Summary
Human neurons engineered from induced pluripotent stem cells (iPSCs) through neurogenin 2 (NGN2) overexpression are widely used to study neuronal differentiation mechanisms and to model neurological diseases. However, the differentiation paths and heterogeneity of emerged neurons have not been fully explored. Here, we used single-cell transcriptomics to dissect the cell states that emerge during NGN2 overexpression across a time course from pluripotency to neuron functional maturation. We find a substantial molecular heterogeneity in the neuron types generated, with at least two populations that express genes associated with neurons of the peripheral nervous system. Neuron heterogeneity is observed across multiple iPSC clones and lines from different individuals. We find that neuron fate acquisition is sensitive to NGN2 expression level and the duration of NGN2-forced expression. Our data reveal that NGN2 dosage can regulate neuron fate acquisition, and that NGN2-iN heterogeneity can confound results that are sensitive to neuron type.
Keywords: NGN2, induced neurons, scRNA-seq, cell fate engineering, neuron fate acquisition
Highlights
-
•
NGN2-iNs are molecularly heterogeneous
-
•
NGN2-iNs subtypes have signatures of central and peripheral nervous system
-
•
Neural fate acquisition is sensitive to the level and duration of NGN2 expression
In this article, Lin, He, and Ebert et al. show that neurons induced through forced expression of NGN2 in iPSCs (NGN2-iNs) present substantial molecular heterogeneity. Using scRNA-seq, they found NGN2-iN subtypes that express genes associated with the peripheral nervous system, and that neural fate acquisition is sensitive to the level and duration of NGN2 expression.
Introduction
Human cell types engineered from induced pluripotent stem cells (iPSCs) through transcription factor overexpression are widely used to study the mechanisms controlling cell fate differentiation, to model human diseases, and to identify potential therapies (Guo and Morris, 2017). Human neurons can be generated through the forced expression of the transcription factor neurogenin 2 (NGN2) with high efficiency and reproducibility (Zhang et al., 2013). These NGN2-induced neurons (NGN2-iNs) functionally mature into morphologically complex and electrophysiological active neurons after approximately 3–4 weeks of co-culture with astrocytes. The NGN2-iN system has been used extensively to understand neuron development and model disease (Lin et al., 2018). However, the characterization of NGN2-iNs so far has generally been limited to functional assays, biomarker expression, and bulk transcriptomics. There is a lack of comprehensive transcriptomic comparison with primary neuron subtypes and it is unclear whether any off-target fate emerges during the differentiation process. Single-cell sequencing methods provide powerful resolution into the heterogeneity of directed differentiation culture systems (Biddy et al., 2018, 2018, 2018; Camp et al., 2018; Karow et al., 2018). Previously, we have used single-cell mRNA sequencing (scRNA-seq) to dissect the differentiation path from mouse embryonic fibroblasts and human pericytes to neurons and identified previously undescribed heterogeneity generated by the overexpression of the pioneer factor ASCL1 (Karow et al., 2018; Treutlein et al., 2016). Here, we set out to characterize NGN2-iNeuron heterogeneity, identify the cell states that are generated during differentiation, and analyze the dynamics of the differentiation process using scRNA-seq.
Results
Heterogeneity of NGN2-induced neurons dissected by scRNA-seq
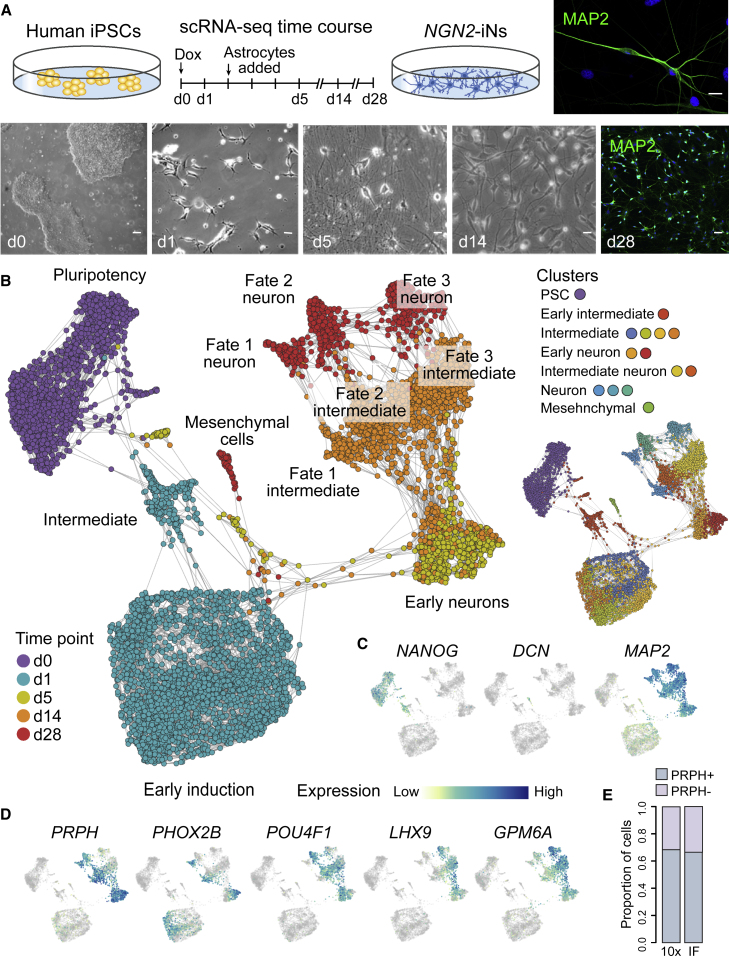
