Summary
Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) is often caused by an adenine to guanine variant at m.3243 (m.3243A>G) of the MT-TL1 gene. To understand how this pathogenic variant affects the nervous system, we differentiated human induced pluripotent stem cells (iPSCs) into excitatory neurons with normal (low heteroplasmy) and impaired (high heteroplasmy) mitochondrial function from MELAS patients with the m.3243A>G pathogenic variant. We combined micro-electrode array (MEA) measurements with RNA sequencing (MEA-seq) and found reduced expression of genes involved in mitochondrial respiration and presynaptic function, as well as non-cell autonomous processes in co-cultured astrocytes. Finally, we show that the clinical phase II drug sonlicromanol can improve neuronal network activity when treatment is initiated early in development. This was intricately linked with changes in the neuronal transcriptome. Overall, we provide insight in transcriptomic changes in iPSC-derived neurons with high m.3243A>G heteroplasmy, and show the pathology is partially reversible by sonlicromanol.
Keywords: MELAS, micro-electrode arrays, neurons, sonlicromanol, iPSC-derived neurons, mitochondria
Highlights
-
•
High m.3243A>G heteroplasmy leads to neuronal network dysfunction
-
•
Expression of genes involved in mitochondrial and synaptic function are affected
-
•
Gene expression in co-cultured rat astrocytes is non-cell autonomously affected
-
•
Sonlicromanol partially rescues the network activity and transcriptome changes
Using human induced pluripotent stem cell-derived neurons with high levels of m.3243A>G heteroplasmy, Klein Gunnewiek et al. report on transcriptome changes underlying the functional neuronal network phenotype and show that sonlicromanol can partially improve both the neuronal network phenotype and the transcriptome changes.
Introduction
Mitochondrial diseases (MDs) predominantly involve tissues with high energy needs, such as the brain (Gorman et al., 2016; Kim et al., 2019; Klein Gunnewiek et al., 2020). The most common MD, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), presents with epilepsy, stroke-like episodes, intellectual and cortical sensory deficits, cognitive decline, muscle weakness, cardiomyopathy, and/or diabetes (El-Hattab et al., 2015). The pathogenic variant underlying most MELAS cases is an adenine to guanine variant at position m.3243 (m.3243A>G) of the MT-TL1 gene (tRNAleu(UUR)), in the mitochondrial genome (mtDNA) (OMIM: 590050) (Goto et al., 1990). Approximately 1:20,000 are clinically affected by this pathogenic variant (Chinnery et al., 2000; Majamaa et al., 1998; Manwaring et al., 2007). The m.3243A>G pathogenic variant affects mitochondrial protein synthesis (King et al., 1992; King and Koga, 1992), likely by reducing amino acid incorporation efficiency during translation of 13 mtDNA-encoded proteins that are part of the oxidative phosphorylation system (OXPHOS) subcomplexes I–V (Sasarman et al., 2008). When the percentage of mutated mtDNA copies (heteroplasmy) in a cell reaches a threshold, normal OXPHOS function is disrupted (Ciafaloni et al., 1992; Kobayashi et al., 1990; Ylikallio and Suomalainen, 2012). OXPHOS reduces oxygen to water using electrons from NADH and FADH2, producing adenine triphosphate (ATP), and serves as the main production site of reactive oxygen species (ROS) (Holmström and Finkel, 2014). OXPHOS complex (I–V) deficiencies lead to a disbalanced cellular redox state (Tito et al., 2016), often with increased ROS production (Distelmaier et al., 2009), distorted mitochondrial signaling, and macromolecule damage (Daiber, 2010).
Classic treatments for mitochondrial disease, such as antioxidants (Garrido-Maraver et al., 2012; Glover et al., 2010) or dietary supplements (Parikh et al., 2015), mainly target symptoms, but none consistently improve patients' strength or quality of life (Pfeffer et al., 2012). Sonlicromanol (KH176), full name (S)-6-hydroxy-2,5,7,8-tetramethyl-N-((R)-piperidin-3-yl)chroman-2-carboxamide hydrochloride, is currently in clinical trial stage IIB (Janssen et al., 2019). As a Trolox derivative (soluble vitamin E), it aims to restore the redox balance and reduce ROS (Beyrath et al., 2018; Koene et al., 2017). This is achieved by modulation of the thioredoxin system/peroxiredoxin enzyme machinery (TrxR-Trx-Prdx system), which reduces H2O2 into water using electrons from NADPH (Beyrath et al., 2018). Rodent studies detected sonlicromanol in muscle and the brain, in a dose-dependent manner and without marked accumulation after frequent daily exposure (Beyrath et al., 2018). Ndufs4−/− mice treated with sonlicromanol lived longer and showed improved performance and gait, with less retinal ganglion cell degeneration (Frambach et al., 2020; Haas et al., 2017). Phase I and II clinical trials demonstrated acceptable safety, pharmacokinetic properties (Koene et al., 2017), and tolerance in humans, and improved alertness, as well as reduced depressive symptoms in patients (Janssen et al., 2019).
Here, we combined neuronal network recordings from microelectrode arrays (MEAs) with transcriptomics data (MEA measurements with RNA sequencing [MEA-seq]) to investigate (1) the mechanisms downstream of the impaired neural bioenergetics, and (2) whether we can reverse neuronal pathology caused by the m.3243A>G pathogenic variant. Our MEA-seq approach revealed reduced expression of genes involved in mitochondrial and presynaptic function linked to neuronal network dysfunction in high m.3243A>G heteroplasmy neurons. This neuronal phenotype non-cell autonomously induced gene expression changes in the healthy co-cultured rat astrocytes. Furthermore, sonlicromanol treatment improved the neuronal network pathology and reversed gene expression changes in a patient-specific manner.
Results
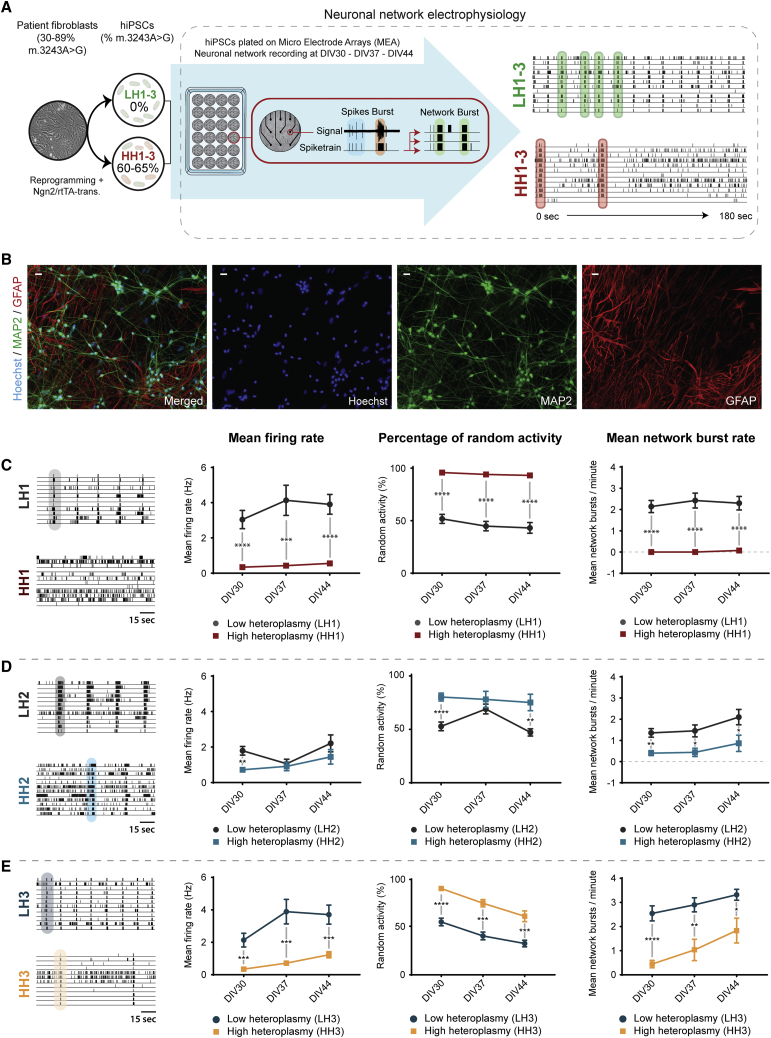
High m.3243A>G heteroplasmy affects neuronal network development
We previously generated induced pluripotent stem cell (iPSC) clones, with a wide range of m.3243A>G heteroplasmy levels on an isogenic background, ranging from 0% to 83%, from three separate individuals (Klein Gunnewiek et al., 2020). We specifically differentiated these isogenic sets of iPSCs with low (0%; LH1–3) and high (±60%; HH1–3) levels of heteroplasmy into glutamatergic neurons via tetracycline-induced Neurogenin-2 (Ngn2) expression (iNeurons) (Figures 1A and S1A). We co-cultured them with rat astrocytes to ensure proper maturation (Figure 1B) and measured the spontaneous neuronal network activity using MEAs, at 30, 37, and 44 days in vitro (DIV). In healthy control networks, the firing and network burst rates remained relatively stable from DIV30–37 onward (Figure S1B), indicators of a functionally “mature” network (Frega et al., 2017). We found that HH lines as a group showed a significantly different mean firing rate (MFR), percentage of random (non-burst) activity (PRS)??, and network burst rate (NBR) (Figure S1C). When we looked at each HH line specifically, HH1, HH2, and HH3 lines all showed a reduced NBR up to DIV44, compared with their isogenic controls (Figures 1C–1E). Furthermore, all three lines displayed an increased PRS and a reduced MFR over time (Figures 1C–1E). Other parameters, such as the burst rate, were not affected (Figures S1D–S1G). For HH2, the differences in the MFR and PRS were less pronounced, especially at DIV37, indicating HH2 neuronal network activity is less affected compared with HH1 and HH3. Nevertheless, the results point to a persistent neuronal network phenotype in the HH iNeurons past DIV30.
