Summary
Activation of NOTCH signaling in human hematopoietic stem/progenitor cells (HSPCs) by treatment with an engineered Delta-like ligand (DELTA1ext-IgG [DXI]) has enabled ex vivo expansion of short-term HSPCs, but the effect on long-term repopulating hematopoietic stem cells (LTR-HSCs) remains uncertain. Here, we demonstrate that ex vivo culture of human adult HSPCs with DXI under low oxygen tension limits ER stress in LTR-HSCs and lineage-committed progenitors compared with normoxic cultures. A distinct HSC gene signature was upregulated in cells cultured with DXI in hypoxia and, after 21 days of culture, the frequency of LTR-HSCs increased 4.9-fold relative to uncultured cells and 4.2-fold compared with the normoxia + DXI group. NOTCH and hypoxia pathways intersected to maintain undifferentiated phenotypes in cultured HSPCs. Our work underscores the importance of mitigating ER stress perturbations to preserve functional LTR-HSCs in extended cultures and offers a clinically feasible platform for the expansion of human HSPCs.
Keywords: Notch, hypoxia, hematopoietic stem cell, endoplasmic reticulum stress, expansion, signaling, pathways, gene therapy, transplantation
Graphical abstract
Highlights
-
•
Superior NOTCH-mediated expansion of human HSPCs in hypoxic cultures
-
•
Culture under hypoxia mitigates ER stress in human HSPCs
-
•
Hypoxia potentiates NOTCH intracellular signaling in cultured human HSPCs
Larochelle and colleagues concurrently activate NOTCH and hypoxic pathways in ex vivo cultures of human adult HSPCs to enable a clinically relevant expansion of cells with long-term repopulating potential after transplantation. They characterize the molecular intersection between the two signaling pathways and demonstrate the important role of low oxygen tension in mitigating ER stress during NOTCH-mediated expansion of human HSPCs.
Introduction
Allogeneic transplantation and autologous ex vivo gene correction strategies for blood disorders are often restricted due to insufficient numbers of hematopoietic stem and progenitor cells (HSPCs) derived from umbilical cord blood (UCB) units, or from bone marrow (BM) and mobilized peripheral blood (MPB) of patients with impaired hematopoiesis. Likewise, due to inefficiencies of current nuclease-based protocols for targeted integration or correction of a therapeutic gene within human HSPCs only rare gene-edited cells are generally available for transplantation. Ex vivo expansion of transplantable hematopoietic stem cells (HSCs) would circumvent these shortcomings. Most efforts to expand human HSCs have focused on cells derived from UCB sources (Delaney et al., 2010; Horwitz et al., 2019; Wagner et al., 2016). However, UCB HSCs are biologically distinct and display a higher BM repopulating potential compared with adult HSCs; conditions developed for expansion of these cells typically show lower efficacy when applied to adult sources of HSCs (Tanavde et al., 2002). Hence, the development of safe and effective expansion strategies for adult HSCs remains a challenging goal in clinical hematology.
Ectopic activation of NOTCH signaling in human HSPCs promotes self-renewal (Kumano et al., 2003; Stier et al., 2002) and has been exploited for their expansion ex vivo (Delaney et al., 2005, 2010, 2016). NOTCH signaling is initiated by engagement of Jagged or Delta-like ligands to one of four NOTCH receptors on neighboring cells, resulting in a series of proteolytic cleavages and release of the intracellular domain (ICD) of NOTCH receptors from the cellular membrane (Artavanis-Tsakonas et al., 1995). The released domains translocate to the nucleus where they form transcriptionally active protein complexes and stimulate NOTCH-regulated genes, such as HES and HEY transcriptional targets. NOTCH activation in cultured human UCB HSPCs by treatment with DELTA1ext-IgG (DXI), an engineered NOTCH Delta-like ligand composed of the extracellular domain of DELTA1 fused to the Fc portion of the human immunoglobulin 1 (IgG1), has enabled clinically significant ex vivo expansion of short-term HSPCs and reduced times to neutrophil recovery following transplantation (Delaney et al., 2005, 2010). However, in this double UCB transplantation system in which only one of the cord units was expanded and the other left unmanipulated, sustained engraftment of the expanded donor graft was generally not observed. Although T cells found only in the unmanipulated UCB fraction may have triggered an immune-mediated rejection of the expanded cells after transplantation, this study also raised the possibility of ineffective expansion of long-term repopulating HSCs (LTR-HSCs) in cultures with DXI.
Recent studies have highlighted how increased proliferative demand triggers ER stress perturbations that collectively impair HSC function in ex vivo culture (Miharada et al., 2014; Milyavsky et al., 2010; Mohrin et al., 2010; Sigurdsson et al., 2016; van Galen et al., 2014, 2018; Xie et al., 2019). Accumulation of misfolded and unfolded proteins within the ER has been identified as an important mediator of ER stress in highly proliferating cells (Walter and Ron, 2011). The cellular ER responds to the burden of improperly assembled proteins by activating the unfolded protein response (UPR). Under mild and transient stress conditions, proteome homeostasis (i.e., proteostasis) is re-established and stress signals promote HSC survival, whereas under strong and persistent stress stimuli, UPR and chaperone activities initially increase in an attempt to mitigate ER stress but cell death through apoptosis eventually ensues when cellular homeostasis can no longer be restored (Tabas and Ron, 2011).
Multipotent HSCs occupy the most hypoxic niches within the BM (Giuntoli et al., 2007; Kubota et al., 2008; Parmar et al., 2007). Their bioenergetic demands are largely met by metabolic pathways based on anaerobic glycolysis. The limited production of reactive oxygen species (ROS) from anaerobic metabolic processes maintains their quiescent state (Bigarella et al., 2014; Jang and Sharkis, 2007). Hypoxia-inducible factors (HIFs) function as master regulators of the cellular response to hypoxic signals (Semenza, 2012). HIF-1 and its paralog HIF-2 consist of an alpha subunit (HIF-1α or HIF-2α) that can heterodimerize with the beta subunit of HIF-1 (HIF-1β), also known as aryl hydrocarbon receptor nuclear translocator (ARNT) (Mohyeldin et al., 2010). Under well-oxygenated conditions, HIF-α proteins are ubiquitinated and targeted for proteasomal degradation, thus preventing formation of functional heterodimers with HIF-1β. Unbound HIF-1β/ARNT can complex with the aryl hydrocarbon receptor (AHR) to induce expression of target genes, such as CYP1A1 and CYP1B1, which promote cellular differentiation (Lindsey and Papoutsakis, 2012). In contrast, HIF-α subunits are stabilized under hypoxia and form heteromers with HIF-1β proteins which are no longer accessible for binding with AHR. Instead, HIF-α/HIF-1β duplexes bind to hypoxia response elements within promoter regions of genes regulating metabolic states of HSCs, resulting in cellular quiescence and protection from oxidative stress (Semenza, 2012; Simsek et al., 2010). Evidence indicates that culture under hypoxia limits ER stress stimuli and may thus offer a simple approach to preserve and expand higher numbers of functional HSCs under highly proliferative conditions, such as those promoted in culture with DXI (Jang and Sharkis, 2007; Rouault-Pierre et al., 2013). Because HSCs display a lower tolerance to cellular stress and an increased propensity to apoptosis compared with downstream progeny, hypoxia may provide a distinct advantage to long-term HSCs subjected to high proliferative pressure (Miharada et al., 2014; Milyavsky et al., 2010; Mohrin et al., 2015).
In addition to the potential role of low oxygen tension in mitigating ER stress, accumulating evidence indicates that microenvironmental cues such as hypoxia can directly modulate NOTCH signaling output in various cell types (Landor and Lendahl, 2017; Poellinger and Lendahl, 2008). Studies in precursor cell lines (Gustafsson et al., 2005) established a molecular mechanism based on a direct interaction between NOTCH1 ICD and HIF-1α. Whether NOTCH and hypoxic signals intersect in human HSCs is not known, but in several other stem cell types the role of hypoxia in maintaining their primitive phenotype and promoting their proliferation requires functional NOTCH signaling (Fraker et al., 2007; Gustafsson et al., 2005; Xu et al., 2013). These observations suggest that culture of HSPCs with DXI under hypoxia may influence the potency and/or duration of NOTCH signaling to promote their expansion ex vivo.
Here, we provide evidence that concurrent activation of NOTCH and hypoxic pathways in ex vivo cultures of human adult HSPCs enables a clinically relevant expansion of LTR-HSCs in xenotransplant models. We demonstrate a NOTCH-hypoxia molecular crosstalk in HSPCs cultured with DXI under hypoxic conditions, and a transcriptomic signature that coheres with a mitigated ER stress response within the HSC compartment compared with counterpart cells from normoxia + DXI cultures.
