set consisted of TB26-9 (5′AGCACTTTGGAAAGAGAG3′) and TB26-10 (5′CCTAAGCTCGAACGTGCC3′), amplifying a 259-bp sequence. The reaction mixtures used in the present series of experiments consisted of 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 0.1% oxtoxynol (Triton X-100), 160 mM (each) deoxynucleoside triphosphates (dATP, dCTP, dTTP, and dGTP), and 1 U of Thermus aquaticus (Taq) DNA polymerase (Takara Taq; Takara Shuzo Co., Otsu, Japan). In a single PCR step, 50 pmol of each outer primer was combined with 5 μl of the prepared sample to a final volume of 50 μl. The tube was covered with mineral oil to prevent evaporation, and PCR was conducted in an automatic thermal cycler (program temperature control system PC-700; Astec, Co., Fukuoka, Japan). The amplification reactions were run for 30 cycles of DNA denaturation at 94°C for 1 min, primer annealing at 55°C for 2 min, and DNA extension at 72°C for 2 min. The final cycle of DNA extension was carried out at 72°C for 10 min. In the nested-PCR step, 1 μl of the first amplification product was added to a new reaction mixture with 50 pmol of each inner primer and reamplified with 30 cycles as described above, except for the primer annealing step. This step was carried out at 63°C for 2 min. To avoid possible contamination of PCR mixtures, all reactions were performed under stringent conditions, as recommended by Kwok and Higuchi (8). Three negative controls, including control reagents and sera from healthy subjects, were run along with the test samples for all reactions. The nested-PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide, and the results were photographed.
Southern blot hybridization.
Gel-fractionated nested-PCR products were transferred onto nylon membranes (Hybond N+; Amersham International, Buckinghamshire, United Kingdom) and then hybridized with a DNA probe specific for T. asahii, TA-p (5′TAGTAGGAATGTGACTTCTCCGGAA3′), or specific for T. mucoides, TM-p (5′ATAGTAGGAATGTAGCTCCCCCGGG3′), labeled with an enhanced chemiluminescence detection system (ECL 3′-oligolabeling and detection system; Amersham). The membranes were washed according to the instructions provided by the manufacturer and exposed to X-ray films for 60 min.
Glucuronoxylomannan antigen assay.
Circulating GXM antigen was detected by a latex agglutination test, the Serodirect “Eiken” Cryptococcus test (Eiken Kagaku Co., Tokyo, Japan). Assays were performed according to the instructions provided by the manufacturer. Briefly, the serum sample was incubated at 56°C with a serum treatment solution for 30 min. After being heated at 95°C for 5 min, the mixture was tested with latex beads.
RESULTS
Specificity of nested PCR for Trichosporon species.
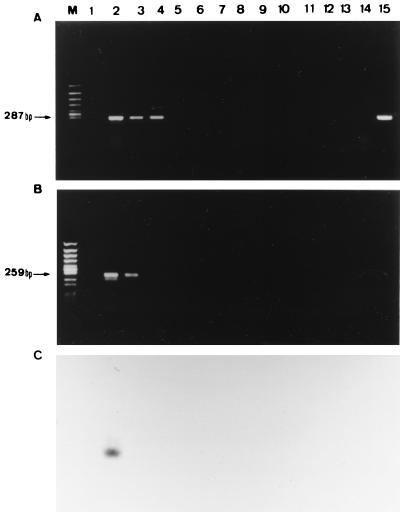
DNA samples from the organisms tested were examined to see whether the primer amplified the same DNA products. Using a single PCR with outer primers, a specific 287-bp fragment was amplified from all Trichosporon tested species (T. asahii, T. mucoides, and T. montevideense) and other basidiomycetous fungi tested but not from other microorganisms. In nested PCR with inner primers, a specific 259-bp fragment was amplified from T. asahii and T. mucoides, but not from other microorganisms, including other basidiomycetous fungi tested (T. montevideense, Cryptococcus neoformans, and so on) (Table 1 and Fig. 1).
FIG. 1.
Specificity of PCR assay with DNA from various pathogenic fungi. (A) Single PCR assay. (B) Nested-PCR assay. (C) Southern blot hybridization with TA-p probe following nested PCR. Lanes: M, molecular size marker (Φ174/HincII digest); 1, distilled water; 2, T. asahii; 3, T. mucoides; 4, T. montevideense; 5, A. fumigatus; 6, A. flavus; 7, A. nidulans; 8, A. niger; 9, A. terreus; 10, C. albicans; 11, C. glabrata; 12, C. parapsilosis; 13, C. tropicalis; 14, P. crustosum; 15, C. neoformans.