Figure 1.
Neuronal network development for LH1-3, and HH1-3 neuronal networks
(A) Patient-derived fibroblasts were reprogrammed to iPSCs, generating low (0%) and high (60%–65%) heteroplasmy clones. These were differentiated into excitatory neurons, co-cultured with rat astrocytes, on MEAs recorded for a 10-min period at DIV30, -DIV37, and -DIV44.
(B) Representative image of iPSC-derived neurons and co-cultured rat astrocytes at 1:1 ratio, stained for MAP2 (green), GFAP (red), and Hoechst (blue) at DIV30 (scale bar, 30 μm).
(C–E) Representative raster plots of MEA recordings, as well as the quantification of the MFR, the PRS, and the NBR, for (C) LH1 (n = 27) and HH1 (n = 27), (D) LH2 (n = 16) and HH2 (n = 14), and (E) LH3 (n = 23) and HH3 (n = 21). Data represent means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, using restricted maximum likelihood model, with Holm-Sidak's correction for multiple comparisons.
See also Figure S1.
Mitochondrial and synaptic gene expression are affected in high m.3243A>G heteroplasmy iNeurons
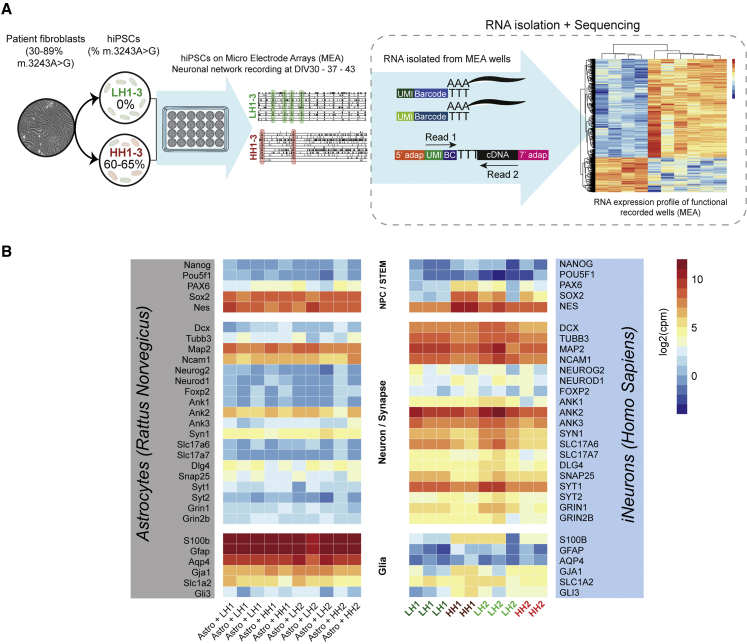
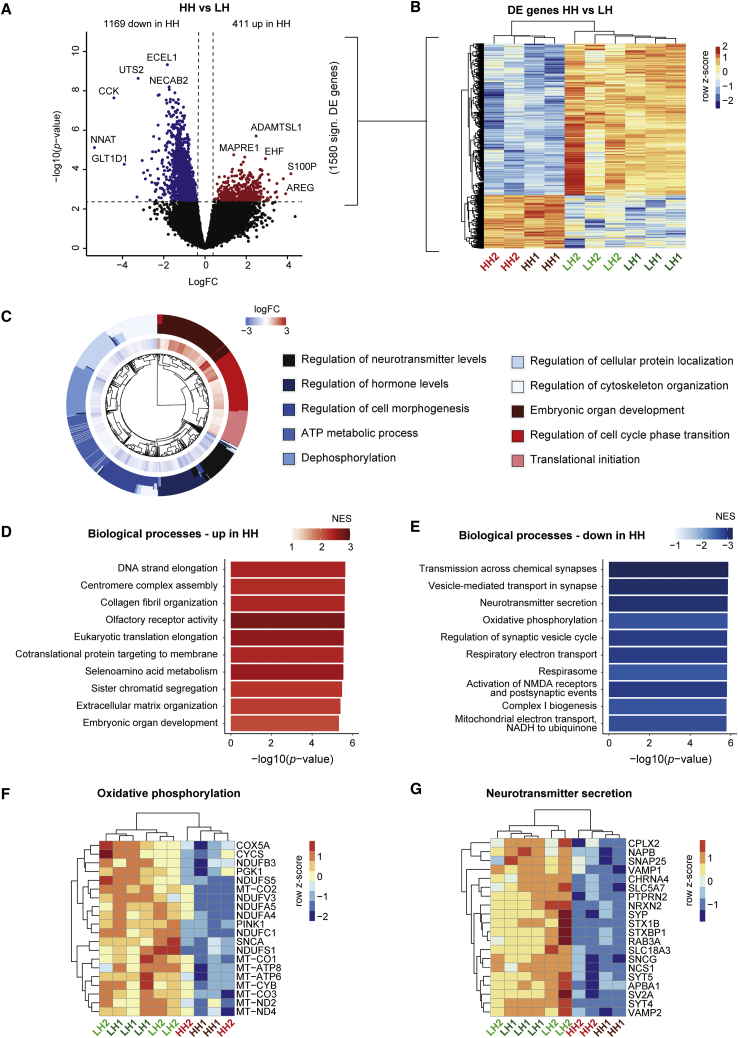
We optimized a bulk RNA sequencing (RNA-seq) method that can be used in combination with MEA experiments (MEA-seq). RNA was isolated from the neuronal co-cultures, directly after network activity was measured on DIV44 (Figure 2A). Reads from the human neurons and rat astrocytes were separated by mapping to a combined human- and rat genome. We confirmed the cell identity based on the expression of genes known to be highly expressed in either neurons (e.g., MAP2) or astrocytes (e.g., GFAP) (Figure 2B). We determined how the impaired neuronal bioenergetics caused by high m.3243A>G heteroplasmy levels affect gene expression. Therefore, we used the gene expression data from HH and LH iNeurons by extracting the reads that uniquely mapped to the human genome. Principal component analysis (PCA) showed that HH samples cluster away from LH samples (Figure S2A). Differential expression (DE) analysis between HH samples (HH1 + HH2) and LH samples (isogenic controls, LH1 + LH2) revealed 1,169 downregulated genes and 411 upregulated genes (adjusted p < 0.05) (Figures 3A and 3B; Table S1). To gain insight into the biological processes that were affected, we performed gene set enrichment analysis (GSEA) on the DE results. We observed enrichment of upregulated genes involved in cell cycle regulation, extracellular matrix organization, protein translation, and embryonic organ development (adjusted p < 0.05) (Figures 3C and 3D; Table S1). Interestingly, several of the top gene sets enriched for downregulated genes represented (pre)synaptic processes and mitochondrial respiration (adjusted p < 0.05) (Figures 3C and 3E; Table S1).
Figure 2.
Expression of stem cell, neuronal progenitor cell, neuronal, and glial cell markers in iNeurons co-cultured with astrocytes
(A) RNA-seq was performed on several representative MEA wells containing iNeurons co-cultured with rat astrocytes.
(B) Heatmap showing gene expression levels of stem cell/NPC genes (top section), neuronal and synaptic genes (middle section), and glial genes (bottom section), including rat homologs (left section, astrocyte samples), and human homologs (right section, iNeuron samples) in LH1 (n = 3), LH2 (n = 3), HH1 (n = 2), and HH2 (n = 2) iNeurons co-cultured with rat astrocytes. For iNeuron gene expression profiles, reads uniquely mapping to the human genome were extracted. For astrocyte gene expression profiles, reads uniquely mapping to the rat genome were extracted. Voom-transformed and batch-corrected counts per million (log2 scale) are shown.
