Summary
Regeneration of skeletal muscle requires resident stem cells called satellite cells. Here, we report that the chromatin remodeler CHD4, a member of the nucleosome remodeling and deacetylase (NuRD) repressive complex, is essential for the expansion and regenerative functions of satellite cells. We show that conditional deletion of the Chd4 gene in satellite cells results in failure to regenerate muscle after injury. This defect is principally associated with increased stem cell plasticity and lineage infidelity during the expansion of satellite cells, caused by de-repression of non-muscle-cell lineage genes in the absence of Chd4. Thus, CHD4 ensures that a transcriptional program that safeguards satellite cell identity during muscle regeneration is maintained. Given the therapeutic potential of muscle stem cells in diverse neuromuscular pathologies, CHD4 constitutes an attractive target for satellite cell-based therapies.
Keywords: Chd4, satellite cells, skeletal muscle, regeneration, lineage maintenance, NuRD, muscle stem cell
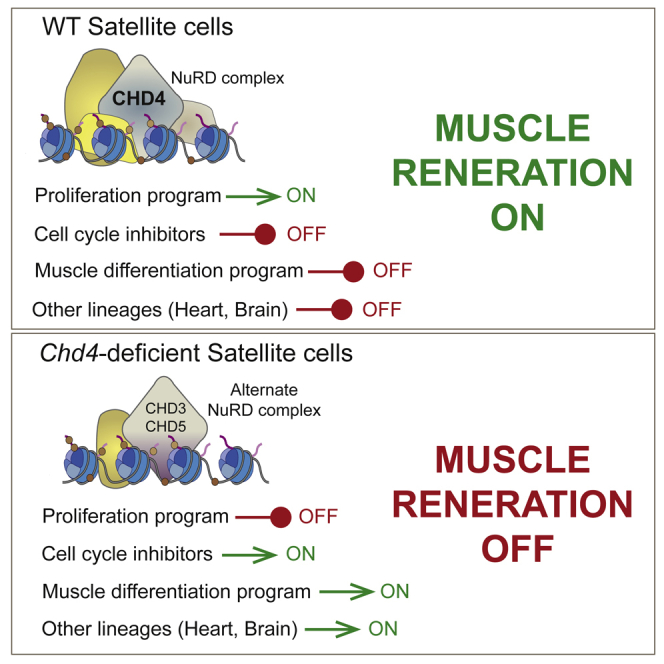
Graphical abstract
Highlights
-
•
CHD4/NuRD regulates satellite cell (SC) fate commitment
-
•
CHD4 deficiency blocks SC proliferation and disrupts skeletal muscle regeneration
-
•
CHD4/NuRD repress myogenic differentiation genes during SC proliferative expansion
-
•
CHD4/NuRD represses genes associated with other fates (brain, heart) in SCs
In this article, Sreenivasan and colleagues show that the chromatin-remodeling complex CHD4/NuRD is required to maintain muscle stem cell (satellite cell) identity. CHD4 represses the skeletal muscle differentiation gene program during satellite cell proliferation and maintains genes from other lineages (brain, heart) silent, being essential for proper skeletal muscle regeneration.
Introduction
Adult skeletal muscles regenerate through a population of muscle-resident Pax7-expressing stem cells (satellite cells [SCs]) that are quiescent in resting conditions. Upon injury, SCs exit quiescence and proliferate, and their progeny either differentiate to form new muscle fibers or self-renew to replenish the quiescent stem cell pool (Yin et al., 2013). SC fate is controlled by an interplay of instructive and responsive cues, which are epigenetically regulated at a cell-autonomous level (Ermolaeva et al., 2018; Faralli et al., 2016; Juan et al., 2011; Robinson and Dilworth, 2018; Schworer et al., 2016; Zhang et al., 2015).
The chromatin-remodeling complex NuRD (nucleosome remodeling and deacetylase) is an epigenetic regulator implicated in various cellular processes, such as cell-cycle progression, DNA damage response, maintenance of genome integrity, lineage commitment of different cell types (including stem cells), and proliferation of cancer cells (Gomez-del Arco et al., 2016; Kashiwagi et al., 2007; Lai and Wade, 2011; O'Shaughnessy and Hendrich, 2013; Ostapcuk et al., 2018; Williams et al., 2004; Xu et al., 2020; Zhang et al., 1998; Zhao et al., 2017). Different NuRD complexes with a diverse assembly of protein components yield distinct outcomes, including transcriptional activation or repression of target genes. These components include the chromodomain-helicase-DNA-binding proteins 3 and 4 (CHD3, CHD4), which both possess helicase/ATPase activity, the class I histone deacetylases 1 and 2 (HDAC1 and HDAC2), the metastasis-associated proteins Mta1/2/3, and the methyl-CpG-binding domain family members MBD2 or MBD3. These core factors can assemble into mutually exclusive CHD4/NuRD-like complexes (Le Guezennec et al., 2006). We previously demonstrated that Chd4 maintains tissue homeostasis and identity of mature cardiac and skeletal muscle (Gomez-del Arco et al., 2016) and plays a decisive role in the de-repression of RIPK3, a key effector of necroptosis (Sreenivasan et al., 2020).
