Abstract
Introduction
The pandemic of a novel coronavirus disease 2019 (COVID-19) caused by a severe acute respiratory coronavirus 2 (SARS-CoV-2) infection has been problematic worldwide. A new SARS-CoV-2 diagnostic test (SmartAmp) was licensed in Japan in July 2021. This method, which enables us to diagnose COVID-19 as well as a gene mutation on the virus, is promising to reduce medical costs and staff labor.
Patients and methods
To analyze the diagnostic accuracy of the SmartAmp assay for diagnosing COVID-19, we performed this retrospective study at our institute during April and May 2021. We compared the results of the SmartAmp assay and real-time reverse transcription-polymerase chain reaction (rRT-PCR) using a saliva sample from individuals suspected as having COVID-19.
Results
Out of 70 samples tested, the SmartAmp assay had 50 (71%) positive and 20 (29%) negative results. Using rRT-PCR as a reference, the diagnostic accuracy displayed a sensitivity of 84%, a specificity of 95%, a positive predictive value of 97.7%, and a negative predictive value of 70.4%. On the other hand, false-negative cases were found in 7 (10%), and there was no significant difference of Ct-value between true positive and false negative cases (Mean Ct-value 25.2 vs. 27.5 cycles, p = 0.226 by Mann-Whitney U test).
Conclusion
The SmartAmp assay is a valuable method to diagnose COVID-19 rapidly. However, the negative predictive value is not high enough to diagnose the disease, so that negative results should be considered for rRT-PCR testing if patients are suspected of having COVID-19.
Keywords: SmartAmp, COVID-19, SARS-CoV-2, Reverse transcription-polymerase chain reaction, Coronavirus
The emergence of a novel coronavirus in Wuhan, China, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the pandemic of COVID-19 (coronavirus disease 2019) cause health and economic crises and reveal human relations problems such as racism and conflict between nations [[1], [2], [3]]. In Japan, we face several problems; the emergence of social distancing enforcers, discrimination against medical staff who provide medical care for the patients with COVID-19, misleading information by the mass media regarding COVID-19 vaccines, and hosting the Tokyo Olympics 2020 during the pandemic of COVID-19. COVID-19 highlights various problems in Japan, which must be overcome. After a vaccination program started in the USA, the number of patients with COVID-19 dramatically decreased (as of 1:07 p.m. CET, 5 July 2021. https://covid19.who.int/ ). On the other hand, Japan is still facing several problems, including a vaccination program that has been delayed compared to other countries, resulting in the possible fifth wave of COVID-19 infections. This will impact the Tokyo Olympics and can lead to a collapse of the medical care system. Although COVID is becoming a community infection, we have no rapid diagnostic method that is easy to handle everywhere. Thus, we have an issue with whether the patient should be isolated or not. While the diagnosis of COVID-19 has been a gold standard by reverse-transcription-polymerase chain reaction (RT-PCR), it is costly and requires a trained laboratory technician and medical equipment to perform, taking 3–4 h per assay [4,5]. With an increase in numbers for COVID-19 testing, although rapid antigen testing has prevailed widely, there is no reliable evidence to support it.
The SmartAmp method is an isothermal DNA amplification technology for rapid detection, which enables us to detect genetic polymorphisms or mutations in 25–45 min after preprocessing under isothermal conditions without the need for DNA isolation or PCR amplification. Some had already reported its utility for detecting a single nucleotide polymorphism (SNP). Besides, the method has prevailed in several fields of medicine [[6], [7], [8]]. SmartAmpSARS-CoV-2 kit (DANAFORM Inc., Tokyo, Japan) was commercialized to diagnose COVID-19 in July 2021 in Japan. We evaluated the efficacy and validity of the SmartAmp assay for the diagnosis of COVID-19. This is the first report documenting that the SmartAmp assay would be a proper diagnostic method for diagnosing COVID-19.
We collected 70 saliva clinical specimens from individuals from April 2021 until May 2021 at Aichi Medical University Hospital and affiliated facilities. All patients were suspected of having COVID-19 based on their clinical symptoms (within nine days from the onset) or met the definition of close contact with COVID-19 patients. Saliva was collected based on the standard protocol in the same manner as the previous study [9]. Using the samples, we performed rRT-PCR as well as SmartAmp assay in diagnosing COVID-19. Then, we analyzed the diagnostic characteristics such as diagnostic accuracy and ROC curves, threshold cycle (Ct) value of the rRT-PCR, and compared the results of the two methods. This study was approved by the Institutional Review Board of Aichi Medical University Hospital (H17-106).