Figure 3.
Gene expression changes in HH versus LH iNeurons
(A) Volcano plot showing DE genes in HH versus LH iNeurons (genes with adjusted p < 0.05 are labeled), with upregulated genes in red (logFC > 0) and downregulated genes in blue (logFC < 0).
(B) Heatmap showing expression of DE genes in HH iNeurons (n = 4) versus LH iNeurons (n = 6). Voom-transformed and batch-corrected counts per million (log2 scale) were scaled per gene.
(C) Circos plot showing gene sets significantly enriched for up- and downregulated genes in HH versus LH iNeurons. LogFC is shown for leading edge genes within each gene set.
(D and E) Bar plot showing 10 gene sets that are among the top gene sets enriched for (D) upregulated or (E) downregulated genes in HH versus LH iNeurons. The −log10(p value) (x axis) and the normalized enrichment score (NES) (color coded: red for positive NES, blue for negative NES) for Reactome pathways and GO terms representing biological processes (BPs) are shown.
(F and G) Heatmap showing top 20 leading edge genes for the gene sets (F) oxidative phosphorylation and (G) neurotransmitter secretion. Voom-transformed and batch-corrected counts were scaled per gene.
See also Figure S2.
Focusing on the mitochondrial and synaptic genes, we observed significant downregulation of several mtDNA genes regulated by MT-TL1, namely MT-ATP6, MT-CO1, MT-CO2, MT-CO3, MT-CYB, MT-ND2, and MT-ND4 (adjusted p < 0.05) (Figure 3F, Table S1). Furthermore, several nuclear mitochondrial genes encoding for complex I and complex V subunits of the electron transport chain were significantly downregulated, e.g., NDUFA4, NDUFA5, NDUFS5, and COX5A (adjusted p < 0.05) (Figure 3F, Table S1). Downregulated synaptic genes were involved in synaptic vesicle formation and fusion with the presynaptic membrane, e.g., SYP, SYT4, SYT5, VAMP1, and VAMP2 (adjusted p < 0.05) (Figure 3G, Table S1). In addition, we observed downregulation of genes linked to epilepsy, such as STXBP1, DNM1, KCNT1, KCNQ3, and SCN2A (adjusted p < 0.05) (Table S1). Overall, the results show that high m.3243A>G heteroplasmy is accompanied by significant expression changes in genes essential for mitochondrial and synaptic function, which likely contribute to the neuronal network dysfunction we observed on MEAs.
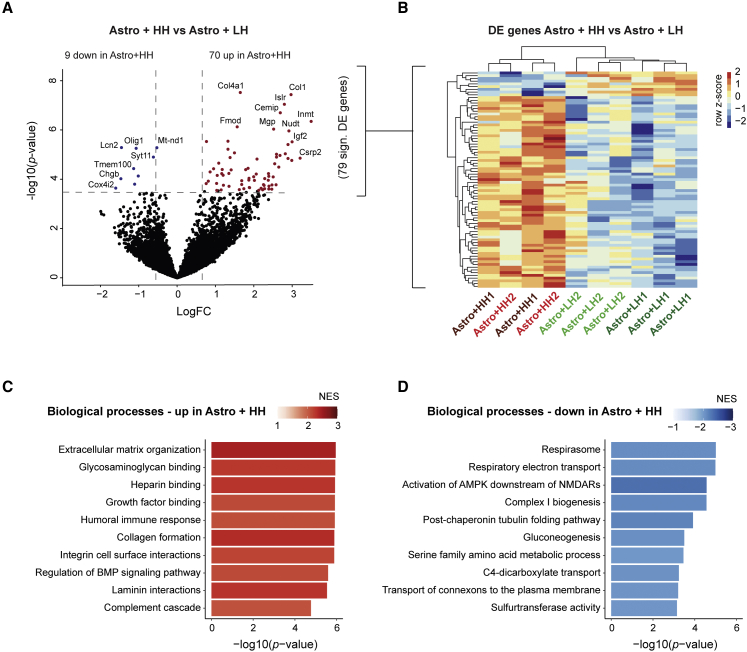
Non-cell autonomous effects of high m.3243A>G neuronal heteroplasmy on astrocytes
The different species of origin (rat astrocytes versus human neurons) allowed us to investigate whether the neuronal m.3243A>G heteroplasmy level non-cell autonomously affected gene expression in astrocytes. Gene expression profiles from astrocytes were obtained from the co-cultured samples by extracting the reads that uniquely mapped to the rat genome. We compared gene expression profiles of astrocytes co-cultured with HH iNeurons (Astro+HH1 and Astro+HH2) with astrocytes co-cultured with LH iNeurons (Astro+LH1 and Astro+LH2). DE analysis revealed 79 significant DE genes (adjusted p < 0.05); 70 upregulated genes and nine downregulated genes (Figures 4A, 4B, and S2B; Table S1). Interestingly, two downregulated genes play key roles in mitochondrial ATP production: Cox4i2 and Mt-nd1. GSEA revealed significant enrichment of upregulated genes in extracellular matrix organization, the immune response, and signaling cascades (adjusted p < 0.05) (Figure 4C; Table S1). The top gene sets enriched for downregulated genes contain genes involved in, e.g., mitochondrial function, amino acid metabolism, and synaptic function (Figure 4D; Table S1). These data reveal that the neuronal m.3243A>G heteroplasmy is accompanied by non-cell autonomous changes in astrocytic gene expression, including genes involved in mitochondrial function. Possibly, the changes in astrocyte gene expression result from the neuronal bioenergetic deficit induced by m.3243A>G heteroplasmy (Klein Gunnewiek et al., 2020), or direct neuronal-astrocytic connections (Sun et al., 2012; Wang and Gerdes, 2015).
Figure 4.
Non-cell autonomous effects of HH iNeurons on astrocyte gene expression
(A) Volcano plot showing DE genes in astrocytes co-cultured with HH iNeurons compared with astrocytes co-cultured with LH iNeurons (genes with adjusted p < 0.05 are labeled), with upregulated genes in red (logFC > 0) and downregulated genes in blue (logFC < 0).
(B) Heatmap showing expression of DE genes in astrocytes co-cultured with HH iNeurons (n = 4) versus astrocytes co-cultured with LH iNeurons (n = 6). Voom-transformed and batch-corrected counts (log2 scale) were scaled per gene.
(C and D) Bar plot showing 10 gene sets that are among the top gene sets enriched for (C) up- or (D) downregulated genes in astrocytes co-cultured with HH iNeurons compared with astrocytes co-cultured with LH iNeurons. The −log10(p value) (x axis) and the NES (color coded: red for positive NES, blue for negative NES) for Reactome pathways and GO terms representing BPs are shown.
See also Figure S2.
Sonlicromanol improves neuronal network dysfunction of high m.3243A>G heteroplasmy iNeurons in a patient-specific manner
Next, we exposed HH neuronal networks to sonlicromanol (KH176) (Beyrath et al., 2018; Haas et al., 2017; Koene et al., 2017) to test its ability to improve the HH neuronal network phenotypes. We treated MEA-grown neuronal networks (HH1–HH3) short term (2 weeks; DIV29–DIV44) and long-term (6 weeks; DIV3–DIV44), with different concentrations of sonlicromanol (0.5 μM, 1 μM, 3 μM, and 5μM + dimethyl sulfoxide [DMSO] vehicle condition), and recorded network activity at DIV30, DIV37, and DIV44 (Figure 5A). We chose these concentrations based on previous work that found improved viability at 0.1–1 μM sonlicromanol (half-maximal effective concentration [EC50] = 0.27 μM) in fibroblasts with complex I subunit pathogenic variants (Beyrath et al., 2018). We included higher concentrations (3 and 5 μM) to reach maximal efficacy in our cell type.