Here, we show that CHD4 is not only an essential component of the muscle regenerative program that sustains efficient expansion of adult muscle stem cells but it is also required to maintain their transcriptional identity.
Results
CHD4 expression in SCs is required for muscle regeneration
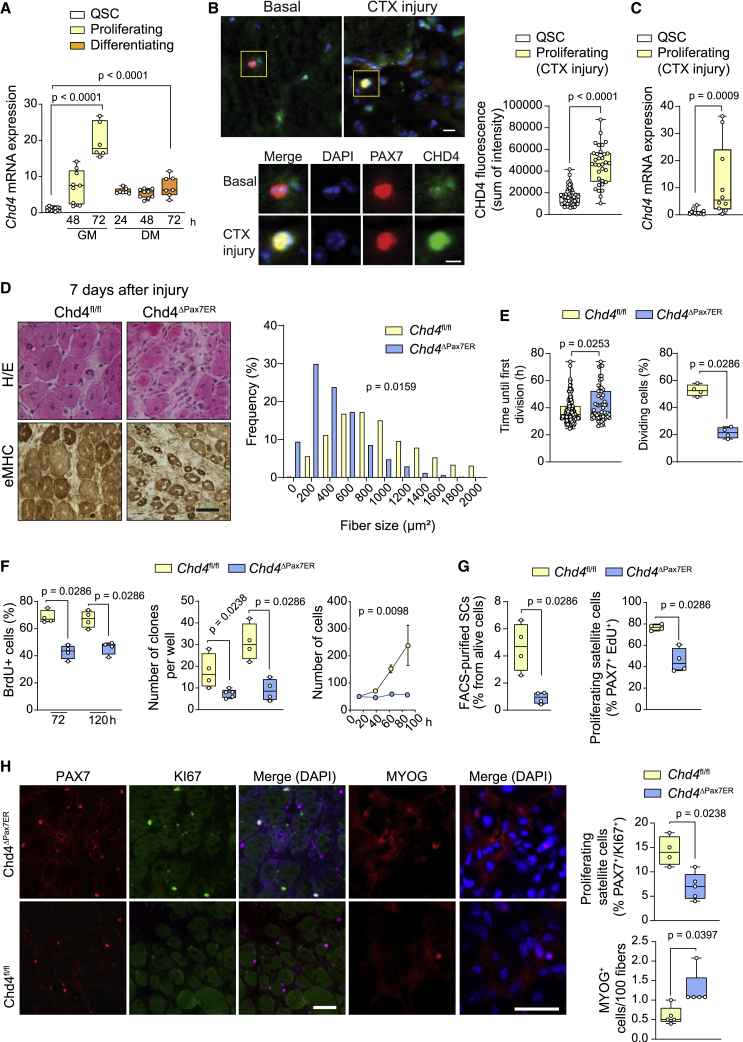
We first assessed Chd4 expression in quiescent SCs (QSCs) isolated from resting muscle of wild-type (WT) mice using a α7-integrin+/CD34+/SCA1−/CD45− fluorescence-activated cell sorting (FACS) protocol and cultured in growth medium for 48/72 h, or differentiation medium for 24/48/72 h. Chd4 mRNA expression was upregulated in proliferating SCs, decreasing during early differentiation stages (Figure 1A). Consistently, in SCs isolated from regenerating muscle 72 h after injury, the peak of the proliferative response, Chd4 expression was strongly induced (Figure 1B) at both transcript and protein levels (Figure 1C).
Figure 1.
Chd4 is required for the proliferative expansion of SCs during muscle regeneration
(A) Chd4 mRNA relative expression in QSCs, and in SCs under proliferative (in growth medium [GM]) or differentiation (in differentiation medium [DM]) conditions at the indicated time points in culture from WT mice. Data were normalized to housekeeping gene Rpl7 with the QSCs set to 1 (from left to right, n = 13, 9, 6, 7, 9, or 7 mice, respectively).
(B) Representative pictures and quantification of Chd4 in SCs (Pax7+) from non-injured muscles from WT mice, or at 72 h after muscle injury with CTX (n = 33 or 60 cells, respectively, from four mice). The sum of the Chd4 fluorescence intensity per cell is represented. Scale bar, 5 μm.
(C) Chd4 mRNA expression in freshly isolated SCs from non-injured muscles and at 72 h after muscle injury with CTX. Data were normalized to housekeeping gene Rpl7, with the QSCs set to 1 (n = 12 or 9 mice, respectively).
(D) Representative pictures of sections of regenerating muscles from TMX-treated Chd4fl/f and Chd4ΔPax7ER mice at day 7 after injury, stained with hematoxylin and eosin and antibodies against eMHC. Scale bar, 10 μm (top), 5 μm (bottom). Frequency distribution of cross-sectional area (μm2) of regenerating fibers from TMX-treated Chd4fl/f and Chd4ΔPax7ER mice (n = 4 and 5 mice, respectively) (bottom).
