Summary
Uterine leiomyoma (LM) is the most common tumor in women. Via its receptor (PGR) expressed in differentiated LM cells, progesterone stimulates paracrine signaling that induces proliferation of PGR-deficient LM stem cells (LSCs). Antiprogestins shrink LM but tumors regrow after treatment cessation possibly due to persisting LSCs. Using sorted primary LM cell populations, we found that the PGR gene locus and its target cistrome are hypermethylated in LSCs, inhibiting the expression of genes critical for progesterone-induced LSC differentiation. PGR knockdown shifted the transcriptome of total LM cells toward LSCs and increased global DNA methylation by regulating TET methylcytosine dioxygenases. DNA methylation inhibitor 5′-Aza activated PGR signaling, stimulated LSC differentiation, and synergized with antiprogestin to reduce tumor size in vivo. Taken together, targeting the feedback loop between DNA methylation and progesterone signaling may accelerate the depletion of LSCs through rapid differentiation and sensitize LM to antiprogestin therapy, thus preventing tumor regrowth.
Keywords: uterine leiomyoma, DNA methylation, progesterone receptor, stem cell regulation
Graphical abstract
Highlights
-
•
PGR and its target genes are hypermethylated in uterine leiomyoma (LM) stem cells.
-
•
PGR regulates pathways critical for LM stem cell differentiation and depletion.
-
•
PGR is necessary for TET expression and global DNA methylation in LM.
-
•
5′-Aza increases PGR level and potentiated antiprogestin effects to shrink LM.
Liu and colleagues find that hypermethylation of PGR and its target genes in leiomyoma stem cells prevents their depletion via differentiation. PGR-regulated TET expression and global DNA methylation in leiomyoma cells. A DNA methylation inhibitor activated PGR signaling, stimulated stem cell depletion, and synergized with antiprogestin to shrink tumors; this combination may be a future treatment for leiomyoma.
Introduction
Uterine leiomyoma (LM) is the most common tumor in reproductive-age women, affecting ~80% of women by age 50 years (Bulun, 2013; Kim et al., 2013). LM can cause significant morbidity, including excessive uterine bleeding, anemia, and labor obstruction and may delay the diagnosis of ovarian cancer (Bulun, 2013). Approximately 250,000 hysterectomies or myomectomies are performed annually in the US to remove LM, costing up to $34.4 billion (Cardozo et al., 2012). No new pharmaceutical therapy has been approved in the US for LM since the 1990s; that treatment, a GnRH agonist, has limited efficacy and significant side effects, with frequent regrowth of tumors. There is an urgent need for new treatment options for LM.
Stem cells are critical for normal tissue and disease development. We identified three molecularly and functionally distinct cell populations in LM with hierarchical differentiation: LM stem-like cells (LSCs, 5%), LM intermediate cells (LICs, 7%), and terminally differentiated LM cells (LDCs, 88%) (Moravek et al., 2017). Progesterone (P4), its receptor (PGR), and LSCs are indispensable for LM growth; however, LSCs are deficient in PGR, suggesting that paracrine interactions between the LM cell populations mediate the response to P4 (Mas et al., 2012; Ono et al., 2013). LM and breast cancer share some similarities in hormone responses (Kim et al., 2013; Yin et al., 2012). PGR signaling plays dual roles in mammary gland, regulating mammary stem cell differentiation and proliferation through paracrine interaction with adjacent differentiated cells (Joshi et al., 2010; Lee et al., 2013). In LM, we observed that P4, via PGR expressed in LDCs, stimulates paracrine signaling that induces the proliferation of PGR-deficient LSCs, leading to tumor growth (Ikhena et al., 2018; Moravek et al., 2017; Ono et al., 2013). Whether PGR regulates LSC differentiation remains unknown.
Epigenetic alterations, such as DNA methylation, play crucial roles in stem cell regulation and disease progression (Ohm et al., 2007; Sheaffer et al., 2014). Previous studies demonstrated dysregulated DNA methylation in LM versus normal myometrium (MM) (Navarro et al., 2012) (Maekawa et al., 2013). We recently demonstrated that LSCs harbor a unique DNA methylome, which suppresses key genes important for differentiation (Liu et al., 2020). In this report, we examined how DNA methylation in LSCs affects PGR function and alters LSC differentiation and disease progression.