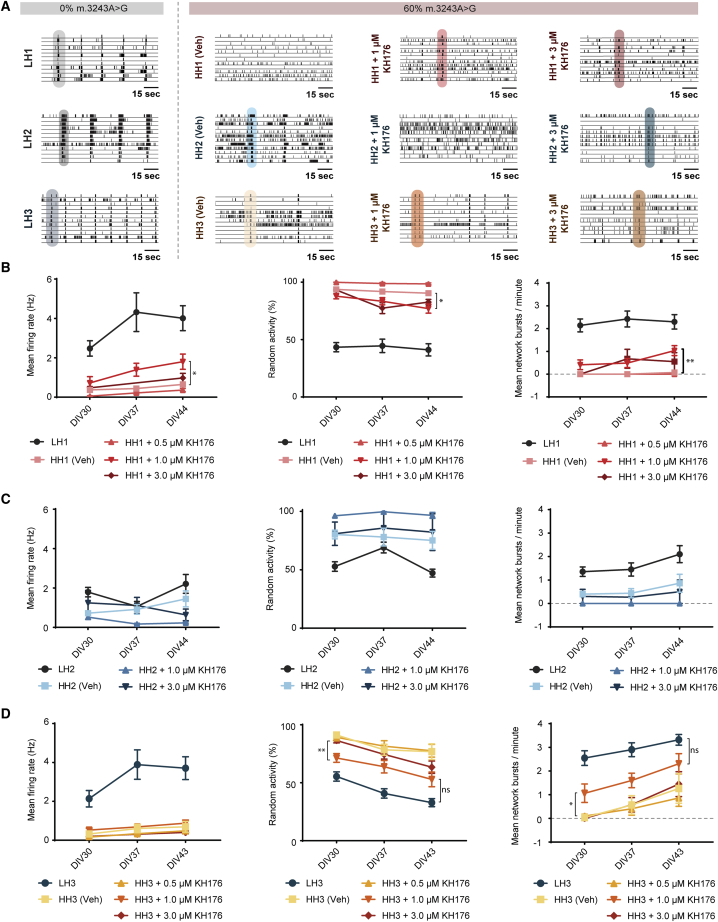
Figure 5.
Neuronal network activity after sonlicromanol (KH176) treatment starting from DIV3, in HH1-3 neuronal networks
(A) Representative raster plots of 2-min neuronal network activity recordings at DIV44 of LH1 (n = 17), HH1 vehicle (n = 12), HH1 + 1 μM sonlicromanol (n = 8), HH1 + 3 μM sonlicromanol (n = 5), LH2 (n = 6), HH2 vehicle (n = 16), HH2 + 1 μM sonlicromanol (n = 4), HH2 + 3 μM sonlicromanol (n = 5), LH3 (n = 16), HH3 vehicle (n = 9), HH3 + 1 μM sonlicromanol (n = 14), and HH3 + 3 μM sonlicromanol (n = 10).
(B–D) Quantification of the overall MFR, PRS, and NBR for (B) set #1 including HH1 + 0.5 μM sonlicromanol (n = 5), (C) set #2, and (D) set #3 including HH3 + 0.5 μM sonlicromanol (n = 13). Data represent means ± SEM. We determined the statistical difference between untreated- and sonlicromanol-treated HH1–3 conditions. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, using restricted maximum likelihood model, with Holm-Sidak's correction for multiple comparisons between treated and untreated samples.
See also Figure S3.
Sonlicromanol treatment of mature HH1 or HH3 networks from DIV29 onward had no significant effect on the key MEA parameters (Figures S3A and S3B). When we started sonlicromanol treatment earlier in neuronal development, at DIV3, we observed improved MEA activity in lines HH1 and HH3, predominantly at the 1 μM concentration (Figures 5B–5D). At DIV44, we found a significant increase in the MFR (p < 0.01) and NBR (p < 0.001), and a decrease in the PRS (p < 0.005), in HH1 + 1 μM sonlicromanol compared with the HH1-vehicle condition (Figure 5B). We report similar but less pronounced improvements in HH1 + 3 μM sonlicromanol (Figure 5B) but observed no differences at 5 μM. The HH3 line showed similar improvements when treated with 1 μM sonlicromanol from DIV3 onward (Figure 5D). Already at DIV30 we observed a significantly reduced PRS (p < 0.01) and increased NBR (p < 0.05) in HH3 + 1 μM sonlicromanol compared with HH3 + vehicle condition. At DIV44, these differences between HH3 + 1 μM and HH3 + vehicle were no longer significant. However, where the HH3 + vehicle still showed a significantly higher PRS and reduced NBR compared with LH3 isogenic controls, the HH3 + 1 μM sonlicromanol did not significantly differ from LH3. This shows sonlicromanol at a concentration of 1 μM improved the neuronal phenotype of HH3 neurons to more closely resemble LH3 isogenic controls. Interestingly, these improvements did not translate to the HH2 line (Figure 5C), suggesting the effects could be patient specific as well as dose dependent.
Next, we investigated whether the sonlicromanol treatment affected neuronal survival, m.3243A>G heteroplasmy levels, and ROS-induced DNA damage. A propidium iodide test showed no cell death in the untreated and 0–5 μM sonlicromanol-treated neurons, confirming the compound is not toxic (Figure S4A). Furthermore, we observed no differences in MAP2-positive cell density between untreated and 0–5 μM sonlicromanol-treated cultures (Figure S4B and S4C), nor did we find changes in m.3243A>G heteroplasmy levels (Figure S4D). Subsequently, we asked whether the sonlicromanol treatment reduced ROS levels, as in other models (Beyrath et al., 2018; Haas et al., 2017). We quantified the level of oxidative stress-induced DNA damage, using an 8-Oxo-2'-deoxyguanosine (8-OXO-dg) staining (Figures S5A–S5D). We observed significantly increased DNA damage in the soma of HH1 (p < 0.001) and HH3 (p < 0.05) iNeurons compared with their LH1 and LH3 isogenic controls (Figures S5B and S5D). However, we found no significant reductions in 8-OXO-dg levels in iNeurons treated with 1–3 μM sonlicromanol, compared with their DMSO vehicle controls. It is possible sonlicromanol targets different forms of ROS (e.g., causing lipid peroxidation), or the 1–3 μM sonlicromanol concentration was insufficient to reduce the DNA damage, since previous work showed the compound is 10 times more potent to restore the redox balance compared with its effects on ROS levels (Beyrath et al., 2018; Haas et al., 2017). These results show the sonlicromanol treatment did not improve neuronal network performance by affecting the network density, nor through overall reductions in m.3243A>G heteroplasmy or the levels of oxidative stress-induced DNA damage.
Together, we showed that, for patients with HH1 and HH3, sonlicromanol was beneficial in restoring some neuronal network activity and synchronicity, when treatment was initiated early in neuronal network development. These effects were dose dependent and patient specific.
Sonlicromanol reverses changes in gene expression in high m.3243A>G heteroplasmy iNeurons in a patient-specific manner
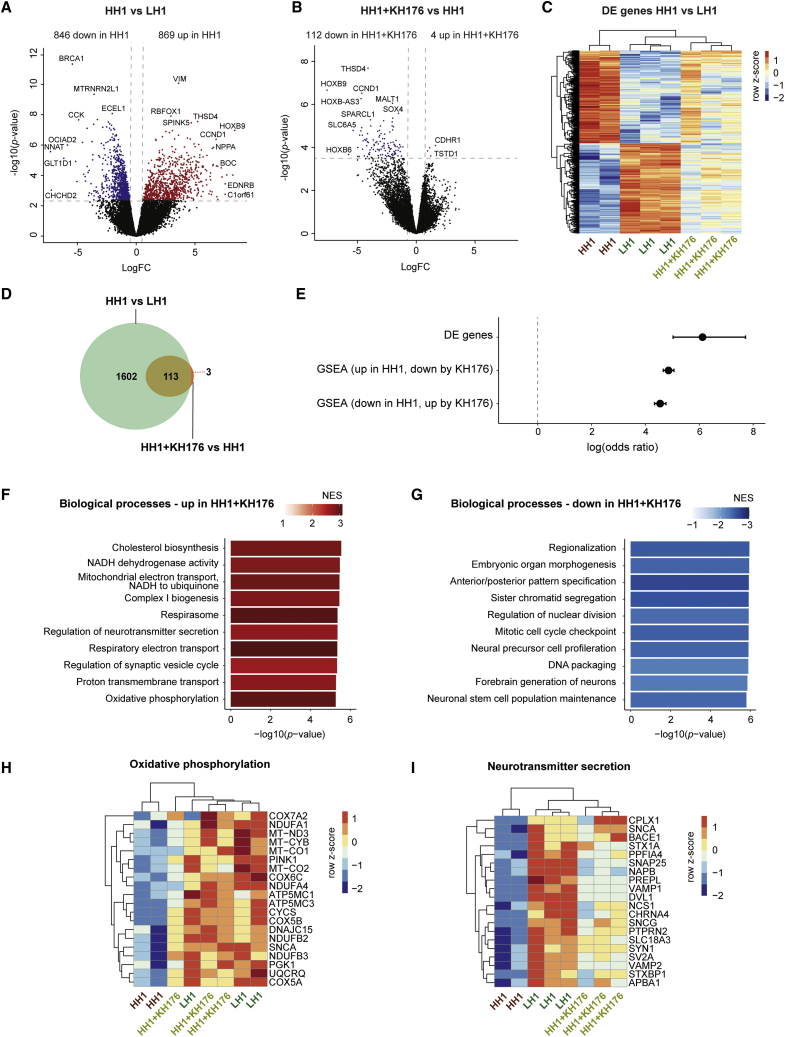
To determine how the sonlicromanol treatment affects gene expression in HH iNeurons, we isolated RNA at DIV44 from HH1 and HH2 iNeurons co-cultured with rat astrocytes on MEAs, with and without treatment of 1 μM sonlicromanol. Again, we extracted reads uniquely mapping to the human genome to investigate the effects of sonlicromanol on the iNeurons specifically. We compared gene expression changes for the first (HH1 and LH1) and second (HH2 and LH2) isogenic sets separately, to determine potential patient-specific treatment-induced gene expression changes. DE analysis for the first isogenic set (HH1 versus LH1) revealed 1,715 significantly DE genes (adjusted p < 0.05); 846 downregulated and 869 upregulated, in HH1 samples (Figures 6A and 6C; Table S2). Sonlicromanol treatment of HH1 (HH1 + KH176) compared with untreated HH1 revealed 116 DE genes (adjusted p < 0.05); 112 downregulated genes and four upregulated genes (Figure 6B and Table S2). Remarkably, 113 of these 116 genes overlapped with genes differentially expressed in HH1 compared with LH1 (odds ratio [OR] = 454, p < 2.2 × 10−16) (Figures 6D and 6E). We observed sonlicromanol-treated HH1 samples cluster toward LH1 samples, showing sonlicromanol reversed gene expression changes affected in HH1 (Figures S2C and 6C).