(E) Average time until first division during live time-lapse microscopy, and percentage of dividing SCs from TMX-treated Chd4fl/f and Chd4ΔPax7ER mice (138 or 58 individual cells from three different mice, respectively).
(F) Percentage of BrdU+ SCs cultured in GM for either 72 or 120 h (left), the number of SC colonies with more than five cells counted throughout 4 days after isolation and culture in GM (middle), and the number of SCs per well after seeding 50 cells/well at time 0 (right), from TMX-treated Chd4fl/f and Chd4ΔPax7ER mice (n = 4 independent experiments).
(G) Relative number of total and proliferating SCs after FACS purification from TMX-treated Chd4fl/f or Chd4ΔPax7ER mice at 3 days after injury (n = 4 mice/group).
(H) Representative pictures and quantification of proliferating (Pax7+/Ki67+; n = 4 mice/group) or differentiating cells (Myog; n = 5 mice/group) in muscles of TMX-treated Chd4fl/f or Chd4ΔPax7ER mice at 7 days after injury. Scale bar, 50 μm.
To assess the specific contribution of Chd4 to SC functions, we generated mice with conditional deletion of Chd4 in SCs (Chd4ΔPax7ER mice) by intercrossing Chd4fl/fl and Pax7CRE-ER mice (Nishijo et al., 2009; Williams et al., 2004). Loss of Chd4 after tamoxifen (TMX) administration was verified by RT-PCR (Figure S1A). In the absence of injury, muscles of Chd4ΔPax7ER and Chd4f/fl (control) mice showed no significant differences in muscle morphology (Figure S1B), and the number of Pax7+ cells was similar, with a tendency to decrease by 90 days after TMX administration (Figure S1C). At 7 days after injury, muscle regeneration in Chd4ΔPax7ER mice was severely impaired, as indexed by the reduced size of myofibers positive for embryonic myosin heavy chain (eMHC), a marker of newly formed myofibers (Figure 1D). The muscles of Chd4ΔPax7ER mice continued to show smaller eMHC+ and centrally nucleated fibers (indicative of regeneration) than control mice at 14 or 21 days after injury (Figures S1D and S1E). This persistent regenerative defect in Chd4ΔPax7ER mice was exacerbated after two rounds of muscle injury (Figure S1F).
CHD4 is essential for efficient SC proliferation during muscle regeneration
We next determined myogenic functions of SCs upon quiescence exit. Compared with Chd4fl/fl, Chd4ΔPax7ER SCs exhibit proliferation arrest as revealed by live time-lapse microscopy (Figure 1E), 5-bromo-2′-deoxyuridine (BrdU) incorporation in growth-promoting conditions (Figure 1F), and clonogenic assays (Figure 1F). Importantly, acute Chd4 deletion ex vivo by adenovirus-CRE (Ad-CRE, compared with Ad-GFP) in SCs isolated from Chd4fl/fl mice (Figure S1G) induced a proliferation arrest, thereby excluding potential indirect effects of long-term TMX-mediated Chd4 deletion (Figure S1G).
Consistently, compared with Chd4fl/fl, the total number and fraction of proliferating (PAX7+/EdU+ or PAX7+/KI67+) SCs from regenerating muscles in vivo were dramatically lower in Chd4ΔPax7ER mice (Figures 1G and 1H), coinciding with an increased number of differentiating (Myogenin, Myog+) SCs (Figure 1H), and a reduced proportion of self-renewed SCs (Figure S1H). Consistently, impairment of self-renewal and regeneration in Chd4ΔPax7ER mice was exacerbated upon repetitive injuries (Figure S1F). Furthermore, regenerating myofibers in Chd4ΔPax7ER mice contained significantly fewer myonuclei (Figure S1I), a feature also observed in in vitro-formed myotubes (Figure S1J).