Results
PGR expression is inhibited by hypermethylation in LSCs
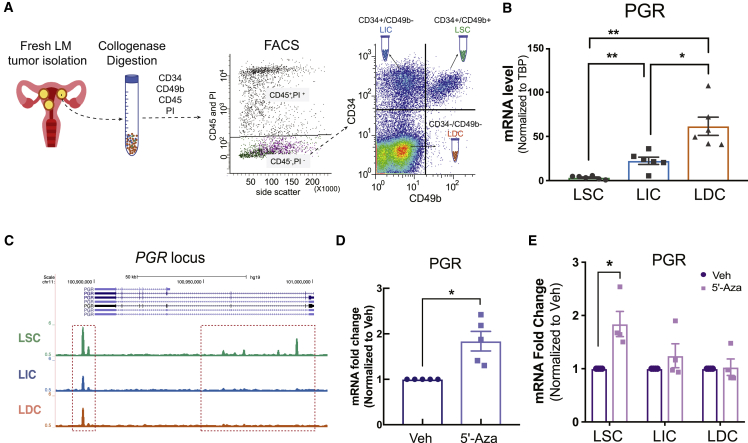
We used fluorescence-activated cell sorting (FACS) to isolate three cell populations from fresh LM tissue: LSCs (CD34+CD49b+), LICs (CD34+CD49b−), and LDCs (CD34−CD49b−) and evaluated the mRNA levels of PGR in each cell population (Figure 1A). PGR mRNA expression was lower in LSCs compared with LICs and LDCs (Figure 1B), whereas the PGR gene was hypermethylated at several intronic regions in LSCs (Figure 1C). To determine whether hypermethylation accounts for the low PGR transcriptional activity in LSCs, we treated total LM cells (non-sorted cells dissociated from LM) and FACS-sorted individual cell populations with vehicle (DMSO) or the DNA methylation inhibitor, 5′-Aza. 5′-Aza increased PGR mRNA levels in total LM cells and LSCs (Figures 1D and 1E). These data suggest that hypermethylation suppresses PGR signaling in LSCs.
Figure 1.
PGR gene expression is inhibited by DNA methylation in LSCs
(A) Schematic showing the workflow for isolation of three LM cell populations from fresh LM tissue.
(B) Real-time qPCR of PGR mRNA levels in each LM population (n = 6 patients, one-way ANOVA, ∗∗p < 0.01, ∗p < 0.05).
(C) Representative genome browser view shows methylation levels of the most significant differentially methylated regions around the PGR gene locus among the three LM cell populations.
(D) Real-time qPCR of PGR mRNA levels in total primary LM cells treated with 5′-Aza (100 nM) or vehicle (DMSO) for 96 h (n = 5 patients, t test, ∗p < 0.05).
(E) Bar graph shows the PGR mRNA fold change in each LM cell population treated with 5′-Aza (96 h, 100 nM) versus vehicle (n = 4 patients, two-way ANOVA, ∗p < 0.05).
PGR mRNA expression has been shown to be lower in MM versus LM (Liu et al., 2019). We analyzed our published MethylCap-seq data (GSE113108) and found that the PGR gene locus was hypermethylated in MM compared with LM, showing a similar pattern observed in LSCs versus LICs/LDCs (Figure S1A). 5′-Aza treatment also induced PGR gene expression in MM cells (Figure S1B), suggesting that 5′-Aza also affects PGR signaling in MM.
PGR cistromes are hypermethylated and associated with repressive gene transcription activity in LSCs
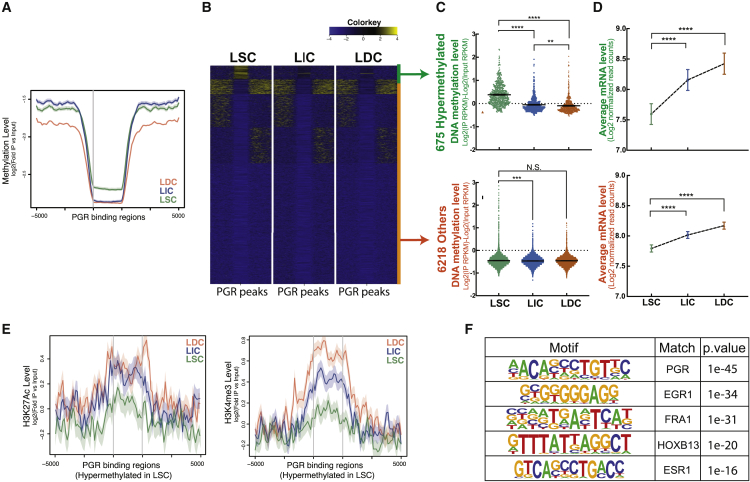
Although PGR plays a critical role in LM development, the complex mechanisms underlying PGR-mediated transcriptional activity, specifically during LSC differentiation, remain unclear (Ishikawa et al., 2010). To evaluate the role of DNA methylation in regulating PGR downstream signaling, we examined the methylation status adjacent to global PGR-binding sites in each LM cell population. We performed PGR chromatin immunoprecipitation sequencing (ChIP-seq), searched for PGR-binding sites that were only present in all 5 LM tissues, and identified 6,893 regions. We then integrated the PGR cistrome with published methylome data and found that, among the three populations, LSCs had the highest overall DNA methylation level in regions at or adjacent to (<1 kb) the 6,893 PGR-binding sites (Figure 2A) (Liu et al., 2020). K-means clustering pinpointed a cohort of PGR-binding regions (675 of 6,893, 9.8%) that were markedly hypermethylated in LSCs compared with LICs and LDCs (Figures 2B and 2C). DNA methylation levels were lower at the other 6,218 PGR-binding regions and showed only small differences among the three cell populations (Figure 2C).