Sensitivity of nested PCR for Trichosporon species.
We used a T. asahii DNA, extracted as described above, to evaluate the sensitivity of nested PCR. Serial 10-fold dilutions of the gene were prepared, and each dilution was then subjected to amplification by PCR. Five hundred femtograms of target DNA was detected by a single PCR, whereas as little as 5 fg was detected by the nested PCR, indicating that the sensitivity of nested PCR was 100 times higher than that of the single PCR. The method involving nested PCR with a gel detection system was as sensitive as the following hybridization method (Fig. 2).
FIG. 2.
Sensitivity of PCR assay with DNA from T. asahii. (A) Single PCR assay. (B) Nested-PCR assay. (C) Southern blot hybridization with TA-p probe following nested PCR. Lanes: M, molecular size marker (Φ174/HincII digest); 1, distilled water; 2, 5 × 10−9 g of DNA; 3, 5 × 10−10 g; 4, 5 × 10−11 g; 5, 5 × 10−12 g; 6, 5 × 10−13 g; 7, 5 × 10−14 g; 8, 5 × 10−15 g; 9, 5 × 10−16 g; 10, 5 × 10−17 g; 11, 5 × 10−18 g.
Southern blot hybridization for Trichosporon species.
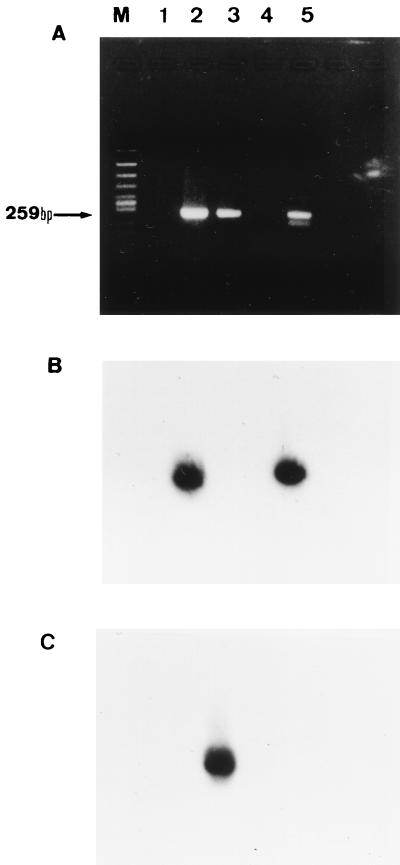
As shown in Fig. 3, we were able to differentiate between T. asahii and T. mucoides by Southern blot hybridization performed after the nested PCR by using DNA probes specific for each of these two species.
FIG. 3.
Southern blot hybridization of the nested-PCR products. (A) Nested PCR. (B) Southern blot hybridization with TA-p probe following nested PCR. (C) Southern blot hybridization with TM-p probe following nested PCR. Lanes: M, molecular size marker (Φ174/HincII digest); 1, distilled water; 2, T. asahii; 3, T. mucoides; 4, T. montevideense; 5, clinical isolate of T. asahii.
Detection of DNA specific for Trichosporon species in clinical samples.
A summary of the results of blood culture, GXM antigen assays, and nested PCR using serum samples of patients with a histological diagnosis of disseminated trichosporonosis is shown in Table 2. The detection sensitivity levels of these three assays were as follows: 72% (8 of 11) for blood cultures, 54% (6 of 11) for the GXM antigen assay, and 64% (7 of 11) for nested PCR. All isolates of Trichosporon species were further identified as T. asahii var. asahii by the technique based on the DNA-DNA homology of Trichosporon species. The results of a Southern blot hybridization that was performed after nested PCR also showed that all seven PCR-positive patients had T. asahii infection (data not shown). All samples from healthy subjects were negative in these three assays. The results of nested PCR became available a few days to a few weeks earlier than those of blood cultures for five subjects (data not shown).
TABLE 2.
Comparison of results of blood culture, nested PCR, and GXM assay with serum samples from patients with disseminated trichosporonosis
| Patient no. | Sex/age (yr)a | Underlying diseaseb | Result byc:
|
||
|---|---|---|---|---|---|
| Blood culture | GXM antigen | Nested PCR | |||
| 1 | M/55 | AML | + | + | + |
| 2 | M/73 | ALL | + | + | + |
| 3 | F/55 | MDS | + | + | + |
| 4 | M/59 | AML | + | + | + |
| 5 | M/73 | AML | + | + | + |
| 6 | M/60 | AML | + | + | − |
| 7 | M/62 | MM | + | − | − |
| 8 | M/71 | CLL | + | − | + |
| 9 | F/53 | CML | − | − | − |
| 10 | M/59 | ML | − | − | + |
| 11 | F/67 | MM | − | − | − |
M, male; F, female.