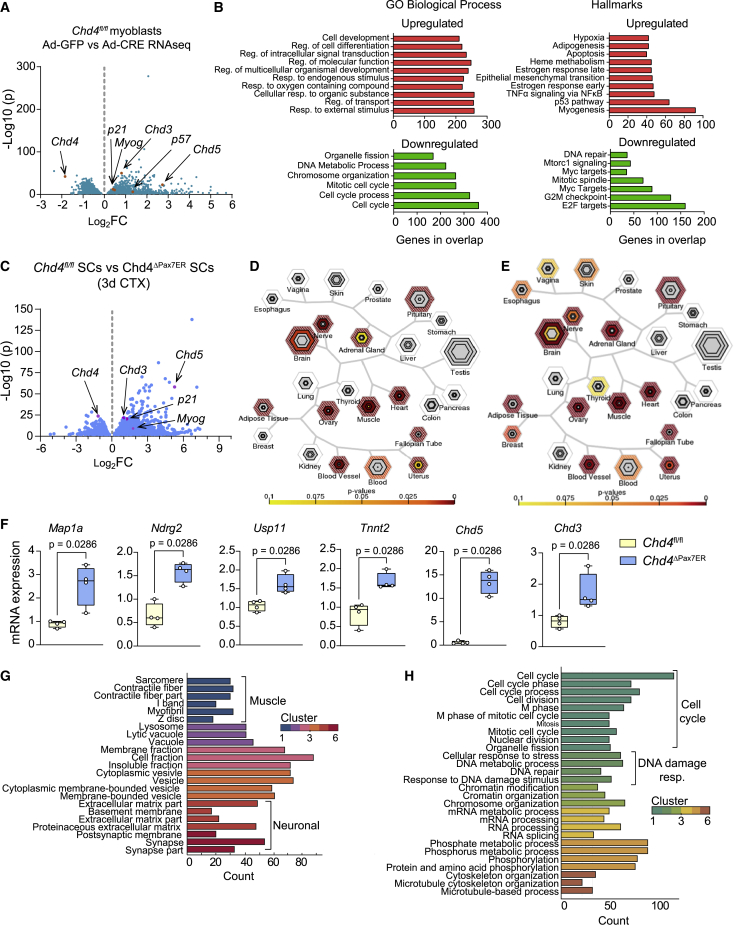
To obtain a molecular understanding of how Chd4 affects SC proliferation, we performed whole-transcriptome analyses of Chd4fl/fl SCs cultured in growth medium for 72 h with acute Ad-CRE-mediated deletion of Chd4, resulting in the upregulation of 1,975 genes, and the downregulation of 1,843 genes (adjusted p < 0.01), compared with Ad-GFP transduced controls (Figure 2A). The upregulated genes included the Chd4 homologs Chd3 and Chd5, suggesting a rapid compensatory attempt to uphold the availability of an alternative NuRD complex. Upregulated gene clusters were involved in cell differentiation toward myogenesis but also other cell lineages, and downregulated gene clusters were involved in cell-cycle and DNA metabolism processes (Figure 2B). Within the upregulated genes, we found increased Myog expression, a member of the myogenic regulatory factor (MRF) family required for muscle-cell differentiation (Figures 2A and S2A), and of Cdkn1c (p57) (Figures 2A and S2B), which has been linked to cell-cycle arrest and is required for the initiation of muscle differentiation (Mademtzoglou et al., 2018; Zhang et al., 1999). In agreement, we confirmed an upregulation of p57 and Myog in SCs isolated from regenerating muscles of Chd4ΔPax7ER mice after injury (Figures S2A and S2B).
Figure 2.
Chd4 controls the expression of cell-cycle regulators and lineage-specific genes in SCs
(A) Volcano plot (−log10(p) versus log2(fold change)) showing RNA-seq results from Chd4fl/fl SCs transduced with Ad-CRE or Ad-GFP (control) (n = 4 independent experiments).
(B) GO analysis of the Biological Process database and Hallmark database of the significantly upregulated and downregulated genes.
(C) Volcano plot showing RNA-seq results from FACS-purified SCs at 3 days after TMX-induced muscle injury, in Chd4fl/fl and Chd4ΔPax7ER mice (n = 4 mice/group).
(D) TSEA analyses comparing gene expression of Chd4fl/fl SCs transduced with Ad-CRE or Ad-GFP.
(E) TSEA analyses of gene expression from SCs purified from TMX-treated Chd4fl/fl or Chd4ΔPax7ER mice at 3 days after muscle injury.
(F) Relative mRNA expression of the indicated genes in Chd4fl/f or Chd4ΔPax7ER mice SCs at 3 days after injury (n = 4 mice/group). Data were normalized to housekeeping gene Rpl7, with the Chd4fl/f set to 1.
(G and H) Functional annotation clustering based on DAVID biological processes of upregulated genes (G) or downregulated genes (H) in FACS-purified SCs, as in (C).
We next investigated Chd4 regulation of p57 due to its important role in regulating growth arrest of adult SCs. CHD4 chromatin immunoprecipitation (ChIP) followed by PCR revealed a direct association to the p57 gene regulatory regions in proliferating SCs (Figure S2C). In agreement with this, lentivirus-mediated knockdown of p57 increased SC proliferation (Figure S2D). To determine the influence of the upregulated p57 expression in SCs of Chd4ΔPax7ER mice in vivo, we used a p57 conditional-mutant allele (Mademtzoglou et al., 2017) to generate Chd4ΔPax7ER;p57ΔPax7ER double-mutant mice. Genetic inactivation of p57 in Chd4-deficient SCs failed to restore SC proliferation or normal muscle regeneration (Figure S2E), and also did not affect the dysregulation of genes from other lineages in vivo (Figure S2F), showing that CHD4 controls SC function through the concurrent regulation of several target genes with distinct biological functions.