Figure 2.
PGR cistromes are hypermethylated and associated with repressive gene transcription activity in LSCs
(A) Average line plots show DNA methylation levels of 6,893 PGR-binding sites in the three LM cell populations (n = 5 patients, 6,893 PGR peaks ± 5,000 bp).
(B) Heatmaps show the DNA methylation level around PGR-binding sites in each LM population. K-means clustering identified PGR-binding sites that were hypermethylated in LSCs versus LICs and LDCs. Yellow indicates hypermethylation and blue indicates hypomethylation.
(C) Dot plots show quantification of DNA methylation levels of PGR-binding sites in each LM population. Top: PGR-binding sites with hypermethylation in LSCs; bottom: other PGR-binding sites (one-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(D) Average dot plots show mRNA levels (RPKM) of PGR target genes in each LM population. Top: genes associated with 675 PGR-binding sites hypermethylated in LSCs; bottom: genes associated with other PGR-binding sites (one-way ANOVA, ∗∗∗∗p < 0.0001).
(E) Average line plots show histone modification levels (left: H3K27Ac level; right: H3K4me3 level) of PGR-binding sites with hypermethylation in LSCs.
(F) Homer motif analysis of PGR-binding sites related to hypermethylation in LSCs.
Using RNA sequencing (RNA-seq), we further evaluated gene transcript levels associated with PGR-binding sites (Liu et al., 2020). Compared with LICs and LDCs, LSCs expressed the lowest levels of genes adjacent to the 675 hypermethylated PGR-targeted regions and other PGR-binding regions (Figure 2D, top and bottom panel, respectively). The expression difference of genes near the 675 hypermethylated PGR-targeted regions was particularly dramatic, possibly due to hypermethylation-related suppression in LSCs. We also found the lowest enrichment of active histone marks (H3K27ac, H3K4me3) associated with these regions in LSCs, confirming the inhibited status of the neighboring genes (Figure 2E) (Liu et al., 2020).
To evaluate transcription factors other than PGR that might be affected by hypermethylation, we conducted motif analysis to identify consensus sequences enriched in the 675 hypermethylated PGR-binding sites. P4 response elements and several other hormone-related transcription factor-binding elements, such as FRA1/AP-1 and ESR1, were highly enriched in the hypermethylated PGR-binding sites in LSCs (Figure 2F), suggesting that the disruption of P4 signaling in LSCs is accomplished by both PGR deficiency and hypermethylation of hormone-response elements.
Next, we investigated whether DNA methylation influences PGR recruitment to its target gene loci to modulate gene transcription. As a proof-of-concept, we selected several candidate genes, including TIMP3, ROR2, GREB1, MYH11, and WNT5A, which showed the lowest gene expression by RNA-seq and harbor hypermethylated PGR-binding sites in LSCs (Liu et al., 2020). These genes have established roles in hormone signaling, stem cell regulation, and carcinogenesis or LM pathogenesis (Cheng et al., 2018; Goikuria et al., 2018; Jackson et al., 2015; Tarfiei et al., 2011; Zhou et al., 2017). We treated primary LM cells with 5′-Aza and evaluated PGR-binding activity at the hypermethylated sites identified in LSCs. 5′-Aza enhanced PGR recruitment to its target gene loci in primary LM cells (Figure S2A). We then examined whether 5′-Aza regulates the expression of these genes and whether PGR is necessary for the regulation. 5′-Aza significantly increased the mRNA levels of these genes (Figure S2B). PGR knockdown not only significantly decreased mRNA levels of the five candidate genes but also completely blocked 5′-Aza-stimulated expression of GREB1, ROR2, and WNT5A, indicating that PGR is critical for their expression (Figure S2B).
