Abstract
Immune checkpoint blockade, an immunotherapy, has been applied in multiple systemic malignancies and has improved overall survival to a relatively great extent; whether it can be applied in breast cancer remains unknown. We endeavored to explore possible factors that may influence immunotherapy outcomes in breast cancer using several public databases. The possible treatment target TNF superfamily member 4 (TNFSF4) was selected from many candidates based on its abnormal expression profile, survival-associated status, and ability to predict immune system reactions. For the first time, we identified the oncogenic features of TNFSF4 in breast carcinoma. TNFSF4 was revealed to be closely related to treatment that induced antitumor immunity and to interact with multiple immune effector molecules and T cell signatures, which was independent of endocrine status and has not been reported previously. Moreover, the potential immunotherapeutic approach of TNFSF4 blockade showed underlying effects on stem cell expansion, which more strongly and specifically demonstrated the potential effects of applying TNFSF4 blockade-based immunotherapies in breast carcinomas. We identified potential targets that may contribute to breast cancer therapies through clinical analysis and real-world review and provided one potential but crucial tool for treating breast carcinoma that showed effects across subtypes and long-term effectiveness.
Subject terms: Cancer, Cell biology, Computational biology and bioinformatics, Genetics, Immunology, Molecular biology, Stem cells, Biomarkers, Molecular medicine, Oncology, Risk factors
Introduction
Breast carcinoma treatments have been evolving for years, accompanied by the emergence of reagents referred to as endocrine therapy and targeted therapy and improvements in chemotherapeutics1,2. However, the development of therapies seems to have encountered a bottleneck, and what approach should be pursued to further prolong patient survival is unclear3–5. Can we draw lessons from the experience of immunotherapy?
Improving cancer-related immune resistance is mainly achieved through immune checkpoint blockade therapy, and multiple immune checkpoint-targeting agents have been identified and clinically applied to treat lung cancer, esophageal cancer, melanoma, etc. Therapeutic targets include but are not limited to PD-L1 (CD274), PD-L2 (PDCD1LG2), CD80, CD86, and CD70, and novel applied therapies have shown promise in lung cancer and melanoma treatment6–10. However, how immune checkpoint blockade therapy acts in the breast cancer treatment process and what checkpoint may be targeted to achieve benefits are totally unknown. Recently, CCR9 was demonstrated to exert strong immunoregulatory effects on T cell responses in multiple tumors, and through inhibiting TCR signaling, CCR9 regulates STAT signaling in T cells, resulting in reduced T helper 1 cytokine secretion. Unlike PD-L1 blockade, inhibition of CCR9 expression on tumor cells facilitates immunotherapeutic effects via tumor-specific T cells in vivo11,12. However, whether and how such immune checkpoints are involved in breast cancer prognosis and therapeutic efficacy remain largely unclear.
Therefore, this study aimed to clarify the clinical significance of immunotherapies, especially the uncertain efficacy of immune checkpoint blockade, in breast cancer patients and to propose more suitable strategies for improving breast cancer treatments by applying our knowledge on immunity.
Results
Screening possible immune functions related to anticancer treatment
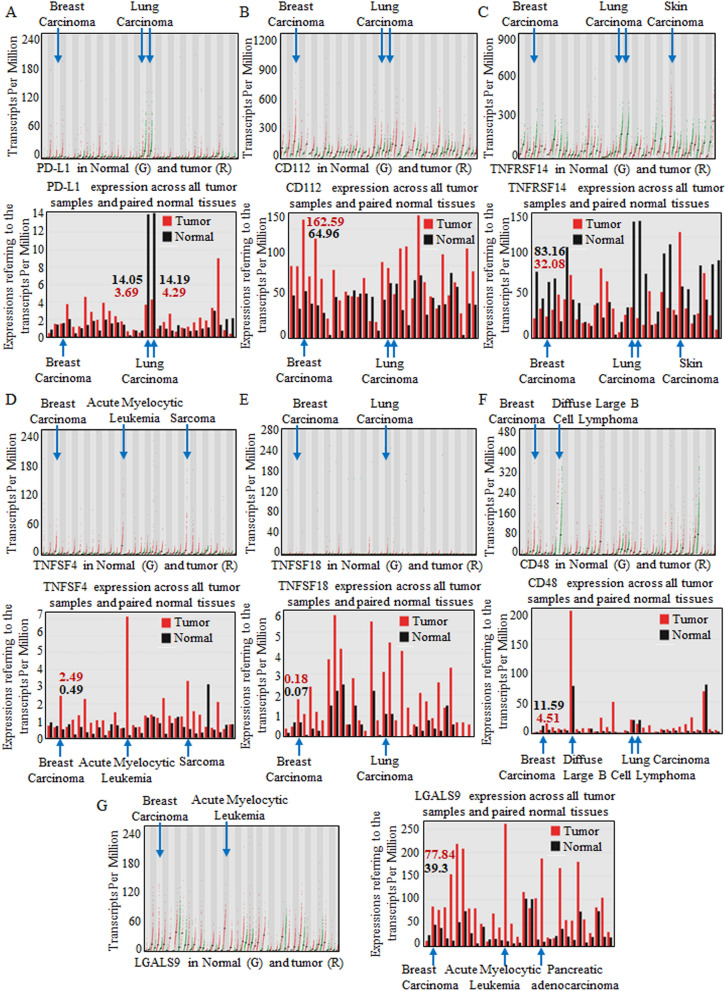
Common immune checkpoints were browsed and screened for possible immunotherapeutic value, and the representative immune checkpoint molecules13–15 ADORA2A, BTLA, Nectin-2 (CD112), CD160, CD244, PD-L1, CD96, CSF1R, CTLA4, HAVCR2, IDO1, IL10, IL10RB, KDR, KIR2DL1, KIR2DL3, LAG3, LGALS9, PDCD1, PDCD1LG2, PVRL2, TGFB1, TGFBR1, TIGIT, VTCN1, TNF Receptor Superfamily Member 14 (TNFRSF14), TNF superfamily member 4 (TNFSF4), and TNF superfamily member 18 (TNFSF18) were all input into the Tumor and Immune System Interaction Database (TISIDB) for potential effect predication in the integrated repository portal for tumor-immune system interactions16. Primarily, PD-L1, CD112, TNFRSF14, TNFSF4, TNFSF18, CD48, and LGALS9 were selected for their significant differential expression patterns, and the patterns are shown in Fig. S1. The paired red and green colors indicate the expression patterns in multiple organs and systems.