CHD4 maintains the transcriptional identity of SCs during muscle regeneration
To gain a deeper mechanistic understanding of the action of CHD4 in SCs in vivo, we performed RNA sequencing (RNA-seq) and transposase-accessible chromatin high-throughput sequencing (ATAC-seq) on SCs isolated from TMX-treated Chd4fl/fl and Chd4ΔPax7ER mice 3 days post injury. RNA-seq analysis identified 1,721 significantly upregulated genes and 1,226 significantly downregulated genes in Chd4ΔPax7ER SCs (Figures 2C), 41% and 47% of which were similarly up- or downregulated in vitro (Figure S2G). In agreement with our previous report, Chd4 loss upregulated Ripk3 (Sreenivasan et al., 2020). Furthermore, comparison of the in vitro and in vivo RNA-seq data confirmed increased expression of p21, a trend toward p57 upregulation, as well as increased expression of the MRFs MyoD and Myog, involved in SC proliferation and early differentiation, and of genes specific to mature skeletal muscle (Figures 2D and 2E). Notably, tissue specific expression analysis (TSEA) further revealed upregulation of genes not related to the skeletal muscle lineage, including brain-related genes such as Map1a, Nrdg2, and Usp11, and heart-related genes such as cardiac troponin T (Tnnt2), which were confirmed by qRT-PCR (Figures 2F and S2H). These observations, together with clustering of altered Gene Ontology (GO) biological process categories via using the database for annotation, visualization and integrated discovery (DAVID) (Figure 2G), strongly indicate that SCs require Chd4 to retain muscle stem cell identity in regenerating muscles, especially during the proliferative phase after injury.
RNA-seq of Chd4-deficient SCs in vivo also identified the downregulation of two important gene clusters: one involved in positive regulation of the cell cycle (e.g., cyclin-dependent kinase 4 [Cdk4]), and one in DNA damage repair (e.g., replication protein A1 [Rpa1]); these may contribute to the observed proliferative phenotype (Figure 2H). Chd4ΔPax7ER SCs also showed changes in the expression of other NuRD complex subunits (e.g., histone deacetylase 1 [Hdac1]), other CHD family genes (e.g., chromodomain helicase DNA-binding protein 5 [Chd5]), histone variants (e.g., histone cluster 1 H1 family member C [Hist1h1c]), and some Polycomb repressive complex 2 (PRC2) components (e.g., enhancer of zeste 2 [Ezh2]) (Figure S3A). These changes are particularly important, pointing to the consequent complexity of dysregulated epigenetic repression occurring in the absence of Chd4, and thereby providing a potential explanation for the expression of genes from diverse lineages.
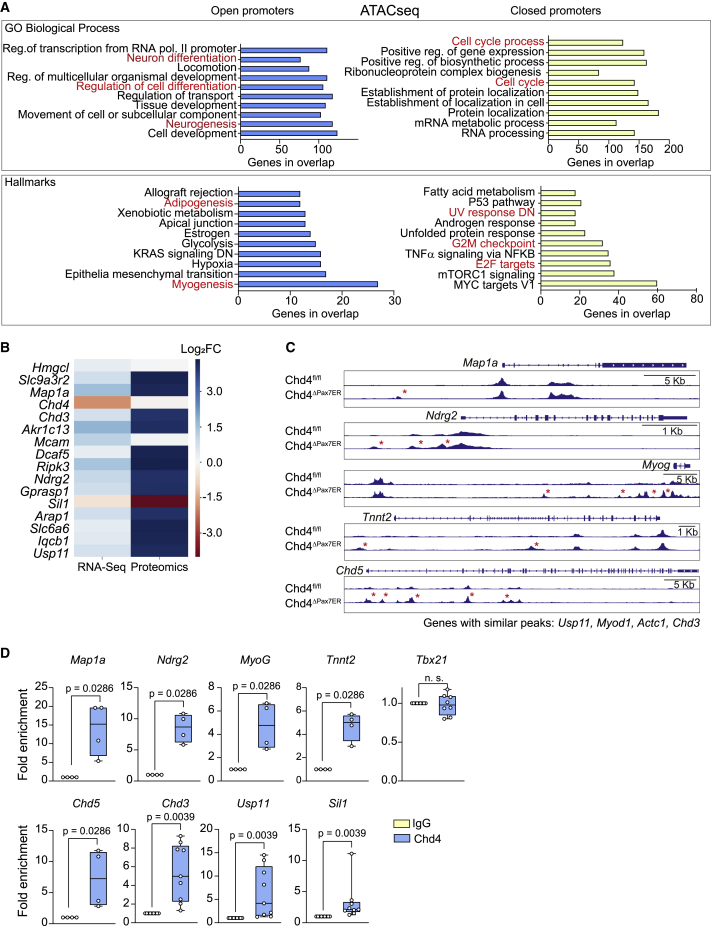
Supporting the transcriptomic data, ATAC-seq analysis of Chd4-deficient cells sorted 3 days after muscle injury showed extensive changes in proximal promoters near transcriptional start sites (TSSs), with 1,600 peaks significantly more open (e.g., with more accessible chromatin) and 1,436 significantly more closed. GO analysis of these changed genes revealed similar categories to those found by RNA-seq. For instance, myogenesis and neuronal differentiation gene promoters were significantly more open, whereas cell-cycle regulation, G2/M checkpoint, and E2F-regulated proliferation-related gene promoters were significantly more closed (Figure 3A).