PGR target genes are involved in LSC differentiation
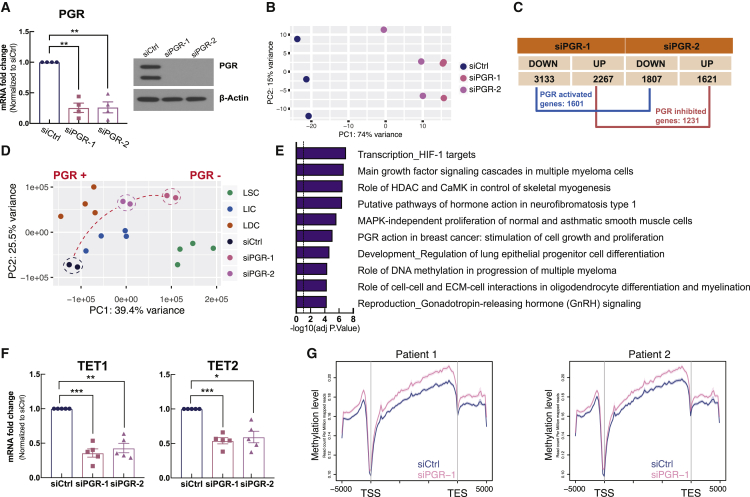
As upregulation of PGR is a hallmark of LSC differentiation to LICs/LDCs (Ono et al., 2013; Yin et al., 2015), we investigated the potential role of PGR signaling in LSC differentiation by relating PGR target gene expression to transcriptional alterations during the LSC transition to LICs/LDCs. To evaluate the impact of PGR on the transcriptome, we performed RNA-seq in total primary LM cells with endogenous PGR knockdown by small interfering RNA (siRNA). Both siPGR-1 and siPGR-2 significantly downregulated PGR mRNA and protein levels (Figure 3A). RNA-seq (on three samples with knockdown efficiency >70%) showed a significant transcriptome shift caused by PGR knockdown, identifying 2,832 genes regulated by both siPGR-1 and siPGR-2, with 1,601 genes downregulated and 1,231 genes upregulated (Figures 3B and 3C). Focusing on the genes previously identified as differentially expressed between LSCs and LICs/LDCs, principal-component analysis (PCA) indicated that, upon PGR knockdown, the transcriptome of total primary LM cells shifted toward that of LSCs (Figure 3D) (Liu et al., 2020). Based on PCA and hierarchical clustering analyses, one outlier that had significantly higher PGR expression than the other two samples (~1.5- and ~3-fold higher before and after knockdown, respectively) was excluded from this integration assay (Figures S3A–S3C). Gene set enrichment analysis was performed to functionally profile the identified PGR-regulated genes; PGR knockdown inhibited genes involved in extracellular matrix (ECM) regulation, a phenotype of differentiated LM cells, and activated genes related to cell cycle and proliferation (Figure S3D).
Figure 3.
PGR target genes are important for LSC differentiation
(A) Left panel: real-time qPCR quantification of PGR mRNA levels in LM cells transfected with two different PGR siRNAs (siPGR-1 and siPGR-2) or scrambled control siRNA (siCtrl) for 96 h (n = 4 patients, one-way ANOVA, ∗∗p < 0.01). Right panel: representative immunoblot shows PGR protein levels after PGR knockdown.
(B) Principal-component analysis (PCA) plot shows transcriptome clustering among siCtrl and siPGR samples (n = 3 patients).
(C) Table shows up- and downregulated genes after siPGR transfections.
(D) PCA plot shows clustering of LM populations (n = 4 patients) and LM total primary cells transfected with siCtrl or siPGRs (n = 2 patients) based on mRNA levels of the 4,357 differentially expressed genes between LSCs and LICs/LDCs.
(E) Pathway and network enrichment analysis (Metacore) of the 651 PGR direct target genes.
(F) Bar graphs show mRNA levels of TET1 and TET2 in siCtrl- and siPGR-transfected primary LM cells (n = 5 patients, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(G) Average line plot comparing DNA methylation levels between siCtrl- and siPGR-1-transfected primary LM cells at the gene body regions (n = 2 patients).
Integrating the PGR tissue ChIP-seq and PGR knockdown RNA-seq data, we found that PGR-binding sites were present near 651 PGR-regulated genes, 436 activated and 215 inhibited, suggesting that these genes are potentially direct targets of PGR (Figure S3E). These genes were highly enriched in functional pathways involved in LM development, including myogenesis, PGR action, progenitor cell differentiation, ECM-cell interaction, and DNA methylation (Figure 3E). These data suggest that PGR may be a critical transcription factor governing the transcriptional shift during LSC differentiation.
Interestingly, “DNA methylation” was identified as a pathway regulated by PGR. Focusing on this pathway, we discovered PGR binds to two key DNA demethylases, TET1 and TET2, and upregulates their expression (Figures S3F and 3F). Transcript levels of TET methylcytosine dioxygenases were found to be lower in LSCs versus LICs and LDCs, which may account for the global hypermethylation identified in the stem cell population (Liu et al., 2020). Given the important role of TETs in actively regulating DNA methylation, these findings prompted us to investigate whether PGR influences DNA methylation in LM cells. We performed MethylCap-seq to determine the global DNA methylation profiles in cells with or without PGR knockdown. Transient PGR knockdown upregulated overall DNA methylation levels around gene body regions in total LM cells (Figure 3G). Together, we found that DNA methylation affects PGR signaling in LSCs and PGR mediates methylation via transcriptional regulation of DNA demethylases, suggesting that DNA methylation and PGR play an integral role in regulating LSC stemness.