Furthermore, PD-L1 (Fig. 1A), CD112 (Fig. 1B), TNFRSF14 (Fig. 1C), TNFSF4 (Fig. 1D), TNFSF18 (Fig. 1E), CD48 (Fig. 1F), and LGALS9 (Fig. 1G) were analyzed for abnormal expression patterns in breast carcinomas. Specifically, CD112, TNFSF4, TNFSF18, and LGALS9 were significantly overexpressed in breast carcinomas, strongly suggesting potent and valuable effects.
Figure 1.
Analyzing candidate immune checkpoint molecules targeted by blockade in carcinomas. Carcinomas throughout the body were enrolled for analysis, and multiple systems showed various immune checkpoint molecule patterns. PD-L1 (A), CD112 (B), TNFRSF14 (C), TNFSF4 (D), TNFSF18 (E), CD48 (F), and LGALS9 (G) were analyzed for abnormal expression in breast carcinomas. In each image, the specific quantitative value is shown at the top, and each dot represents the expression in samples. The bar graphs below were used for comparison, and bar height bar represents the median expression of the specific tumor type or normal tissue. Specifically, CD112, TNFSF4, TNFSF18, and LGALS9 were relatively overexpressed in breast carcinomas.
Immunotherapeutic targets participated in multiple breast carcinogenic processes by interacting with key cancer-stimulating factors
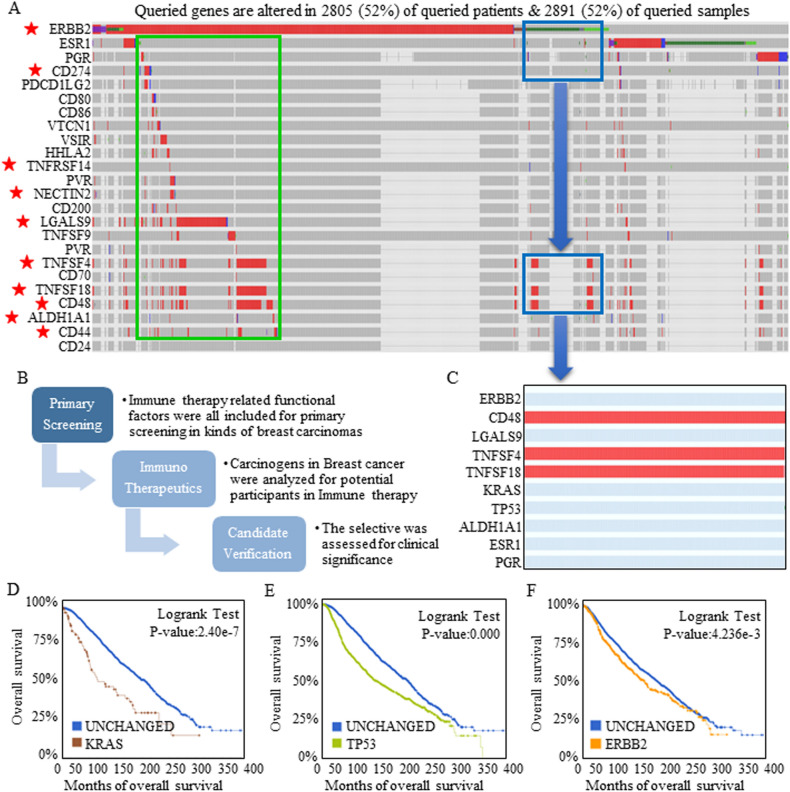
The heterogenetic features of breast cancer cause breast carcinomas with different hormone receptor statuses to benefit from different and specific treatment strategies. As a double-edged sword, a specific agent will not function in another kind of breast carcinoma, and to determine the potential of immune checkpoint blockade therapies, native expression signatures and correlations were studied using cBioPortal and Gene Expression Profiling Interactive Analysis 2 (GEPIA2). An expression heat-map was used to visualize the expression patterns of the candidates in red bars, and the selected representative genes described in the first part of Result section are labeled with a red star (Fig. 2A). The analysis flowchart is shown in Fig. 2B. All the enrolled factors were input and studied for potential functional correlations (Fig. S2). Specifically, the main oncogenes ERBB2, KRAS, and TP53 were analyzed for their intrinsic connections in breast carcinomas, and promising correlations between TNFSF4 and ERBB2, KRAS, and TP53 were confirmed (Fig. S3, Table 1).
Figure 2.
Selection of the most representative immune checkpoint molecules targeted by blockade. Expression signatures and correlations were studied using cBioPortal and GEPIA2. (A) Expression heat-map showing the expression of each candidate in a red bar, and the most representative genes are labeled with a red star. The red bars indicate actual data points, and the gray bars indicate that no specific data were available. (B) The analysis flowchart is shown. (C) Nearly all the immunotherapeutic targets showed expanded expression intervals referring to ESR1 and PGR, and TNFSF4, TNFSF18, and CD48 showed increased expression in breast carcinomas without ERBB2, ESR1, and PGR expression. Amplification or mutation of KRAS (D), TP53 (E), or ERBB2 (F) indicated worse predicted survival.