Figure 3.
Chd4 maintains the transcriptional identity of SCs during muscle regeneration
(A) GO analysis of the genes with differential open (left) and closed (right) chromatin from SCs obtained from TMX-treated Chd4fl/fl or Chd4ΔPax7ER mice at 3 days after muscle injury (n = 4 mice/group). Data were analyzed using gene set enrichment analysis (GSEA) (MySigDB 6.2 Database). Reg, regulation; pol, polymerase. All categories represented obtained p < 0.0001.
(B) Color map (coded for Log2(fold change)) representing targets commonly found to be differentially expressed (p < 0.01) in the RNA-seq and protein mass spectrometry of Chd4fl/fl SCs transduced with Ad-CRE or Ad-GFP (n = 4 independent experiments). Data were analyzed using DESeq2 (RNA-seq) or MaxQuant (mass spectrometry).
(C) Genome browser tracks of ATAC-seq data at the Map1a, Ndrg2, Myog, Tnnt2, and Chd5 loci from SCs obtained from TMX-treated Chd4fl/fl and Chd4ΔPax7ER mice at 3 days after muscle injury (overlayed tracks of n = 4 mice).
(D) ChIP-qPCR of Chd4 protein binding to the indicated gene loci. The Tbx21 gene was used as control. Data were normalized to immunoglobulin G (IgG), which was set to 1 (n = 4 independent experiments).
Integration of proteomic and transcriptomic analyses of Chd4-deficient SCs (Sreenivasan et al., 2020), identified 18 commonly upregulated or downregulated proteins, including CHD3 and the neural proteins NDRG2, USP11, and MAP1a (Figure 3B). More importantly, Chd4ΔPax7ER SCs displayed inappropriate accessibility of lineage genes including Map1a, Ndrg2, Myog, Tnnt2, and Chd5 (Figure 3C). CHD4 binding to these loci was confirmed by ChIP-qPCR (Figure 3D), indicating direct repression.
CHD4 functions in SCs are intrinsic to the NuRD complex
We analyzed the interaction of other core NuRD subunits (MTA1 and HDAC1) in the absence of Chd4. Interestingly, the binding of MTA1 to HDAC1 was significantly reduced but not completely abolished (Figure S3B), indicating that CHD4 functions to promote the association of the NuRD complex to cognate binding sites in SCs. In agreement with this, histone acetylation was increased at the promoters of Chd4-regulated genes (Figure S3C), indicating reduced HDAC activity of the CHD4/NuRD complex in those promoters. ChIP-PCR analysis of HDAC1 on its cardinal target p21 gene locus (Mal et al., 2001) showed that the absence of Chd4 impaired HDAC1 recruitment (Figure S3D). Consistently, inhibition of HDAC1 in WT cells with romidepsin (Furumai et al., 2002) induced upregulation of the Map1a and Ndrg2 genes, similarly to Chd4 loss (Figure S3E).
As Chd4 deletion upregulated the expression of Chd3, we asked if an alternate CHD3/NuRD complex exists and, if so, whether it could act redundantly. Notably, Chd3 knockdown reduced the expression of Chd4, and vice versa (Figure S3F), pointing to a cross-regulation of both genes in SCs. Knockdown of either Chd3 or Chd4 resulted in a similar reduction of SC proliferation (Figures 1E–1G and S3F). Interestingly, the reduction of SC proliferation by single knockdown of Chd4 or Chd3 was not increased upon knockdown of both Chd4 and Chd3 (Figure S3F). These results show that CHD4's actions in SCs are integral to the NuRD complex and that the CHD4 and CHD3 complexes may be present in SCs with overlapping functionalities.
Discussion
This study dissects novel functional roles of the epigenetic modifier CHD4 in SCs during skeletal muscle regeneration, including cell identity and cellular expansion. Our results revealed that Chd4 loss breaks satellite lineage confinement and allows the expression of genes associated with other fates, indicating that its function is required for strict control of muscle lineage plasticity during regeneration.
