Summary
The health and homeostasis of skeletal muscle are preserved by a population of tissue-resident muscle stem cells (MuSCs) that maintain a state of mitotic and metabolic quiescence in adult tissues. The capacity of MuSCs to preserve the quiescent state declines with aging and metabolic insults, promoting premature activation and stem cell exhaustion. Sestrins are a class of stress-inducible proteins that act as antioxidants and inhibit the activation of the mammalian target of rapamycin complex 1 (mTORC1) signaling complex. Despite these pivotal roles, the role of Sestrins has not been explored in adult stem cells. We show that SESTRIN1,2 loss results in hyperactivation of the mTORC1 complex, increased propensity to enter the cell cycle, and shifts in metabolic flux. Aged SESTRIN1,2 knockout mice exhibited loss of MuSCs and a reduced ability to regenerate injured muscle. These findings demonstrate that Sestrins help maintain metabolic pathways in MuSCs that protect quiescence against aging.
Keywords: satellite cells, aging, reactive oxygen species, oxidative stress, mTORC1, metabolism, regeneration, RNA sequencing
Highlights
-
•
Sestrin deficiency alters mTORC1 signaling in MuSCs
-
•
In young mice, Sestrin knockout alters metabolism of MuSCs
-
•
Sestrin deficiency accelerates age-dependent loss and dysfunction of MuSCs
Age-dependent changes in metabolism supportive of quiescence are underexplored in adult stem cells. Aguilar and colleagues show that loss of Sestrins, a class of stress-inducible proteins with antioxidant functions, upregulates mTORC1 activity in muscle stem cells (MuSCs) and promotes premature activation and exhaustion of the MuSC compartment. Loss of Sestrins with aging exacerbates these effects, resulting in reduced protection against accumulated metabolic insults.
Introduction
Tissue-resident stem cells are crucial regulators of homeostasis that resist perpetual activation through mitotic quiescence, a reversible restraint of entry into the G0 phase of the cell cycle. After injury, tissue-specific stem cells leave the quiescent state, activate, and proliferate, generating either committed progenitors that differentiate to repair tissue or return toward quiescence to replenish the stem cell pool (Cheung and Rando, 2013). An excellent example of this process is in skeletal muscle, a post-mitotic tissue supported by muscle stem cells (MuSCs) (Lepper et al., 2011), also known as satellite cells, that reside between the basal lamina and sarcolemma. After activation, myogenic progenitors (myoblasts) fuse with themselves and existing myofibers to regenerate and repair damaged tissue (Tierney and Sacco, 2016). MuSC quiescence is critically determined both by internal mechanisms, such as chromatin state (Beerman and Rossi, 2015; Shcherbina et al., 2020) and external communication with the extracellular matrix (Xin et al., 2016) and other cell types (Feige et al., 2018). Defects in the balance between quiescence and activation have been observed in cachexia (Baracos et al., 2018), sarcopenia (Chakkalakal et al., 2012), and in diseases, such as Duchenne muscular dystrophy (Chang et al., 2016; Feige et al., 2018), highlighting a clinical need to understand the molecular mechanisms by which healthy MuSCs regulate quiescence and signaling cues that promote activation (Yang et al., 2021).
A critical yet underexplored element of quiescence regulation in adult stem cells is the maintenance of metabolic homeostasis. Quiescent MuSCs utilize fatty acid oxidation (FAO) in quiescence but increases in stressors, such as reactive oxygen species (ROS), DNA damage (Cheung and Rando, 2013; Sperka et al., 2012), and inflammation, shift metabolism away from FAO toward glycolysis, which in turn prompts MuSCs to activate and leave the quiescent state (Ryall et al., 2015). Several genes influence quiescence and FAO, including the forkhead box O (FOXO) family of transcription factors, which maintains bioenergetic demands (Eijkelenboom and Burgering, 2013) and nutrient consumption in MuSCs by inhibiting oxidative stress and activation of the mammalian target of rapamycin complex (mTORC) signaling through the phosphatidylinositide 3-kinase (PI3K)/AKT pathway (Briata et al., 2012; Serra et al., 2007; Touil et al., 2013; Yu and Cui, 2016). Age-induced deterioration of quiescence and hyperactivation of both the PI3K/AKT and mTORC pathways leads to increased stem cell turnover and impaired self-renewal (Chakkalakal et al., 2012; García-Prat et al., 2016; Ho et al., 2017). Inhibitors of mTORC activity (Liu and Sabatini, 2020), such as the tuberous sclerosis proteins 1 and 2 (TSC1/2) complex (Rodgers et al., 2014), and the AMP-activated protein kinase (AMPK) complex (Theret et al., 2017), have both been shown to regulate MuSC metabolism and longevity. Another mTORC inhibitor called Sestrin (Lee et al., 2010a), a set of conserved proteins encoded by Sesn1, Sesn2, and Sesn3 in mammals, mediate metabolic responses to oxidative stress by physically activating AMPK signaling and indirectly inhibiting mTORC1 pathways (Kim et al., 2015; Lee et al., 2013, 2016). In skeletal muscle, only Sesn1 and Sesn2 are expressed (Kim et al., 2020; Li et al., 2019) and Sestrin-mediated inhibition of mTORC1 and AKT pathways upregulates autophagy and downregulates anabolic pathways to maintain proteostasis and organelle quality, ultimately preserving muscle mass and force (Segalés et al., 2020). While Sestrins are implicated in muscle pathophysiology and regulate mTORC1 and aging, their role in MuSCs and stem cells in general remains unexplored.
Here, we utilized SESTRIN1,2 knockout (SKO) murine models to evaluate the influence of Sestrins on MuSC functions. Muscles of SKO mice resulted in hyperactivation of the mTORC1 complex in MuSCs, even in the absence of regenerative stimuli, accumulation of ROS, and premature entry into the cell cycle. Despite the alterations in basal stem cell status, application of a muscle injury to SKO mice revealed highly similar regenerative trajectories as wild-type (WT) matched controls. We used RNA sequencing (RNA-seq) and metabolic flux modeling to show that SKO MuSCs displayed modifications to FAO and other amino acid metabolic fluxes, which impinge on mTORC1 and reduce ability to maintain quiescence. To probe deeper into this result, we aged SKO mice and observed an accelerated age-dependent loss of MuSCs. Repeated muscle injuries to aged SKO mice showed increases in fibrosis and smaller cross-sectional areas of myofibers indicating reductions in regenerative potential. These studies demonstrate a unique role for Sestrins as arbiters of MuSC metabolism and protection against aging.