Table 1.
The analysis tested in 276 pairs between the 24 tracks in the OncoPrint to reveal the connections among immunotherapies related genes.
| A | B | A Not B | B Not A | Both | Log2 | P value | q value |
|---|---|---|---|---|---|---|---|
| TNFSF4 | TNFSF18 | 5 | 4 | 70 | > 3 | < 0.001 | < 0.001 |
| TNFSF18 | CD48 | 11 | 40 | 63 | > 3 | < 0.001 | < 0.001 |
| TNFSF4 | CD48 | 14 | 42 | 61 | > 3 | < 0.001 | < 0.001 |
| CD274 | PDCD1LG2 | 3 | 1 | 20 | > 3 | < 0.001 | < 0.001 |
| ERBB2 | TP53 | 369 | 1345 | 441 | 1.409 | < 0.001 | < 0.001 |
| PVR | NECTIN2 | 3 | 9 | 12 | > 3 | < 0.001 | < 0.001 |
| ERBB2 | LGALS9 | 120 | 9 | 27 | > 3 | < 0.001 | < 0.001 |
| TNFSF9 | CD70 | 1 | 3 | 7 | > 3 | < 0.001 | < 0.001 |
| HHLA2 | CD200 | 6 | 3 | 6 | > 3 | < 0.001 | < 0.001 |
| KRAS | TP53 | 47 | 1717 | 69 | 1.501 | < 0.001 | < 0.001 |
| CD80 | CD200 | 2 | 5 | 4 | > 3 | < 0.001 | < 0.001 |
| CD86 | CD200 | 7 | 5 | 4 | > 3 | < 0.001 | < 0.001 |
| CD274 | TP53 | 10 | 1013 | 27 | 2.355 | < 0.001 | < 0.001 |
| CD80 | CD86 | 3 | 8 | 3 | > 3 | < 0.001 | < 0.001 |
| CD80 | HHLA2 | 3 | 9 | 3 | > 3 | < 0.001 | < 0.001 |
| NECTIN2 | CD48 | 12 | 94 | 9 | 2.902 | < 0.001 | 0.001 |
| CD86 | HHLA2 | 8 | 9 | 3 | > 3 | < 0.001 | 0.003 |
| PVR | CD48 | 8 | 96 | 7 | > 3 | < 0.001 | 0.004 |
| CD44 | TP53 | 10 | 332 | 19 | 1.984 | < 0.001 | 0.005 |
| VSIR | TP53 | 5 | 338 | 13 | 2.421 | < 0.001 | 0.011 |
| CD200 | TP53 | 1 | 343 | 8 | > 3 | < 0.001 | 0.012 |
| PDCD1LG2 | TP53 | 7 | 484 | 14 | 2.138 | < 0.001 | 0.012 |
| VTCN1 | TNFRSF14 | 27 | 19 | 3 | > 3 | 0.001 | 0.015 |
| CD274 | PVR | 18 | 12 | 3 | > 3 | 0.003 | 0.031 |
| HHLA2 | TP53 | 3 | 342 | 9 | 2.614 | 0.004 | 0.041 |
| ALDH1A1 | CD44 | 10 | 26 | 3 | > 3 | 0.004 | 0.048 |
The close connections among the candidate immune checkpoints, and the connections among checkpoints and malignancy stimulators of either KRAS, TP53, ERBB2, ALDH1A1, CD44, were all calculated and exhibited. The close connections strongly suggested the crucial roles of candidate immune checkpoints in therapy response, and also suggested the potential effects they may exert.
In general, nearly all the immunotherapeutic targets showed expanded expression intervals in ESR1+ and PGR+ cases, the existence of which demonstrated a better response to standard (“chemotherapeutic”) and anti-hormone treatments, indicating that immunotherapy may compensate for current deficiencies. More importantly and interestingly, TNFSF4, TNFSF18, and CD48 all showed increased expression in breast carcinomas bearing no ERBB2, ESR1, or PGR expression, strongly suggesting that immunotherapy may show efficacy in all types of breast cancer, and a partially enlarged drawing is shown in Fig. 2C. In detail, TNFSF4, TNFSF18 and CD48 showed aberrant expression in breast cancer cases with different genetic backgrounds, including those without KRAS (Fig. 2D), TP53 (Fig. 2E), or ERBB2 (Fig. 2F) expression and activation; these three factors dominated the survival prognosis and led to short predicted survival.