We generated a stable iPSC line expressing NGN2 that can be induced by doxycycline (Dox) and drives the differentiation toward iNeurons (Zhang et al., 2013). We then performed scRNA-seq (10× Genomics) at multiple time points during directed differentiation (Figure 1A). After filtering the data of astrocytes, multiplets, and cells with insufficient unique molecular identifiers (UMIs), a total of 6,764 cells (day 0 [d0], 1,412 cells; d1, 2,688 cells; d5, 524 cells; d14, 1,515 cells; d28, 625 cells) were included in the analysis. We combined all time course data and reconstructed the differentiation path of NGN2-iNs (Figures 1B–1D). Surprisingly, we found at least four transcriptionally distinct cell populations at the d28 time point. One population is marked by DCN/COL5A1 and we interpret this cluster as an off-target mesenchymal population (Figure 1C). In addition, we observed three different neuronal clusters that express high levels of pan-neuronal genes (MAP2, NCAM1) yet are molecularly distinct (Figure 1C). Two clusters have high expression of PRPH, an intermediate neurofilament that is highly expressed in neurons of the peripheral nervous system and some central nervous system regions that have neural projections toward peripheral structures (Yuan et al., 2012). These two PRPH+ clusters segregate into a PHOX2B+ cluster and a POU4F1+ cluster (Figure 1D). The other neuronal cluster is marked by GPM6A expression, which is expressed throughout both the central nervous system (CNS) and the spinal cord during mouse development (Figure 1D) (Diez-Roux et al., 2011).
Figure 1.
Diverse subpopulations emerge during NGN2-directed neuron differentiation into iNs from human iPSCs
(A) Schematic of scRNA-seq time course experiment and representative images from human iPSCs differentiating into NGN2-iNs. Cells were analyzed with scRNA-seq (10× Genomics) at multiple time points during differentiation. Immunohistochemical staining of NGN2-iNs at d28 with MAP2 (green) and DAPI (blue). Scale bars, 10 μm.
(B) SPRING embedding shows the developmental relationships of 409B2-derived NGN2-iNs with cells colored by time points (left) or cluster (right).
(C) Expression feature plots stem cell, mesenchymal, and neural marker genes.
(D) Expression feature plots of NGN2-iN clusters.
(E) Proportion of PRPH+ and PRPH− cells quantified using scRNA-seq (10×) or immunofluorescence.
See also Figures S1A and S1B.
We characterized the neural identity and presence of molecular heterogeneity in our NGN2-iN culture using immunofluorescence of TUBB3 and PRPH (Figures S1A and S1B). The percentage of PRPH+ cells was quantified to compose 67% of neural cells, comparable with the percentage estimated by scRNA-seq (Figures 1E and S1B). We examined the presence of common makers that were used to characterize NGN2-iNs and how they overlap with PRPH expression (Figures S1C–S1E). Most NGN2-iNs and PRPH+ cells express CUX1 and VGlut1, but not GAD1/2, supporting their cortical excitatory feature as reported previously (Zhang et al., 2013). However, unlike PRPH and other identified cluster markers, common neural markers are not able to resolve the heterogeneity in our dataset. We noticed that the percentage of PRPH+ cells in our dataset is higher than previous reports (Chen et al., 2020; Nickolls et al., 2020; Schörnig et al., 2021). This can be due to differences in protocol, the particular readout of neural identity used in the previous reports, or thresholds for assigning positive staining from immunohistochemistry (Figure S2). Together, these data suggest that NGN2-iNs generated from our protocol are comparable with other published reports of induced neurons resulting from NGN2 overexpression, and differences in the method of readout (selected markers versus whole transcriptome) can influence the interpretations of heterogeneity.
