Abstract
Background
The cycle of feeding and fasting is fundamental to life and closely coordinated with changes of metabolic programs. During extended starvation, ketogenesis coupled with fatty acid oxidation in the liver supplies ketone bodies to extrahepatic tissues as the major form of fuel. In this study, we demonstrated that PAQR9, a member of the progesterone and adipoQ receptor family, has a regulatory role on hepatic ketogenesis.
Methods
We analyzed the phenotype of Paqr9-deleted mice. We also used biochemical methods to investigate the interaction of PAQR9 with PPARα and HUWE1, an E3 ubiquitin ligase.
Results
The expression of Paqr9 was decreased during fasting partly depending on PPARγ. The overall phenotype of the mice was not altered by Paqr9 deletion under normal chow feeding. However, fasting-induced ketogenesis and fatty acid oxidation were attenuated by Paqr9 deletion. Mechanistically, Paqr9 deletion decreased protein stability of PPARα via enhancing its poly-ubiquitination. PAQR9 competed with HUWE1 for interaction with PPARα, thus preventing ubiquitin-mediated degradation of PPARα.
Conclusion
Our study reveals that PAQR9 impacts starvation-mediated metabolic changes in the liver via post-translational regulation of PPARα.
Keywords: PAQR9, PPARα, HUWE1, Starvation, Ketogenesis, Fatty acid oxidation
1. Introduction
The cycle of feeding and fasting is the most fundamental physiological change occurring in all forms of life, including humans. During starvation, a delicate set of metabolic programs are initiated in order to meet the fuel needs of the body [1]. During the early phase of starvation, glucose generated by glycogen breakdown serves as the major form of fuel to the body. Later on, gluconeogenesis in the liver is initiated to provide glucose. During extended starvation, ketogenesis in the liver supplies ketone bodies as the major form of fuel for extrahepatic tissues, especially the brain [1]. Ketone bodies are formed from acetyl-CoA, a product of mitochondrial β-oxidation from fatty acids that are derived from lipolysis in adipose tissues during fasting. The changes in metabolic programs in the liver during starvation are controlled by the orchestrated actions of a set of transcription factors, including glucocorticoid receptor (GR), cAMP responsive element binding protein 1 (CREB1), peroxisome proliferator activated receptor alpha (PPARα), and CCAAT/enhancer binding protein beta (CEBPβ) [2,3].
PPARα plays a critical role in regulating both fatty acid oxidation (FAO) and ketogenesis [[4], [5], [6]]. PPARα-null mice show dysregulation of FAO in response to fasting [7,8]. The expression of rate-limiting enzymes of ketogenesis and fatty acid oxidation as well as fasting-responsive genes such as Hmgcs2, Cpt1, and Fgf21 are mainly regulated at the transcription level by PPARα [[9], [10], [11]]. Transcriptional activation of PPARα is initiated via binding with various fatty acids or fatty acid–derived compounds [4]. Other transcription factors such as GR also influence the functions of PPARα [3]. Additionally, PPARα can be regulated at the posttranscriptional level via ubiquitin-mediated degradation. In particular, HUWE1, an E3 ligase, was identified to directly bind with PPARα and cause PPARα protein degradation by a proteasome-mediated pathway [12].
PAQR9 is a member of the progesterone and adipoQ receptor (PAQR) family [13]. PAQR9 was originally called mPRε and reported to function by coupling to G-proteins, serving as an integral membrane progesterone receptor in neurons [14]. However, when PAQR9 is ectopically expressed in mammalian cells, it is mainly localized in ER but not the plasma membrane [15]. It was later found that PAQR9 plays a role in the regulation of the protein quality control of mislocalized membrane proteins by affecting their interaction with BAG6 [15]. However, the in vivo function of PAQR9 is currently unknown. We report here that the expression of Paqr9 is altered by fasting in mice. Furthermore, we found that PAQR9 plays a role in fasting-induced ketogenesis and FAO in the liver by regulating the protein stability of PPARα, thus revealing an in vivo function of PAQR9 during starvation.