Evaluation of the cluster of TNFSF4, TNFSF18, CD48 and LGALS9 as therapeutic immune therapeutic targets
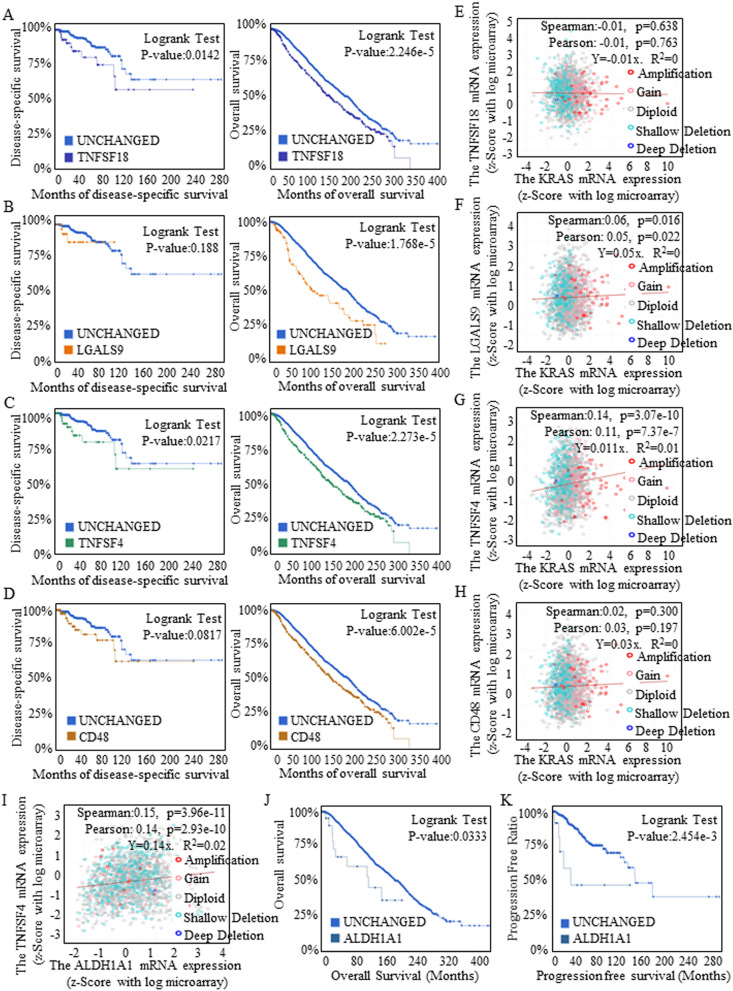
The clinical significance of a cluster including TNFSF18 (Fig. 3A), LGALS9 (Fig. 3B), TNFSF4 (Fig. 3C) and CD48 (Fig. 3D) was also analyzed for the value of these molecules as immunotherapeutic targets, and TNFSF4 and TNFSF18 showed greater significance in predicting disease-free survival and overall survival (OS). KRAS greatly stimulated carcinogenesis and determined the anticancer treatment response. We further explored the correlations of KRAS with TNFSF18 (Fig. 3E), LGALS9 (Fig. 3F), TNFSF4 (Fig. 3G) and CD48 (Fig. 3H), and the positive correlation between TNFSF4 and KRAS strongly suggested the prospect of achieving therapeutic effects by using TNFSF4 as a target for manipulation or blockade.
Figure 3.
Clinical evaluations of potential therapeutic candidates. Data were acquired from the combined study cohorts of BREAST (METABRIC 2016), BREAST CANCER (MSK 2018), and BREAST INVASIVE CARCINOMA BREAST (TCGA PANCAN 2018). The clinical significance of the cluster including TNFSF18 (A), LGALS9 (B), TNFSF4 (C) and CD48 (D) was analyzed for the value of these molecules as immunotherapeutic targets. The correlations between KRAS and TNFSF18 (E), LGALS9 (F), TNFSF4 (G) or CD48 (H) were analyzed, and the positive correlation between TNFSF4 and KRAS strongly suggested the prospective therapeutic effects of using TNFSF4 as target for manipulation or blockade. (I) A close correlation between TNFSF4 and ALDH1A1 expression was identified. Stem cells with a positive ALDH1A1 phenotype had a shorter survival time (J) and shorter progression-free time (K).
Groups of stem cells are considered the root of cancer recurrence due to their steady status and superior renewal ability. We previously identified stem-like ALDH1A1+ cells in breast cancer groups2,17,18, and in this study, we noticed a close correlation between TNFSF4 and ALDH1A1 expression (Fig. 3I), the latter of which indicated shorter survival (Fig. 3J) and progression-free times (Fig. 3K). These results strongly suggested that oncogenic TNFSF4 may be an effective immunotherapeutic target, and the proposed survival benefits may also be achieved by repressing stem cell expansion, fully assuring a positive outcome for anti-TNFSF4 treatment. The other candidates, CD112, TNFRSF14, and PD-L1, failed to be involved upon further analysis, as indicated by their negative associations with survival (Fig. S4).
Probable mechanism by which TNFSF4 functions
The immune system becomes suppressed as carcinomas become aggressive, and when immune function inhibitors are applied (immune blockade approaches), active immune cells begin to infiltrate and execute cellular functions (Fig. 4A). All types of cancer cells, including the heterogenetic subtype cells known as cancer stem cells (CSCs), can be eliminated by immune attack. To investigate the crucial role of the selected immune target, infiltrating lymphocyte functions and connective functional factors were analyzed for insights into the underlying mechanisms. Both ALDH1A1 overexpression (Fig. 4B) and TNFSF4 (Fig. 4C) overexpression correlated with more active lymphocytes. However, infiltrating immune cells were suppressed by highly expressed immune inhibitors on cells with increased ALDH1A1 (Fig. 4D) or TNFSF4 expression (Fig. 4E). These results indicated that TNFSF4 can potentially reactivate the immune response and partially function by precisely neutralizing stem cells.
Figure 4.
TNFSF4-based immunotherapy may intersect with cancer stem cell signature repression. (A) Schematic figure illustrating the immune response. the immune system becomes suppressed when a carcinoma becomes aggressive. Later, when immune function inhibitors are blocked, active immune cells begin to infiltrate and exert cytotoxic activities against all tumor cells. Spearman correlations between TNFSF4 and immunoinhibitory factors (Y axis) across human cancers (X axis). The items in the column are listed in sequence as follows: ADORA2A, BTLA, CD160, CD244, CD274, CD96, CSF1R, CTLA4, HAVCR2, IDO1, IL10, IL10RB, KDR, KIR2DL1, KIR2DL3, LAG3, LGALS9, PDCD1, PDCD1LG2, PVRL2, TGFB1, TGFBR1, TIGIT, and VTCN1. The infiltrating lymphocyte functions and connective functional factors were analyzed, and both ALDH1A1 overexpression (B) and TNFSF4 overexpression (C) were correlated with more lymphocyte infiltration. Spearman correlations between TNFSF4 and kinds of lymphocytes (Y axis) across human cancers (X axis). The items in the column are listed in sequence as follows: ADORA2A, BTLA, CD160, CD244, CD274, CD96, CSF1R, CTLA4, HAVCR2, IDO1, IL10, IL10RB, KDR, KIR2DL1, KIR2DL3, LAG3, LGALS9, PDCD1, PDCD1LG2, PVRL2, TGFB1, TGFBR1, TIGIT, and VTCN1. However, infiltrating immune cells were suppressed by highly expressed immune inhibitors in cells with increased ALDH1A1 (D) or TNFSF4 expression (E). These results indicated that TNFSF4 blockade treatment could potentially reactivate the immune response and partially function through precisely inhibiting stem cells.