In support of this notion, Chd4 loss induced upregulation of genes corresponding to multiple lineages in proliferating SCs, both in vitro and during muscle regeneration, including the brain and neural proteins NDGR2, USP11, and MAP1A at both the transcript and the protein level. Notably, Chd4 depletion also triggered the upregulation of Chd5, which encodes a neural-specific chromatin remodeler in the same CHD family as CHD3 and CHD4. CHD5 activates neuronal genes required for terminal neuronal differentiation (Egan et al., 2013; Potts et al., 2011). Therefore, it is tempting to speculate that CHD5 could be responsible for de-repression of neural genes, in an NuRD-dependent or -independent manner. Also, Chd4 loss significantly upregulated the expression of cardiac-specific genes, in agreement with our previous studies showing inappropriate expression of cardiac sarcomeric genes in Chd4-deficient mature skeletal muscles (Gomez-del Arco et al., 2016). Importantly, chromatin accessibility and chromatin binding assays confirmed that Map1a, Ndrg2, Myog, Tnnt2, and Chd5 are direct CHD4 targets, which explains the loss of their expression in the absence of Chd4. Chd4 depletion also changed the gene expression profiles of the Chd4 homolog Chd3 and epigenetic modulators Ezh2 and Suz12. The cross-regulation of Chd4 and Chd3 in SCs indicates that several NuRD complexes control SC proliferation, even though the CHD3 and CHD4 proteins form distinct NuRD complexes with specific target genes (Hoffmeister et al., 2017). This functional substitution of CHD4 by either CHD3 or CHD5 may contribute to the observed stem cell lineage-loss phenotype. The biological implication of these changes upon Chd4 depletion in SCs remains to be determined, despite reports of Ezh2 controlling SC identity (Juan et al., 2011). The results presented here show that CHD4 exerts a tight control on muscle stem cell fate commitment and lineage fidelity, acting as a master transcriptional repressor of non-muscle lineage genes and of differentiation-specific myogenic genes during the proliferative expansion of SCs. These CHD4-regulated processes are essential for the regenerative functions of muscle stem cells.
Experimental procedures
Mice
The satellite cell-specific Chd4 transgenic mouse model (Chd4ΔPax7ER) was generated by crossing mice that express CreERT2 from the Pax7 promoter (Pax7CRE-ER) with Chd4-floxed mice (Chd4fl/fl)) to generate Chd4ΔPax7ER mice. p57 conditional mice were used to generate Chd4ΔPax7ER/p57ΔPax7ER mice. Genetic ablation was induced by injection of 100 μL of tamoxifen (TMX) (T5648, Sigma-Aldrich, 20 mg/mL in corn oil) intraperitoneally for four consecutive days. Chd4fl/fl mice were used as control for the TMX treatment. Regeneration of skeletal muscle was induced by intramuscular injection of cardiotoxin (CTX; Latoxan, 10−5 M). The Catalan Government approved the work protocols, following applicable legislation. Both male and female mice were used in each experiment unless stated otherwise. Live colonies were maintained and genotyped as per Jackson Laboratories' guidelines and protocols. Mice were housed together, health was monitored daily for sickness symptoms, and euthanized immediately at the clinical endpoint when recommended by veterinary and biological services staff members.
SC isolation culture, immunohistochemistry, and cellular assays
A detailed description is provided in supplemental experimental procedures.
RNA-seq and ATAC-seq, ChIP, and RT-qPCR
A detailed description is provided in supplemental experimental procedures.
Statistical analysis
Data were analyzed using GraphPad Prism 8.0. The sample size (n) of each experimental group is described in each corresponding figure legend, and all experiments were repeated at least with three biological replicates. Data presented as mean ± SD. p < 0.05 was considered statistically significant. Mann-Whitney U test (independent samples, two sided) was used for pairwise comparisons among groups at each time point.
Data and code availability
RNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE179683.
Author contributions
Conceptualization, K.S., A.R., J.K., A.L.S., E.P., and P.M.-C.; formal analysis, K.S., A.R., and E.P.; investigation, K.S., A.R., D.M., J.S., I.E., and A.I.; writing –review & editing, K.S., A.R., J.K., A.L.S., E.P., and P.M.-C.; funding acquisition, T.B. and P.M.-C.; resources, F.R., P.G.-d.A., and J.M.R.; visualization, A.L.S. and E. P.; supervision, J.K., A.L.S., E.P., and P.M.-C.
Acknowledgments
We thank C. Keller for the Pax-CRE-ER mouse line; V. Lukesova, L. Ortet, E. Andrés, and A. Navarro for technical contributions; J. López for bioinformatics; J. Martín-Caballero (PRBB Animal Facilities); O. Fornas (UPF/CRG FACS Facility); A. Dopazo (CNIC-Genomics Facility); E. Rebollo (IBMB-CSIC Molecular Imaging Platform). The authors acknowledge funding from MINECO-Spain (grant no. RTI2018-096068), ERC-AdG-741966, LaCaixa-HEALTH-HR17-00040, MWRF, MDA, UPGRADE-H2020-825825, AFM, and DPP-Spain to P.M.C. P.G.-d.A. was supported by MINECO-Spain (grant no. SAF2016-77816-P); French ANR Labex REVIVE (grant no. ANR-10-LABX-73) to F.R. A.R. was supported by a DCEXS-UPF, BIST Fellowship. Fundació la Marató de TV3 supported A.L.S. (grant no. 202033) and P.M.C. (grant no. 202021) and the María-de-Maeztu-Program for Units of Excellence to UPF (grant no. MDM-2014-0370) and the Severo-Ochoa-Program for Centers of Excellence to CNIC (grant no. SEV-2015-0505).
Published: August 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.07.022.
Contributor Information
Antonio L. Serrano, Email: antonio.serrano@upf.edu.
Eusebio Perdiguero, Email: eusebio.perdiguero@upf.edu.
Pura Muñoz-Cánoves, Email: pura.munoz@upf.edu.