Transcriptome trajectory analysis along the path of NGN2-iN development
We further analyzed NGN2-iN developmental trajectories after additional integration and clustering of all time course data (Figures S3A and S3B). NGN2 induction resulted in major gene expression changes early on in programming (d0, d1, and d5), likely driven by the immediate downstream targets of NGN2. Based on the observation of rapid transition from iPSCs to cells committed to a neuronal fate (Figures 1B and S3B), we hypothesized that directed differentiation bypasses early transitional states that are usually observed in vivo to reach neuronal states. To test the hypothesis, we ordered NGN2-iNs in pseudotime based on transcriptome similarities and compared the resulting trajectories with development of neurons in brain organoids (Figures S3C and S3D) (Kanton et al., 2019). We observed an escape from the early developmental stages from pluripotency directly into neural precursor stages, skipping multiple intermediate stages, including neuroectoderm and neuroepithelium induction, supporting a more direct differentiation model (Figure S3D).
To investigate the molecular events underlying the dramatic developmental changes, we identified 3,231 genes with significant expression changes along the course of NGN2-iN development and segregated them into six clusters with their expression peak at different stages (Figure S3E). We performed functional enrichment analysis on these differentially expressed (DE) genes and recovered gene ontology terms related to neural development (Figure S3F; Table S1). We cross-referenced DE genes with annotated transcription factors (TFs) (Hu et al., 2019) to identify potential drivers of gene expression changes during NGN2-iN development (Figure S1G). We focused on TFs that changed from 6 to 12 h to d1 after Dox induction and constructed a gene-regulatory network (Aibar et al., 2017), incorporating transcription factor binding site prediction in promoters with TF-target co-expression (Figure S3H). NGN2 was predicted to connect with some TFs with the highest centrality in the constructed regulatory network, including POU5F1, HES6, and SOX11, supporting its role in driving direct reprogramming from iPSCs to induced neurons.
While neural induction begins before d1, the NGN2-iN heterogeneity emerges later as the expression of NGN2-iN subtype markers were detected after d1 (pseudotime Pt ∼0.4) (Figures S3B and S3I). Interestingly, we found that PHOX2B and POU4F1 had divergent expression from the beginning of their activation, while POU4F1 and GPM6A bifurcated later at d5 (pseudotime Pt ∼0.75) (Figures S3J and S3K). PRPH, on the other hand, was detected only after the PHOX2B and POU4F1 bifurcation, suggesting that it was activated independently in PHOX2B- and POU4F1-expressing cells (Figure S3I).
NGN2-iN heterogeneity is commonly detected
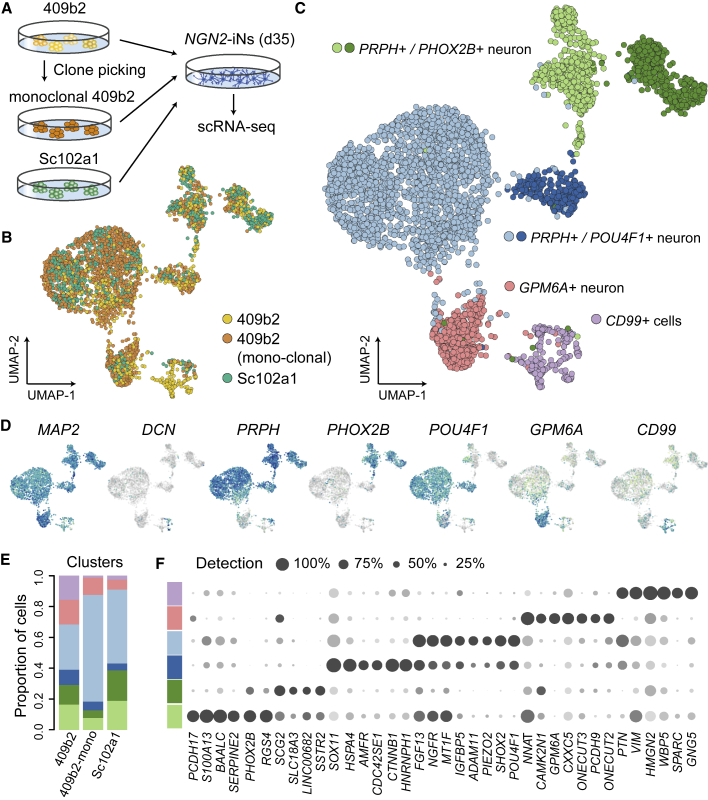
We next determined if the NGN2-iN heterogeneity results from heterogeneous iPSC populations used to induce iNs, or if the heterogeneity was specific to the particular iPSC line. We established a single iPSC clone (409B2 monoclonal) from the parent 409B2 line and additionally generated a polyclonal NGN2-inducible line from another individual (Sc102a1). We induced these lines and analyzed the resulting transcriptomes at d35 of differentiation (Figure 2A). We found that significant heterogeneity is still observed in an integrated analysis of the scRNA-seq data, with six molecularly distinct clusters containing cells from each of the starting iPSC lines. We note that the three main neural subtypes and off-target cells are all observed for each line (Figures 2B–2E). We searched for the top DE genes in the six clusters, and observed distinct gene expression patterns for each cluster (Figure 2F; Table S2). These data suggest that NGN2-based neural reprogramming is intrinsically heterogeneous, independent of the purity of the starting cell population, and that the heterogeneity is detected in neurons from multiple iPSC lines.