Stem cell signatures predicted the response to immunotherapy
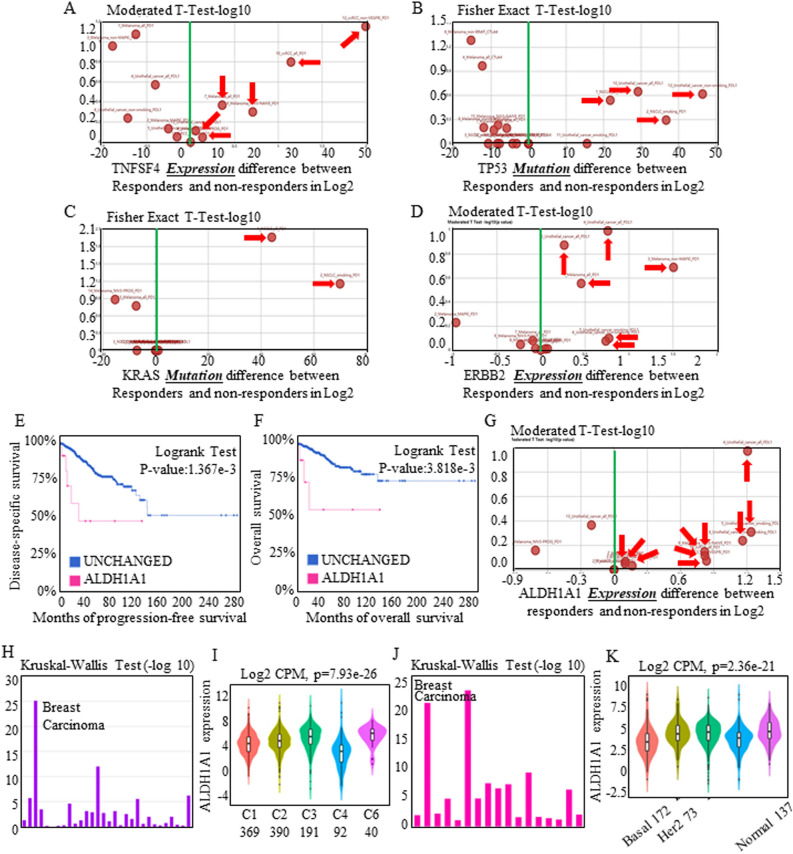
Immunotherapy responses and effects were believed to be correlated with lymphocyte activation and infiltration, and we first assessed the effects of immunotherapies in multiple systems. Treatments targeting TNFSF4 tended to produce relatively good outcomes (Fig. 5A). Additionally, TNFSF4-associated TP53 (Fig. 5B), KRAS (Fig. 5C), and ERBB2 (Fig. 5D) all indicated a relatively good immunotherapy response, which further supported the crucial position of TNFSF4. Red-labeled text refers to one specific study, and detailed study information can be reviewed by searching certain PMID numbers in TISIDB.
Figure 5.
TNFSF4 blockade therapies could be assessed in the stem cell signature-associated mode. The transcriptomic and genomic profiles of pretreated tumor biopsies from responders and non-responders treated with anti-PDL1 and anti-PD1 antibodies were enrolled for analysis. In total, the responders tended to exhibit higher TNFSF4 expression levels, and each study could be reviewed by searching for a certain PMID number (red labeling). Assessment of real-world immunotherapeutic effects indicated that effects related to TNFSF4 (A) tended to imply better outcomes and that TNFSF4-associated TP53 (B), KRAS (C), and ERBB2 (D) all indicated better immunotherapy response. Increased ALDH1A1 expression indicated shorter disease-specific survival (E) and overall survival (F), and ALDH1A1 surprisingly correlated with higher therapeutic response ratios (G) through clinical data assessment. (H,I) ALDH1A1 was analyzed for its roles in predicting immunotherapy response, and the 5 subgroups C1 (N = 369), C2 (N = 390), C3 (N = 191), C4 (N = 92), and C6 (N = 40) were involved in assessing functional aspects. ALDH1A1 expression dominated in all the subtypes, participating in multiple immune reaction processes. (J,K) Associations between ALDH1A1 expression and molecular subtypes across human cancers were also identified, and the signature of increasing ALDH1A1 expression tended to be found in all kinds of breast carcinomas. Specifically, C1 represents wound healing, C2 represents IFN-gamma dominant, C3 represents inflammatory, C4 represents lymphocyte depleted, C5 represents immunologically quiescent and is not shown, and C6 represents TGF-b dominant.
TNFSF4 and ALDH1A1 were confirmed to participate in immune-activating therapies, and TNFSF4 treatment predicted a therapeutic response. Immunotherapy consistently repressed tumor growth once the immune system was activated, and from the above results, we further found that TNFSF4-associated therapy might also influence stem cell expansion. Increased ALDH1A1 expression indicated shorter disease-specific survival (Fig. 5E) and overall survival (Fig. 5F), and in clinical assessments, ALDH1A1 surprisingly correlated with higher therapeutic response ratios (Fig. 5G), which has not ever been reported or analyzed.