As for the SmartAmp assay, we collected saliva samples from individuals who were suspected of having COVID-19 and transferred the samples to ReproCELL Inc. (Kanagarwa, Japan) to outsource the SmartAmp assay. rRT-PCR was performed by using BD MAX system (a fully integrated, automated platform that performs nucleic acid extraction and real-time PCR) (Japan Becton Dickinson and Company, Japan). The re-suspended saliva was centrifuged at 500×g for 1 min, and the volumes of 750 μL supernatant fluid were assayed on the BD MAX system using the BD SARS-CoV-2 reagents for BD MAX System. In these reagents, the primer and double-quencher probe sets were based on the United States Centers for Disease Control and Prevention (US CDC) assay for specific detection of SARS-CoV-2 by amplifying two unique regions of the N gene (i.e., N1 and N2), and the human RNase P gene as an internal control.
Statistical analyses were performed using GraphPad Prism version 9.1.0 for Mac, GraphPad Software, La Jolla California USA.) P-values<0.05 were considered statistically significant.
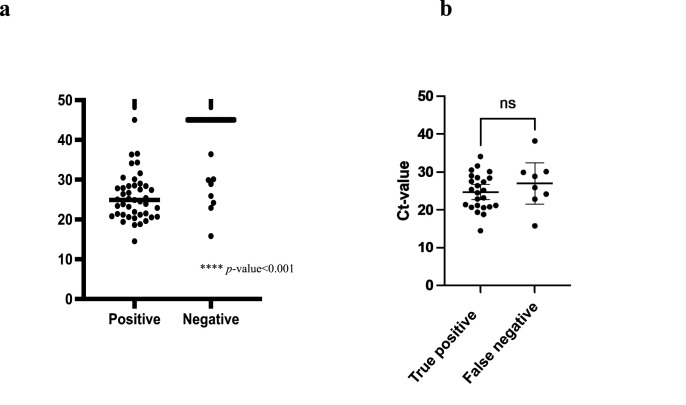
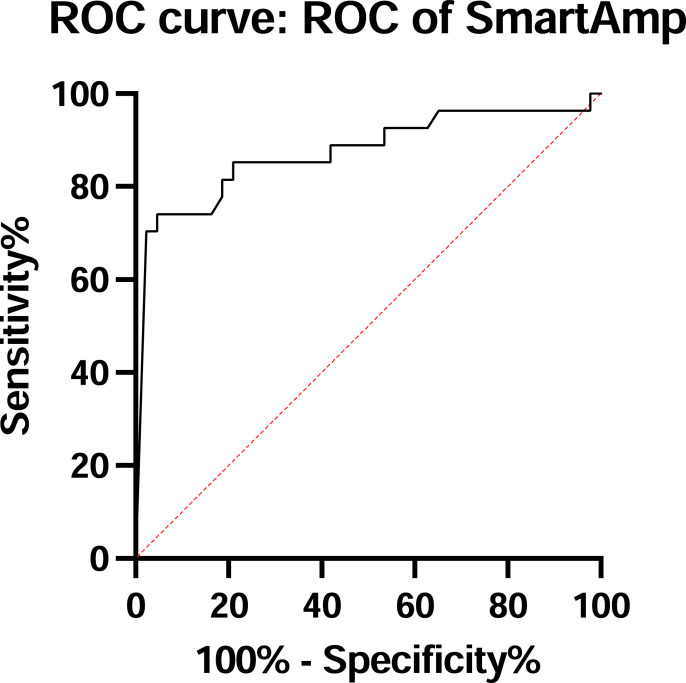
A total of 70 saliva samples were tested. The rRT-PCR revealed 50 (71%) positive and 20 (29%) negative results, while The SmartAmp assay showed 43 (61%) positive and 27 (39%) negative cases. We had one false-positive (positive-SmartAmp assay, but negative-rRT-PCR) and seven false-negative (negative-SmartAmp assay, but positive-rRT-PCR). A false-positive sample exhibited a 40 Ct-value. False-negative samples revealed the median Ct-value of rRT-PCR was 27.6 cycles (range 15.8–38.2 cycles). Comparing the mean Ct-value between the positive and negative samples, the mean Ct-value in the positive samples was lower than in the negative ones (25.6 vs. 39.6 cycles, p < 0.001 by Mann-Whitney U test) (Fig. 1 a). Using the result of rRT-PCR as a reference, the area under the receiver-operating characteristic (AUROC) curve is 0.891 [p < 0.001, 95% confidence interval (CI) 0.8–0.981] as shown in Fig. 2 . SmartAmp assay had a sensitivity of 84%, a specificity of 95%, a positive predictive value of 97.7%, and a negative predictive value of 70.4%, as shown in Table 1 . As for a gene mutated expression on true positive samples, N501Y and E484K were seen in 35 (83%) and 7 (17%), respectively (data not shown).