Figure 2.
NGN2-iN neuron diversity is recapitulated in multiple iPSC lines
(A–C) (A) scRNA-seq was performed on d35 NGN2-iNs from polyclonal 409B2, monoclonal 409B2, and polyclonal Sc102a1 iPSCs. (B and C) UMAP embedding of Seurat 3.0 integrated scRNA-seq data, with cells colored by cell source (B) or cluster annotated by marker genes (C).
(D) Feature plots showing the expression of marker genes.
(E) Stacked bar plot showing proportions of clusters in each sample.
(F) Dot plot of marker gene expression patterns and detection rates across clusters.
Molecular features of NGN2-iN subpopulations
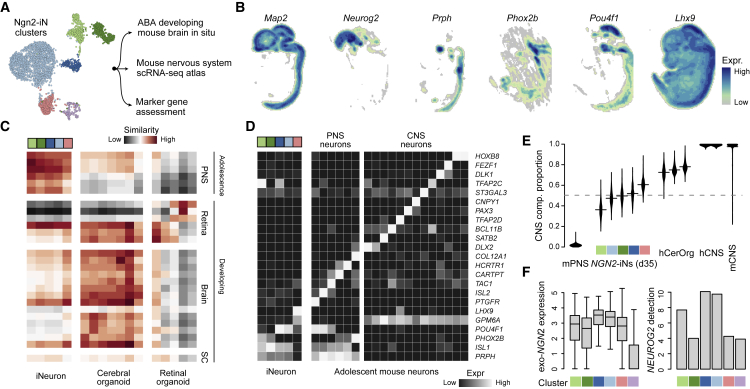
We next analyzed the molecular signatures that distinguished each NGN2-iN cluster. We compared the signatures with in situ hybridization (Ravasi et al., 2010) data from the Allen Developing Mouse Brain Atlas, and with single-cell transcriptome atlases containing primary neural cells (Figure 3A). We also assessed the expression of neurotransmitters and other markers of neuron specialization. Based on whole-embryo mouse ISH data (Thompson et al., 2014), we find that Ngn2 is expressed in progenitor zones in the developing telencephalon and many other brain structures (Figures 3B and S4). Prph is expressed in the neural retina, trigeminal nerve, and nuclei within the gray horn of the spinal cord. Phox2b is expressed in rhombencephalon/brain stem neurons as well as neurons in the peripheral nervous system (PNS). Pou4f1 is expressed in the retina, mesencephalon derivatives, trigeminal nerve, and gray horn nuclei. We next compared each NGN2-iN cluster with PNS and CNS neurons from primary reference cell atlases (Clark et al., 2019; La Manno et al., 2020; Zeisel et al., 2018) (Figure 3C). Unlike neurons in the iPSC-derived cerebral and retinal organoids, the NGN2-iN clusters did not show specific transcriptomic similarity to any CNS neuron subtypes (Figures 3C and S4D). Some NGN2-iN clusters were relatively similar to PNS neurons, especially the PRPH+ clusters, although they did not specify any of the PNS neuron subtypes. We explored if NGN2-iN expressed markers of primary neuron subtypes (Figures 3D and S4E). There is no clear in vivo neuron population as the counterpart of any NGN2-iN cluster. We deconvoluted the ratio of PNS/CNS identity for each cluster and found that all NGN2-iNs have mixed signatures of CNS- and PNS-derived neurons without a clearly established identity (Figure 3E). We find that the GPM6A+ cluster has more CNS features while the PRPH+ clusters have a biased PNS signature, in line with GPM6A and PRPH showing high expression in CNS and PNS neurons, respectively. Altogether, our data suggested that NGN2-iNs have a mixture of neuronal signatures, and we were not able to establish a clear identity of NGN2-iN populations. We note that this lack of in vivo counterpart could be due to incomplete reference cell atlases, as well as discrepancies between human and mouse neurons.