Associations between ALDH1A1 expression and immune subtypes across human cancers were included for analysis, and ALDH1A1 expression dominated in all the subtypes (Fig. 5H), participating in multiple immune reaction processes (Fig. 5I). Associations between ALDH1A1 expression and molecular subtypes across human cancers were also identified (Fig. 5J), and a signature of increasing ALDH1A1 levels tended to be observed in all kinds of breast carcinoma (Fig. 5K), indicating the universal therapy-responsive role of ALDH1A1 in achieving better outcomes.
Preliminary exploration of clinical significance
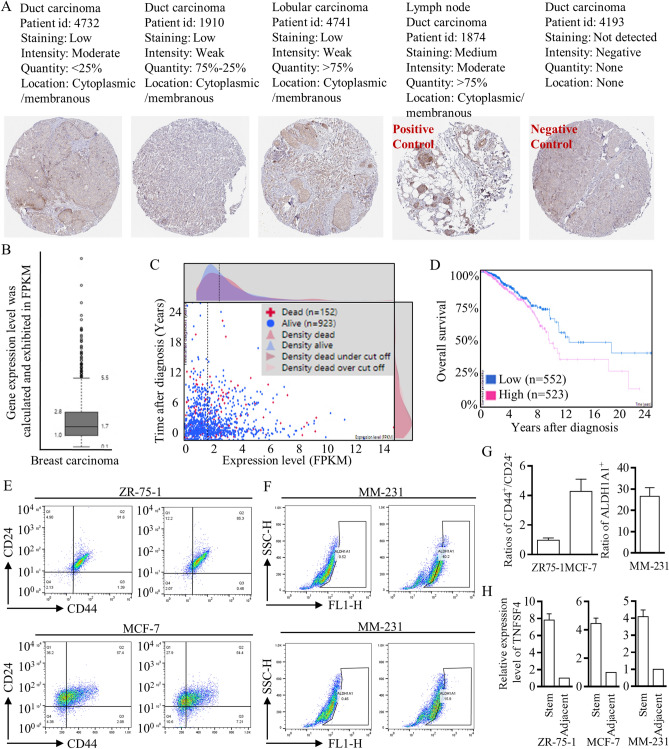
Immunohistochemical (IHC) staining images were acquired from “The Human Protein Atlas” project (funded by the Knut & Alice Wallenberg Foundation), and representative images were arranged referring to different groups in the public dataset (Fig. 6A). High expression of TNFSF4 was universally identified in breast carcinoma (Fig. 6B) and pointed to poorer survival outcomes (Fig. 6C,D). To further verify the potential effects of TNFSF4 on stem cell renewal, we isolated stem cells and identified higher TNFSF4 expression in the CD44+/24− cell group with the luminal A/B phenotype (Fig. 6E) and in the ALDH1A1+ cell group with the basal-like phenotype (Fig. 6F). For the first time, we derived solid results through database screening and bench-to-clinic exploration; the results strongly suggested the value of TNFSF4-based immunotherapy (Fig. 6G,H).
Figure 6.
Exploration of the putative clinical roles of TNFSF4. (A) IHC staining images are shown to clarify different expression patterns (left to right, in sequence, < 25%, 25–75%, and > 75%). A lymph node slide was set as a positive control, and an unstained slide was set as a negative control. (B) High RNA expression of TNFSF4 was universally identified in breast carcinoma, with testing and calculation based on FPKM, and the cutoff line is labeled, which was used for clinical predictions. (C,D) Higher TNFSF4 expression pointed to poorer survival outcomes. (E,F) Flow cytometry with FACSAria sorting was applied to isolate stem cells from ZR75-1, MCF-7, and MM-231 cells. (G,H) Stem cells with a CD44+/24− or ALDH1A1+ phenotype were identified and isolated, and the TNFSF4 expression patterns in different cell lines were checked to illustrate the increased expression.
Discussion
The treatment approaches for breast carcinoma are constantly evolving in accordance with newly confirmed molecular and biological findings, which lead to reconfiguration of treatment methods. Shifting treatment strategies may expand benefits across various carcinomas; however, immunotherapy is not yet effective in most malignancies. Several specific contributions have been identified for breast carcinogenesis, and related therapies that target or relieve the malignant process have greatly improved patient lives. Currently, endocrine therapy, anti-Her-2 therapy, and improved chemotherapy all prolong survival outcomes. We must confront the fact that basal-like breast carcinoma, which is always identified as triple-negative carcinoma, is without any endocrine factors or precise targets for treatment. As an example, immunotherapy has been used across the squamous, adenocarcinoma, and small cell types of lung cancer, indicating universal inhibition. To explore potential effective immune targets, we first input all immunotherapy-related functional factors, and after screening in all kinds of breast carcinomas, several candidates were selected, and their correlations with aggressive breast carcinogens were analyzed to evaluate the possible effects of targeted therapy. To finally identify the most promising immunotherapy candidate, the selected factors were assessed for clinical significance (Fig. 2B).
TNFSF4 belongs to the tumor necrosis factor ligand family and functions in T cell-antigen-presenting cell (APC) interactions, mediating the adhesion of activated T cells to target cells. TNFSF4 can bind to TNFRSF4 and collaboratively stimulate T cell proliferation and cytokine production. There are only limited studies exploring the possible functions of TNFSF419,20, and the roles of TNFSF4 in breast carcinoma are unknown. We found that TNFSF4 was highly expressed in all kinds of breast carcinomas, and its aberrant overexpression was associated with shorter overall survival and disease-free survival. Importantly, TNFSF4 was further revealed to be closely related to antitumor immunity, including multiple immune effector molecules and T cell signatures, as presented and illustrated.