Results
NOTCH-mediated ex vivo expansion of human CD34+ progenitor cells is enhanced by culture under hypoxic conditions
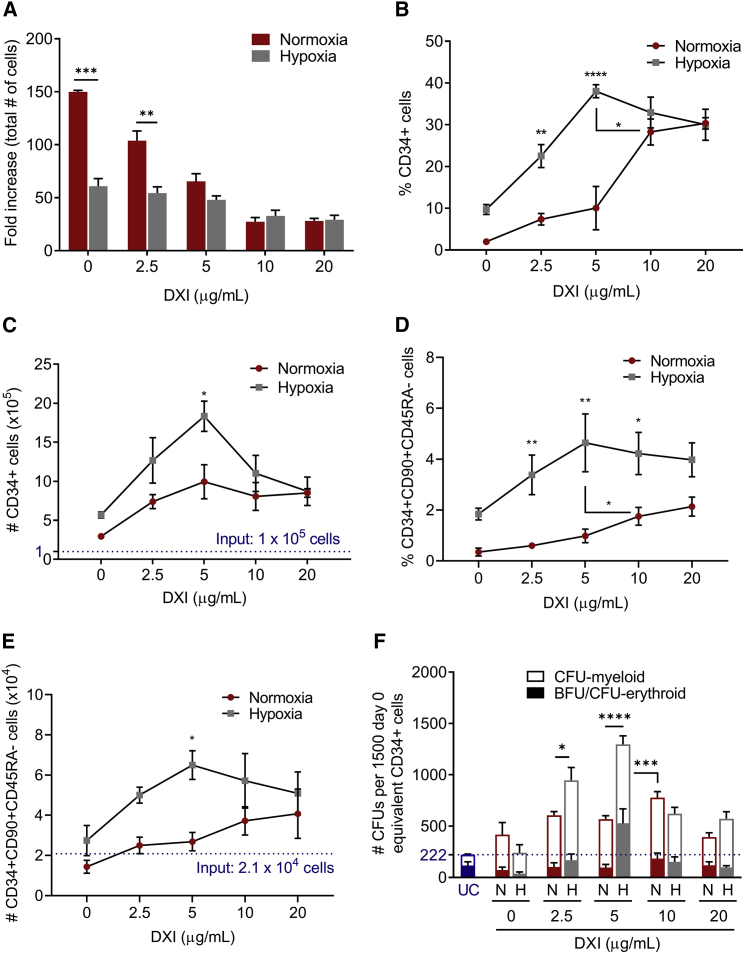
We first compared the global effect of normoxia and hypoxia on cellular growth, survival, and differentiation in the presence or absence of DXI. A total of 1 × 105 human MPB CD34+ cells were cultured for 21 days under normoxic (21% O2) or hypoxic (1%–2% O2) conditions in vessels coated with fibronectin alone or combined with increasing concentrations of DXI (2.5, 5, 10, and 20 μg/mL). After expansion, cells were counted and characterized by flow cytometry and functional assays. Low oxygen tension alone markedly reduced total cell numbers compared with normoxic cultures. In both normoxia and hypoxia, addition of DXI further decreased total cell counts in a density-dependent manner compared with control cultures without DXI (Figure 1A). The overall diminished cell production in hypoxia was not due to increased apoptosis (Figure S1A) or differentiation block of a specific hematopoietic lineage, although some skewing toward erythroid production was observed at all DXI concentrations compared with normoxic cultures (Figures S1B–S1F). The reduced total cell numbers at higher ligand densities in both hypoxia and normoxia were also not associated with significant differences in the percentages of cells that underwent apoptosis (Figure S1A) or lineage-specific inhibition of differentiation (Figures S1B–S1F). We observed decreased monocytic (CD14) differentiation at higher DXI concentrations in normoxic and hypoxic cultures, but the overall contribution of this lineage was low and alone cannot account for the observed decrease in total cell counts (Figure S1B). From these data, we infer that concurrent activation of NOTCH and hypoxia pathways in human CD34+ cells decreased the rate of cellular proliferation in a DXI-density-dependent manner, with minimal impact on cell survival or skewing of lineage differentiation.
Figure 1.
NOTCH-mediated ex vivo expansion of human CD34+ progenitor cells is enhanced by culture under hypoxic conditions
Human CD34+ cells were cultured under normoxia (21% O2) or hypoxia (1%–2% O2) in vessels coated with fibronectin alone or combined with increasing concentrations of DXI. After 21 days in culture, cells were counted and characterized by flow cytometry and colony-forming unit (CFU) assays.
(A) Total cell numbers (n = 4 donors).
(B) Percentages of CD34+ cells (n = 4 donors).
(C) Absolute CD34+ cell numbers (n = 4 donors).
(D) Percentages of CD34+CD90+CD45RA− cells (n = 4 donors).
(E) Absolute CD34+CD90+CD45RA− cell numbers (n = 4 donors).
(F) Numbers of myeloid and erythroid CFUs per 1,500 day-0 equivalent CD34+ cells plated (n = 6–9 technical replicates, from three donors).
In (A), (C), (E), and (F), blue horizontal dotted lines represent corresponding data from uncultured cells. Data are displayed as mean ± SEM; two-way ANOVA was used. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S1.
Next, we evaluated the relative effect of normoxia and hypoxia on the CD34+ progenitor compartment after expansion for 21 days with or without DXI. In the absence of NOTCH ligand, residual percentages (Figure 1B) and absolute numbers (Figure 1C) of CD34+ cells were generally poor, albeit better in hypoxic than normoxic cultures. Compared with control groups (no DXI), cells cultured with NOTCH ligand showed a marked increase in percentages (Figure 1B) and numbers (Figure 1C) of CD34+ cells at all tested concentrations of ligand in both normoxia and hypoxia. We observed a peak 18.3-fold expansion of CD34+ cells in hypoxic cultures plated with 5 μg/mL DXI relative to baseline cell input, whereas normoxic conditions increased CD34+ cell numbers up to 10-fold at the same or higher concentrations of NOTCH ligand (Figure 1C). A ligand-density-dependent expansion of HSC-enriched populations of CD34+CD90+CD45RA− cells was also observed, with maximum detection of these cells at DXI concentrations of 10–20 μg/mL in normoxia or 5 μg/mL in hypoxia (Figures 1D and 1E). In clonogenic progenitor assays, growth of myeloid (CFU-G, CFU-M, and CFU-GM) and erythroid (BFU-E and CFU-E) colonies also increased at all DXI densities under both normoxic and hypoxic conditions relative to colony counts derived from the same number of nonexpanded CD34+ cells (Figure 1F). At the prime concentrations of DXI in normoxia (10 μg/mL) and hypoxia (5 μg/mL), hypoxic cultures enabled a superior rise (1.7-fold) in total progenitor numbers compared with cells cultured under normal oxygen tension (Figure 1F). Collectively, our data indicate that NOTCH-mediated ex vivo expansion of human MPB CD34+ progenitor cells is enhanced by culture under hypoxic conditions, albeit minimally (<2-fold), when comparing optimized densities of ligand for each culture condition.