Results
Sestrin loss upregulates mTORC1 in unstimulated MuSCs
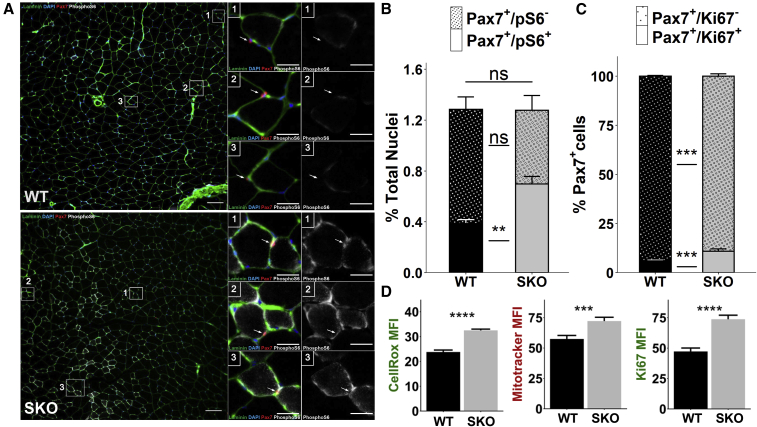
To examine the role that Sestrins play in regulating MuSCs, we isolated hindlimb muscles from 2-month-old uninjured C57BL6J (WT) and previously generated SKO mice (Kim et al., 2020; Li et al., 2019). Comparisons of myofiber size and fiber typing revealed no significant differences for SKO muscles (Figures S1A–S1C). No statistically significant change in the total number of PAX7+ MuSCs was observed between WT and SKO quadriceps (Figures 1A and 1B). However, in situ staining of PAX7 and the phosphorylated form of ribosomal protein S6 (pS6), an mTORC1 target, in quadriceps muscle revealed a substantial increase in the fraction of pS6+/PAX7+ MuSCs for SKO muscles compared with WT controls (Figures 1A and 1B). The increase in pS6+ in SKO MuSCs is associated with hyperactivation of mTORC1 and is in line with previous studies of TSC1,2 KO mice (Rodgers et al., 2014) and our data showing that Sestrins are negative regulators of mTORC1 (Kim et al., 2015, 2020). Given mTORC1 contributes to MuSC activation (Rodgers et al., 2014), we reasoned that SKO MuSCs would display reduced quiescence. To further determine if SKO MuSCs displayed reductions in quiescence, we performed in situ co-staining of Ki67 and PAX7 (Figures 1C and S1D). In line with our observations of enhanced mTORC1 activity in SKO MuSCs, we observed an increased number of Ki67+/PAX7+ cells in SKO muscles compared with WT.
Figure 1.
Loss of SESTRIN1,2 induces hyperactivation of mTORC1 signaling in MuSCs
(A) Representative in situ immunohistochemical images of whole quadriceps muscle sections from wild-type (WT, top) and SESTRIN1,2−/− (SKO, bottom) mice stained for DAPI (blue), PAX7 (red), laminin (green), and phospho-S6 (pS6, white), a marker of mTORC1 activation. Scale bars, 100 μm. Magnified images show PAX7+ cells for each condition. Arrows mark PAX7+ cells. Scale bars, 25 μm.
(B) Quantification of pS6+ (plain) and pS6− (stippled) PAX7+ cells as a percentage of all nuclei in whole WT and SKO quadriceps sections (WT, n = 3 mice; SKO, n = 3 mice). Statistical comparisons are two-sided Mann-Whitney U tests.
(C) Quantification of Ki67+ (plain) and Ki67− (stippled) PAX7+ cells in WT and SKO quadriceps muscle sections (WT, n = 4 mice; SKO, n = 4 mice). Statistical comparisons are two-sided Mann-Whitney U tests.
(D) Quantification of mean fluorescence intensity (MFI) from CellRox, MitoTracker, and Ki67 in WT and SKO MuSCs fixed immediately after isolation. Statistical comparisons are two-sided, unpaired Student's t tests. All data are shown as mean ± SEM. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figures S1 and S2.
Sestrin loss reduces ability to maintain MuSC quiescence
To probe further if Sestrin loss and associated increases in mTORC1 signaling influenced MuSC quiescence, WT and SKO hindlimb muscles (tibialis anterior, extensor digitorum longus, gastrocnemius, and quadriceps) were extracted and MuSCs isolated using fluorescent-activated cell sorting (FACS) (Cerletti et al., 2008) with both negative (SCA-1−, CD45−, MAC-1−, TER-119−) and positive surface markers (CXCR4+, β1-integrin+) (Figure S2A). Consistent with our in situ results, freshly isolated SKO MuSCs displayed decreased PAX7 expression (Figures S2B and S2C) with higher levels of ROS (CellROX; Figures 1D and S2D) and mitochondria (MitoTracker; Figures 1D and S2E) compared with WT MuSCs (Figures 1D, S2D, and S2E). SKO MuSCs also displayed increases in proliferation (as measured by Ki67; Figures 1D and S2E) but similar levels of MYOD (Figures S2B and S2C), suggesting stronger resistance to activation in WT than SKO MuSCs. Culture of both types of MuSCs in activating conditions for 72 h showed similar levels of PAX7, but increased MYOD expression in SKO MuSCs (Figures S2F and S2G). Given that autophagy is critical for MuSC activation (García-Prat et al., 2016), and that Sestrins suppress autophagic degradation (Lee et al., 2013), we performed immunofluorescence (IF) staining of p62/sequestosome (SQSTM) on freshly isolated and MuSCs cultured for 72 h. We observed no differences between WT and SKO MuSCs for p62 (Figures S2H–S2K), and no differences in total AMPKα1 levels between WT and SKO MuSCs were observed at either time point (Figures S2L–S2O). Integrating these results suggests that Sestrins influence MuSCs in homeostasis, but do not significantly affect activation dynamics.