DNA methylation inhibitor 5′-Aza enhances the therapeutic efficacy of antiprogestin in LM
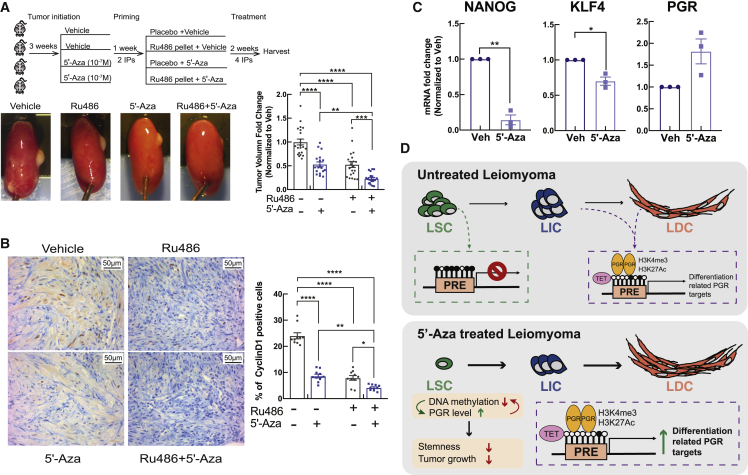
As demonstrated above, 5′-Aza upregulated PGR expression in LSCs. RU486 is a P4 antagonist that competes with P4 for PGR binding and alters receptor conformation (Allan et al., 1992). We tested whether 5′-Aza alters the inhibitory effect of RU486 on the growth of existing LM tumors using an in vivo mouse xenograft model. Primary LM cell xenografts were established for 3 weeks, then mice were given vehicle (PBS) or 5′-Aza twice per week followed by treatment with or without RU486 (Figure 4A). 5′-Aza or RU486 alone partially decreased LM growth, but the combination treatment completely blocked LM growth. The combined treatment also significantly decreased the number of cyclin D1-positive cells compared with the vehicle and RU486-only groups (Figure 4B). RU486 but not 5′-Aza treatment increased cell density in the xenografted tumors (Figure S4A), consistent with our previous report that RU486 partially shrinks LM via reduction in ECM volume and/or cell size (Ishikawa et al., 2010). 5′-Aza treated xenografts showed significantly lower NANOG and KLF4 mRNA levels and slightly higher PGR mRNA expression (Figure 4C). These findings indicate that 5′-Aza treatment synergizes with antiprogestin to reduce tumor size, at least in part by decreasing LSC stemness and tumor cell proliferation.
Figure 4.
DNA methylation inhibitor 5′-Aza sensitizes LM to antiprogestin treatment
(A) Top: workflow of in vivo xenograft mouse experiments using primary LM cells. Bottom left: representative pictures of regenerated tumors under each treatment. Bottom right: tumor volume quantification (n = 3 patient tissues; vehicle group kidney graft n = 22; RU486 group kidney graft, n = 21; 5′-Aza group kidney graft, n = 18; RU486 + 5′-Aza group kidney graft, n = 20, two-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(B) Left panel, representative images of immunohistochemical staining of Cyclin D1; right panel, percentage of Cyclin D1-positive cells among total cells counted at 40× magnification (n = 10 fields from three patients, two-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
(C) Bar graphs show mRNA levels of NANOG, KLF4, and PGR in xenograft tumors treated with vehicle or 5′-Aza (n = 3 patients, t test, ∗p < 0.05, ∗∗p < 0.01).
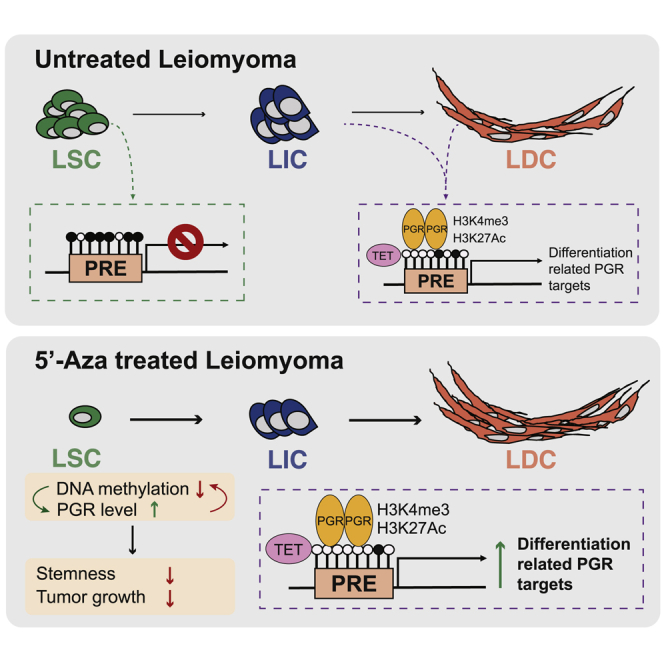
(D) Schematic of the proposed role of PGR signaling and DNA methylation interaction in the regulation of LSC stemness and LM growth.