NOTCH and hypoxia pathways cooperate in human HSPCs
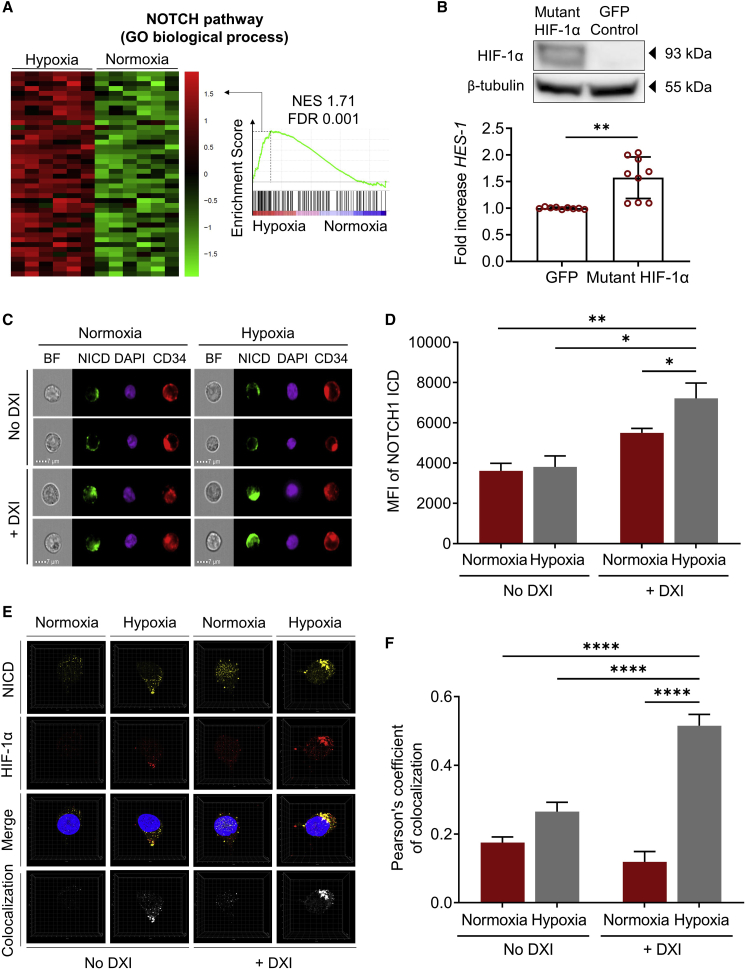
Evidence that hypoxia can directly modulate NOTCH signaling output in various cellular systems and our observation that lower densities of DXI were generally required to support the expansion of CD34+ cells under hypoxic conditions suggested that molecular regulators of the hypoxia pathway may influence the potency and/or duration of NOTCH signaling in HSPCs. To test this hypothesis, we first cultured human MPB CD34+ cells under normoxic or hypoxic conditions for 24 h with or without DXI, and compared expression of the primary NOTCH target gene HES-1 by qRT-PCR. We observed a global increase in HES-1 gene expression in cells cultured under hypoxia compared with normoxia (Figure S2A). Consistent with the known regulation of HIF-1α by hypoxia at the protein level, mRNA expression of HIF-1α was comparable between normoxic and hypoxic conditions (Figure S2B). We then undertook a global transcriptomic analysis of purified CD34+ cells treated for 24 h in normoxic or hypoxic cultures in wells coated with DXI at the optimized densities (10 μg/mL in normoxia; 5 μg/mL in hypoxia). We observed several differentially expressed genes between each experimental group (Figure S2C and Table S1; false discovery rate [FDR] < 0.05; fold change > 1.5 or < −1.5). The ranked gene lists were evaluated using gene set enrichment analysis (GSEA) against open-access expression databases for both hypoxia and NOTCH pathways. When comparing the hypoxia pathway for cells cultured with DXI under normoxic or hypoxic settings, we observed the expected enrichment of this gene set in the hypoxia group (Figure S2D and Table S2). Analysis of gene sets implicated in NOTCH signaling revealed increased activity of the NOTCH pathway in the hypoxia + DXI group relative to the normoxia + DXI control (Figure 2A and Table S2), notwithstanding the lower concentration of DXI used in hypoxic cultures.
Figure 2.
NOTCH and hypoxia pathways cooperate in human HSPCs
(A) Heatmap of leading edge subset (left) and enrichment plot (right) showing relative expression of genes associated with the NOTCH pathway in CD34+ cells cultured in normoxia (21% O2) or hypoxia (1%–2% O2) with optimized densities of DXI. NES, normalized enrichment score; FDR, false discovery rate.
(B) Top: western blot analysis confirming exogenous HIF-1α expression in human CD34+ cells cultured under normoxic conditions with DXI after transduction with a mutant HIF-1α-expressing lentiviral vector. A control group transduced with a GFP-expressing lentiviral vector is shown for comparison. β-Tubulin was used as loading control. Bottom: HES-1 gene expression measured by qRT-PCR (n = 9 technical replicates, from three donors). Results indicate the fold increase in HES-1 levels normalized to a GFP-transduced control group.
(C) Representative ImageStream images of human CD34+ cells cultured under normoxic or hypoxic conditions in the absence or with optimized concentrations of DXI. BF, bright-field; NICD, NOTCH1 intracellular domain (ICD); DAPI, 4′,6-diamidino-2-phenylindole.
(D) Quantification of the mean fluorescence intensity (MFI) of NOTCH1 ICD in 10,000 cells/condition.
(E) Representative high-resolution images of individual cells assessed by confocal microscopy. The bottom panel displays a colocalization channel generated with Imaris imaging software.
(F) Quantification of NOTCH1 ICD and HIF-1α signal colocalization in individual cells for each condition by the Pearson's correlation coefficient (n = 20 cells/condition).
In (B), (D), and (F), data are displayed as mean ± SEM. Two-sided (B and F) and one-sided (D) unpaired t test were used. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001. See also Figures S2 and S3; Table S2.
To determine whether the impact of hypoxia on NOTCH signaling was linked to the specific recruitment of HIF-1α, we measured HES-1 expression in CD34+ cells transduced with a lentiviral vector expressing a mutant HIF-1α (P402A/P564A) known to resist proteasome degradation (Yan et al., 2007), and cultured these cells under normoxic conditions with DXI (Figure 2B, top panel). Notably, after 48 h of normoxic culture, HES-1 expression was significantly increased in mutant HIF-1α-overexpressing cells compared with a GFP-transduced control group (Figure 2B, bottom panel). Together, these results suggest an HIF-1α-mediated cooperation between NOTCH and hypoxia pathways in human HSPCs, analogous to observations in other stem/progenitor cell populations (Gustafsson et al., 2005).
Because activation of NOTCH receptors ultimately leads to the liberation of NOTCH ICDs, we next queried whether this step in the NOTCH signaling cascade might be modulated by hypoxia. We used ImageStream to compare protein levels of NOTCH1 and NOTCH2 ICD in human CD34+ cells cultured under normoxic or hypoxic conditions in the absence or with previously optimized concentrations of DXI. When examining NOTCH1, we first confirmed that the ICDs were readily detected within the nucleus upon addition of the ligand in normoxia and hypoxia (Figure 2C). Importantly, the signal intensity of NOTCH1 ICD significantly increased in hypoxic cultures compared with control groups (Figures 2C and 2D), indicating that hypoxia affects signaling mediated by the ICD of NOTCH1 receptor. However, no differences in ICD fluorescence intensities were observed in experiments examining the effect of hypoxia on NOTCH2 signaling (Figures S3A and S3B).
Because NOTCH1 ICD and HIF-1α were previously shown to cooperate by direct physical association in other cellular systems, we examined the localization of their fluorescence signals by confocal microscopy within human CD34+ cells cultured for 24 h under normoxia or hypoxia, with or without DXI. Similar to findings by ImageStream, we observed enhanced NOTCH1 ICD signal intensities under hypoxic conditions (Figure 2E, NICD). Importantly, this approach also revealed increased spatial overlap of NOTCH1 ICD with HIF-1α after concomitant activation of NOTCH and hypoxia pathways in culture compared with control groups (Figure 2E, colocalization). Statistical quantification of colocalization in individual cells for each condition by the Pearson's correlation coefficient confirmed an increased co-occurrence of NOTCH1 ICD and HIF-1α in HSPCs cultured under hypoxia with DXI relative to control groups (Figure 2F). In contrast, hypoxic culture did not enhance colocalization of NOTCH2 ICD and HIF-1α (Figure S3C). Taken together, these results support a molecular mechanism by which HIF-1α enhances signaling mediated by the ICD of NOTCH1 in human HSPCs.