Sestrin deficiency does not alter injury responses of MuSCs in young mice
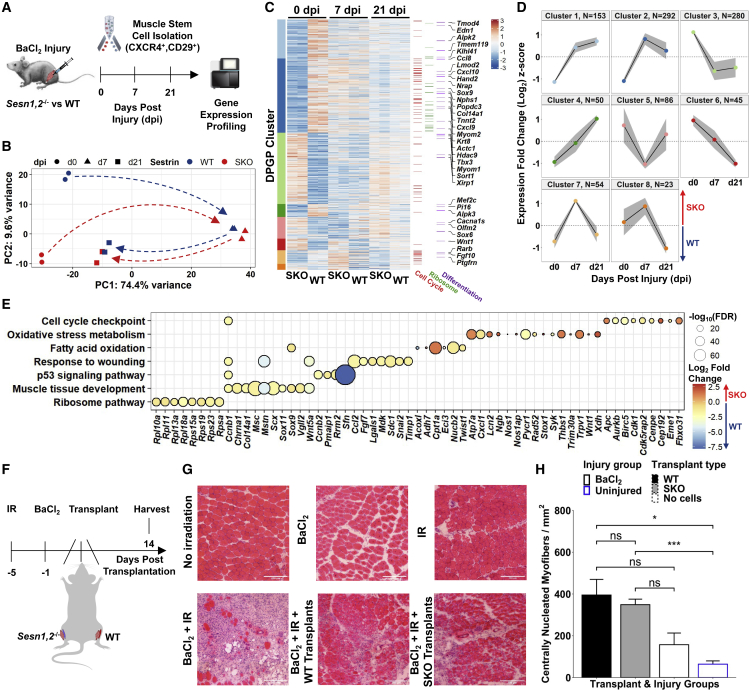
To glean deeper insights into the consequences of alterations in homeostasis from SESTRIN1,2 KO, WT and SKO hindlimb muscles (tibialis anterior, gastrocnemius, and quadriceps) were injured through BaCl2 injections. MuSCs were isolated with FACS before and after injury (0, 7, and 21 days post injury [dpi]), and submitted for RNA-seq (Aguilar et al., 2016) (Figure 2A). Gene expression profiles from WT and SKO samples demonstrated strong agreement between biological replicates at each time point (Spearman > 0.97) (Figure S3A) and principal-component analysis (PCA) revealed that, while WT and SKO MuSCs were transcriptionally dissimilar before injury, they migrated along similar regeneration trajectories (Figure 2B). A total of 982 differentially expressed genes were identified across all time points, many of which (~90%) were uniquely expressed at 0 dpi (Figures 2C and S3B). Dirichlet process Gaussian process (DPGP) clustering revealed time-based differences in enriched gene ontology terms and KEGG pathways (Figures 2D, S3C, and S3D). Visualizing the DPGP clustered genes in PCA space showed that the temporal dynamics of clusters 1, 2, 4, and 7 were predominantly driven by genes that were differentially expressed as a result of injury (Figure S3E). Consistent with Sestrins contributing to variations in metabolism, genes in cluster 1 that were upregulated in WT before injury were enriched for terms related to fatty acid derivative metabolism (e.g., Cyp2s1, Fabp5) and restraint of activation (e.g., Mstn, Snai2). Cluster 7 was enriched for genes comprising cell-cycle regulation terms (e.g., Cdk1, Ccna2, Ccnb1, Cdc25c), and summing these results further suggests that SKO MuSCs display alterations in metabolism and reductions in the ability to retain quiescence. To probe further into changes in homeostasis, we evaluated differentially expressed genes at 0 dpi. This analysis revealed that SKO MuSCs contained upregulated genes for cell-cycle checkpoints and oxidative stress metabolism, while WT MuSCs upregulated genes related to ribosomal activity and inhibition of muscle tissue development (Figure 2E). Given the variation in metabolic genes between SKO and WT MuSCs, we used genome-scale metabolic modeling to assess the relationship between 3,744 metabolic reactions, 2,766 metabolites, 1,496 metabolic genes, and 2,004 metabolic enzymes (Duarte et al., 2007; Shcherbina et al., 2020). The model predicted that uninjured SKO MuSCs upregulated metabolic flux through acetyl CoA synthetase, and glycine, serine, and threonine metabolism (Table S1). Given that mTORC1 is regulated by acetyl CoA (Son et al., 2019), these results are consistent with over-activated mTORC1 in SKO MuSCs and increases in expression of acetyl-transferases and deacetylases (Figure S4F). Collectively, the observed changes in expression show that knocking out Sesn1,2 does not significantly alter muscle regeneration but rather drives alterations in metabolism that contribute to reduced quiescence in MuSCs.
Figure 2.
Sesn1,2 Knockout MuSCs show robust regenerative responses despite having strongly altered basal-level transcriptomes
(A) Schematic diagram of BaCl2 injury model in the hindlimbs of WT and SKO mice followed by MuSC isolation via FACS and RNA-seq profiling before and during regeneration.
(B) Principal-component analysis (PCA) of MuSC replicates from WT and SKO mice.
(C) Standardized heatmap (Z scores) of 982 differentially expressed genes (SKO over WT) from WT and SKO MuSCs grouped by Dirichlet process Gaussian process (DPGP) clusters. Genes that participate in the cell cycle (Buettner et al., 2015), the Ribosome KEGG pathway (mmu03010), and the muscle cell differentiation gene ontology (GO) term (GO: 0042692) are marked on the right.
(D) DPGP mixture model-based clustering of mean gene expression time series Z scores (gray bars are 2× standard deviation and black line is cluster mean).
(E) Gene set terms and KEGG pathways at 0 dpi plotted against constituent genes.
(F) Schematic diagram for SKO and WT MuSC transplantation experiment. The hindlimbs of young mice (5-month-old males) were irradiated 5 days before transplantation and TAs were injured with an intramuscular BaCl2 injection or left uninjured 1 day before transplantation. Alternating TAs received either SKO cells (n = 4 mice), WT cells (n = 4 mice), or no cells (n = 3 mice and 4 mice for injured and uninjured controls, respectively) during transplantation. TAs were harvested 2 weeks post-transplantation.
(G) Representative H&E images of TA cross-sections following no injury, only BaCl2 injury, or only irradiation (top), and following a combination of BaCl2 and irradiation followed by no cell transplantation, WT cell transplantation, or SKO cell transplantation (bottom). Scale bars, 200 μm.
(H) Quantification of centrally nucleated myofibers per total cross-sectional area (mm2) between transplant groups. Bar plot fill colors indicate transplant type and border colors indicate injury type. Statistical comparisons are two-sided, unpaired Student’s t tests. All data are shown as mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001. See also Figure S3 and Table S1.
To rule out if changes in the muscle microenvironment from loss of Sestrins impacted the regenerative activity of MuSCs, we performed transplantation of MuSCs from WT and SKO muscles. Hindlimbs of WT young (5 months) mice were irradiated (18 Gy), followed by injury via BaCl2 injection into tibialis anterior (TA) muscles (Figure 2F). One day after muscle injury, ~18,000 FACS-sorted MuSCs from SKO and WT muscles were transplanted into recipients and muscle regeneration evaluated 14 days later. Irradiated limbs that received muscle injury displayed considerable degeneration and fibrosis after muscle injury (Boldrin et al., 2012) (Figure 2G). Both WT and SKO transplanted MuSCs improved regeneration as assessed by the number of myofibers with centrally located nuclei, but no significant difference between the two types of transplanted MuSCs was observed (Figure 2H). These results confirm that loss of Sestrins in myofibers does not impact muscle regeneration through MuSCs, and that WT and SKO MuSCs display comparable regenerative potential.