AML, acute myelogenic leukemia; ALL, acute lymphoblastic leukemia; MDS, myelodysplastic syndrome; MM, multiple myeloma; CLL, chronic lymphoblastic leukemia; CML, chronic myelogenic leukemia; ML, malignant lymphoma.
+, positive; −, negative.
For the screening of PCR inhibitors in samples, Trichosporon DNA-negative samples were tested to detect human genomic DNA (β-globin DNA) by nested PCR according to the methods described by Saiki et al. (15), and these samples were positive for β-globin DNA. On the basis of these results, it was considered that there were no PCR inhibitors in these samples (data not shown).
DISCUSSION
Deep-seated trichosporonosis is a lethal opportunistic infection associated with morbid severe conditions in immunocompromised patients, disseminating to various organs, such as the lungs, kidneys, digestive tract, adrenal gland, and central nervous system (19). Even though disseminated trichosporonosis is resistant to antifungal agents such as amphotericin B (21), combination therapy consisting of a recombinant human G-CSF and an azole antifungal agent (in particular, fluconazole) may be effective (14). However, due to the lack of rapid and reliable methods for early diagnosis of disseminated trichosporonosis, it has been difficult to institute therapy at the early stages of the disease. Disseminated trichosporonosis was previously diagnosed by mycological, histopathological, or serodiagnostic tests. However, since Trichosporon species colonize and proliferate in the oral cavity, digestive tract, or urinary tract or on the skin, detection of Trichosporon species in the sputum, feces, and urine is unreliable. Nonetheless, when a Trichosporon species is detected in an abscess or aseptic specimens, such as blood, spinal fluid, or pleural effusion, the detection of such fungi is clinically significant. However, a long period of time is needed before the results of the diagnostic tests mentioned above become available. Because of these delays in diagnosis, trichosporonosis is sometimes diagnosed after the patient has died. Even though trichosporonosis can be diagnosed by an immunohistological assay using monoclonal antibodies, invasive examinations such as biopsy cannot often be performed in patients with severe underlying diseases. Furthermore, serological diagnosis also has certain disadvantages. For example, Trichosporon species and C. neoformans are known to share common antigens. While a latex agglutination test is used for the diagnosis of cryptococcosis, patients with deep-seated trichosporonosis are also found positive by this test (9, 10). Consequently, trichosporonosis and cryptococcosis cannot be distinguished by this assay. Furthermore, (1→3)-β-d-glucan, a common component of the cell wall of fungi, can be detected in patients with disseminated trichosporonosis (11), but since this compound can be detected in patients with almost all deep-seated mycotic diseases, except for mucormycosis (12), assays for the detection of (1→3)-β-d-glucan lack specificity for trichosporonosis. Trichosporon cutaneum was reported as the causative agent of disseminated trichosporonosis, but due to the molecular evolution classification system, based on DNA-DNA homology, introduced by Gueho et al. in 1992, T. cutaneum was reclassified into 20 species (4). In the past, serum types I and II were classified as the causative agents of deep-seated mycosis, but after the new classification, they were identified as T. mucoides and T. asahii, respectively (17). The new primers developed in the present study were capable of specifically detecting these two species of Trichosporon. Therefore, unlike the GXM antigen assay, the nested-PCR assay distinguished between clinically important Trichosporon species and other basidiomycetous fungi, including T. montevideense (serum type III, which is not a usual causative agent of deep-seated mycosis).
Furthermore, the two DNA probes (TA-p and TM-p) used in the present study distinguished T. asahii from T. mucoides. Trichosporon species are resistant to certain antifungal agents, such as amphotericin B (21). However, Anaissie et al. reported that the activity of amphotericin B varied, with some strains exhibiting a good in vivo response and others showing resistance to this agent, suggesting this heterogeneity is based on differences among species of Trichosporon (1). Therefore, implementation of molecular techniques used in previous studies (4, 17) and the present investigation would be also helpful for the development of new antifungal agents against certain species of Trichosporon.