Concomitant activation of NOTCH and hypoxia pathways enhances ex vivo expansion of human LTR-HSCs
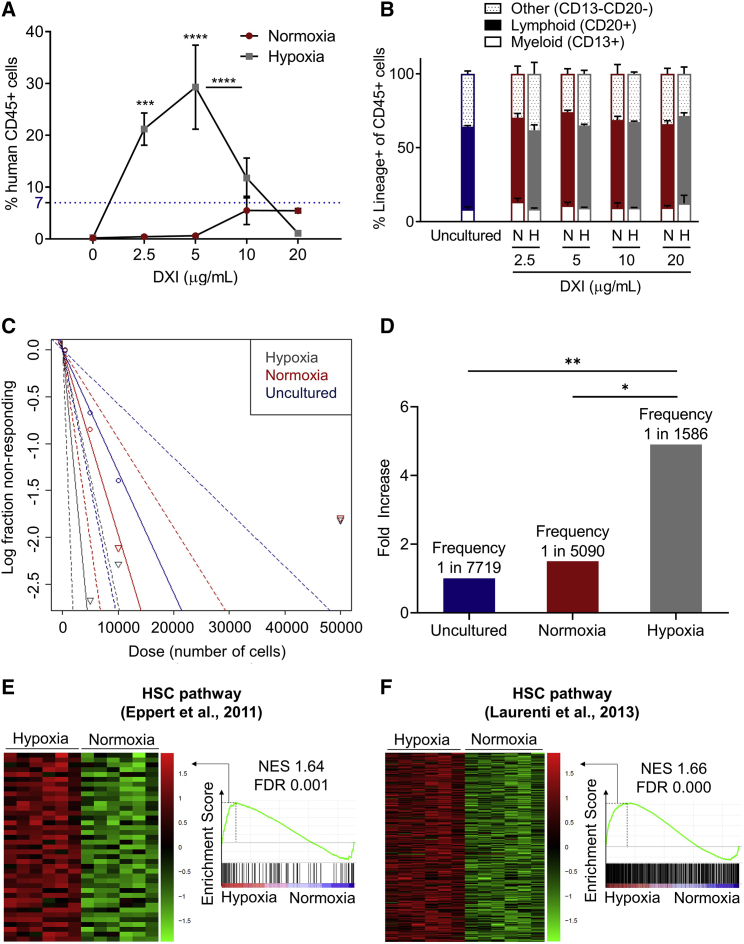
To address whether LTR-HSCs can be expanded in our culture system, we quantified human cell engraftment at 4 months after transplantation of NOD-Scid-IL2rynull (NSG) mice with 1 × 105 unmanipulated MPB CD34+ cells or the 21-day expanded progeny of the same starting cell number for each culture condition. In contrast to previous studies utilizing HSPCs derived from UCB sources (Delaney et al., 2005, 2010), MPB CD34+ cells cultured in normoxia with 10 or 20 μg/mL DXI resulted in levels of engraftment in NSG mice comparable but not superior to uncultured cells, and lower densities of DXI were insufficient to maintain cells with repopulating activity (Figure 3A). Notably, we observed a significant increase (∼5-fold) in human cell engraftment when hypoxia was used in combination with 2.5 or 5 μg/mL DXI compared with unexpanded CD34+ cells or HSPCs cultured in normoxia at optimal density (10 μg/mL) of DXI (Figure 3A). Multilineage engraftment was detected in all groups in proportions similar to engraftment derived from uncultured cells (Figure 3B).
Figure 3.
Concomitant activation of NOTCH and hypoxia pathways enhances ex vivo expansion of human LTR-HSCs
(A) Percentages of human CD45+ cells in the BM of NSG mice 4 months after transplantation of 1 × 105 uncultured CD34+ cells (blue horizontal dotted line) or the 21-day expanded progeny of the same starting cell number for normoxic and hypoxic culture conditions in the absence or with increasing concentrations of DXI (n = 3–4 mice/group).
(B) Lineage distribution of human CD45+ cells in the BM of NSG mice. N, normoxia; H, hypoxia (n = 3–4 mice/group).
(C) Semilogarithmic plot of LTR-HSC frequency 4 months after transplantation of uncultured CD34+ cells or the 21-day expanded progeny of the same starting cell number for normoxic or hypoxic culture conditions in the presence of optimized concentrations of DXI. Solid lines indicate the best-fit linear model for each dataset. Dotted lines represent 95% confidence intervals.
(D) Summary of LTR-HSC frequency and fold increase in LTR-HSC frequency after 21 days of expansion in normoxia or hypoxia with optimized concentrations of DXI relative to uncultured CD34+ cells. p values were measured by extreme limiting dilution analysis.
(E and F) Heatmaps of leading edge subset (left) and enrichment plots (right) showing the relative expression of genes associated with HSC pathways (E, Eppert et al., 2011; F, Laurenti et al., 2013) in CD34+ cells cultured in normoxia or hypoxia with optimized densities of DXI. NES, normalized enrichment score; FDR, false discovery rate.
In (A) and (B), data are displayed as mean ± SEM. Two-way ANOVA was used unless otherwise specified. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Table S2.
To elucidate how hypoxia improved engraftment of CD34+ cells cultured in the presence of DXI compared with normoxia, we speculated that either the homing potential of repopulating cells, their absolute numbers, or both properties were enhanced by hypoxia. We first measured expression of CXCR4, a chemokine receptor regulated via HIF-dependent pathways and well known to mediate homing of HSPCs within the BM after transplantation (Kahn et al., 2004; Schioppa et al., 2003; Speth et al., 2014; Staller et al., 2003). Cultures of CD34+ cells from eight independent donors were established under normoxic or hypoxic conditions in wells coated with DXI at previously optimized concentrations. No statistically significant difference in CXCR4 expression was seen in CD34+ cells after 21 days of culture under normoxia or hypoxia (31.7% versus 35.8%, p = 0.19). Similar results were obtained when CXCR4 expression was compared in a more purified CD34+CD90+CD45RA− cell population (37.4% versus 38.8%, p = 0.55), indicating that upregulation of CXCR4 was likely not the primary mechanism by which hypoxia augmented engraftment of DXI expanded cells.
Next, we performed limiting dilution analyses to compare the frequency of LTR-HSCs within the CD34+ cell compartment at baseline and after 21 days of normoxic or hypoxic cultures supplemented with the optimized concentrations of DXI (Figures 3C and 3D). Engrafting cells in uncultured CD34+ cells were measured at the expected frequency of 1 in 7,719 (Huntsman et al., 2015). When analyzed at 4 months post transplantation, a limited (1.5-fold) increase in frequency (1 in 5,090) was obtained from normoxic cultures relative to uncultured cells. In contrast, the frequency of long-term repopulating cells (1 in 1,586) was 4.9-fold higher (p = 0.007) in hypoxic cultures compared with uncultured cells and 4.2-fold higher (p = 0.045) than in the normoxia group (Figures 3C and 3D).
To corroborate these findings molecularly, we queried the ranked gene lists from our previously established RNA sequencing (RNA-seq) dataset (see Figure 2A, Table S2, and Figures S2C and S2D) using GSEA against published gene expression signature databases of LTR-HSCs (Eppert et al., 2011; Laurenti et al., 2013). Consistent with functional transplantation assays, we found that HSC gene expression signatures were significantly upregulated in cells cultured with DXI in hypoxia compared with cells exposed to DXI under normoxic conditions (Figures 3E and 3F; Table S2). Collectively, these data show that hypoxia supports a superior ex vivo expansion of human LTR-HSCs compared with normoxia at optimized densities of DXI ligand.
Downregulation of ER stress pathways by hypoxia during DXI-mediated expansion of human LTR-HSCs
To further clarify how hypoxia improved NOTCH-mediated expansion of LTR-HSCs, we performed single-cell RNA-seq (scRNA-seq) of sorted MPB CD34+ cells treated with optimized DXI densities under normoxic or hypoxic conditions. In total, after quality control 1,532 and 2,709 single CD34+ cell libraries from normoxic and hypoxic cultures, respectively, were retained for further analyses. Summary statistics are shown in Figure S4A.