Sestrin loss decreases MuSC number and function in middle age
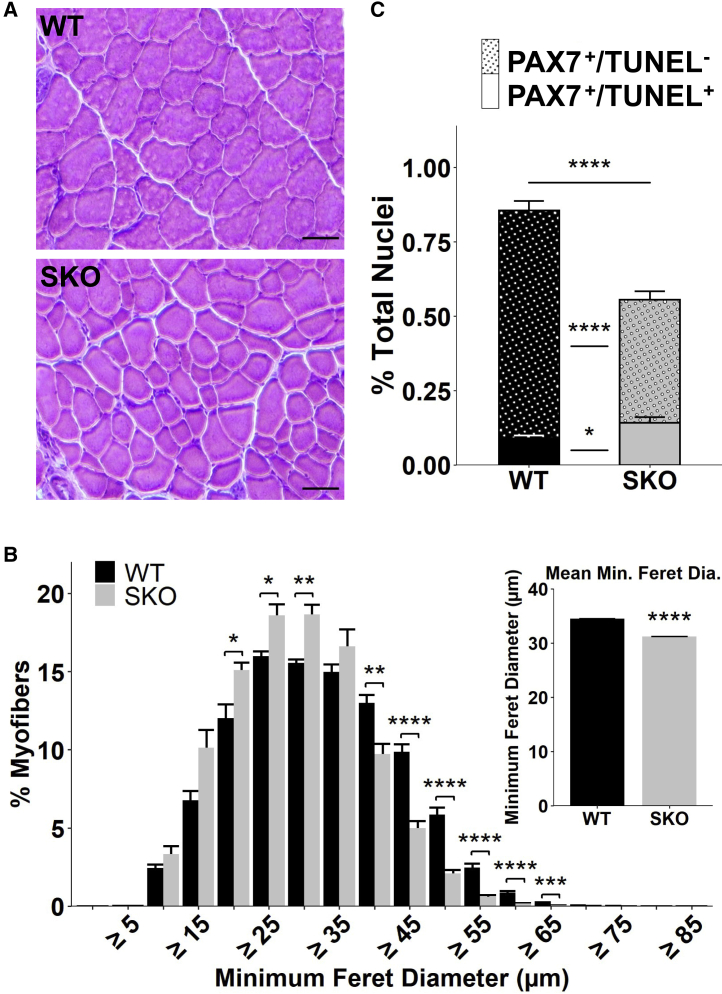
One way to evaluate changes in metabolism and long-term homeostasis of MuSCs is through natural aging, whereby increases in oxidative damage and negative changes in proteostasis promote constitutive MuSC activation and exhaustion (Chakkalakal et al., 2012; García-Prat et al., 2016). We therefore examined gene expression datasets obtained from young and aged MuSCs before and after injury (Shcherbina et al., 2020) and observed that aged MuSCs exhibit reductions in Sesn1 expression before and during regeneration (Figure S4A). To probe deeper into these observations, we aged SKO and WT mice to middle age (14 months), when hyperactivated mTORC1 mice from TSC1,2 KO demonstrated accelerated aging phenotypes (Castets et al., 2019). We extracted hindlimb muscles from middle-aged SKO and WT mice and observed smaller myofibers (Figures 3A and 3B) in addition to comparable numbers of centrally nucleated fibers and fiber types for SKO muscles compared with WT (Figures S4B–S4D). Consistent with TSC1,2 KO mice (Haller et al., 2017; Rodgers et al., 2014), middle-aged SKO MuSCs displayed a loss in the number of total PAX7+ MuSCs, increased fractions of TUNEL+/PAX7+ MuSCs (Figure 3C) and increased numbers of pS6+/PAX7+ MuSCs (Figures S4E and S4F) compared with WT. We reasoned that the loss of MuSCs may be the result of changes in autophagy but did not observe variations in p62 levels between SKO and WT muscles (Figures S4G–S4I). These results suggest loss of Sestrins and persistent activation of mTORC1 promotes premature loss in the total number of PAX7+ MuSCs.
Figure 3.
Sustained mTORC1 activation from loss of SESTRIN1,2 in aging induces loss of MuSCs
(A) Representative images of H&E staining in TA muscle sections from middle-aged mice (14 months). Scale bars, 100 μm.
(B) Size distributions of WT and SKO myofibers in TA muscle sections from middle-aged SKO and WT mice (WT, n = 5 mice; SKO, n = 3 mice). Inset shows mean size per condition. Statistical comparisons are two-sided, unpaired Student’s t tests with Holm multiple testing correction for the histogram.
(C) Quantification of TUNEL+ (plain) and TUNEL− (stippled) PAX7+ cells as a fraction of total nuclei in TA muscle sections from 14-month-old SKO and WT mice (WT, n = 4 mice; SKO, n = 4 mice). Statistical comparisons are two-sided Mann-Whitney U tests. All data are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S4.
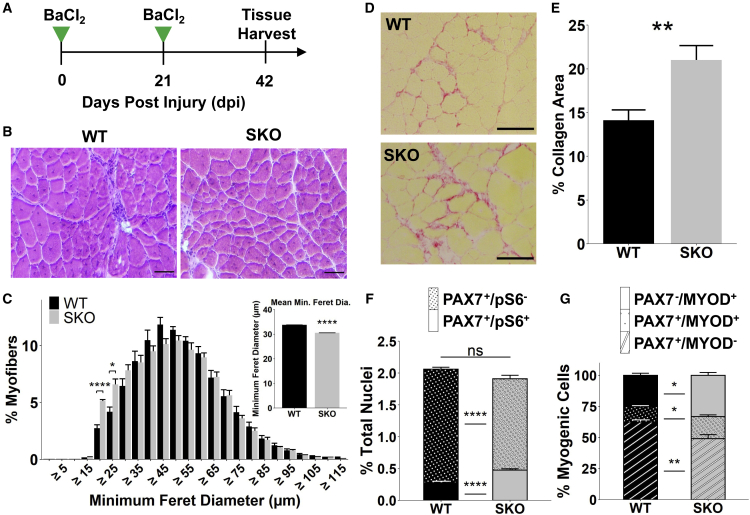
To further examine if changes in the regenerative capacity of MuSCs are modified with loss of Sestrins during aging, we performed repetitive injuries by injecting BaCl2 into the TA muscle (injuries spaced 21 days apart between injections) (Figure 4A). After repetitive injuries, SKO muscles displayed reductions in fiber size (Figures 4B and 4C), enhanced collagen deposition (Figures 4D and 4E), and significant loss and gain of type IIx and type IIb fiber types, respectively, consistent with previous observations of myofiber transitions in aging muscle (Crupi et al., 2018; Ham et al., 2020; Liu et al., 2017) (Figures S4J and S4K). To determine if MuSCs from SKO muscle retained increased mTORC1 activity after response to injury, we assessed the fraction of pS6+/PAX7+ MuSCs and consistent with uninjured MuSCs, observed increased numbers of pS6+/PAX7+ MuSCs compared with WT (Figure 4F). To further determine if MuSCs remained activated, we performed co-staining of MYOD and PAX7 in situ and observed an increase of the fraction of PAX7+/MYOD+ and PAX7−/MYOD+ cells, as well as a decrease in PAX7+/MYOD− cells in SKO muscles compared with WT (Figures 4G and S4L). Summing these results demonstrates that long-term Sestrin deficiency produces an attenuated regenerative response owing to a reduced MuSC population.