Figure 6.
Effects of sonlicromanol (KH176) treatment on gene expression in HH iNeurons for isogenic set 1 (HH1)
(A and B) Volcano plot showing DE genes in (A) HH1 compared with LH1 iNeurons or (B) HH1+KH176 compared with untreated HH1 iNeurons (genes with adjusted p < 0.05 are labeled), with upregulated genes in red (logFC > 0) and downregulated genes in blue (logFC<0).
(C) Heatmap showing expression of DE genes in HH1 versus LH1, for LH1 (n = 3), HH1 (n = 2), and HH1+KH176 (n = 3) samples. Voom-transformed and batch-corrected counts per million (log2 scale) were scaled per gene.
(D) Venn diagram showing overlap between DE genes in HH1 versus LH1, and DE genes in HH1+KH176 versus HH1.
(E) The log(odds ratio) is shown for overlap between DE genes in HH1 versus LH1 and HH1+KH176 versus HH1, and for overlap between gene sets enriched for upregulated genes in HH1 versus LH1 and gene sets enriched for downregulated genes in HH1+KH176 versus LH1, and vice versa. Error bars represent the 95% confidence interval.
(F and G) Bar plot showing 10 gene sets that are among the top gene sets enriched for (F) upregulated or (G) downregulated genes in HH1+KH176 versus HH1. The −log10(p value) (x axis) and the NES (color coded: red for positive NES, blue for negative NES) for Reactome pathways and GO terms representing BPs are shown.
(H and I) Heatmap showing top 20 leading edge genes in HH1+KH176 versus HH1 for the gene sets (H) oxidative phosphorylation, and (I) neurotransmitter secretion, for LH1, HH1, and HH1+KH176 samples. Voom-transformed and batch-corrected counts were scaled per gene.
See also Figures S2+ S6.
We performed GSEA on DE genes from HH1 + KH176 versus HH1 and HH1 versus HL1 to investigate what biological processes are reversed by sonlicromanol treatment (adjusted p < 0.05) (Table S2). We first confirmed there was a high overlap between gene sets upregulated in HH1 and downregulated by sonlicromanol treatment, and vice versa (OR = 129 and OR = 94, respectively; p < 2.2 × 10−16) (Figure 6E). We observed enrichment of upregulated genes in HH1 iNeurons treated with sonlicromanol for gene sets representing mitochondrial respiration and (pre)synaptic function (Figure 6F), and enrichment of downregulated genes in gene sets representing, e.g., chromatid segregation and embryonic organ development (Figure 6G), which were affected in HH lines (Figures 3D and 3E). Genes that were partly normalized in HH lines after sonlicromanol treatment were genes encoding for members of the electron transport chain, including MT-CO1, MT-CO2, MT-CYB, and COX5A (unadjusted p < 0.1) (Figure 6H). In addition, we identified genes important for synaptic vesicle release that were upregulated and showed a trend in the direction of the LH lines (unadjusted p < 0.05), including SNAP25, SYT4, VAMP1, VAMP2, STXBP1, SCNA, and SYN1 (Figure 6I). Sonlicromanol's positive effect on synaptic gene expression likely results from improved mitochondrial function and could contribute to the increased neuronal activity observed in sonlicromanol-treated HH1 iNeurons.
Finally, we analyzed the RNA-seq data for the second isogenic set, which did not show neuronal network improvements after sonlicromanol treatment. A comparison of gene expression profiles from HH2 to LH2 iNeurons identified 857 DE genes (adjusted p < 0.05); 197 upregulated genes and 660 downregulated genes (Figures S6A and S2D and Table S3). Interestingly, we identified no significant DE genes when comparing sonlicromanol treated with untreated HH2 samples (Table S3). A heatmap of HH2 versus LH2 DE genes revealed HH2 iNeurons treated with sonlicromanol clustered together with HH2 (untreated) samples, whereas LH2 samples clustered away, confirming the compound had minor to no effects on expression of genes affected in HH2 iNeurons (Figure S6B).
Overall, these results illustrate those significant functional improvements observed after sonlicromanol treatment were accompanied by improved transcriptomic changes, showing increased expression of genes involved in mitochondrial and synaptic function.
Discussion
We combined MEA recordings with transcriptomics to study the biological processes underlying the neuronal network phenotype linked to high levels (60%) of m.3243A>G heteroplasmy. This revealed downregulation of mtDNA and nuclear genes involved in mitochondrial ATP production, and genes involved in (pre)synaptic processes, in line with decreased neuronal network activity, and previously observed bioenergetic and single-cell presynaptic deficits (Klein Gunnewiek et al., 2020). Sonlicromanol treatment, started early in neuronal development, resulted in improved neuronal network activity in two patient-derived iNeuron lines (HH1 and HH3) and the upregulation of genes involved in mitochondrial respiration and (pre)synaptic function.
Synaptic function, and specifically the presynaptic vesicle cycle, rely on local ATP synthesis (Rangaraju et al., 2014). We previously linked high m.3243A>G heteroplasmy to reduced OPXHOS, less Synapsin 1/2 puncta, and less mitochondria at presynaptic sites (Klein Gunnewiek et al., 2020). Here, this was accompanied by downregulation of mtDNA genes encoding for subunits of OXPHOS complexes I, III, IV, and V; nuclear genes coding for complex I; and various genes linked to mitochondrial disease (Table S1) (Rahman and Rahman, 2018; Thompson et al., 2020). Furthermore, key synaptic genes were downregulated, such as SYN1 and SYN2 (Synapsin 1/2), GRIA4, GRIN1/3A, and VAMP1/2 (Table S1), as well as presynaptic Ca2+ sensor synaptotagmins (SYT1/4/5/6/7), linked to presynaptic release probabilities (Kwon et al., 2016). Combined, these findings suggest the high m.3243A>G heteroplasmy affects a wide range of mitochondrial and presynaptic processes, not merely dysfunctional ATP production. Reduced expression of STXBP1 and other epilepsy-related genes stand out, as epilepsy occurs frequently in MELAS patients. STXBP1 is linked to infantile epileptic encephalopathy and epilepsy (Pavone et al., 2012), and STXBP1 knockdown reduces sEPSC frequency (not amplitude) in human iNeurons (Zhang et al., 2013), as we previously observed in HH iNeurons (Klein Gunnewiek et al., 2020). Future work using excitatory-inhibitory neuronal co-cultures could further elucidate these findings.
Sonlicromanol, a compound targeting mitochondrial function, improved both neuronal network function and changes in gene expression, representing mitochondrial respiration and synaptic function. In a phase II clinical trial, sonlicromanol improved mood and alertness, often found in patients suffering from m.3243A>G related and other MDs (Janssen et al., 2019). We provide molecular and functional evidence for potential positive effects of sonlicromanol on these neuronal-specific deficits. We observed no toxicity at any sonlicromanol concentrations, nor any effects of treatment on neuronal survival or m.3243A>G heteroplasmy levels, suggesting the drug intrinsically improves neuronal function. ROS-induced DNA damage, as seen in MELAS (Katayama et al., 2009) and following stroke (Mizukoshi et al., 2006), was not improved by sonlicromanol treatment, but this could be due to insufficiently high sonlicromanol concentrations or a different ROS adduct type that is affected by sonlicromanol (e.g., lipid peroxidation).