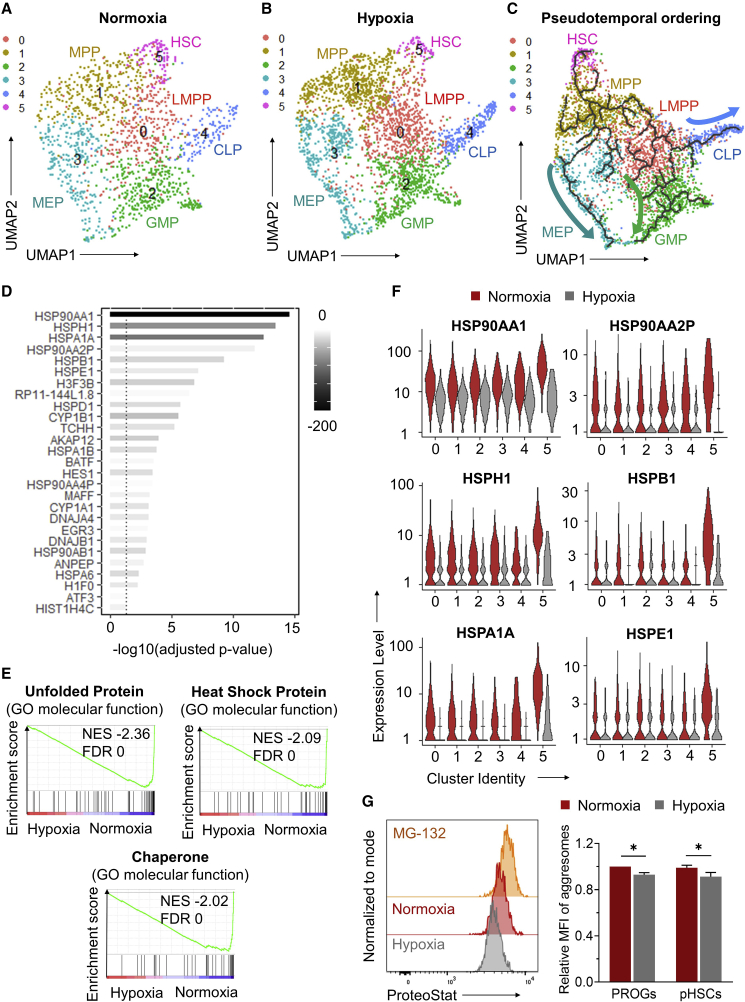
To impute a pattern of HSPC differentiation, we clustered and visualized transcriptome sequencing data of single CD34+ cells from each experimental group in two-dimensional uniform manifold approximation and projection (UMAP). We identified six distinct cell clusters with a comparable distribution of cells per cluster between normoxic (Figure 4A) and hypoxic cultures (Figure 4B). By associating cluster-specific transcripts with defined lineage transcriptome signatures (Eppert et al., 2011; Laurenti et al., 2013; Laurenti and Gottgens, 2018; Tirosh et al., 2016; Zhao et al., 2017), CD34+ cell clusters could be computationally assigned to define hematopoietic subpopulations, including lymphoid-primed multipotent progenitors (cluster 0), multipotent progenitors (cluster 1), granulocyte-monocyte progenitors (GMP, cluster 2), megakaryocytic-erythroid progenitors (MEP, cluster 3), common lymphoid progenitors (CLP, cluster 4), and a single HSC population (cluster 5) (Figures 4A, 4B, and S4B). Pseudotemporal ordering was used to reconstruct hematopoiesis. By mapping the cell type profile from UMAP, we developed a trajectory of differentiation from HSCs to MEP and to myeloid (GMP) and lymphoid progenitors (CLP) (Figure 4C). Thus, we were able to deconvolute the heterogeneous CD34+ cell population at single-cell level, and reconstructed a trajectorial pattern of hematopoiesis.
Figure 4.
Downregulation of ER stress pathways by hypoxia during DXI-mediated expansion of human LTR-HSCs
(A and B) UMAP visualization of scRNA-seq data for CD34+ cells treated in vessels coated with optimized densities of DXI under normoxic (A) or hypoxic (B) conditions. A total of six cell clusters were identified and annotated as lymphoid-primed multipotent progenitors (LMPP, cluster 0), multipotent progenitors (MPP, cluster 1), granulocyte-monocyte progenitors (GMP, cluster 2), megakaryocytic-erythroid progenitors (MEP, cluster 3), common lymphoid progenitors (CLP, cluster 4), and hematopoietic stem cells (HSC, cluster 5).
(C) Reconstruction of the hematopoietic hierarchy pseudotime ordering by Monocle in the integrated dataset.
(D) List of 27 differentially expressed genes (DEGs) (all downregulated) identified by comparing the transcriptome of HSC cluster 5 from normoxic and hypoxic cultures. These genes represent the overlap between two lists of DEGs identified using two independent statistical methods, Wilcoxon rank sum (Figure S4C) and DESeq2 (Figure S4D).
(E) Enrichment plots showing the relative expression of genes associated with unfolded protein, heat-shock protein, and chaperone pathways. NES, normalized enrichment score; FDR, false discovery rate.
(F) Violin plots showing expression levels of the top downregulated genes identified in (D).
(G) Quantification of protein aggregates (aggresomes) in human CD34+CD38+ progenitors (PROGs) and phenotypically defined CD34+CD38−CD90+CD45RA− LTR-HSC populations (pHSCs) cultured with optimized DXI densities under normoxic or hypoxic conditions, as measured using a ProteoStat staining approach. Left: representative flow-cytometry histograms; the proteasome inhibitor MG-132 was used as positive control to define a population of ProteoStathigh population. Right: summary of relative mean fluorescence intensity (MFI) of aggresomes (n = 9 technical replicates, from three independent donors). Data are displayed as mean ± SEM. One-way repeated-measures ANOVA was used. ∗p ≤ 0.05.
See also Figures S4 and S5; Tables S3, S4, and S5.
To assess the relative impact of normoxia and hypoxia on the HSC compartment, we performed GSEA of cells within HSC cluster 5 for each culture condition. A total of 27 differentially expressed genes (all downregulated) were identified in HSCs exposed to hypoxia + DXI compared with control cultures (Figures 4D, S4C, and S4D; Tables S3 and S4). Consistent with the known molecular crosstalk between hypoxic and AHR signaling pathways (Lindsey and Papoutsakis, 2012), culture with DXI under hypoxia downregulated expression of two AHR target genes (i.e., CYP1A1 and CYP1B1), providing an objective validation of the scRNA-seq dataset (Figure 4D). Pathways indicative of cellular ER stress, including UPR, heat-shock protein (HSP), and chaperone systems, were most significantly downregulated in hypoxia-treated cells relative to normoxic cultures (Figure 4E and Table S5). When the top differentially expressed genes in HSC cluster 5 were examined across all cell clusters, we observed a more prominent upregulation of these genes within transcriptionally defined HSCs exposed to normoxia relative to more mature progenitor populations in clusters 0–4 (Figure 4F, red violin plots). Notably, hypoxia lessened the cellular stress response in both progenitors and HSCs, but the mitigation was more apparent in the HSC population (Figure 4F, gray violin plots).
To independently confirm these findings, we performed qRT-PCR of the top differentially expressed ER stress response genes (see Figure 4D) from MPB CD34+CD38+ hematopoietic progenitors and HSC-enriched CD34+CD38− populations treated with optimized DXI densities under normoxic or hypoxic conditions. Consistent with the scRNA-seq analysis, we observed an upregulation of ER stress response genes in LTR-HSC-enriched populations exposed to normoxia relative to more mature hematopoietic progenitors, and hypoxia lessened ER stress response more prominently in LTR-HSC-enriched populations (Figure S5). To assess the impact of NOTCH signaling inhibition on ER stress response, we also incubated cells from each culture condition with LY411575, a small-molecule inhibitor of γ-secretase-mediated proteolytic cleavage of NOTCH ICD. Blockade of NOTCH ICD cleavage and release by LY411575 did not reverse downregulation of ER stress response mediated by hypoxia during ex vivo culture with DXI (Figure S5).
To further corroborate gene expression analysis of ER stress response pathways, we compared the burden of improperly assembled proteins in human HSPCs cultured with optimized DXI densities under normoxic or hypoxic conditions. Accumulation of protein aggregates (i.e., aggresomes) was previously shown to correlate with activation of ER stress response in HSPCs (Sigurdsson et al., 2016). In agreement with transcriptomic data, we found that hypoxia lowered aggresome formation in both CD34+CD38+ progenitors and phenotypically defined CD34+CD38−CD90+CD45RA− LTR-HSC populations (pHSCs), as measured using a ProteoStat staining approach (Figure 4G). From these data, we infer that hypoxia curtailed ER stress response pathways during DXI-mediated expansion of LTR-HSCs.