Discussion
We demonstrated crosstalk between P4 signaling and DNA methylation that regulates LSC stemness and LM growth (Figure 4D). The PGR gene locus and its genome-wide cistrome were hypermethylated in LSCs, repressing gene expression fundamental for LSC differentiation. The demethylating reagent 5′-Aza not only stimulated PGR expression but also increased recruitment of PGR to its target gene loci to activate expression. PGR upregulated the TET methylcytosine dioxygenase, and PGR depletion by siRNA led to global hypermethylation in LM cells. Furthermore, 5′-Aza increased the efficacy of RU486 in shrinking existing LM in a xenograft mouse model. These observations suggest a potential feedback loop between DNA methylation and P4 signaling to induce LSC differentiation and sensitize LM to antiprogestin treatment.
In the mammalian genome, DNA methylation is crucial for the regulation of differentiation-related gene expression in many self-renewing tissues (Sen et al., 2010). We recently reported that the LSC genome is hypermethylated, especially at the loci of genes crucial for differentiation (Liu et al., 2020). Demethylation decreased LSC numbers and inhibited their colony-forming ability, likely by stimulating LSC differentiation. Our finding that the PGR gene locus is hypermethylated in LSCs, which inhibits PGR expression, is consistent with previous findings in endometrial cancer and endometriosis (Sasaki et al., 2001; Wu et al., 2006). In LSCs, we identified several tissue-specific methylation sites around the PGR locus, with the most significant hypermethylation in the intronic region (Figure 1C). The hypermethylation landscape in LSCs interfered with the interaction of PGR with its cistrome and suppressed PGR target gene expression, further blocking P4 signaling in LSCs.
In the complex biology of LM, P4 and PGR play dual roles (Joshi et al., 2010; Lee et al., 2013). P4/PGR in LICs/LDCs induces paracrine signaling that acts on PGR-deficient LSCs to stimulate self-renewal and proliferation (Moravek et al., 2017; Ono et al., 2013). Our current findings suggest another intriguing role of PGR: stem cell differentiation. PGR target genes are enriched in pathways critical for LSC differentiation and LM development, some of which were inhibited by hypermethylation in LSCs. Knockdown of PGR shifted the transcriptome of total primary LM cells toward that of LSCs. Our data also indicate that PGR signaling can influence LSC differentiation by affecting the DNA methylation process. At a single-gene level, Verde et al. (2018) recently reported that PGR maintains ESR1 expression by preserving low DNA methylation at the ESR1 promoter in breast cancer cells. We found that PGR regulates TET1 and TET2 expression via binding to their promoter regions (Figures S3F) and that transient PGR depletion upregulated DNA methylation levels around gene body regions in LM cells. Together with a previous study showing that TETs are expressed at low levels in LSCs, these findings suggest that PGR activation may contribute to global DNA methylation loss during LSC differentiation (Liu et al., 2020).
Notably, cotreatment of an LM xenograft mouse model with RU486 and 5′-Aza, but not either reagent alone, was able to almost completely shrink LM tumors. Therefore, it is reasonable to suggest that targeting the feedforward interaction between DNA methylation and PGR signaling may be a potential strategy to accelerate LSC differentiation and increase susceptibility to antiprogestin, leading to tumor eradication. However, as a demethylating agent, 5′-Aza may have toxic effects (Christman, 2002). Using a similar treatment regimen used by other publications showing no obvious adverse effects, we observed a slight body weight loss in mice treated with 5′-Aza (Figure S4B) (Zorn et al., 2007). Our findings that 5′-Aza increased PGR gene expression in MM cells and that PGR-binding sites were hypermethylated in MM versus LM, also suggest that 5′-Aza treatment may affect PGR signaling pathway activity and function in normal MM (Liu et al., 2019). Thus, we will continue investigating LSC differentiation to identify more specific druggable targets to shrink existing LM and prevent regrowth.
These findings not only reveal crosstalk between epigenetic and hormonal regulation during LSC differentiation but also suggest a possible treatment strategy in which LSCs may be sensitized to antiprogestin by stimulating differentiation. By dissecting the complex interactions between P4 action and DNA methylation during LM stem cell differentiation, we provide new insights into the mechanisms behind the epigenomic regulation of hormone-dependent tumor growth.
Experimental procedures
A comprehensive description of the methodology is included in the supplemental information. All antibodies and primers used in this manuscript are listed in Tables S1–S3.