Intriguingly, astrocyte gene expression was affected by the presence of HH iNeurons. Genes involved in metabolism and tubulin formation were downregulated in astrocytes co-cultured with HH iNeurons. Naturally, neuron activity facilitates astrocyte maturation and expression of astrocyte genes, potentially through Notch signaling Hasel et al. (2017). Both neuronal Notch ligands and astrocyte Notch1/2 receptors facilitate astrocytic glutamate uptake, and although Notch1/2 expression is unaffected in astrocytes co-cultured with HH iNeurons, the HH iNeurons do show reduced expression of Notch ligands JAG2 and DLK2. Aberrant Notch signaling has also been observed in MELAS organoids (Winanto et al., 2020), and could present an additional treatment target.
Our study has several limitations. A small patient sample size prevents us from drawing definitive conclusions about the utility of sonlicromanol to treat the neurological symptoms of MELAS. This is further confounded by the significant inter-patient variability in the results, the use of rodent as opposed to human astrocytes, and the inherent variance of our analytical methods. Testing sonlicromanol's efficacy in a larger cohort of individuals with MELAS is warranted to increase clinical trial readiness. Although the HH1–3 lines have similar heteroplasmy levels, it is unclear whether other nuclear factors might influence treatment response. This could contribute to a specific, individual response to sonlicromanol. If so, the efficacy of sonlicromanol should be assessed in, e.g., MEA-based in vitro platforms to test individual variations in drug response. This could refine enrollment (only responders to sonlicromanol would be enrolled) and could ultimately increase the success of future clinical trials. Furthermore, personalized testing like “N = 1” clinical trials (Schork, 2015) might be more suited for MELAS and mitochondrial disease, where average performance scores could blur promising findings. More advanced co-culture setups, using human astrocytes and other neuronal subtypes, will improve predictive value of our model.
By combining electrophysiological and transcriptomics data we disentangled several neuronal features of MELAS, advancing the understanding of the impact of m.3243A>G heteroplasmy on the human nervous system. Our findings on neuronal responses to sonlicromanol treatment confirm the heterogeneity and individualized nature of patient response to treatment, as well as the need for treatments tailored to the needs of the individual mitochondrial disease.
Experimental procedures
iPSC generation and culture
iPSC reprogramming and karyotyping for lines LH1–3 and HH1–3 have been described previously (Klein Gunnewiek et al., 2020; Perales-Clemente et al., 2016). Fibroblasts of MELAS subjects with the pathogenic variant m.3243A>G in MT-TL1 (tRNALeu(UUR)) were reprogrammed (Takahashi and Yamanaka, 2006), generating clones with 0% m.3243A>G heteroplasmy (LH1–3) and one clone with 60%–65% m.3243A>G heteroplasmy (HH1–3). Lines LH2+3 and HH2+3 were generous gifts from Ester Perales-Clemente and Timothy Nelson. All lines were previously karyotyped and were tested for m.3243A>G heteroplasmy levels using droplet digital PCR (ddPCR). All iPSCs were discarded after 15 passages post initial heteroplasmy measurement, to ensure heteroplasmy levels did not decrease. iPSCs were passaged 1–2 per week; medium was changed every 2–3 days. Collecting patient material and establishing hiPSCs have all been performed according to locally (Radboudumc) IRB protocols.
Neuronal differentiation
Neuronal differentiation was described previously (Frega et al., 2017; Klein Gunnewiek et al., 2020). iPSCs were derived into upper-layer, excitatory cortical neurons by overexpressing Neurogenin 2 (Ngn2). rtTA/Ngn2-positive iPSCs were plated as single cells at DIV0 onto 24-well multi-electrode arrays (Multichannel Systems, MCS GmbH, Reutlingen, Germany), coated with 50 μg/mL poly-L-ornithine hydrobromide (PLO; Sigma-Aldrich #P3655-10MG) and 5 μg/mL human recombinant laminin 521 (BioLamina #LN521-02) in E8 basal medium (Gibco #A1517001) supplemented with 1% penicillin/streptomycin (Pen/Strep; Sigma-Aldrich P4333), 1% RevitaCell (Thermo Fisher Scientific #A2644501), and 4 μg/mL doxycycline (Sigma-Aldrich #D9891-5G) to drive Ngn2 expression, at 20,000 cells per well for LH1+2, and 30,000 cells per well for HH1+2, to ensure similar mature neuron cell density. At DIV1, medium was changed to DMEM/F12 (Gibco #11320-074) supplemented with 1% Pen/Strep, 4 μg/mL doxycycline, 1% N-2 supplement (Gibco #17502-048), 1% MEM non-essential amino acid solution (NEAA; Sigma-Aldrich #M7145), 10 ng/mL human recombinant brain-derived neurotrophic factor (BDNF) (Promokine #C66212), 10 ng/mL human recombinant NT-3 (Promokine #C66425). We added primary cortical rat astrocytes (isolated as described by Frega et al., 2017) in a 1:1 ratio on DIV2 and changed 100% of medium at DIV3, to Neurobasal (Gibco #21103-049), supplemented with 20 μg/mL B-27 (Gibco #0080085SA), 1% GlutaMAX (Gibco), 1% Pen/Strep, 4 μg/mL doxycycline, 10 ng/mL human recombinant NT3, and 10 ng/mL human recombinant BDNF. At DIV3 only we added 2 μM cytosine β-D-arabinofuranoside (Ara-C; Sigma-Aldrich C1768-100MG) to remove proliferating cells. From DIV5 to DIV44, 50% of the medium was refreshed every 2 days, supplemented with 2.5% fetal bovine serum (FBS; Sigma-Aldrich #F2442-500ML) from DIV9 onward.
MEA recordings
Following a 10-min acclimatization period (37°C; 5% CO2), we recorded 10-min periods of spontaneous activity of LH1–3 and HH1–3 neuronal networks at DIV30, DIV37, and DIV44, using the 24-well MEA system (Multichannel Systems, MCS GmbH, Reutlingen, Germany), at a 10-KHz sampling rate, high-pass filter (i.e., Butterworth, 100-Hz cutoff frequency), and ±4.5 SD noise threshold. Spike trains were extracted using Multiwell Analyzer. A custom script for MATLAB (The Mathworks, Natick, MA) extracted parameters describing the network activity (Bologna et al., 2010), using thresholds to determine the MFR, mean burst rate (MBR), and NBR, as per Klein Gunnewiek et al. (2020) and Mossink et al. (2021).
Sonlicromanol (KH176) was provided by Khondrion as a powder for reconstitution in DMSO (Beyrath et al., 2018). DMSO was used as vehicle. Sonlicromanol was added at 500 nM, 1 μM, 3 μM, or 5 μM, from either DIV3 or from DIV29, up to DIV44, during every medium change.
Immunocytochemistry
iNeurons were washed with ice-cold DPBS (Gibco #14190-094) and fixated with 4% paraformaldehyde/4% sucrose (v/v), and permeabilized (DPBS, 0.2% Triton X-100, Sigma-Aldrich #9002-93-1). Cells were DPBS washed three times and incubated in blocking buffer to prevent a specific antibody binding (DPBS, 5% normal horse serum, 5% normal goat serum, 5% normal donkey serum, 0.1% bovine serum albumin [BSA], 1% glycine, 0.4% Triton, and 0.1% lysine, all from Sigma-Aldrich) for 1 h at room temperature. Primary antibodies were diluted 1:1,000 in blocking buffer, and were incubated overnight at 4°C. Next, cells were DPBS washed three times and incubated with secondary antibodies, diluted 1:1,000 in blocking buffer, for 1 h at room temperature. After three DPBS washing steps, Hoechst (Thermo Fischer Scientific #H3570) diluted 1:10,000 in DPBS was added for 10 min at room temperature. After one last DPBS wash, the coverslips were embedded in mounting medium (DAKO #S3023). Primary antibodies: mouse anti-8-OXO-dg (1:100; R&D Systems 4354-MC-050), guinea pig anti-MAP2 (1:1,000; Synaptic Systems 188,004), and rabbit anti-GFAP (1:1,000; Abcam AB7260). Secondary antibodies: goat anti-mouse Alexa Fluor 488 (1:1,000, Invitrogen A-11029), and goat anti-guinea pig Alexa Fluor 568 (1:1,000, Invitrogen). We imaged on a Zeiss Axio Imager Z1 with apotome, using the same settings for all batches and groups, at a resolution of 1,024 × 1,024 at 40× magnification. We imaged 15 random areas, and quantified the MAP2-positive cells per surface area, using ImageJ software (Schneider et al., 2012).
RNA-seq
RNA-seq was performed on human iPSC-derived neurons from LH (LH1+2) samples, HH (HH1+2) samples, and HH1+2 samples treated with 1 μM sonlicromanol. In all conditions, iNeurons were co-cultured with rat astrocytes. RNA was isolated after measuring network activity of the neurons on MEAs at DIV44, from two to three biological replicates per condition. LH and HH samples were distributed across two MEA batches. HH samples treated with sonlicromanol were included only in the second batch. RNA was isolated with the Quick-RNA Microprep kit (Zymo Research, R1051) according to manufacturer's instructions. RNA quality was checked using Agilent's Tapestation system (RNA High Sensitivity ScreenTape and Reagents, 5067–5579/80). RIN values ranged between 6.4 and 9.1. Library preparation was performed using a published single-cell RNA-seq protocol (Cao et al., 2017), adapted for bulk RNA-seq experiments.