Downregulation of apoptotic pathways by hypoxia during DXI-mediated expansion of human LTR-HSCs
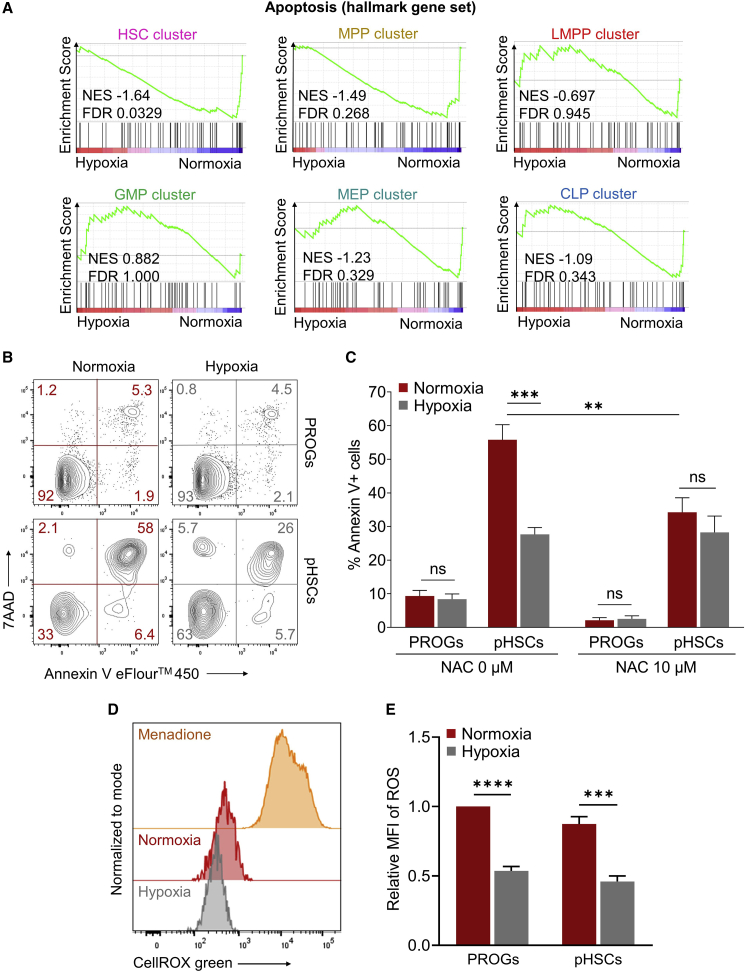
To investigate whether decreased ER stress response in hypoxic cultures resulted in improved survival of LTR-HSCs more than progenitors, we first evaluated the ranked gene lists from each scRNA-seq cell cluster using GSEA against the Hallmark Collection of the Molecular Signatures Database, which includes gene sets linked to apoptosis (Liberzon et al., 2015). A significant (FDR < 0.05) hypoxia-mediated decrease in apoptosis was observed exclusively within the HSC cluster (Figure 5A).
Figure 5.
Reduced apoptosis and ROS levels by hypoxia during DXI-mediated expansion of human LTR-HSCs
(A) Enrichment plots showing the relative expression of genes associated with the apoptosis pathway for all six cell clusters identified by scRNA-seq of CD34+ cells treated in vessels coated with optimized densities of DXI under normoxic or hypoxic conditions. NES, normalized enrichment score; FDR, false discovery rate.
(B) Representative flow-cytometry plots obtained by dual annexin V/7AAD immunostaining of human CD34+ cells treated as described in (A).
(C) Summary of percentages of annexin V+ cells in CD34+ cell subpopulations comprising hematopoietic CD34+CD38+ progenitors (PROGs) or phenotypically defined CD34+CD38−CD90+CD45RA− LTR-HSC populations (pHSCs) (n = 3 independent donors). Addition of N-acetyl-L-cysteine (NAC) ROS scavengers in culture rescued pHSC apoptotic death under normoxic conditions.
(D) Quantification of ROS levels in human PROGs and pHSCs after culture for 24 h with optimized densities of DXI under normoxic or hypoxic conditions using a CellROX green assay. Representative flow-cytometry plots are shown. Cells treated with menadione were included as a positive control.
(E) Summary of relative mean fluorescence intensity (MFI) of ROS (n = 3 independent donors).
Data are displayed as mean ± SEM. Ordinary one-way ANOVA was used. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; ns, not significant.
To independently confirm these findings, we next treated CD34+ cells with optimized DXI densities under normoxic or hypoxic conditions for 24 h and measured levels of apoptosis by annexin V staining assay in subpopulations composed of hematopoietic progenitors or highly enriched in LTR-HSCs. In both normoxic and hypoxic groups, the majority (>90%) of progenitor cells were viable (i.e., annexin V−/7AAD−). In contrast, the proportion of LTR-HSC-enriched populations that underwent apoptosis (i.e., annexin V+) rose markedly in normoxic cultures (55.8% ± 4.5%, mean ± standard error of the mean [SEM]), and a significantly lower fraction of this population (27.7% ± 2.0%, mean ± SEM) underwent apoptosis in hypoxia (Figures 5B and 5C).
Because ER stress-mediated apoptosis can be exacerbated by increased ROS production and vice versa, we also quantified ROS molecules in normoxic and hypoxic cultures. We confirmed the expected lower ROS levels in the hypoxia group (Figures 5D and 5E) and showed rescue of pHSC apoptotic death in normoxia by addition of N-acetyl-L-cysteine (NAC) ROS scavengers in culture (Figure 5C). Taken together, these results indicate that hypoxia enables DXI-mediated expansion of LTR-HSCs by mitigating ER stress, ROS production, and apoptosis associated with increased cellular proliferation under normoxic ex vivo culture conditions.
Discussion
In this study we provide functional, phenotypic, and molecular evidence that ex vivo culture of human adult CD34+ cells under hypoxic conditions enables a superior DXI-mediated expansion of hematopoietic cells with long-term lymphoid and myeloid repopulating capacity compared with normoxic cultures. Our data suggest a two-pronged mechanism by which optimal ectopic activation of NOTCH signaling in human HSCs enhances their self-renewal, and culture under hypoxia mitigates ER stress triggered by the increased proliferative demand, as evidenced by reduced UPR and HSP chaperone activity, lower ROS levels, and decreased formation of aggresomes, resulting in improved survival of expanding HSCs.
A key finding emerging from our work is the distinct influence of hypoxia on LTR-HSCs and progenitors during expansion with DXI. As previously shown in alternative culture systems (Milyavsky et al., 2010; Mohrin et al., 2010; Sigurdsson et al., 2016; van Galen et al., 2014; Xie et al., 2019), activation of ER stress and apoptotic pathways under normoxic conditions was more apparent in cell populations enriched in LTR-HSCs than in hematopoietic progenitors. Importantly, the benefits provided by low-oxygen cultures were most notable in the primitive HSC compartment, consistent with a recent investigation suggesting that, under lower stress conditions, pro-survival integrated stress responses are uniquely activated in HSCs to restore cellular homeostasis and ensure their long-term persistence (van Galen et al., 2018). Other approaches have also been proposed to confer protection against ER stress, including addition or enforced expression of chaperones (Miharada et al., 2014; Sigurdsson et al., 2016; van Galen et al., 2014) and transient pharmacological inhibition of de novo sphingolipid synthesis in HSPCs (Xie et al., 2019), but their utility in combination with DXI-mediated NOTCH activation for LTR-HSC expansion will require direct testing.
Further investigation is needed to fully understand the mechanism by which hypoxia limits activation of ER stress pathways in HSPCs, and most notably in LTR-HSCs treated with DXI. Our data suggest that the stabilized alpha subunits of HIF-1 and HIF-2 regulators in hypoxia likely play a critical role by averting extensive ROS formation known to simulate ER stress and trigger apoptosis by activation of the UPR pathway (Rouault-Pierre et al., 2013). The importance of thwarting excessive ROS production for optimal expansion of LTR-HSCs was recently highlighted in a study using three-dimensional culture of human HSPCs in a degradable zwitterionic hydrogel that promoted extensive CD34+ cell proliferation by suppressing ROS levels (Bai et al., 2019). Likewise, prevention of ER stress by ectopic expression of the RNA binding protein Dppa5 (Miharada et al., 2014) or the mitochondrial HSP Mortalin (Tai-Nagara et al., 2014) maintained low cytosolic ROS levels and improved HSC function compared with untreated controls. Alternative strategies to scavenge or limit production of ROS within CD34+ cell populations, including supplementation of culture media with common antioxidant agents (e.g., NAC) (Hu et al., 2014), pharmacological inhibitors of mitochondrial permeability transition pore (e.g., cyclosporin A) (Mantel et al., 2015), or stabilizers of HIF alpha subunits (e.g., dimethyloxylalylglycine) (Speth et al., 2014), may be useful in combination with NOTCH signaling activation to expand LTR-HSCs. However, cellular toxicity associated with these small molecules is often limiting (Bai et al., 2019; Broxmeyer, 2016).