Figure 3.
Molecular signatures of NGN2-iN compared with primary neuronal cell types mouse reference atlases
(A) NGN2-iN subpopulation signatures were compared with diverse reference atlases.
(B) Spatial expression patterns of selected markers as maximum intensity projections across sagittal sections in the embryonic day 13.5 mouse brain from the Allen Developing Mouse Brain Atlas.
(C) Transcriptomic similarities between NGN2-iNs, other iPSC-derived neurons, and primary neurons represented as Pearson correlations between expression profiles. SC, spinal cord.
(D) Average expression of various marker genes of primary neuron subtypes in NGN2-iN clusters and primary mouse PNS and CNS neuron subtypes.
(E) Proportions of the estimated CNS component in NGN2-iNs, cerebral organoid neurons, and human/mouse primary mature PNS/CNS neurons.
(F) Expression of exogenous (left) and endogenous (right) NGN2 in different NGN2-iN clusters. The boxes show the lower and upper quartiles of the distributions. the bars extend to the min/max or 1.5x interquartile range.
The expression level of reprogramming factors could affect the outcome of reprogramming (Sommer et al., 2012) and lead in part to the heterogeneity of NGN2-iNs. We thus analyzed the relationship between NGN2 expression and the molecular identity of corresponding NGN2-iNs. The NGN2 expression level and proportion of NGN2-expressing cells was indeed lower in the off-target cluster (CD99+), in line with previous studies that failed reprogramming is linked to silenced reprogramming factors (Treutlein et al., 2016) (Figure 3F). Among the successfully reprogrammed neural clusters, we observed variable expression levels of NGN2 and proportion of NGN2-expressing cells, prompting us to examine whether NGN2 dosage affects the NGN2-iN reprogramming heterogeneity.
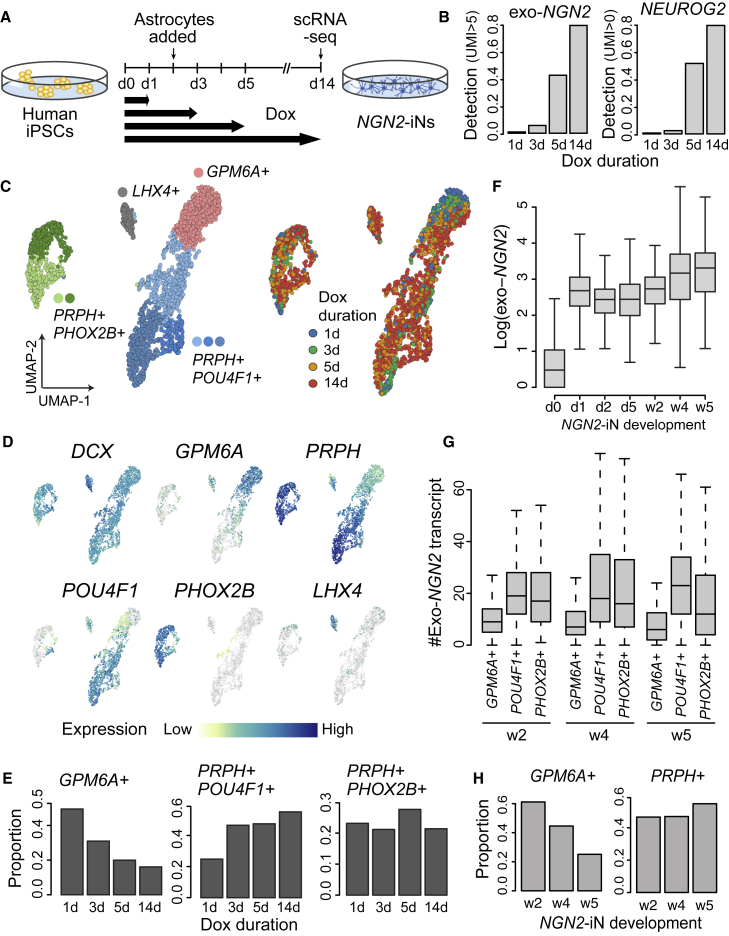
Duration of NGN2 induction affects NGN2-iN subtype configuration
We manipulated NGN2 expression by shortening the duration of Dox treatment and analyzed the resulting cells at d14 using scRNA-seq (Figure 4A). A total of 2,767 cells (d1, 311 cells; d3, 378 cells; d5, 727 cells; d14, 1,351 cells) were included in the analysis. As expected, the expression level of exogenous NGN2 is correlated with the duration of Dox treatment (Figure 4B). Endogenous NGN2 expression is also positively correlated with Dox treatment duration, likely as a result of positive autoregulation (Figure 4B) (Ejarque et al., 2013). Each of the previously identified major clusters were detected in all samples; however, the duration of Dox treatment affects the proportion of samples among each cluster (Figures 4C–4E). Specifically, we found that the GPM6A+ population was enriched in samples with shorter Dox treatment, while the PRPH+/POU4F1+ population was more abundant in samples with increased Dox treatment (Figure 4E). Given the observation that NGN2 expression could affect NGN2-iN fate specification, we revisited the time course data for NGN2-iN development. The expression of NGN2 remained nearly consistent during NGN2-iN development, with a slight increase of NGN2 expression at weeks 4 and 5 (Figures 4F and S3B). Interestingly, the expression level of NGN2 is consistently lower in GPM6A+ cells, independent of the duration of development (Figure 4G). Supporting this data, the proportion of GPM6A+ cells also decreased with longer NGN2-iN culture (Figure 4H). This could imply that prolonged expression of NGN2 potentially steers NGN2-iN fate away from CNS lineages. Altogether, these data show that the duration of NGN2 induction impacts the proportion of neuron subtypes that emerge in this single factor reprogramming paradigm.