For each sample, 25 ng total RNA was used as input for RNA-seq library preparation. In short, an anchored oligo-dT primer was used for reverse transcription, followed by second-strand synthesis and subsequent removal of excess primers using Exonuclease I (NEB, M0293). cDNA samples were pooled per sets of eight, randomized across three pools, and a 1.2× Ampure XP beads clean-up was performed (Beckman Coulter, A63881). Next, tagmentation was performed using TDE1 enzyme (Illumina, 15027865), followed by a 2.0× beads clean-up. PCR amplification was performed for 15 cycles using the NEBNext High-Fidelity 2X PCR Master Mix (NEB, M0541), followed by a 0.8× beads clean-up. Gel extraction was performed to select for products between 200 and 1,000 bp. cDNA concentrations of the final libraries were measured by Qubit dsDNA HS Assay kit (Invitrogen, Q32854). Product size distributions were visualized using Agilent's Tapestation system (D5000 ScreenTape and Reagents, 5067–5588/9). Libraries were sequenced on the NextSeq 500 platform (Illumina) using a V2 75 cycle kit (read 1, 18 cycles; read 2, 52 cycles; index 1, 10 cycles). A full description of the RNA-seq library preparation can be found in the supplemental information.
RNA-seq data pre-processing
Base calls were converted to fastq format and demultiplexed using Illumina's bcl2fastq conversion software (v.2.16.0.10) tolerating one mismatch per library barcode. Reads were filtered for a valid unique molecular identifier (UMI) and sample barcode, tolerating one mismatch per barcode. Trimming was performed using Trimmomatic (version 0.33) (Bolger et al., 2014). Trimmed reads were mapped to a combined human (GRCh38.p12) and rat (Rnor_6.0) reference genome using STAR (Dobin et al., 2013) (version 2.5.1b), with default settings (--runThreadN 1, --outReadsUnmapped None, --outFilterType Normal, --outFilterScoreMin 0, --outFilterMultimapNmax 10, --outFilterMismatchNmax 10, --alignIntronMin 21, --alignIntronMax 0, --alignMatesGapMax 0, --alignSJoverhangMin 5, --alignSJDBoverhangMin 3, --sjdbOverhang 100). Uniquely mapped reads (mapping quality of 255) were extracted and read duplicates were removed using the UMI-tools software package (Smith et al., 2017). Raw reads from BAM files were further processed to generate count matrices with HTSeq (Anders et al., 2015) (version 0.9.1) using a combined Gencode GRCh38.p12 (release 29, Ensembl 94) and rat (Rnor_6.0) reference transcriptome, to separate counts from human neurons and rat astrocytes.
RNA-seq data analysis
Raw counts from count tables were transformed to counts per million (cpm) using edgeR version 3.26.8 (R package) (Robinson et al., 2009). Transcripts with a cpm > 2 in at least two samples were included. Counts were voom-transformed (log2-transformation on cpm values), and corrected for MEA batch effect and genetic background for DE analysis using limma version 3.40.6 (R package) (Ritchie et al., 2015). Genes with a Benjamini-Hochberg (BH)-corrected p < 0.05 were considered to be significantly differentially expressed between two conditions. GSEA was performed using fgsea version 1.10.1 (R package) (Korotkevich and Sukhov, 2019), using a DE gene list ranked on the t statistic. Enrichment of genes was tested in Gene Ontology (GO) terms (C5 collection), Reactome pathways (C2 canonical pathways sub-collection), and in NRF2 transcription factor target (TFT) gene sets (C3 TFT sub-collection) from the Molecular Signatures Database (MSigDB) using msigdbr version 7.1.1 (R package) (Dolgalev, 2019). Gene sets with a BH-corrected p < 0.05 were considered to be significantly enriched for up- or downregulated genes. A detailed description can be found in the supplemental information.
ddPCR to measure MT-TL1 m.3243A>G heteroplasmy
This was described in detail previously (Klein Gunnewiek et al., 2020). In short, ddPCR was performed on DNA extracted from neurons at DIV44, using ddPCR primers custom synthesized to amplify the mitochondrial MT-TL1 m.3243 region. Data analysis was performed with QuantaSoft Analysis Pro version 1.0.596 (Bio-Rad).
Statistical analysis
Analysis was done using unpaired t tests, one-way analysis of variance with Bonferroni post hoc correction, one-way repeated measures ANOVA with sequential post hoc Bonferroni corrections, or Kolmogorov-Smirnov test, and restricted maximum likelihood model with Holm-Sidak's correction for multiple comparisons, where appropriate, using GraphPad Prism 6 (GraphPad Software). p values of p < 0.05, were deemed significant. Sample sizes were based on our previous experiences in the calculation of experimental variability. The number of wells (n) used are reported for each experiment.
Data and code availability
The GEO accession number for the RNA-seq data in this paper is GSE154825.
Author contributions
T.M.K.G. and A.H.A.V. performed the experiments and analyzed the data. T.M.K.G., A.H.A.V., T.K., and N.N.K. conceived the hypothesis and designed the experiments. I.P., M.H., and C.S. assisted in experiments and technical optimization. H.R., J.B., and J.S. provided sonlicromanol and assistance. T.M.K.G. and A.H.A.V. drafted the manuscript. All authors edited the draft manuscript.
Conflict of interests
J.B. and H.R. are full-time employees of the SME Khondrion (www.khondrion.com). J.S. is the founding CEO of Khondrion. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Eva Morava and David Cassiman of the University Hospital Leuven, and Ester Perales-Clemente and Timothy Nelson (Mayo Clinic, RC, MN) for their generous donation of iPSCs. We thank the department of Molecular Developmental Biology at the Radboud Institute of Molecular Life Sciences for RNA library sequencing. We thank Khondrion for permission to use the sonlicromanol. This work was made possible by the generosity of the Marriott family (to T.K.) and supported by the Tjalling Roorda Foundation (to T.M.K.G.), Stichting Stofwisselingskracht (project number 2017-20 to T.K. and N.N.K.), Netherlands Organization for Health Research and Development ZonMw grant 91217055 (to N.N.K.), ERA-NET NEURON DECODE! grant (NWO) 013.18.001 (to N.N.K.), and Epilepsiefonds WAR 18-02 (to N.N.K.).
Published: July 29, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.002.
Contributor Information
Tamas Kozicz, Email: kozicz.tamas@mayo.edu.
Nael Nadif Kasri, Email: n.nadif@donders.ru.nl.