2. Materials and methods
2.1. Reagents and antibodies
Cycloheximide (CHX), polybrene, polyethylenimine (PEI), puromycin, and Oil Red O were from Sigma–Aldrich (St. Louis, MO, U.S.A.), MG132 was from BD Biosciences (Franklin Lakes, NJ, U.S.A.), and Polyjet, Lipo2000 and TRIzol reagent were from Invitrogen (Carlsbad, CA, U.S.A.). RIPA buffer and PCR 2X mix were from Yeasen (Shanghai, China). Rapamycin was from MCE (NJ, USA). AICAR was from CSNpharm (Chicago, MI, USA). Sodium caprylate was from Sangon (Shanghai, China). Antibodies against Flag tag and α-tubulin were from Sigma–Aldrich; antibodies against Myc tag and mouse IgG were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.); antibodies against GFP, HUWE1, HSP90, CREB, phosphorylated CREB, and TP53 were from Cell Signaling Technology (Boston, MA, USA); antibodies against HA and GAPDH were from ABclonal (Boston, MA, U.S.A.); antibodies against PAQR9, PPARα, Alexa Fluor 546 goat anti-mouse IgG, and Alexa Fluor 546 goat anti-rabbit IgG were from Abcam (Cambridge, U.K.); antibody against ubiquitin was from Santa Cruz Biotechnology (TX,U.S.A.).
2.2. Animal studies
The Paqr9 deletion mice were developed in the Model Animal Research Center of Nanjing University (Nanjing, Jiangsu, China). All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences with approval number SINH-2020-CY-1. Mice were maintained on a 12-hour light/dark cycle at 25 °C. Primary hepatocytes were isolated as described [12]. Mouse blood ketone bodies were measured with a kit purchased from Abbott Diabetes Care (Chicago, MI, USA).
2.3. Plasmids, cell transfection, and lentivirus
Plasmids of Flag-tagged PPARα, differently tagged full-length PAQR9 overexpression, and Paqr9 shRNA constructs were reported previously [12,15]. PPARγ was cloned from the cDNA of MCF7 cells by PCR and inserted into vector 3 × Flag. Various lengths of Paqr9 promoter were cloned from the genomic DNA of HeLa cells by PCR and inserted into vector pGL.3-basic. Vector pRL-TK was used to express renilla luciferase for luciferase assay control. pMD2. G and psPAX2 plasmids were used for lentivirus packaging. HEK293T cells were transfected with PEI, HeLa cells were transfected with Polyjet, and Hep3B cells were transfected with Lipo2000. Paqr9-deleted Hep3B cells were generated by lentivirus transfection and screened with puromycin. Lentivirus was packaged with the psPAX2-pMD2. G system. In brief, each 10-mm dish of HEK293T cells was transfected with 7.5 μg of psPAX2, 3 μg of pMD2. G, and 10 μg of shRNA-containing plasmid. The cell culture medium was collected 48 h later and directly used to infect Hep3B cells for 48 h with 4 μg/mL polybrene, then treated with 5 μg/mL puromycin. HUWE1 knockout plasmid was generated by CRISPR-Cas9 system with LentiCRISPRV2 containing a sequence of sgRNA 5′-CTGTAGCTGGTTTTCAGTG-3'. PPARα knockout was performed with the same method containing a sequence of sgRNA 5′- GCACCATCTGGTCGCGATGG -3'. The HUWE1-and PPARα-deleted Hep3B cells were generated by lentivirus transfection and screened with puromycin.
2.4. Cell fractionation
Hep3B cells were first lysed with membrane lysis (20 mM Tris, 137 mM NaCl, 5 mM EDTA, 1% NP40, and 10% glycerinum) at 4 °C for 15 min. The supernatant was collected after centrifugation at 4 °C for 10 min (12,000 rpm) for detection of proteins in the cytoplasm. The sediment was washed twice by PBS and then lysed with RIPA at 4 °C for 30 min, followed by centrifugation at 4 °C for 10 min (12,000 rpm). The supernatant was then used to detect proteins in the nucleus.