Figure 4.
Loss of SESTRIN1,2 in aging muscle impairs regeneration and promotes MuSC loss following injury
(A) Schematic of BaCl2 double injury model.
(B) Representative images of H&E staining in injured TA muscle sections from middle-aged mice (14 months). Scale bars, 100 μm.
(C) Size distribution of WT and SKO myofibers after injury (WT, n = 4 mice; SKO, n = 3 mice). Statistical comparisons are two-sided, unpaired Student’s t tests with Holm multiple testing correction for the histogram. Inset is mean myofiber size per condition.
(D) Representative images of Picrosirius red-stained collagen in whole injured TA muscle sections. Muscle fibers are shown in yellow. Scale bars, 100 μm.
(E) Collagen area fraction in whole TA muscle sections as determined by Picrosirius red staining. Statistical comparison is a two-sided, unpaired Student’s t test (WT, n = 3 mice; SKO, n = 4 mice).
(F) Quantification of pS6+ (plain) and pS6− (stippled) PAX7+ cells as percentages of all nuclei in whole, injured TA muscle sections. Statistical comparisons are two-sided Mann-Whitney U tests (WT, n = 3 mice; SKO, n = 3 mice).
(G) Quantification of PAX7+/MYOD− (striped), PAX7+/MYOD+ (stippled), and PAX7−/MYOD+ (plain) cells as percentages of all nuclei in whole, injured TA muscle sections (WT, n = 3 mice; SKO, n = 3 mice). Statistical comparisons are two-sided Mann-Whitney U tests. All data are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. See also Figure S4.
Discussion
Tissue homeostasis is critically dependent on the maintenance and activity of stem cells, which in turn are regulated by metabolic status. A focal point of metabolic signaling is the nutrient-sensitive protein complex, mTORC1, which balances protein biosynthesis and autophagy to meet anabolic demands (Wullschleger et al., 2006). Regulation of mTORC1 (Haller et al., 2017) is essential to preventing stem cell overactivation (Meng et al., 2018) and maintaining quiescence (Nieto-González et al., 2019), and loss of mTORC1 inhibition results in the depletion of hematopoietic stem cells (Lee et al., 2010b). Sestrins are a unique class of metabolic regulators that titrate mTORC1 signaling and act as oxidative stress sensors, but their roles in adult stem cells have yet to be evaluated. Our results show that Sestrins contribute to MuSC homeostasis by inhibiting mTORC1 and maintaining metabolism supportive of quiescence. The loss of Sestrins resulted in upregulation of genes related to oxidative stress metabolism and a reduced capacity to resist activation. Persistent increases in oxidative stress from hyperactivation of mTORC1 have been shown to promote an aged phenotype in muscle (Tang et al., 2019) and prime MuSCs for activation through accelerated cell-cycle entry and increased mitochondrial activity (Rodgers et al., 2014). Our studies are consistent with these observations and demonstrate that Sestrins fine-tune the basal metabolism of MuSCs via stress-inducible genes with antioxidant functions, such as nuclear factor (erythroid-derived 2)-like 2 (Nrf2) (Ho et al., 2016), and other ROS-sensitive factors, such as p53 and the FOXO family (Lee et al., 2013). Given that hyperactivated mTORC1 promotes mitochondrial biogenesis and oxidative phosphorylation, a source of ROS generation and electron leakage (Filomeni et al., 2015), our results suggest that Sestrins balance mTORC1 and the cellular redox state in MuSCs.
The regeneration of muscle is mediated through MuSC actions, but the ability of MuSCs to repair tissue and self-renew declines in aging (Blau et al., 2015). This is due in part to the accumulation of DNA damage (Sousa-Victor et al., 2014), metabolic reprogramming in response to increases in oxidative stress (Pala et al., 2018), and altered chromatin packaging (Shcherbina et al., 2020). Our results demonstrate that Sestrin-deficient muscle displays comparable regenerative capacity in young mice (2–3 months), which gives way to pathological aging in middle age (14 months) due to MuSC loss that is compounded by impaired regenerative capacity independent of changes in autophagic flux. These results may be driven by alterations in mitochondria and energy production. Previously, we observed that Sestrin loss resulted in the attenuation of mitochondrial biogenesis and maximal respiratory capacity (Kim et al., 2020). Both are pivotal for proper muscle repair via MuSCs, and interruptions to mitochondrial generation and bioenergetics can be detrimental to MuSC differentiation. We posit that Sestrins provide a control layer for MuSC mitochondria that protects against age-associated insults and is diminished in aging, resulting in imbalanced ROS levels (Bigarella et al., 2014). In support of this, both SESTRIN1 and 2 exhibited decreased expression in aged MuSCs compared with young MuSCs (Shcherbina et al., 2020).
Integrating our data suggests that Sestrins are important regulators of MuSC metabolism and the quiescent state. Several studies have shown that mTORC1 hyperactivation results in stem cell loss (Haller et al., 2017; Nieto-González et al., 2019) and interrupted maintenance of the quiescent state, but none have yet demonstrated this behavior through Sestrins. Accordingly, Sestrin-dependent changes may be strong targets for restoring MuSC metabolism in aging that expand our understanding of metabolic regulation in stem cells across lifespan.