Figure 4.
Neuronal fate specification is sensitive to NGN2 dosage
(A) Schematic for the doxycycline (Dox) treatment duration experiment.
(B) Detection rates for cells expressing exogenous NGN2 (>5 UMI) and endogenous NGN2 (>0 UMI) from each sample.
(C) UMAP embedding of NGN2-iN cells from each sample. scRNA-seq data were integrated using cluster similarity spectrum-based integration (He et al., 2020).
(D) UMAP plots colored by marker gene expression. DCX marks the neural lineages. LHX4 marks the identity of the off-target cluster.
(E) Proportion of cells per Dox treatment time point in each of the three neural clusters.
(F) Expression level of exogenous NGN2 from the time course experiment presented in Figure S1A. The boxes show the lower and upper quartiles of the distributions. the bars extend to the min/max or 1.5x interquartile range.
(G) Numbers of exogenous NGN2 transcripts in the three neural clusters from different time points of NGN2-iN development. The boxes show the lower and upper quartiles of the distributions. the bars extend to the min/max or 1.5x interquartile range.
(H) Proportion of cells from different time points of NGN2-iN development.
Discussion
Cell fate engineering of neural subtypes from human iPSCs using defined TFs provides extraordinary new inroads into disease modeling and therapy screening using human cells. Methods to rapidly generate mature human neurons are exciting and transformative for these endeavors. It has been established that the NGN2-iN protocol is able to reprogram stem cells to general neural fates, with less heterogeneity and higher consistency across multiple stem cell lines compared with traditional reprogramming strategies mediated by small-molecule inhibitors/activators. However, our analysis suggests that the emergent neuron population is heterogeneous, with the heterogeneity being consistent across different cell lines. We are unable to assign the neuron populations to a particular identity with high confidence. We note that this may be due to the fact that current single-cell and spatial transcriptome reference atlases are incomplete. However, without a specific matrix and guiding molecules it may be expected that neurons are not able to establish the molecular profile observed in vivo with high precision. Our data show that multiple NGN2-iN subpopulations are more similar to neurons of the PNS than CNS, and it is unclear if this culture paradigm is indicative of CNS functionality. Modifications of the NGN2-iN protocol by adding developmental patterning factors to the culture medium can steer neuron differentiation to a desired path (Nehme et al., 2018). Our data support a continued effort into identifying combinatorial transcription factor overexpression systems (Ravasi et al., 2010) and medium conditions that can support precise neuron cell type engineering. Furthermore, comprehensive human nervous system reference cell atlases are required to understand the identity of cell states that emerge in in-vitro-engineered neuron systems. Single-cell genomics and comparisons with high-dimensional reference atlases should become a field gold standard to assess the heterogeneity and precision of in vitro engineered neurons.
Experimental and computational procedures
Cell culture
All cells described in this work were incubated at 37°C, 5% CO2, and 90% humidity unless otherwise stated. 409B2 (RIKEN BRC Cell Bank), Sc102a1 (System Biosciences) stem cells, and corresponding rtTA/NGN2-derivatives were cultured in standard feeder-free conditions in mTeSR1 (STEMCELL Technologies) on plates coated with Matrigel (Corning). Primary cortical rat astrocytes (Gibco) were cultured in high-glucose DMEM containing 10% fetal calf serum and 1% pen/strep on plates coated with poly-D-lysine (Sigma-Aldrich). Fresh medium was added to the astrocytes every 4–5 days and passaged once a week with trypsin-EDTA digestion at a standard ratio of 1:2. Astrocytes were used up to passage 10, with passage 0 being the culture of initial isolation. rtTA/NGN2 double-positive stem cell lines were generated and differentiated into NGN2-iNs as described previously (Frega et al., 2017).