Our results with clinical samples from 11 patients with histologically diagnosed disseminated trichosporonosis showed that 7 patients (64%) were positive for nested PCR. In two patients who had positive blood cultures but who were negative for nested PCR (patients 6 and 7), T. asahii was first detected by a blood culture that was conducted just before death, while serum samples for PCR assays were collected 4 or 16 days before the blood culture. Therefore, the time difference in sample collection might have played an important role in the false-negative test. In patients 9 and 11, blood culture, GXM antigen assay, and nested PCR were all negative. It is possible that these results may be due to the presence of only a small number of causative fungi below the threshold level of these assays. This assumption is based on the fact that these two patients were treated with a drip infusion of amphotericin B or fluconazole for more than 10 days before samples were obtained for these diagnostic tests.
The new nested-PCR assay was performed on more than one occasion with samples from eight patients, and the assay detected the causative fungi a few days to a few weeks earlier than blood culture in five patients (data not shown). Therefore, our nested-PCR assay is useful for the early diagnosis of disseminated trichosporonosis.
In a clinical situation, Trichosporon species were also recognized to be the causative agents of summer-type hypersensitivity pneumonia and white piedra. In addition, these fungi are considered to colonize at the catheter sites. We had a chance to test samples from two patients with summer-type hypersensitivity pneumonia. These samples were negative for Trichosporon DNA by nested PCR (data not shown). Because the Trichosporon organisms usually cannot be isolated from the blood of patients with summer-type hypersensitivity pneumonia, the organisms are not considered to exist in such patients’ blood. Therefore, the sera from these patients should be negative for Trichosporon DNA. In this study, we could not test the samples from patients with white piedra and catheter site colonization, because we did not have serum from such patients. Recently, Walsh et al. reported cases of development of disseminated aspergillosis from primary cutaneous aspergillosis (22). There is a possibility that white piedra also advances to disseminated trichosporonosis, so we think that, in patients with white piedra, the PCR might be positive. On the other hand, we consider that PCR should be positive with sera from patients with venous catheter site colonization because of the existence of the components, including DNA, of Trichosporon in the blood vessels. Therefore, the PCR method might be positive with the serum of some patients with nondisseminated trichosporonosis. However, we believe that the results of the nested PCR should provide very helpful information for diagnosing disseminated trichosporonosis when the other clinical data are combined.
In conclusion, we have described the development of a novel PCR assay for the detection of clinically important species of Trichosporon in serum samples from patients with disseminated trichosporonosis. Detection of DNA specific for Trichosporon species with this method requires only a serum sample and can be repeated several times during the course of the disease. Further studies are necessary to prospectively evaluate our new PCR with a large clinical population sample.
ACKNOWLEDGMENTS
We thank Takako Shinoda, Department of Microbiology, Meiji College of Pharmacy, for performing identification of Trichosporon species by a technique based on DNA-DNA homology. We also thank Kiyoko Arita, Second Department of Internal Medicine, Oita Medical University, for preparing fungal strains and F. G. Issa, Department of Medicine, University of Sydney, for assistance with reading and editing the manuscript.
REFERENCES
- 1.Anaissie E J, Hachem R, Karyotakis N C, Gokaslan A, Dignani M C, Stephens L C, Tin-U C K. Comparative efficacies of amphotericin B, triazoles, and combination of both as experimental therapy of murine trichosporonosis. Antimicrob Agents Chemother. 1994;38:2541–2544. doi: 10.1128/aac.38.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grauer M E, Bokemeyer C, Bautsch W, Freund M, Link H. Successful treatment of a Trichosporon beigelii septicemia in granulocytopenic patients with amphotericin B and granulocyte colony-stimulating factor. Infection. 1994;22:238–286. doi: 10.1007/BF01739918. [DOI] [PubMed] [Google Scholar]