In conclusion, TNFSF4 is a potential immunotherapy target due to its aberrant expression pattern in breast carcinoma, and this protein was even found to be overexpressed in carcinomas without ERBB2, ESR1, or PGR1 amplification and those without either KRAS or TP53 mutation and amplification. TNFSF4, together with other candidates, was included for evaluation, and the positive correlations with immune function inhibitors and lymphocytes drew much attention. In the clinical case analysis, TNFSF4-targeted therapy might show the best response to therapy, and interestingly, TNFSF4 also perturbed stem cell expansion, a signature that is critical for long-term recurrence and response to therapy. Therefore, for the first time, we aimed to identify the key factors that may contribute to breast cancer therapies and provide one potential but crucial approach for treating all types of breast carcinoma with long-term effectiveness.
Methods and materials
Database availability and integrated analysis
To analyze immune checkpoint-related prognoses in breast cancer, breast cancer genomics-related datasets in The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) and International Cancer Genome Consortium (ICGC, https://icgc.org) were individually collected and subsequently subjected to bioinformatic analysis with the web servers GEPIA2 (http://gepia2.cancer-pku.cn)21,22, cBioPortal for Cancer Genomics (http://www.cbioportal.org)23,24, and TISIDB (http://cis.hku.hk/TISIDB)25,26. In detail, GEPIA2 was used to calculate prognostic indexes, including differential expression, pathological stage, gene correlations, and patient survival; cBioPortal was used to visualize and compare gene alterations; and TISIDB was used to explore correlations between the abundances of immunomodulators and the expression of the investigated genes.
Tumor-infiltrating lymphocytes were analyzed using TISIDB27 and the immune-related signatures of 28 tumor-infiltrating lymphocyte types from Charoentong’s study (https://www.cell.com/cell-reports/fulltext/S2211-1247(16)31709-0), which can be viewed on the download page. The relative abundances of tumor-infiltrating lymphocytes were inferred by using gene set variation analysis (GSVA) based on the gene expression profile. Overall survival and disease-free survival analyses were performed using the Kaplan–Meier method with a 50% cutoff to separate the low- and high-expression groups. The log-rank test, also known as the Mantel–Cox test, was used for the hypothesis test. The Cox proportional hazard ratio (HR) and 95% confidence interval were also included in survival plots. A P value < 0.05 was considered statistically significant.
Clinical data analysis
The cutoff (FPKM) was set based on the FPKM value of each gene. When applying survival curve analysis, patients were classified into two groups, and the association between prognosis (survival) and gene expression (FPKM) was examined. The best expression cutoff referred to the FPKM value that yielded the maximal difference with regard to survival between two groups at the lowest log-rank P value. The best expression cutoff was selected based on survival analysis. Median expression referred to the median FPKM value calculated based on the gene expression (FPKM) data from all patients in the dataset. The median follow-up time referred to the median time (years) after diagnosis with a specific type of cancer based on clinical data from a public dataset.
Stem cell isolation and RNA quantification
The breast cancer cell lines MCF-7, ZR75-1, and MM-231 were purchased from American Type Culture Collection and maintained in DMEM (Gibco) or RPMI 1640 medium (HyClone). Total mRNA was reverse transcribed into cDNA by using an RT-PCR kit (AT301 TransGen Biotech), and real-time quantitative PCR (RT-qPCR) was performed with a CFX96 Real-Time PCR Detection System (Bio-Rad)17,18. For analysis of the expression of the breast cancer stem cell (BCSC) markers CD44 and CD24, cells in different treatment groups were collected and washed with PBS twice. Then, the cells were incubated with anti-CD44-FITC (clone G44-26) and anti-CD24-PE (clone ML5) antibodies (Invitrogen) at 4 °C for 30 min. After two washes, the samples were analyzed using a flow cytometer (FACSCalibur, BD Biosciences)28,29. An LSRII flow cytometer (BD Biosciences Pharmingen, San Diego, USA) was used to analyze and separate CSCs based on cell labeling and fluorescence-activated cell sorting. The activated ALDEFLUOR reagent and DEAM purchased from STEMCELL Technologies (Vancouver, BC, Canada) were used to isolate aldehyde dehydrogenase (ALDH+) cells (stem-like cells). The percentages of ALDH1+ stem cells in different groups were analyzed5,18.
Statistical significance and classification
The Spearman method was used to analyze pairwise gene expression correlations, and a P value < 0.05 was considered statistically significant. The correlation degree was identified from the absolute value of the correlation coefficient, and the detailed classifications were as follows: ≤ 0.4, weak; 0.41–0.60, moderate; 0.61–0.80, strong; and 0.81–1.0, very strong. The cooccurrence and mutual exclusivity of genetic alterations between genes of interest and each immune checkpoint molecule were determined from the log2 odds ratio, P value, and Q value. A Q value < 0.05 was considered statistically significant. The investigated immune inhibitors were collected according to Charoentong’s study27,30,31, and each Spearman correlation between the investigated genes and a distinct immune inhibitor in an individual cancer type was integrated into the indicated heat-map. Log-rank P values for Kaplan–Meier plots represents results from an analysis of the correlation between an mRNA expression level and patient survival.
Ethics approval and consent to participate
All procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards for retrospective studies. We confirmed that the IHC results of the human participants were generated in “The Human Protein Atlas” project (www.proteinatlas.org), and the representative images were arranged referring to different groups in the public dataset.
Consent for publication
The authors declare that each author has approved this article for publication.
Supplementary Information
Acknowledgements
We very much appreciate Mallory Ellis and Dr. Leslie Robinson for their careful revision of this manuscript and language polishing.