Tissue collection
Northwestern University’s institutional review board approved the use of human tissue. After receiving verbal informed consent, MM and LM tissues were obtained from 40 premenopausal women undergoing myomectomy or hysterectomy (age 38 ± 9 years, range 29–47 years). Patients receiving hormone treatment within 6 months before surgery were excluded. Tissues were dissociated and cells were isolated as described previously (Yin et al., 2015).
Data and materials availability
Data from PGR ChIP-seq, MethylCap-seq, and RNA-seq following PGR knockdown have been deposited in the GEO under the accession code Database: GSE148257. Methylome, transcriptome, and histone modification data for each LM cell population were previously published and deposited in the GEO under the accession code Database: GSE138051.
Author contributions
S.L., P.Y., and S.E.B. conceptualized and designed the study. S.L. performed most of the experiments and data analysis with help from P.Y., A.J.D., H.Z., S.A.K., and J.S.C.V. J.X. and Y.D. contributed to data analyses. S.L. wrote the manuscript under the supervision of P.Y. and S.E.B. All authors commented on the manuscript and approved its content.
Acknowledgments
We acknowledge the Northwestern University Flow Cytometry Facility, NUSeq Core, and Pathology Core Facility, which are co-funded by a National Cancer Institute Cancer Center grant CA060553. Flow Cytometry Cell Sorting was performed on a BD FACSAria SORP system, purchased through the support of NIH 1S10OD011996-01. This work was supported by the National Institutes of Health grants P01-HD057877 and P50-HD098580 (to S.E.B. and P.Y.).
Published: August 12, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.013.
Supplemental information
References
- Allan G.F., Tsai S.Y., Tsai M.J., O'Malley B.W. Ligand-dependent conformational changes in the progesterone receptor are necessary for events that follow DNA binding. Proc. Natl. Acad. Sci. U S A. 1992;89:11750–11754. doi: 10.1073/pnas.89.24.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulun S.E. Uterine fibroids. N. Engl. J. Med. 2013;369:1344–1355. doi: 10.1056/NEJMra1209993. [DOI] [PubMed] [Google Scholar]
- Cardozo E.R., Clark A.D., Banks N.K., Henne M.B., Stegmann B.J., Segars J.H. The estimated annual cost of uterine leiomyomata in the United States. Am. J. Obstet. Gynecol. 2012;206:211 e211–219. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Michalski S., Kommagani R. Role for growth regulation by Estrogen in Breast Cancer 1 (GREB1) in hormone-dependent cancers. Int. J. Mol. Sci. 2018;19:2543. doi: 10.3390/ijms19092543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman J.K. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Goikuria H., Freijo M.D.M., Vega Manrique R., Sastre M., Elizagaray E., Lorenzo A., Vandenbroeck K., Alloza I. Characterization of carotid smooth muscle cells during phenotypic transition. Cells. 2018;7:23. doi: 10.3390/cells7030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikhena D.E., Liu S., Kujawa S., Esencan E., Coon J.S.t., Robins J., Bulun S.E., Yin P. RANKL/RANK pathway and its inhibitor RANK-Fc in uterine leiomyoma growth. J. Clin. Endocrinol. Metab. 2018;103:1842–1849. doi: 10.1210/jc.2017-01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ishi K., Serna V.A., Kakazu R., Bulun S.E., Kurita T. Progesterone is essential for maintenance and growth of uterine leiomyoma. Endocrinology. 2010;151:2433–2442. doi: 10.1210/en.2009-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson H.W., Waterhouse P., Sinha A., Kislinger T., Berman H.K., Khokha R. Expansion of stem cells counteracts age-related mammary regression in compound Timp1/Timp3 null mice. Nat. Cell Biol. 2015;17:217–227. doi: 10.1038/ncb3118. [DOI] [PubMed] [Google Scholar]
- Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- Kim J.J., Kurita T., Bulun S.E. Progesterone action in endometrial cancer, endometriosis, uterine fibroids, and breast cancer. Endocr. Rev. 2013;34:130–162. doi: 10.1210/er.2012-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Gallego-Ortega D., Ledger A., Schramek D., Joshi P., Szwarc M.M., Cho C., Lydon J.P., Khokha R., Penninger J.M., et al. Progesterone drives mammary secretory differentiation via RankL-mediated induction of Elf5 in luminal progenitor cells. Development. 2013;140:1397–1401. doi: 10.1242/dev.088948. [DOI] [PubMed] [Google Scholar]