Experimental procedures
MuSC isolation via FACS
For tissue collection, mice were anesthetized with 3% isoflurane, then euthanized by cervical dislocation, bilateral pneumothorax, and removal of the heart. Hindlimb muscles (tibialis anterior, gastrocnemius, and quadriceps) of WT and Sestrin double-KO (DKO) mice were quickly harvested using sterile surgical tools and placed in separate plastic Petri dishes containing cold PBS. Using surgical scissors, muscle tissues were minced and transferred into 50 mL conical tubes containing 20 mL of digest solution (2.5 U/mL dispase II and 0.2% [~5,500 U/mL] collagenase type II in DMEM medium per mouse). Samples were incubated on a rocker placed in a 37°C incubator for 90 min with manual pipetting the solution up and down to break up tissue every 30 min using a fetal bovine serum (FBS)-coated 10 mL serological pipette. Once the digestion was completed, 20 mL of F10 medium containing 20% heat-inactivated FBS was added into each sample to inactivate enzyme activity. The solution was then filtered through a 70 μm cell strainer into a new 50 mL conical tube and centrifuged again at 350 × g for 5 min. The pellets were re-suspended in 6 mL of staining medium (2% heat-inactivated FBS in Hank's buffered salt solution) and divided into separate FACS tubes. The FACS tubes were centrifuged at 350 × g for 5 min and supernatants discarded. The cell pellets were then re-suspended in 200 μL of staining medium and antibody cocktail containing Sca-1:APC (1:400), CD45:APC (1:400), CD11b:APC (1:400), Ter119:APC (1:400), CD29/B1-integrin:PE (1:200), and CD184/CXCR-4:biotin (1:100) and incubated for 30 min on ice in the dark. Cells and antibodies were diluted in 3 mL of staining solution, centrifuged at 350 × g for 5 min, and supernatants discarded. Pellets were re-suspended in 200 μL staining solution containing PECy7:streptavidin (1:100) and incubated on ice for 20 min in the dark. Again, samples were diluted in 3 mL staining solution, centrifuged, supernatants discarded, and pellets re-suspended in 200 μL staining buffer. Live cells were sorted from the suspension via addition of 1 μg of propidium iodide stain into each experimental sample and all samples were filtered through 35 μm cell strainers before the FACS. Cell sorting was done using a BD FACSAria III Cell Sorter (BD Biosciences, San Jose, CA) and APC-negative, PE/PECy7 double-positive MuSCs were sorted into staining solution for immediate processing.
MuSC culture and IF staining
PAX7, MYOD, p62, and AMPKα1 staining in MuSCs (freshly isolated and after 3 days of culture)
Ninety-six-well plates were coated in either Cell-Tak (3.5 μg/cm2, Fisher Scientific no. C354240) for immediate attachment and fixation of cells or 0.5% gelatin (Sigma-Aldrich, no. G2500) solution for 3 days of culture in myoblast growth medium (80% Ham’s F10, 20% FBS, 1% penicillin/streptomycin solution, 20 ng/mL basic fibroblast growth factor) at 37.5°C and 5.0% CO2. MuSCs were isolated as described above from two WT mice and two DKO mice (all male, aged 3–4 months). A portion of the isolated MuSCs was re-suspended in 1× PBS, seeded in Cell-Tak-coated wells at a density of 7,800 cells/cm2, and briefly allowed to attach before fixation with methanol at −20°C for 10 min. The remainder of the MuSCs were re-suspended in myoblast growth medium and seeded in gelatin-coated wells also at a density of 7,800 cells/cm2. The medium was replenished every 24 h and cells were similarly fixed with methanol after 3 days of culture.
Cells were blocked and permeabilized with 0.3% Triton X-100 (Acros Organics no. A0376210) and 1% BSA (Fisher Scientific no. BP9703-100) in 1× PBS. Cells were then incubated overnight at 4°C with either a combination of AF488-conjugated anti-PAX7antibody (1:100) and AF647-conjugated anti-MYODantibody (1:50) in 0.2% BSA in 1× PBS, anti-AMPKα1 (1:1,000) primary antibody in 0.2% BSA in 1× PBS, or anti-p62 (1:800) primary antibody in 1% BSA in 1× PBS. Wells stained for AMPKα1 or p62 were incubated with an AF647 goat anti-rabbit secondary antibody for 1 h at room temperature, protected from the light. All stains were performed in duplicate for each genotype and time point. Nuclei were stained with 1 μg/mL DAPI in 1× PBS for 10 min at room temperature. Images (20× magnification) were acquired on a Zeiss Axio Vert.A1 inverted microscope with a Colibri 7 LED light source and an AxioCam MRm camera. Images were subsequently analyzed in FIJI. Regions of interest (ROIs) were generated by thresholding on the DAPI image to identify nuclei. The average fluorescent intensities of each stain within these ROIs were recorded. The average PAX7, MYOD, AMPKα1, and p62 signals were compared between SKO and WT cells using a two-sample Student’s t test with the significance level set to α = 0.05.
MitoTracker DeepRed, CellRox green, and Ki67 staining in freshly isolated MuSCs
Ninety-six-well plates were coated in 0.5% gelatin (Sigma-Aldrich, no. G2500) solution before seeding MuSCs isolated via FACS (two WT, two DKO, all female, aged 3–4 months). Cells were re-suspended in myoblast growth medium and seeded at a density of 7,800 cells/cm2. Plates were incubated at 37°C for 3 h to allow the cells to attach before replacing the medium with MitoTracker DeepRed (diluted to 500 nM in pre-warmed medium) or CellRox Green Reagent (diluted 1:500 in pre-warmed medium). Cells were incubated at 37°C with their respective live cell stains for 30 min before aspirating off all medium and fixing in 4% paraformaldehyde in 1× PBS for 10 min at room temperature. Cells were then permeabilized in 0.1% Triton X-100 in 1× PBS for 15 min at room temperature and blocked for 1 h at room temperature in 1% BSA, 0.5% goat serum, 22.52 mg/mL glycine in PBST (0.1% Tween 20 in PBS). After three washes with 1× PBS, cells were then stained with PE-conjugated Ki67 antibody at a dilution of 1:50 in 1% BSA in PBST overnight at 4°C, protected from light. Afterward, cells were washed three times with PBST and nuclei were stained with 1 μg/mL DAPI in PBS for 10 min at room temperature. All stains were performed in duplicate across all samples. MitoTracker and Ki67 images were captured on a Zeiss Axio Vert.A1 inverted microscope with a Colibri 7 LED light source and an AxioCam MRm camera at a 40× magnification. CellRox images were captured on a Nikon A1si confocal microscope at a magnification of 10×. ROIs were generated by thresholding on the DAPI image to identify nuclei in FIJI, followed by measurements of average fluorescent intensity within each ROI. A two-sample t test with the significance level set to α = 0.05.
Preparation of RNA-seq libraries and sequencing
MuSCs were FACS sorted directly into TRIzol and snap frozen in liquid nitrogen. Samples were subsequently thawed, and RNA was extracted using a QIAGEN miRNeasy Micro Kit as per the manufacturer’s instructions. The integrity of the isolated RNA was verified using a Bioanalyzer (Agilent 2100) and 1–10 ng of high-quality RNA (RNA integrity number, RIN > 8) was used to produce cDNA libraries using the Smart-Seq v.4 protocol (Clontech) as per the manufacturer’s instructions. cDNAs were prepared into sequencing libraries using 150 pg of full-length cDNA amplicons (Nextera XT DNA Library Preparation Kit, Illumina) with dual index barcodes. Barcoded cDNA libraries were pooled into a single tube and sequenced on a NextSeq (Illumina) using 76 bp single-ended reads.