Fig. 1.
shows the comparison of threshold cycle values of rRT-PCR between true positive and negative cases (Fig. 1a), and between true positive and false negative cases (Fig. 1b) by SmartAmp assay.
Fig. 2.
Shows the area under receiver-operating characteristic curve for the diagnosis of COVID-19, using the result of rRT-PCR as a reference.
Table 1.
Diagnostic accuracy of RT-SmartAmp assay for the diagnosis of COVID-19.
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | YI |
|---|---|---|---|---|
| All cases (n = 70) | ||||
| 84 | 95 | 97.7 | 70.4 | 0.79 |
| Cases showing Ct-value≥27 (n = 39) | ||||
| 78.9 | 95 | 93.8 | 82.6 | 0.74 |
COVID-19, coronavirus disease 2019; Ct, threshold cycle; PPV, positive predictive value; NPV, negative predictive value; YI, Youden index.
The results showed that the Smart-Amp assay is effective and valid for diagnosing COVID-19 using saliva specimens. Moreover, obtaining saliva samples is safer and more accessible than nasopharyngeal samples to prevent secondary transmission from patients to medical staff [3,5,6,9]. Although the gold standard diagnostic testing for COVID-19 is rRT-PCR, SmartAmp assay has several advantages in managing the COVID-19 patients. This method can reduce medical costs, showing a lower cost of 1980 Japanese yen (JPY), which is about 18 USD per sample [7], while PCR is more costly, showing about 250–300 USD per sample in Japan [5]. It takes 25–45 min after preprocessing to diagnose the disease, which is much shorter than rRT-PCR. We currently have an issue regarding the emergence of gene mutations on SARS-CoV-2, contributing to a reduction in the efficacy of vaccination [10,11]. It might be helpful to detect a gene mutation for surveillance and general practice, even though there is no evidence that a gene mutation affects the prognosis among the patients [[12], [13], [14]].
As for the diagnostic values, the diagnostic accuracy of the testing displayed a high positive predictive value. On the other hand, false-negative cases were found in 7 (10%), showing a low negative predictive value of 70.4%, as shown in Table 1. We previously reported that the automated quantitative CLEIA antigen test by saliva sample is helpful for the diagnosis of COVID-19. The diagnostic accuracy of the testing in patients with Ct ≧ 27 cycles is lower than those with Ct < 27 cycles [5]. The SmartAmp assay can show a false-negative result, even though the Ct-value ranged 15.8 to 38.2 cycles. Comparing the Ct-values between true positive and false negative cases, there was no significant difference between the two groups (Mean Ct-value 25.2 vs. 27.5 cycles, p = 0.226 by Mann-Whitney U test as shown in Fig. 1b). Therefore, clinicians should be aware that the diagnostic method is helpful, but its value is limited. Otherwise, we might miss false-negative cases that can be super spreaders in our society. Although using a saliva sample is an excellent tool to diagnose COVID-19 early, we should be aware that saliva has several factors to inhibit a reaction of amplifying RNA [15]. Thus, collecting a saliva sample for the SmartAmp method should be proper for avoiding an error result. Besides, further study regarding the correlation between the disease onset and diagnostic accuracy should be needed.
There are several limitations to this study. First, we tested only for SARS-CoV-2 from saliva samples and not for co-infection with any other viruses. Viral co-infection could happen, even though it is infrequent. Second, there was no clinical information about the patients. Therefore, we could not evaluate any correlation between clinical manifestations and Ct-value.