- Liu S., Yin P., Kujawa S.A., Coon J.S., Okeigwe I., Bulun S.E. Progesterone receptor integrates the effects of mutated MED12 and altered DNA methylation to stimulate RANKL expression and stem cell proliferation in uterine leiomyoma. Oncogene. 2019;38:2722–2735. doi: 10.1038/s41388-018-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yin P., Xu J., Dotts A.J., Kujawa S.A., Coon V.J., Zhao H., Hilatifard A.S., Dai Y., Bulun S.E. Targeting DNA methylation depletes uterine leiomyoma stem-cell enriched population by stimulating their differentiation. Endocrinology. 2020:bqaa143. doi: 10.1210/endocr/bqaa143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa R., Sato S., Yamagata Y., Asada H., Tamura I., Lee L., Okada M., Tamura H., Takaki E., Nakai A., et al. Genome-wide DNA methylation analysis reveals a potential mechanism for the pathogenesis and development of uterine leiomyomas. PLoS One. 2013;8:e66632. doi: 10.1371/journal.pone.0066632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A., Cervello I., Gil-Sanchis C., Faus A., Ferro J., Pellicer A., Simon C. Identification and characterization of the human leiomyoma side population as putative tumor-initiating cells. Fertil. Steril. 2012;98:741–751.e6. doi: 10.1016/j.fertnstert.2012.04.044. [DOI] [PubMed] [Google Scholar]
- Moravek M.B., Yin P., Coon J.S.t., Ono M., Druschitz S.A., Malpani S.S., Dyson M.T., Rademaker A.W., Robins J.C., Wei J.J., et al. Paracrine pathways in uterine leiomyoma stem cells involve insulin-like growth factor 2 and insulin receptor A. J. Clin. Endocrinol. Metab. 2017;102:1588–1595. doi: 10.1210/jc.2016-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Yin P., Monsivais D., Lin S.M., Du P., Wei J.J., Bulun S.E. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS One. 2012;7:e33284. doi: 10.1371/journal.pone.0033284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm J.E., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W., et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Yin P., Navarro A., Moravek M.B., Coon J.S.t., Druschitz S.A., Serna V.A., Qiang W., Brooks D.C., Malpani S.S., et al. Paracrine activation of WNT/beta-catenin pathway in uterine leiomyoma stem cells promotes tumor growth. Proc. Natl. Acad. Sci. U S A. 2013;110:17053–17058. doi: 10.1073/pnas.1313650110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Dharia A., Oh B.R., Tanaka Y., Fujimoto S., Dahiya R. Progesterone receptor B gene inactivation and CpG hypermethylation in human uterine endometrial cancer. Cancer Res. 2001;61:97–102. [PubMed] [Google Scholar]
- Sen G.L., Reuter J.A., Webster D.E., Zhu L., Khavari P.A. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer K.L., Kim R., Aoki R., Elliott E.N., Schug J., Burger L., Schubeler D., Kaestner K.H. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarfiei G., Noruzinia M., Soleimani M., Kaviani S., Mahmoodinia Maymand M., Farshdousti Hagh M., Pujol P. ROR2 promoter methylation change in osteoblastic differentiation of mesenchymal stem cells. Cell J. 2011;13:11–15. [PMC free article] [PubMed] [Google Scholar]
- Verde G., De Llobet L.I., Wright R.H.G., Quilez J., Peiro S., Le Dily F., Beato M. Unliganded progesterone receptor governs estrogen receptor gene expression by regulating DNA methylation in breast cancer cells. Cancers (Basel) 2018;10:371. doi: 10.3390/cancers10100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Strawn E., Basir Z., Halverson G., Guo S.W. Promoter hypermethylation of progesterone receptor isoform B (PR-B) in endometriosis. Epigenetics. 2006;1:106–111. doi: 10.4161/epi.1.2.2766. [DOI] [PubMed] [Google Scholar]
- Yin P., Ono M., Moravek M.B., Coon J.S.t., Navarro A., Monsivais D., Dyson M.T., Druschitz S.A., Malpani S.S., Serna V.A., et al. Human uterine leiomyoma stem/progenitor cells expressing CD34 and CD49b initiate tumors in vivo. J. Clin. Endocrinol. Metab. 2015;100:E601–E606. doi: 10.1210/jc.2014-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Roqueiro D., Huang L., Owen J.K., Xie A., Navarro A., Monsivais D., Coon J.S.t., Kim J.J., Dai Y., et al. Genome-wide progesterone receptor binding: cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells. PLoS One. 2012;7:e29021. doi: 10.1371/journal.pone.0029021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Kipps T.J., Zhang S. Wnt5a signaling in normal and cancer stem cells. Stem Cells Int. 2017;2017:5295286. doi: 10.1155/2017/5295286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn C.S., Wojno K.J., McCabe M.T., Kuefer R., Gschwend J.E., Day M.L. 5-Aza-2′-deoxycytidine delays androgen-independent disease and improves survival in the transgenic adenocarcinoma of the mouse prostate mouse model of prostate cancer. Clin. Cancer Res. 2007;13:2136. doi: 10.1158/1078-0432.CCR-06-2381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.