Abbreviations
- TNFRSF14
TNF receptor superfamily member 14
- TNFSF4
TNF superfamily member 4
- TNFSF18
TNF superfamily member 18
- CD274
PD-L1
- DCD1LG2
PD-L2
Author contributions
K.L.: bioscientific experiments and study design. L.M.: bioscientific experiments and cell culture. Y.S.: experimental tests and RNA/protein tests. X.L.: paper drafting, study design, and statistical analysis. H.R.: paper drafting and statistical analysis. S.T.: paper drafting, study design, statistical analysis, and image quality control. X.S.: study design, statistical analysis, database screening, figure preparation, and reference cross-checking.
Funding
This work was supported by the National Science Foundation for Young Scientists of China (Grant No. 81602597, to Xin Sun), the Foundation Research Project of Shaanxi Province (2021SF-117, referred to Xin Sun), and The Natural Science Basic Research Program of Shaanxi (No. 2018JM7017, to Xin Sun).
Data availability
The data and relevant supporting materials related to the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in Figure 6, panel E, where the labeling of the flow cytometry antibody gate and thresholds was incorrect. Additionally, the manually added scale number and the gates data have been deleted.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kai Li, Lei Ma and Ye Sun.
Change history
8/1/2023
A Correction to this paper has been published: 10.1038/s41598-023-39545-0
Contributor Information
Shou-Ching Tang, Email: stang2@umc.edu.
Xin Sun, Email: dr_sun_endeavour@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98131-4.
References
- 1.Chen S-S, et al. Predicting the survival of triple-negative breast cancer in different stages: A SEER population based research referring to clinicopathological factors. Cancer Invest. 2020;38:549–558. doi: 10.1080/07357907.2020.1831010. [DOI] [PubMed] [Google Scholar]
- 2.Wang M, et al. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019;52:e12534. doi: 10.1111/cpr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokgun PE, Tokgun O, Kurt S, Tomatir AG, Akca H. MYC-driven regulation of long non-coding RNA profiles in breast cancer cells. Gene. 2019;714:143955. doi: 10.1016/j.gene.2019.143955. [DOI] [PubMed] [Google Scholar]
- 4.Wang, C. et al. Long noncoding RNA EMS connects c-Myc to cell cycle control and tumorigenesis. [DOI] [PMC free article] [PubMed]
- 5.Huang G, et al. TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life Sci. 2019;232:116630. doi: 10.1016/j.lfs.2019.116630. [DOI] [PubMed] [Google Scholar]
- 6.Arman K, Möröy T. Crosstalk between MYC and lncRNAs in hematological malignancies. Front. Oncol. 2020;10:25. doi: 10.3389/fonc.2020.579940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W-Y, et al. The Wnt-β-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J. Cell Sci. 2013;126:2877–2889. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 9.Chang W, et al. Hormonal suppression of stem cells inhibits symmetric cell division and gastric tumorigenesis. Cell Stem Cell. 2020;26:739–754.e738. doi: 10.1016/j.stem.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, et al. Lung cancer pathogenesis and poor response to therapy were dependent on driver oncogenic mutations. Life Sci. 2021;265:118797. doi: 10.1016/j.lfs.2020.118797. [DOI] [PubMed] [Google Scholar]
- 11.Khandelwal N, et al. A high-throughput RNAi screen for detection of immune-checkpoint molecules that mediate tumor resistance to cytotoxic T lymphocytes. EMBO Mol. Med. 2015;7:450–463. doi: 10.15252/emmm.201404414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha A, et al. Programmed death ligand-1 expression on donor T cells drives graft-versus-host disease lethality. J. Clin. Invest. 2016;126:2642–2660. doi: 10.1172/jci85796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren XY, et al. Immune landscape of the B7 and TNFR families in oral squamous cell carcinoma. Chin. J. Dent. Res. 2020;23:109–117. doi: 10.3290/j.cjdr.a44747. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Tang T, Wang X, Bai X, Liang T. Calreticulin couples with immune checkpoints in pancreatic cancer. Clin. Transl. Med. 2020;10:36–44. doi: 10.1002/ctm2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickert D, et al. Circular RNA profiling distinguishes medulloblastoma groups and shows aberrant RMST overexpression in WNT medulloblastoma. Acta Neuropathol. 2021;141:975–978. doi: 10.1007/s00401-021-02306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, et al. CCAT1 stimulation of the symmetric division of NSCLC stem cells through activation of the Wnt signalling cascade. Gene Ther. 2018;25:4–12. doi: 10.1038/gt.2017.98. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–89. doi: 10.1038/cgt.2016.3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol. Cancer. 2020;19:25. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao G, et al. FBXW7 suppresses epithelial-mesenchymal transition and chemo-resistance of non-small-cell lung cancer cells by targeting snai1 for ubiquitin-dependent degradation. Cell Prolif. 2018;51:e12473. doi: 10.1111/cpr.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie F, et al. CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol. Cancer. 2021;20:25. doi: 10.1186/s12943-021-01359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401. doi: 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Z, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol. Cancer. 2020;19:25. doi: 10.1186/s12943-020-01179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, et al. β-Catenin/Lin28/let-7 regulatory network determines type II alveolar epithelial stem cell differentiation phenotypes following thoracic irradiation. J. Radiat. Res. 2021;62:119–132. doi: 10.1093/jrr/rraa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru B, et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 28.Xiao G, et al. MiR-129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem-like cells. Oncotarget. 2017;8:103261–103273. doi: 10.18632/oncotarget.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, et al. MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget. 2015;6:32944–32954. doi: 10.18632/oncotarget.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, et al. Genomic investigation of co-targeting tumor immune microenvironment and immune checkpoints in pan-cancer immunotherapy. NPJ Precis. Oncol. 2020;4:29. doi: 10.1038/s41698-020-00136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charoentong P, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and relevant supporting materials related to the findings of this study are available from the corresponding author upon reasonable request.