scRNA-seq library preparation and sequencing
To prepare scRNA-seq libraries from NGN2-iN single-cell suspensions, Chromium Single-cell 3′ Reagent Kits (10× Genomics, Pleasanton, CA, USA) were applied according to the manufacturer’s instructions. The Chromium Single-cell 3′ Reagent Kits v.2 was employed on NGN2-iN generated from 409B2 time course experiments, monoclonal 409B2 and Sc102a1 iPSCs with approximately 3,000 cells loaded per lane on a 10x microfluidic chip device. Chromium Single-cell 3′ Reagent Kit v.3 was used on Dox treatment duration experiments with nearly 8,000 cells loaded per lane. Quantification and quality control of the 10× library was carried out on a Bioanalyzer (Agilent) using high-sensitivity DNA chips. Libraries prepared from the 10× v.2 kit and v.3 kit were respectively sequenced on the Illumina HiSeq 2500 and Illumina NovaSeq S1 platform.
Data analysis of the scRNA-seq experiments
Cell Ranger was used to demultiplex raw base call files to FASTQ files, align reads to the reference genome and transcriptome with the default alignment parameters, demultiplex human and mouse cells, and generate the count matrices for the human cells. Seurat (v.3.1) was then applied to the human scRNA-seq data for further preprocessing. The scRNA-seq data of all 409b2 cells from iPSCs to NGN2-iN was integrated with cluster similarity spectrum (CSS) (He et al., 2020). The scRNA-seq data of NGN2-iN cells at day 35 of samples from the 409b2 and Sc102a1 human iPSC lines was integrated with Seurat. Generation of UMAP embeddings, clustering, and pseudotime analysis was done on the integrated spaces. Marker genes of different NGN2-iN populations were identified as genes with BH corrected p < 0.01 and expression fold change >1.2. The benchmark of NGN2-iN populations was done by comparisons with the adolescent mouse nervous system atlas (Zeisel et al., 2018), the developing mouse brain (La Manno et al., 2020), and the developing mouse retina (Clark et al., 2019). The NGN2-iN scRNA-seq data with varied Dox treatment durations was processed similarly and integrated with CSS. Details of the computational analysis are described in the supplemental experimental procedures.
Data and code availability
The accession number for the processed scRNA-seq data and computational codes reported in this paper is ArrayExpress E-MTAB-10632.
Author contributions
M. Schörnig established the NGN2 iPSC lines. M. Schörnig and S.E. generated iN cultures with assistance from A.W. S.E. and W.H. established the selective single-cell dissociation. S.E. generated the time course and d35 scRNA-seq NGN2-iN scRNA-seq data with support from M. Schörnig, M. Santel, and W.H. M. Schörnig, H.-C.L., and M.T.N., with support from S.E. and A.W., generated IHC data. N.N.K. provided guidance to establish the NGN2 iPSC lines. E.T. provided guidance for IHC and iN culture. H.C.L. generated the NGN2 induction time course scRNA-seq data with support from S.E. and M. Santel. Z.H., S.E., and H.C.L. analyzed the scRNA-seq data. H.C.L., Z.H., S.E., B.T., and J.G.C. designed the study and wrote the manuscript.
Conflict of interests
B.T. is a member of the Editorial Board of Stem Cell Reports.
Acknowledgments
We thank the Camp and Treutlein labs for helpful discussions. J.G.C. and B.T. are supported by the Chan Zuckerberg Initiative DAF (grant no. CZF2019-002440), an advised fund of Silicon Valley Community Foundation. J.G.C. is supported by the European Research Council (Anthropoid-803441) and the Swiss National Science Foundation (project grant 310030_84795). B.T. is supported by the European Research Council (Organomics-758877, Braintime-874606), the Swiss National Science Foundation (project grant 310030_192604) and the National Center of Competence in Research Molecular Systems Engineering. H.C.L. is supported by the Human Frontier Science Program (LT000399/2020-L).
Published: August 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.006.
Contributor Information
J. Gray Camp, Email: jarrettgrayson.camp@unibas.ch.
Barbara Treutlein, Email: barbara.treutlein@bsse.ethz.ch.