Beyond its role in mitigating ER stress, hypoxia may also play a central role in DXI-mediated expansion of human HSPCs by potentiating NOTCH intracellular signals in cultured cells. The intersection between NOTCH and hypoxia signaling mechanisms has been increasingly recognized in various physiological and disease contexts (Landor and Lendahl, 2017; Poellinger and Lendahl, 2008), and this crosstalk is important to maintain undifferentiated states in several stem cell populations (Fraker et al., 2007; Gustafsson et al., 2005; Xu et al., 2013). Here, we demonstrate that a molecular link also exists between NOTCH signaling and the cellular hypoxic response in human HSPCs. This finding expands on previous reports describing the integration of other intracellular signaling systems in HSPCs, such as NOTCH and Wnt pathways known to converge for the maintenance and self-renewal of HSCs (Duncan et al., 2005; Moore, 2005). Consistent with previous investigations (Gustafsson et al., 2005; Villa et al., 2014), we found that NOTCH1 ICD and HIF-1α are central in the convergence point between the two signaling pathways in HSPCs. The contribution of NOTCH1 ICD is underscored by the increased detection of its signal under hypoxic conditions in flow cytometry and confocal microscopy imaging. The enhanced NOTCH1 ICD activity under hypoxia could result from increased cleavage at the cellular membrane, perhaps via activation of proteolytic enzymes mediating this process (Villa et al., 2014) or from increased domain stability as suggested by pulse-chase experiments in other cellular systems (Gustafsson et al., 2005). The participation of HIF-1α in the NOTCH-hypoxia convergence is highlighted in our study by the upregulation of HES-1 gene expression under normal oxygen tension when a normoxia-resistant form of HIF-1α was upregulated in culture, and by the observed colocalization of HIF-1α and NOTCH1 ICD inferred by metrics based on Pearson's coefficient of correlation. Direct physical association between HIF-1α and NOTCH1 ICD was previously confirmed by coimmunoprecipitation of in vitro translated proteins (Gustafsson et al., 2005), but this pull-down assay is impractical in primary human HSPCs due to the low abundance of both proteins in these cells. Although HIF-1α and the ICD of NOTCH1 likely interact in HSPCs, the precise role and fate of this complex remain unclear. Upregulation of genes involved in NOTCH signaling observed under hypoxia in our study suggests that HIF-1α complexed with NOTCH1 ICD could be recruited onto NOTCH-responsive gene promoters, as shown in previous chromatin immunoprecipitation studies (Gustafsson et al., 2005). Our work also indicates that NOTCH2 is unlikely to be implicated in the intersection of NOTCH and hypoxic signaling in human HSCs. However, it is probable that other constituents of each pathway (e.g., NOTCH3, HIF-2α, Factor Inhibiting HIF-1α) participate in this crosstalk to maintain undifferentiated states in HSPC populations.
In conclusion, we provide new molecular and functional evidence to support the use of DXI in combination with hypoxia to facilitate the expansion of human LTR-HSCs and, to a lesser extent, more mature hematopoietic progenitors. Our findings underscore the importance of incorporating strategies to mitigate ER stress in HSC expansion protocols. Collectively, data presented in this study have implications for fundamental investigations exploring ex vivo expansion of human HSPCs, and for genetic and cellular therapeutics requiring increased cell doses to improve clinical efficacy.
Experimental procedures
See supplemental experimental procedures for a full description of methods.
Cell culture
G-CSF (granulocyte-colony stimulating factor) mobilized human CD34+ cells from healthy donors were thawed and resuspended at a concentration of 1 × 105 cells/mL for 21-day cultures or 5 × 105 cells/mL for short-term cultures in StemSpan Serum-Free Expansion Medium II (STEMCELL Technologies) supplemented with 50 ng/mL of human recombinant Stem Cell Factor, Fms-like tyrosine kinase 3 ligand, and thrombopoietin (PeproTech). Cells were cultured in vessels coated with fibronectin alone or combined with DXI under normoxic (21% O2) or hypoxic (1%–2% O2) conditions with 5% CO2 at 37°C.
Transplantation of human CD34+ HSPCs into NSG mice
Female NSG mice (Jackson Laboratory) were sublethally irradiated (280 cGy) 24 h before tail-vein injection. Animals were housed and handled in accordance with the guidelines set by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council (DHHS publication no. NIH 85-23), and the protocol was approved by the Animal Care and Use Committee of the NHLBI.
Statistics
Data are reported as mean ± SEM. Statistical significance (p < 0.05) was assessed using one-way or two-way ANOVA with Tukey's multiple comparison test or an unpaired Student’s t test. Statistical analyses for bulk and scRNA-seq experiments were performed within the RStudio IDE. All other statistical analyses comparing the different experimental groups were performed using GraphPad Prism 8 software.
Author contributions
Conceptualization, D.A., J.F.F., H.H., R.H.S., and A.L.; in vitro assays, D.A., J.F.F., H.H., P.S.C., and A.C.; NSG mouse transplant assay, J.F.F. and P.S.C.; ImageStream/confocal microscopy, H.H.; RNA-seq preparation, D.A., H.H., L.J.A., and Y.L.; bulk RNA-seq analysis, D.A., F.S., M.P., and A.L.; scRNA-seq analysis, D.A., S.C., L.J.A., and A.L.; Seahorse assay, J.T.; formal analysis, D.A., J.F.F., H.H., S.C., F.S., M.P., and A.L.; funding acquisition, A.L., Y.L., and M.P.; project administration, A.L.; original draft, D.A., H.H., and A.L.; review and editing, D.A., J.F.F., H.H., S.C., F.S., L.J.A., P.S.C., A.C., J.T., Y.L., M.P., R.H.S., and A.L.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
The authors thank all members of the Larochelle lab for helpful discussions; Irwin D. Bernstein MD, Carrie Stein, and Sommer K. Castro at the Fred Hutchinson Cancer Research Center for providing DXI and valuable advice; Keyvan Keyvanfar, Philip McCoy PhD, and the NHLBI Flow Cytometry Core Facility for assisting with flow cytometry and ImageStream analyses; Christian Combs PhD and the Light Microscopy Core Facility for support with confocal microscopy; David Stroncek MD and the NIH Department of Transfusion Medicine and Cell Processing Section staff for apheresis, selection, and cryopreservation of human CD34+ cells; Richard Gustafson RN and the outpatient clinic nursing staff for recruiting normal volunteers and providing G-CSF administration teaching to healthy subjects; Temeri Wilder-Kofie DVM, James Hawkins DVM, and Building 50 Animal Facility staff for excellent animal care. This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health, USA (Z99 HL999999, ZIA HL006218).
Published: August 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.08.001.
Supplemental information
Data and code availability
The data generated for this study have been deposited at the Gene Expression Omnibus under accession codes GEO: GSE157465 and GSE157321.