Supplemental information
References
- Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyrath J., Pellegrini M., Renkema H., Houben L., Pecheritsyna S., Van Zandvoort P., Van Den Broek P., Bekel A., Eftekhari P., Smeitink J.A.M. KH176 safeguards mitochondrial diseased cells from redox stress-induced cell death by interacting with the thioredoxin system/peroxiredoxin enzyme machinery. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-24900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna L.L., Pasquale V., Garofalo M., Gandolfo M., Baljon P.L., Maccione A., Martinoia S., Chiappalone M. Investigating neuronal activity by SPYCODE multi-channel data analyzer. Neural Networks. 2010;23:685–697. doi: 10.1016/j.neunet.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery P.F., Johnson M.A., Wardell T.M., Singh-Kler R., Hayes C., Brown D.T., Taylor R.W., Bindoff L.A., Turnbull D.M. The epidemiology of pathogenic mitochondrial DNA mutations. Ann. Neurol. 2000;48:188–193. [PubMed] [Google Scholar]
- Ciafaloni E., Ricci E., Shanske S., Moraes C.T., Silvestri G., Hirano M., Simonetti S., Angelini C., Donati M.A., Garcia C., et al. MELAS: clinical features, biochemistry, and molecular genetics. Ann. Neurol. 1992;31:391–398. doi: 10.1002/ana.410310408. [DOI] [PubMed] [Google Scholar]
- Daiber A. Redox signaling (cross-talk) from and to mitochondria involves mitochondrial pores and reactive oxygen species. Biochim. Biophys. Acta. 2010;1797:55–56. doi: 10.1016/j.bbabio.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Distelmaier F., Koopman W.J.H., Van Den Heuvel L.P., Rodenburg R.J., Mayatepek E., Willems P.H.G.M., Smeitink J.A.M. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain. 2009;132:833–842. doi: 10.1093/brain/awp058. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgalev Igor. 2019. Msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format. [Google Scholar]
- El-Hattab A.W., Adesina A.M., Jones J., Scaglia F. MELAS syndrome: clinical manifestations, pathogenesis, and treatment options. Mol. Genet. Metab. 2015;116:4–12. doi: 10.1016/j.ymgme.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Frambach S.J.C.M., van de Wal M.A.E., van den Broek P.H.H., Smeitink J.A.M., Russel F.G.M., de Haas R., Schirris T.J.J. Effects of clofibrate and KH176 on life span and motor function in mitochondrial complex I-deficient mice. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:1–12. doi: 10.1016/j.bbadis.2020.165727. [DOI] [PubMed] [Google Scholar]
- Frega M., van Gestel S.H.C., Linda K., van der Raadt J., Keller J., Van Rhijn J.-R., Schubert D., Albers C.A., Nadif Kasri N. Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. J. Vis. Exp. 2017;119:1–10. doi: 10.3791/54900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Maraver J., Cordero M.D., Moñino I.D., Pereira-Arenas S., Lechuga-Vieco A.V., Cotán D., De La Mata M., Oropesa-Ávila M., De Miguel M., Bautista Lorite J., et al. Screening of effective pharmacological treatments for MELAS syndrome using yeasts, fibroblasts and cybrid models of the disease. Br. J. Pharmacol. 2012;167:1311–1328. doi: 10.1111/j.1476-5381.2012.02086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover E.I., Martin J., Maher A., Thornhill R.E., Moran G.R., Tarnopolsky M.A. A randomized trial of coenzyme Q10 in mitochondrial disorders. Muscle Nerve. 2010;42:739–748. doi: 10.1002/mus.21758. [DOI] [PubMed] [Google Scholar]
- Gorman G.S., Chinnery P.F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D.R., Zeviani M., Turnbull D.M. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- Goto Y.I., Nonaka I., Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- Haas R. De, Das D., Gara A., Renkema H.G., Greu R., Van Den Broek P., Perti J., Collin R.W.J.J., Willems P., Beyrath J., et al. Therapeutic effects of the mitochondrial ROS-redox modulator KH176 in a mammalian model of Leigh disease. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-09417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel P., Dando O., Jiwaji Z., Baxter P., Todd A.C., Heron S., Márkus N.M., McQueen J., Hampton D.W., Torvell M., et al. Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat. Commun. 2017;8:1–17. doi: 10.1038/ncomms15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–422. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- Janssen M.C.H., Koene S., De Laat P., Hemelaar P., Pickkers P., Spaans E., Beukema R., Beyrath J., Groothuis J., Verhaak C., et al. The KHENERGY Study: safety and efficacy of KH176 in mitochondrial m. 3243A>G spectrum disorders. Clin. Pharmacol. Ther. 2019;105:101–111. doi: 10.1002/cpt.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Maeda K., Iizuka T., Hayashi M., Hashizume Y., Sanada M., Kawai H., Kashiwagi A. Accumulation of oxidative stress around the stroke-like lesions of MELAS patients. Mitochondrion. 2009;9:306–313. doi: 10.1016/j.mito.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Kim Y., Vadodaria K.C., Lenkei Z., Kato T., Gage F.H., Marchetto M.C., Santos R. Mitochondria, metabolism, and redox mechanisms in psychiatric disorders. Antioxid. Redox Signal. 2019;31:275–317. doi: 10.1089/ars.2018.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M., Koga Y. Mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA (Leu (UUR)) mutation associated with mitochondrial myopathy, encephalopathy. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.P., Koga Y., Davidson M., Schon E.A. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Gunnewiek T.M., Van Hugte E.J.H., Frega M., Guardia G.S., Foreman K.B., Panneman D., Mossink B., Linda K., Keller J.M., Schubert D., et al. m.3243A>G-induced mitochondrial dysfunction impairs human neuronal development and reduces neuronal network activity and synchronicity. Cell Rep. 2020;31:1–16. doi: 10.1016/j.celrep.2020.107538. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Momoi M.Y., Tominaga K., Momoi T., Nihei K., Yanagisawa M., Kagawa Y., Ohta S. A point mutation in the mitochondrial tRNALeu(UUR) gene in melas (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) Biochem. Biophys. Res. Commun. 1990;173:816–822. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- Koene S., Spaans E., Van Bortel L., Van Lancker G., Delafontaine B., Badilini F., Beyrath J., Smeitink J. KH176 under development for rare mitochondrial disease: a first in man randomized controlled clinical trial in healthy male volunteers. Orphanet J. Rare Dis. 2017;12:1–12. doi: 10.1186/s13023-017-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V. Fast gene set enrichment analysis. BioRxiv. 2019;10:1–29. [Google Scholar]
- Kwon S.K., Sando R., Lewis T.L., Hirabayashi Y., Maximov A., Polleux F. LKB1 regulates mitochondria-dependent presynaptic calcium clearance and neurotransmitter release properties at excitatory synapses along cortical axons. PLoS Biol. 2016;14:1–27. doi: 10.1371/journal.pbio.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majamaa K., Moilanen J.S., Uimonen S., Remes A.M., Salmela P.I., Kärppä M., Majamaa-Voltti K.A.M., Rusanen H., Sorri M., Peuhkurinen K.J., et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am. J. Hum. Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwaring N., Jones M.M., Wang J.J., Rochtchina E., Howard C., Mitchell P., Sue C.M. Population prevalence of the MELAS A3243G mutation. Mitochondrion. 2007;7:230–233. doi: 10.1016/j.mito.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Mizukoshi G., Katsura K., Watanabe M., Katayama Y. Evaluation of therapeutic efficacy of free radical scavenger in patients with ischemic stroke. Cereb. Blood Flow Metab. 2006;18:53–60. [Google Scholar]
- Mossink B., van Rhijn J.-R., Wang S., van Hugte E., Linda K., Bak J., Verboven A., Selten M., Anania A., Jansen S., et al. Cadherin-13 is a critical regulator of GABAergic modulation in human stem cell derived neuronal networks. Mol. Psychiatry. 2021 doi: 10.1038/s41380-021-01117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S., Goldstein A., Koenig M.K., Scaglia F., Enns G.M., Saneto R., Anselm I., Cohen B.H., Falk M.J., Greene C., et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet. Med. 2015;17:689–701. doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavone P., Spalice A., Polizzi A., Parisi P., Ruggieri M. Ohtahara syndrome with emphasis on recent genetic discovery. Brain Dev. 2012;34:459–468. doi: 10.1016/j.braindev.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Perales-Clemente E., Cook A.N., Evans J.M., Roellinger S., Secreto F., Emmanuele V., Oglesbee D., Mootha V.K., Hirano M., Schon E.A., et al. Natural underlying mtDNA heteroplasmy as a potential source of intra-person hi PSC variability. EMBO J. 2016;35:1979–1990. doi: 10.15252/embj.201694892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G., Majamaa K., Turnbull D.M., Thorburn D., Chinnery P.F. Treatment for mitochondrial disorders. Cochrane Database Syst. Rev. 2012;4:1–42. doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman J., Rahman S. Mitochondrial medicine in the omics era. Lancet. 2018;391:2560–2574. doi: 10.1016/S0140-6736(18)30727-X. [DOI] [PubMed] [Google Scholar]
- Rangaraju V., Calloway N., Ryan T.A. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156:825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:1–13. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasarman F., Antonicka H., Shoubridge E.A. The A3243G tRNA Leu (UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Hum. Mol. Genet. 2008;17:3697–3707. doi: 10.1093/hmg/ddn265. [DOI] [PubMed] [Google Scholar]
- Schork N.J. Personalized medicine: time for one-person trials. Nature. 2015;520:609–611. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang Y., Zhang J., Tu J., Wang X.J., Su X.D., Wang L., Zhang Y. Tunneling-nanotube direction determination in neurons and astrocytes. Cell Death Dis. 2012;3:e438. doi: 10.1038/cddis.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thompson K., Collier J.J., Glasgow R.I.C., Robertson F.M., Pyle A., Blakely E.L., Alston C.L., Oláhová M., McFarland R., Taylor R.W. Recent advances in understanding the molecular genetic basis of mitochondrial disease. J. Inherit. Metab. Dis. 2020;43:36–50. doi: 10.1002/jimd.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito D.V., Cracan V., Goodman R.P., Peng J., Grabarek Z., Mootha V.K. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–236. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gerdes H.H. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015;22:1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winanto, Khong Z.J., Soh B.S., Fan Y., Ng S.Y. Organoid cultures of MELAS neural cells reveal hyperactive Notch signaling that impacts neurodevelopment. Cell Death Dis. 2020;11:1–8. doi: 10.1038/s41419-020-2383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E., Suomalainen A. Mechanisms of mitochondrial diseases. Ann. Med. 2012;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pak C.H., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for the RNA-seq data in this paper is GSE154825.