Supplemental information
References
- Aibar S., Gonzalez-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddy B.A., Kong W., Kamimoto K., Guo C., Waye S.E., Sun T., Morris S.A. Single-cell mapping of lineage and identity in direct reprogramming. Nature. 2018;564:219–224. doi: 10.1038/s41586-018-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Wollny D., Treutlein B. Single-cell genomics to guide human stem cell and tissue engineering. Nat. Methods. 2018;15:661–667. doi: 10.1038/s41592-018-0113-0. [DOI] [PubMed] [Google Scholar]
- Chen M., Maimaitili M., Habekost M., Gill K.P., Mermet-Joret N., Nabavi S., Febbraro F., Denham M. Rapid generation of regionally specified CNS neurons by sequential patterning and conversion of human induced pluripotent stem cells. Stem Cell Res. 2020;48:101945. doi: 10.1016/j.scr.2020.101945. [DOI] [PubMed] [Google Scholar]
- Clark B.S., Stein-O'Brien G.L., Shiau F., Cannon G.H., Davis-Marcisak E., Sherman T., Santiago C.P., Hoang T.V., Rajaii F., James-Esposito R.E., et al. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102:1111–1126 e1115. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I., et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejarque M., Cervantes S., Pujadas G., Tutusaus A., Sanchez L., Gasa R. Neurogenin3 cooperates with Foxa2 to autoactivate its own expression. J. Biol. Chem. 2013;288:11705–11717. doi: 10.1074/jbc.M112.388173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frega M., van Gestel S.H., Linda K., van der Raadt J., Keller J., Van Rhijn J.R., Schubert D., Albers C.A., Nadif Kasri N. Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. J. Vis. Exp. 2017;119:54900. doi: 10.3791/54900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Morris S.A. Engineering cell identity: establishing new gene regulatory and chromatin landscapes. Curr. Opin. Genet. Dev. 2017;46:50–57. doi: 10.1016/j.gde.2017.06.011. [DOI] [PubMed] [Google Scholar]
- He Z., Brazovskaja A., Ebert S., Camp J.G., Treutlein B. CSS: cluster similarity spectrum integration of single-cell genomics data. Genome Biol. 2020;21:224. doi: 10.1186/s13059-020-02147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchis-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D., et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Karow M., Camp J.G., Falk S., Gerber T., Pataskar A., Gac-Santel M., Kageyama J., Brazovskaja A., Garding A., Fan W., et al. Direct pericyte-to-neuron reprogramming via unfolding of a neural stem cell-like program. Nat. Neurosci. 2018;21:932–940. doi: 10.1038/s41593-018-0168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Siletti K., Furlan A., Gyllborg D., Vinsland E., Langseth C.M., Khven I., Johnsson A., Nilsson M., Lönnerberg P., et al. Molecular architecture of the developing mouse brain. bioRxiv. 2020 doi: 10.1101/2020.07.02.184051. [DOI] [PubMed] [Google Scholar]
- Lin Y.T., Seo J., Gao F., Feldman H.M., Wen H.L., Penney J., Cam H.P., Gjoneska E., Raja W.K., Cheng J., et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–1154.e47. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S., et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls A.R., Lee M.M., Espinoza D.F., Szczot M., Lam R.M., Wang Q., Beers J., Zou J., Nguyen M.Q., Solinski H.J., et al. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 2020;30:932–946 e937. doi: 10.1016/j.celrep.2019.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N., et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schörnig M., Ju X., Fast L., Ebert S., Weigert A., Kanton S., Schaffer T., Kasri N.N., Treutlein B., Peter B.M., et al. Comparison of induced neurons reveals slower structural and functional maturation in humans than in apes. eLife. 2021;10:e59323. doi: 10.7554/eLife.59323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Christodoulou C., Gianotti-Sommer A., Shen S.S., Sailaja B.S., Hezroni H., Spira A., Meshorer E., Kotton D.N., Mostoslavsky G. Residual expression of reprogramming factors affects the transcriptional program and epigenetic signatures of induced pluripotent stem cells. PLoS One. 2012;7:e51711. doi: 10.1371/journal.pone.0051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.L., Ng L., Menon V., Martinez S., Lee C.K., Glattfelder K., Sunkin S.M., Henry A., Lau C., Dang C., et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A., Sim S., Neff N.F., Skotheim J.M., Wernig M., et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A., Sasaki T., Kumar A., Peterhoff C.M., Rao M.V., Liem R.K., Julien J.P., Nixon R.A. Peripherin is a subunit of peripheral nerve neurofilaments: implications for differential vulnerability of CNS and peripheral nervous system axons. J. Neurosci. 2012;32:8501–8508. doi: 10.1523/JNEUROSCI.1081-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J., Haring M., Braun E., Borm L.E., La Manno G., et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the processed scRNA-seq data and computational codes reported in this paper is ArrayExpress E-MTAB-10632.