RNA-seq data processing and analysis
Gene expression estimation
Single-end RNA-seq data were trimmed using Flexbar (v.3.5.0) and pseudo-aligned to the mouse reference genome (GRCm38.p6) using Kallisto (v.0.46.1). Reads averaged 41.75M per sample. The full Kallisto command was as follows:
kallisto quant -b 100 --single -l 300 -s 30 -i [mm10.idx] -t 45 -o [output folder] [trimmed FASTQ]
Differential gene expression
The estimated transcript abundances were summarized to gene-level count matrices using tximport and genes containing at least one read were retained. Differentially expressed genes in treated (KO) samples relative to untreated controls (WT) at each time point (days 0, 7, and 21) were identified using DESeq2 in R with a design formula: Count ~ group, with group = {day + treatment}, day = {0,7,21}, and treatment = {WT,KO}. Surrogate variable analysis was performed on the rlog-transformed count matrix using the SVA package with a null model of rlog(Count) ~ 1 and a design matrix of rlog(Counts) ~ group. Contributions from the surrogate variable were quantified and removed from the rlog-transformed count matrix for downstream analyses. Pairwise Spearman correlation analysis was performed between all replicates, and replicates that had r < 0.9 with other replicates were excluded from further analysis. Pairwise contrasts were examined to find differentially expressed genes between SKO versus WT on each day post injury. Differentially expressed genes were selected using a false discovery rate cutoff of 0.01 and a log2 fold change cutoff of 1.
Time series clustering of differential genes
Genes that were differentially expressed on at least 1 dpi were pooled and submitted for time-clustering using the DPGP algorithm. To prepare inputs for the algorithm, regularized log-transformed counts were averaged across biological replicates on each day post injury and the fold change in averaged counts was calculated between SKO versus WT. The resulting fold changes for each gene were standardized across time points using a Z score transformation. DPGP clustering was performed using the default parameters. The full command was as follows:
DP_GP_cluster.py -i [fold change z-scores] -o./[output file prefix]
DPGP assigned each gene a unique time-dependent cluster based on similar expression dynamics, and clusters that exhibited similar temporal dynamics were manually combined into a single cluster. Log-fold Z scores were plotted as a function of time for each cluster and a heatmap of differentially expressed genes grouped by DPGP cluster was plotted using the ComplexHeatmap package in R.
Additional experimental details on reagents, cell culture, expression analysis, IF staining and quantitation, animals, transplantation, and injury assays can be found in the supplemental information. All procedures, including maintenance of animals, were approved by the Institutional Animal Care and Use Committee and were in accordance with the US National Institutes of Health.
Data and code availability
The accession number for the RNA-seq dataset reported in this study is GEO: GSE162191.
Author contributions
B.A.Y., P.F., J.C.-M., A.C., L.A.B., M.K., and I.L. performed the experiments. B.A.Y. and C.A.A. analyzed the data. S.V.B., J.H.L., and C.A.A. designed the experiments. B.A.Y. and C.A.A. wrote the manuscript with additions from other authors.
Acknowledgments
The authors thank Kanishka de Silva, and the University of Michigan DNA Sequencing Core, for assistance with sequencing. The authors also thank Anna Shcherbina for insights into bioinformatics analysis, and members of the Aguilar, Lee, and Brooks laboratories. Research reported in this publication was partially supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number P30 AR069620 (to C.A.A. and S.V.B.), the 3M Foundation (to C.A.A.), American Federation for Aging Research Grant for Junior Faculty (to C.A.A.), the Department of Defense and Congressionally Directed Medical Research Program W81XWH2010336 (to C.A.A.), the University of Michigan Geriatrics Center and National Institute on Aging under award number P30 AG024824 (to C.A.A. and S.V.B.), National Institute on Aging P01 AG051442 (to S.V.B.), and the National Institute of Biomedical Imaging and Bioengineering Training Award T32 EB005582 (to B.A.Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Published: August 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.014.
Supplemental information
References
- Aguilar C.A., Pop R., Shcherbina A., Watts A., Matheny R.W., Cacchiarelli D., Han W.M., Shin E., Nakhai S.A., Jang Y.C., et al. Transcriptional and chromatin dynamics of muscle regeneration after severe trauma. Stem Cell Rep. 2016;7:983–997. doi: 10.1016/j.stemcr.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracos V.E., Martin L., Korc M., Guttridge D.C., Fearon K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Primer. 2018;4:1–18. doi: 10.1038/nrdp.2017.105. [DOI] [PubMed] [Google Scholar]
- Beerman I., Rossi D.J. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16:613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigarella C.L., Liang R., Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau H.M., Cosgrove B.D., Ho A.T.V. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015;21:854–862. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrin L., Neal A., Zammit P.S., Muntoni F., Morgan J.E. Donor satellite cell engraftment is significantly augmented when the host niche is preserved and endogenous satellite cells are incapacitated. STEM CELLS. 2012;30:1971–1984. doi: 10.1002/stem.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P., Lin W.J., Giovarelli M., Pasero M., Chou C.F., Trabucchi M., Rosenfeld M.G., Chen C.Y., Gherzi R. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 2012;19:478–487. doi: 10.1038/cdd.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- Castets P., Rion N., Théodore M., Falcetta D., Lin S., Reischl M., Wild F., Guérard L., Eickhorst C., Brockhoff M., et al. mTORC1 and PKB/Akt control the muscle response to denervation by regulating autophagy and HDAC4. Nat. Commun. 2019;10:3187. doi: 10.1038/s41467-019-11227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M., Jurga S., Witczak C.A., Hirshman M.F., Shadrach J.L., Goodyear L.J., Wagers A.J. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal J.V., Jones K.M., Basson M.A., Brack A.S. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N.C., Chevalier F.P., Rudnicki M.A. Satellite S. Trends Mol. Med. 2016;22:479–496. doi: 10.1016/j.molmed.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crupi A.N., Nunnelee J.S., Taylor D.J., Thomas A., Vit J.-P., Riera C.E., Gottlieb R.A., Goodridge H.S. Oxidative muscles have better mitochondrial homeostasis than glycolytic muscles throughout life and maintain mitochondrial function during aging. Aging. 2018;10:3327–3352. doi: 10.18632/aging.101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., Vo T.D., Srivas R., Palsson B.Ø. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A., Burgering B.M.T. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- Feige P., Brun C.E., Ritso M., Rudnicki M.A. Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell. 2018;23:653–664. doi: 10.1016/j.stem.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J., Rebollo E., Ruiz-Bonilla V., Gutarra S., Ballestar E., Serrano A.L., et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- Haller S., Kapuria S., Riley R.R., O’Leary M.N., Schreiber K.H., Andersen J.K., Melov S., Que J., Rando T.A., Rock J., et al. mTORC1 activation during repeated regeneration impairs somatic stem cell maintenance. Cell Stem Cell. 2017;21:806–818.e5. doi: 10.1016/j.stem.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham D.J., Börsch A., Lin S., Thürkauf M., Weihrauch M., Reinhard J.R., Delezie J., Battilana F., Wang X., Kaiser M.S., et al. The neuromuscular junction is a focal point of mTORC1 signaling in sarcopenia. Nat. Commun. 2020;11:4510. doi: 10.1038/s41467-020-18140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A., Cho C.-S., Namkoong S., Cho U.-S., Lee J.H. Biochemical basis of sestrin physiological activities. Trends Biochem. Sci. 2016;41:621–632. doi: 10.1016/j.tibs.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.T., Warr M.R., Adelman E.R., Lansinger O.M., Flach J., Verovskaya E.V., Figueroa M.E., Passegué E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Ro S.H., Kim M., Park H.W., Semple I.A., Park H., Cho U.S., Wang W., Guan K.L., Karin M., et al. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep09502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Sujkowski A., Namkoong S., Gu B., Cobb T., Kim B., Kowalsky A.H., Cho C.-S., Semple I., Ro S.-H., et al. Sestrins are evolutionarily conserved mediators of exercise benefits. Nat. Commun. 2020;11:190. doi: 10.1038/s41467-019-13442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Budanov A.V., Park E.J., Birse R., Kim T.E., Perkins G.A., Ocorr K., Ellisman M.H., Bodmer R., Bier E., et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Budanov A.V., Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013;18:792–801. doi: 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Cho U.S., Karin M. Sestrin regulation of TORC1: is sestrin a leucine sensor? Sci. Signal. 2016;9:re5. doi: 10.1126/scisignal.aaf2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Nakada D., Yilmaz O.H., Tothova Z., Joseph N.M., Lim M.S., Gilliland D.G., Morrison S.J. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T.A., Fan C.M. An absolute requirement for pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Huang Y., Semple I., Kim M., Zhang Z., Lee J.H. Cardioprotective roles of sestrin 1 and sestrin 2 against doxorubicin cardiotoxicity. Am. J. Physiol.-Heart Circ. Physiol. 2019;317:H39–H48. doi: 10.1152/ajpheart.00008.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Klose A., Forman S., Paris N.D., Wei-LaPierre L., Cortés-Lopéz M., Tan A., Flaherty M., Miura P., Dirksen R.T., et al. eLife Sciences Publications Limited; 2017. Loss of Adult Skeletal Muscle Stem Cells Drives Age-Related Neuromuscular Junction Degeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D., Frank A.R., Jewell J.L. mTOR signaling in stem and progenitor cells. Development. 2018;145 doi: 10.1242/dev.152595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-González J.L., Gómez-Sánchez L., Mavillard F., Linares-Clemente P., Rivero M.C., Valenzuela-Villatoro M., Muñoz-Bravo J.L., Pardal R., Fernández-Chacón R. Loss of postnatal quiescence of neural stem cells through mTOR activation upon genetic removal of cysteine string protein-α. Proc. Natl. Acad. Sci. 2019;116:8000–8009. doi: 10.1073/pnas.1817183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala F., Girolamo D.D., Mella S., Yennek S., Chatre L., Ricchetti M., Tajbakhsh S. Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J. Cell Sci. 2018;131 doi: 10.1242/jcs.212977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.-R., et al. mTORC1 controls the adaptive transition of quiescent stem cells from G 0 to G Alert. Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall J.G., Cliff T., Dalton S., Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17:651–662. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J., Perdiguero E., Serrano A.L., Sousa-Victor P., Ortet L., Jardí M., Budanov A.V., Garcia-Prat L., Sandri M., Thomson D.M., et al. Sestrin prevents atrophy of disused and aging muscles by integrating anabolic and catabolic signals. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-13832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C., Palacios D., Mozzetta C., Forcales S.V., Morantte I., Ripani M., Jones D.R., Du K., Jhala U.S., Simone C., et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbina A., Larouche J., Fraczek P., Yang B.A., Brown L.A., Markworth J.F., Chung C.H., Khaliq M., Silva K., Choi J.J., et al. Dissecting murine muscle stem cell aging through regeneration using integrative genomic analysis. Cell Rep. 2020;32:107964. doi: 10.1016/j.celrep.2020.107964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S.M., Park S.J., Lee H., Siddiqi F., Lee J.E., Menzies F.M., Rubinsztein D.C. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 2019;29:192–201.e7. doi: 10.1016/j.cmet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Victor P., Gutarra S., García-Prat L., Rodriguez-Ubreva J., Ortet L., Ruiz-Bonilla V., Jardí M., Ballestar E., González S., Serrano A.L., et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- Sperka T., Wang J., Rudolph K.L. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Tang H., Inoki K., Brooks S.V., Okazawa H., Lee M., Wang J., Kim M., Kennedy C.L., Macpherson P.C.D., Ji X., et al. mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell. 2019;18:e12943. doi: 10.1111/acel.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theret M., Gsaier L., Schaffer B., Juban G., Ben Larbi S., Weiss-Gayet M., Bultot L., Collodet C., Foretz M., Desplanches D., et al. AMPKα1-LDH pathway regulates muscle stem cell self-renewal by controlling metabolic homeostasis. EMBO J. 2017;36:1946–1962. doi: 10.15252/embj.201695273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney M.T., Sacco A. Satellite cell heterogeneity in skeletal muscle homeostasis. Trends Cell Biol. 2016;26:434–444. doi: 10.1016/j.tcb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touil Y., Zuliani T., Wolowczuk I., Kuranda K., Prochazkova J., Andrieux J., Le Roy H., Mortier L., Vandomme J., Jouy N., et al. The PI3K/AKT signaling pathway controls the quiescence of the low-rhodamine123-retention cell compartment enriched for melanoma stem cell activity. Stem Cells. 2013;31:641–651. doi: 10.1002/stem.1333. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xin T., Greco V., Myung P. Hardwiring stem cell communication through tissue structure. Cell. 2016;164:1212–1225. doi: 10.1016/j.cell.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.A., Westerhof T.M., Sabin K., Merajver S.D., Aguilar C.A. Engineered tools to study intercellular communication. Adv. Sci. 2021;8:2002825. doi: 10.1002/advs.202002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.S.L., Cui W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq dataset reported in this study is GEO: GSE162191.