We conclude that the SmartAmp assay is a valuable method for the diagnosis of COVID-19. However, the negative predictive value is not reliable enough to diagnose the disease, so that negative results should be considered for performing rRT-PCR on patients if they are suspected of having COVID-19.
Author contributions
NA, AN and HM designed this study. NA drafted this manuscript. AN, DS, HS, YK, NM, TO, AY, SC, IK, and HO performed rRT-PCR. NA, DS, WO, AN, HK, MH,YS, AS and YY contributed to the data collection and data analysis. HM supervised the study. All of co-authors reviewed, and edited the final manuscript.
Declaration of competing interest
H. Mikamo received grant support from Asahi Kasei Pharma Corporation, Shionogi & Co. Ltd., Daiichi Sankyo Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., Pfizer Japan Inc. and FUJIFILM Toyama Chemical Co., Ltd., payment for lectures from Astellas Pharma Inc., MSD K·K., Daiichi Sankyo Co., Ltd., Sumitomo Dainippon Pharma Co., Ltd., MIYARISAN Pharmaceutical Co., Ltd. Becton, Dickinson and Company Japan, and FUJIFILM Toyama Chemical Co. Ltd. The other authors declare that they have no conflicts of interest.
Acknowledgments
We are grateful for the diligent and thorough critical reading of our manuscript by Dr. Yoshihiro Ohkuni, Chief Physician, Taiyo and Mr. John Wocher, Advisor, Kameda Medical Center (Japan).
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai N., Sakanashi D., Nakamura A., Kishino T., Kato H., Hagihara M., et al. Clinical manifestations and radiological features by chest computed tomographic findings of a novel coronavirus disease-19 pneumonia among 92 patients in Japan. J Microbiol Immunol Infect. 2020;S1684–1182(20) doi: 10.1016/j.jmii.2020.07.011. 30168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C.C., Wang C.Y., Ko W.C., Hsueh P.R. In vitro diagnostics of coronavirus disease 2019: technologies and application. J Microbiol Immunol Infect. 2020;54:164–174. doi: 10.1016/j.jmii.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asai N., Sakanashi D., Ohashi W., Nakamura A., Kawamoto Y., Miyazaki N., et al. Efficacy and validity of automated quantitative chemiluminescent enzyme immunoassay for SARS-CoV-2 antigen test from saliva specimen in the diagnosis of COVID-19. J Infect Chemother. 2021;27:1039–1042. doi: 10.1016/j.jiac.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa T., Hayashizaki Y. Clinical SNP detection by the SmartAmp method. Methods Mol Biol. 2013;1015:55–69. doi: 10.1007/978-1-62703-435-7_3. [DOI] [PubMed] [Google Scholar]
- 7.Enokida Y., Shimizu K., Atsumi J., Lezhava A., Tanaka Y., Kimura Y., et al. Rapid detection of SNP (c.309T>G) in the MDM2 gene by the Duplex SmartAmp method. PLoS One. 2013;4 doi: 10.1371/journal.pone.0060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai Y., Kimura Y., Lezhava A., Kanamori H., Usui K., Hanami T., et al. One-step detection of the 2009 pandemic influenza A(H1N1) virus by the RT-SmartAmp assay and its clinical validation. PLoS One. 2012;1 doi: 10.1371/journal.pone.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakanashi D., Asai N., Nakamura A., Miyazaki N., Kawamoto Y., Ohno T., et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J Infect Chemother. 2021;27:126–129. doi: 10.1016/j.jiac.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyakawa K., Jeremiah S.S., Ohtake N., Matsunaga S., Yamaoka Y., Nishi M., et al. Rapid quantitative screening assay for SARS-CoV-2 neutralizing antibodies using HiBiT-tagged virus-like particles. J Mol Cell Biol. 2021;12:987–990. doi: 10.1093/jmcb/mjaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021:6538. doi: 10.1126/science.abg3055. eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021 May;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 14.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;7858:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uribe-Alvarez C., Lam Q., Baldwin D.A., Chernoff J. Low saliva pH can yield false positives results in simple RT-LAMP-based SARS-CoV-2 diagnostic tests. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250202. [DOI] [PMC free article] [PubMed] [Google Scholar]