- 4.Gueho E, de Hoog G S, Billon-Grand G, Christen R, Batenburg van der Vegte W H. Contributions to a revision of genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto A, Yamakami Y, Kamberi P, Yamagata E, Karashima R, Nagaoka H, Nasu M. Comparison of PCR, (1→3)-β-d-glucan and galactomannan assays in sera of rats with experimental invasive aspergillosis. J Clin Lab Anal. 1998;12:257–262. doi: 10.1002/(SICI)1098-2825(1998)12:5<257::AID-JCLA1>3.0.CO;2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herent P, Stynen D, Hernando F, Fruit J, Poulain D. Retrospective evaluation of two latex agglutination tests for detection of circulating antigens during invasive candidosis. J Clin Microbiol. 1992;30:2158–2164. doi: 10.1128/jcm.30.8.2158-2164.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes A R, Cannon R D, Shepherd M G, Jenkinson H F. Detection of Candida albicans and other yeasts in blood by PCR. J Clin Microbiol. 1994;32:228–231. doi: 10.1128/jcm.32.1.228-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 9.McManus E J, Jones J M. Detection of a Trichosporon beigelii antigen cross-reactive with Cryptococcus neoformans capsular polysaccharide in serum from a patient with disseminated Trichosporon infection. J Clin Microbiol. 1985;21:681–685. doi: 10.1128/jcm.21.5.681-685.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melcher G P, Reed K D, Rinaldi M G, Lee J W, Pizzo P A, Walsh T J. Demonstration of a cell wall antigen cross-reacting with cryptococcal polysaccharide in experimental disseminated trichosporonosis. J Clin Microbiol. 1991;29:192–196. doi: 10.1128/jcm.29.1.192-196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazaki T, Kohno S, Mitsutake K, Maesaki S, Tanaka K, Hara K. (1→3)-β-d-Glucan in culture fluid of fungi activates factor G. A limulus coagulation factor. J Clin Lab Anal. 1995;9:334–339. doi: 10.1002/jcla.1860090509. [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki T, Kohno S, Mitsutake K, Maesaki S, Tanaka K-I, Ishikawa N, Hara K. Plasma (1→3)-β-d-glucan and fungal antigenemia in patients with candidemia, aspergillosis, and cryptococcosis. J Clin Microbiol. 1995;33:3115–3118. doi: 10.1128/jcm.33.12.3115-3118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson T F, Miniter P, Patterson J E, Rappaport J M, Andriole V T. Aspergillus antigen detection in the diagnosis of invasive aspergillosis. J Infect Dis. 1995;171:1553–1558. doi: 10.1093/infdis/171.6.1553. [DOI] [PubMed] [Google Scholar]
- 14.Perparim K, Hashimoto A, Nasu M. Efficacy of recombinant human granulocyte-colony stimulating factor alone and in combination with anti-fungal agents against disseminated trichosporonosis in neutropenic mice. J Infect Chemother. 1996;2:232–239. doi: 10.1007/BF02355120. [DOI] [PubMed] [Google Scholar]
- 15.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Sugita T, Nishikawa A, Shinoda T, Kume H. Taxonomic position of deep-seated, mucosa-associated and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J Clin Microbiol. 1995;33:1368–1370. doi: 10.1128/jcm.33.5.1368-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanner D C, Weinstein M P, Fedorciw B, Jono K L, Thorpe J J, Reller L B. Comparison of commercial kits for detection of cryptococcal antigen. J Clin Microbiol. 1994;32:1680–1684. doi: 10.1128/jcm.32.7.1680-1684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tashiro T, Nagai H, Kamberi P, Goto Y, Kikuchi H, Nasu M, Akizuki S. Disseminated Trichosporon beigelii infection in patients with malignant disease, immuno-histochemical study and review. Eur J Microbiol Infect Dis. 1994;13:218–224. doi: 10.1007/BF01974540. [DOI] [PubMed] [Google Scholar]
- 20.Tokimatsu I, Tashiro T, Nasu M. Early diagnosis and monitoring of human cytomegalovirus pneumonia in patients with adult T-cell leukemia by DNA amplification in serum. Chest. 1995;107:1924–1927. doi: 10.1378/chest.107.4.1024. [DOI] [PubMed] [Google Scholar]
- 21.Walsh T J, Melcher G P, Rinaldi M G, Lecciones J, McGough D A, Kelly P, Lee J, Callender D, Rubin M, Pizzo P A. Trichosporon beigelii, an emerging pathogen resistant to amphotericin B. J Clin Microbiol. 1990;28:1616–1622. doi: 10.1128/jcm.28.7.1616-1622.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh T J. Primary cutaneous aspergillosis—an emerging infection among immunocompromised patients. Clin Infect Dis. 1998;27:453–457. doi: 10.1086/514718. [DOI] [PubMed] [Google Scholar]
- 23.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamakami Y, Hashimoto A, Yamagata E, Kamberi P, Karashima R, Nagai H, Nasu M. Evaluation of PCR for detection of DNA specific for Aspergillus species in sera of patients with various forms of pulmonary aspergillosis. J Clin Microbiol. 1998;36:3619–3623. doi: 10.1128/jcm.36.12.3619-3623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]