References
- Artavanis-Tsakonas S., Matsuno K., Fortini M.E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Bai T., Li J., Sinclair A., Imren S., Merriam F., Sun F., O'Kelly M.B., Nourigat C., Jain P., Delrow J.J., et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat. Med. 2019;25:1566–1575. doi: 10.1038/s41591-019-0601-5. [DOI] [PubMed] [Google Scholar]
- Bigarella C.L., Liang R., Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer H.E. Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfus. Apher. Sci. 2016;54:364–372. doi: 10.1016/j.transci.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney C., Heimfeld S., Brashem-Stein C., Voorhies H., Manger R.L., Bernstein I.D. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat. Med. 2010;16:232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney C., Milano F., Cicconi L., Othus M., Becker P.S., Sandhu V., Nicoud I., Dahlberg A., Bernstein I.D., Appelbaum F.R., et al. Infusion of a non-HLA-matched ex-vivo expanded cord blood progenitor cell product after intensive acute myeloid leukaemia chemotherapy: a phase 1 trial. Lancet Haematol. 2016;3:e330–e339. doi: 10.1016/S2352-3026(16)30023-0. [DOI] [PubMed] [Google Scholar]
- Delaney C., Varnum-Finney B., Aoyama K., Brashem-Stein C., Bernstein I.D. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106:2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Rattis F.M., DiMascio L.N., Congdon K.L., Pazianos G., Zhao C., Yoon K., Cook J.M., Willert K., Gaiano N., et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Fraker C.A., Alvarez S., Papadopoulos P., Giraldo J., Gu W., Ricordi C., Inverardi L., Dominguez-Bendala J. Enhanced oxygenation promotes beta-cell differentiation in vitro. Stem Cells. 2007;25:3155–3164. doi: 10.1634/stemcells.2007-0445. [DOI] [PubMed] [Google Scholar]
- Giuntoli S., Rovida E., Gozzini A., Barbetti V., Cipolleschi M.G., Olivotto M., Dello Sbarba P. Severe hypoxia defines heterogeneity and selects highly immature progenitors within clonal erythroleukemia cells. Stem Cells. 2007;25:1119–1125. doi: 10.1634/stemcells.2006-0637. [DOI] [PubMed] [Google Scholar]
- Gustafsson M.V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J.L., Poellinger L., Lendahl U., Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Horwitz M.E., Wease S., Blackwell B., Valcarcel D., Frassoni F., Boelens J.J., Nierkens S., Jagasia M., Wagner J.E., Kuball J., et al. Phase I/II study of stem-cell transplantation using a single cord blood unit expanded ex vivo with nicotinamide. J. Clin. Oncol. 2019;37:367–374. doi: 10.1200/JCO.18.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Cheng H., Gao Y., Shi M., Liu Y., Hu Z., Xu J., Qiu L., Yuan W., Leung A.Y., et al. Antioxidant N-acetyl-L-cysteine increases engraftment of human hematopoietic stem cells in immune-deficient mice. Blood. 2014;124:e45–e48. doi: 10.1182/blood-2014-03-559369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntsman H.D., Bat T., Cheng H., Cash A., Cheruku P.S., Fu J.F., Keyvanfar K., Childs R.W., Dunbar C.E., Larochelle A. Human hematopoietic stem cells from mobilized peripheral blood can be purified based on CD49f integrin expression. Blood. 2015;126:1631–1633. doi: 10.1182/blood-2015-07-660670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y.Y., Sharkis S.J. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J., Byk T., Jansson-Sjostrand L., Petit I., Shivtiel S., Nagler A., Hardan I., Deutsch V., Gazit Z., Gazit D., et al. Overexpression of CXCR4 on human CD34+ progenitors increases their proliferation, migration, and NOD/SCID repopulation. Blood. 2004;103:2942–2949. doi: 10.1182/blood-2003-07-2607. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Takubo K., Suda T. Bone marrow long label-retaining cells reside in the sinusoidal hypoxic niche. Biochem. Bioph Res. Commun. 2008;366:335–339. doi: 10.1016/j.bbrc.2007.11.086. [DOI] [PubMed] [Google Scholar]
- Kumano K., Chiba S., Kunisato A., Sata M., Saito T., Nakagami-Yamaguchi E., Yamaguchi T., Masuda S., Shimizu K., Takahashi T., et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- Landor S.K.J., Lendahl U. The interplay between the cellular hypoxic response and Notch signaling. Exp. Cell Res. 2017;356:146–151. doi: 10.1016/j.yexcr.2017.04.030. [DOI] [PubMed] [Google Scholar]
- Laurenti E., Doulatov S., Zandi S., Plumb I., Chen J., April C., Fan J.B., Dick J.E. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Gottgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553:418–426. doi: 10.1038/nature25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database Hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S., Papoutsakis E.T. The evolving role of the aryl hydrocarbon receptor (AHR) in the normophysiology of hematopoiesis. Stem Cell Rev. Rep. 2012;8:1223–1235. doi: 10.1007/s12015-012-9384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C.R., O'Leary H.A., Chitteti B.R., Huang X., Cooper S., Hangoc G., Brustovetsky N., Srour E.F., Lee M.R., Messina-Graham S., et al. Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell. 2015;161:1553–1565. doi: 10.1016/j.cell.2015.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miharada K., Sigurdsson V., Karlsson S. Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Rep. 2014;7:1381–1392. doi: 10.1016/j.celrep.2014.04.056. [DOI] [PubMed] [Google Scholar]
- Milyavsky M., Gan O.I., Trottier M., Komosa M., Tabach O., Notta F., Lechman E., Hermans K.G., Eppert K., Konovalova Z., et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Mohrin M., Bourke E., Alexander D., Warr M.R., Barry-Holson K., Le Beau M.M., Morrison C.G., Passegue E. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M., Shin J.Y., Liu Y.F., Brown K., Luo H.Z., Xi Y.N., Haynes C.M., Chen D. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A., Garzon-Muvdi T., Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Moore M.A.S. Converging pathways in leukemogenesis and stem cell self-renewal. Exp. Hematol. 2005;33:719–737. doi: 10.1016/j.exphem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Parmar K., Mauch P., Vergilio J.A., Sackstein R., Down J.D. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc. Natl. Acad. Sci. U S A. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poellinger L., Lendahl U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr. Opin. Genet. Dev. 2008;18:449–454. doi: 10.1016/j.gde.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Rouault-Pierre K., Lopez-Onieva L., Foster K., Anjos-Afonso F., Lamrissi-Garcia I., Serrano-Sanchez M., Mitter R., Ivanovic Z., de Verneuil H., Gribben J., et al. HIF-2alpha protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell. 2013;13:549–563. doi: 10.1016/j.stem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Schioppa T., Uranchimeg B., Saccani A., Biswas S.K., Doni A., Rapisarda A., Bernasconi S., Saccani S., Nebuloni M., Vago L., et al. Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 2003;198:1391–1402. doi: 10.1084/jem.20030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson V., Takei H., Soboleva S., Radulovic V., Galeev R., Siva K., Leeb-Lundberg L.M., Iida T., Nittono H., Miharada K. Bile acids protect expanding hematopoietic stem cells from unfolded protein stress in fetal liver. Cell Stem Cell. 2016;18:522–532. doi: 10.1016/j.stem.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Simsek T., Kocabas F., Zheng J., Deberardinis R.J., Mahmoud A.I., Olson E.N., Schneider J.W., Zhang C.C., Sadek H.A. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth J.M., Hoggatt J., Singh P., Pelus L.M. Pharmacologic increase in HIF1alpha enhances hematopoietic stem and progenitor homing and engraftment. Blood. 2014;123:203–207. doi: 10.1182/blood-2013-07-516336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P., Sulitkova J., Lisztwan J., Moch H., Oakeley E.J., Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Stier S., Cheng T., Dombkowski D., Carlesso N., Scadden D.T. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- Tabas I., Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai-Nagara I., Matsuoka S., Ariga H., Suda T. Mortalin and DJ-1 coordinately regulate hematopoietic stem cell function through the control of oxidative stress. Blood. 2014;123:41–50. doi: 10.1182/blood-2013-06-508333. [DOI] [PubMed] [Google Scholar]
- Tanavde V.M., Malehorn M.T., Lumkul R., Gao Z.G., Wingard J., Garrett E.S., Civin C.I. Human stem-progenitor cells from neonatal cord blood have greater hematopoietic expansion capacity than those from mobilized adult blood. Exp. Hematol. 2002;30:816–823. doi: 10.1016/s0301-472x(02)00818-4. [DOI] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Kreso A., Mbong N., Kent D.G., Fitzmaurice T., Chambers J.E., Xie S., Laurenti E., Hermans K., Eppert K., et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- van Galen P., Mbong N., Kreso A., Schoof E.M., Wagenblast E., Ng S.W.K., Krivdova G., Jin L.Q., Nakauchi H., Dick J.E. Integrated stress response activity marks stem cells in normal hematopoiesis and leukemia. Cell Rep. 2018;25:1109–1117.e5. doi: 10.1016/j.celrep.2018.10.021. [DOI] [PubMed] [Google Scholar]
- Villa J.C., Chiu D., Brandes A.H., Escorcia F.E., Villa C.H., Maguire W.F., Hu C.J., de Stanchina E., Simon M.C., Sisodia S.S., et al. Nontranscriptional role of Hif-1alpha in activation of gamma-secretase and notch signaling in breast cancer. Cell Rep. 2014;8:1077–1092. doi: 10.1016/j.celrep.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J.E., Jr., Brunstein C.G., Boitano A.E., DeFor T.E., McKenna D., Sumstad D., Blazar B.R., Tolar J., Le C., Jones J., et al. Phase I/II trial of StemRegenin-1 expanded umbilical cord blood hematopoietic stem cells supports testing as a stand-alone graft. Cell Stem Cell. 2016;18:144–155. doi: 10.1016/j.stem.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Xie S.Z., Garcia-Prat L., Voisin V., Ferrari R., Gan O.I., Wagenblast E., Kaufmann K.B., Zeng A.G.X., Takayanagi S.I., Patel I., et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell. 2019;25:639–653.e7. doi: 10.1016/j.stem.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Liu H., Qu F., Fan J., Mao K., Yin Y., Liu J., Geng Z., Wang Y. Hypoxia inhibits the differentiation of mesenchymal stem cells into osteoblasts by activation of Notch signaling. Exp. Mol. Pathol. 2013;94:33–39. doi: 10.1016/j.yexmp.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Yan Q., Bartz S., Mao M., Li L., Kaelin W.G., Jr. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell Biol. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Gao S., Wu Z., Kajigaya S., Feng X., Liu Q., Townsley D.M., Cooper J., Chen J., Keyvanfar K., et al. Single-cell RNA-seq reveals a distinct transcriptome signature of aneuploid hematopoietic cells. Blood. 2017;130:2762–2773. doi: 10.1182/blood-2017-08-803353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated for this study have been deposited at the Gene Expression Omnibus under accession codes GEO: GSE157465 and GSE157321.