Supplemental information
References
- Egan C.M., Nyman U., Skotte J., Streubel G., Turner S., O'Connell D.J., Rraklli V., Dolan M.J., Chadderton N., Hansen K., et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Ermolaeva M., Neri F., Ori A., Rudolph K.L. Cellular and epigenetic drivers of stem cell ageing. Nat. Rev. Mol. Cell Biol. 2018;19:594–610. doi: 10.1038/s41580-018-0020-3. [DOI] [PubMed] [Google Scholar]
- Faralli H., Wang C., Nakka K., Benyoucef A., Sebastian S., Zhuang L., Chu A., Palii C.G., Liu C., Camellato B., et al. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R., Matsuyama A., Kobashi N., Lee K.H., Nishiyama M., Nakajima H., Tanaka A., Komatsu Y., Nishino N., Yoshida M., Horinouchi S. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- Gomez-del Arco P., Perdiguero E., Yunes-Leites P.S., Acin-Perez R., Zeini M., Garcia-Gomez A., Sreenivasan K., Jimenez-Alcazar M., Segales J., Lopez-Maderuelo D., et al. The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab. 2016;23:881–892. doi: 10.1016/j.cmet.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Hoffmeister H., Fuchs A., Erdel F., Pinz S., Grobner-Ferreira R., Bruckmann A., Deutzmann R., Schwartz U., Maldonado R., Huber C., et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017;45:10534–10554. doi: 10.1093/nar/gkx711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan A.H., Derfoul A., Feng X., Ryall J.G., Dell'Orso S., Pasut A., Zare H., Simone J.M., Rudnicki M.A., Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M., Morgan B.A., Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007;134:1571–1582. doi: 10.1242/dev.001750. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Wade P.A. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X., Vermeulen M., Brinkman A.B., Hoeijmakers W.A., Cohen A., Lasonder E., Stunnenberg H.G. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mademtzoglou D., Alonso-Martin S., Chang T.H., Bismuth K., Drayton-Libotte B., Aurade F., Relaix F. A p57 conditional mutant allele that allows tracking of p57-expressing cells. Genesis. 2017;55 doi: 10.1002/dvg.23025. [DOI] [PubMed] [Google Scholar]
- Mademtzoglou D., Asakura Y., Borok M.J., Alonso-Martin S., Mourikis P., Kodaka Y., Mohan A., Asakura A., Relaix F. Cellular localization of the cell cycle inhibitor Cdkn1c controls growth arrest of adult skeletal muscle stem cells. eLife. 2018;7 doi: 10.7554/eLife.33337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A., Sturniolo M., Schiltz R.L., Ghosh M.K., Harter M.L. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijo K., Hosoyama T., Bjornson C.R., Schaffer B.S., Prajapati S.I., Bahadur A.N., Hansen M.S., Blandford M.C., McCleish A.T., Rubin B.P., et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy A., Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem. Soc. Trans. 2013;41:777–782. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapcuk V., Mohn F., Carl S.H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thoma N.H., et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–743. doi: 10.1038/s41586-018-0153-8. [DOI] [PubMed] [Google Scholar]
- Potts R.C., Zhang P., Wurster A.L., Precht P., Mughal M.R., Wood W.H., 3rd, Zhang Y., Becker K.G., Mattson M.P., Pazin M.J. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.C.L., Dilworth F.J. Epigenetic regulation of adult myogenesis. Curr. Top Dev. Biol. 2018;126:235–284. doi: 10.1016/bs.ctdb.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Schworer S., Becker F., Feller C., Baig A.H., Kober U., Henze H., Kraus J.M., Xin B., Lechel A., Lipka D.B., et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature. 2016;540:428–432. doi: 10.1038/nature20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan K., Ianni A., Kunne C., Strilic B., Gunther S., Perdiguero E., Kruger M., Spuler S., Offermanns S., Gomez-del Arco P., et al. Attenuated epigenetic suppression of muscle stem cell necroptosis is required for efficient regeneration of dystrophic muscles. Cell Rep. 2020;31:107652. doi: 10.1016/j.celrep.2020.107652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.J., Naito T., Arco P.G., Seavitt J.R., Cashman S.M., De Souza B., Qi X., Keables P., Von Andrian U.H., Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Xu N., Liu F., Wu S., Ye M., Ge H., Zhang M., Song Y., Tong L., Zhou J., Bai C. CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer. 2020;20:262. doi: 10.1186/s12885-020-06762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wong C., Liu D., Finegold M., Harper J.W., Elledge S.J. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Gunther S., Looso M., Kunne C., Kruger M., Kim J., Zhou Y., Braun T. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat. Commun. 2015;6:7140. doi: 10.1038/ncomms8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhao H., Han Z., Liu X., Gu J., Tang F., Wei G., Jin Y. The chromatin remodeler Chd4 maintains embryonic stem cell identity by controlling pluripotency- and differentiation-associated genes. J. Biol. Chem. 2017;292:8507–8519. doi: 10.1074/jbc.M116.770248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE179683.