2.5. Cell culture and treatment
HEK293T cells, HeLa cells, Hep3B cells, and primary hepatocytes were cultured in high-glucose DMEM (4.5 g/L) with 100 units/mL penicillin/streptomycin and 10% FBS at 37 °C with 5% CO2. Cells for fasting experiments were washed twice with PBS and then cultured in DMEM with 1 g/L glucose and 2% FBS. 50 nM rapamycin was used for mTOR inhibition and 1 mM AICAR was used for AMPK activation. For in vitro ketogenesis assay, Hep3B cells or hepatocytes were cultured in 12-well plates and treated with DMEM containing 500 μM sodium caprylate for 24 h. The medium was then collected for β-hydroxybutyric acid measurement with a kit purchased from mlbio, Shanghai, China. For lipid accumulation analysis, hepatocytes were treated with 1 g/L glucose and 250 μM sodium caprylate for 36 h, then fixed with 4% PFA for 15 min. 0.5% Oil Red O isopropanol solution was diluted to 0.3% with water and used to stain the cells for 30 min, followed by washing with water. Images of the staining were acquired by OLYMPUS IX73.
2.6. RNA isolation and RT-qPCR
Total RNA of cells and mouse tissues were lysed by TRIzol reagent and purified according to the manufacturer's instructions. RNA was reverse-transcribed with FastQuant RT Kit (Tiangen, Shanghai, China) to obtain cDNA. Real-time PCR was performed with the SYBR Green PCR system (TOYOBO, Tokyo, Japan) with an ABI QuantStudio6 system. The sequence of the primers is shown in Supplemental Table 1.
2.7. Western blotting
Tissues and cultured cells were lysed in RIPA buffer with fresh protease inhibitors (MCE) and phosphatase inhibitors (Sigma–Aldrich), and the supernatant was collected after centrifugation at 4 °C for 20 min at 12,000 rpm. For ubiquitination assay, the cells were treated with or without 10 μM MG132 for 6 h to block proteasomal degradation of proteins before cell lysis. To analyze protein degradation rate, the cells were treated with cycloheximide (CHX) for various times. Co-immunoprecipitation and Western blotting were performed as previously reported [12].
2.8. Dual-luciferase reporter assay
Different lengths of Paqr9 promoter were cloned by PCR into pGL.3-basic plasmids. The plasmids of the promoter and pRL-TK were co-transfected into HEK293T with a ratio of 20:1, and plasmids of transcription factors were co-expressed at the same time. 24 h after the transfection, the cells were lysed and then used to analyze luciferase activity using a kit purchased from Promega (Madison, WI, USA).
2.9. Chromatin immunoprecipitation and ChIP-quantitive PCR (ChIP-qPCR)
ChIP and ChIP-qPCR were carried out as previously described [16].
2.10. Immunofluorescence studies
HeLa cells were cultured on glass coverslips and transfected with different plasmids. 24 h after transfection, the cells were fixed with 4% paraformaldehyde for 10 min and then permeabilized with 0.1% Triton X100 in PBS for 10 min. The cells were blocked in 3% bovine serum albumin in PBS for 30 min, then incubated with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature. The nuclei were stained with Hoechst 33,342 (Molecular Probes) in PBS for 10 min. Fluorescence images were acquired with ZEISS (LSM880).
2.11. Frozen section and Oil Red O staining
Livers were isolated from the mice and fixed overnight with 4% paraformaldehyde (PFA) for paraffin embedding. Frozen sectioning and Oil Red O staining were performed by Servicebio (Wuhan, Hubei, China). The images of the slides were acquired with an Olympus BX51 microscope.
2.12. Statistical analysis
All data are shown as mean ± SEM. The results were analyzed with Student's t-test for comparison between two groups or ANOVA for comparison of more than two groups. Values of P < 0.05 were considered statistically significant.
3. Results
3.1. Paqr9 expression is regulated by fasting and refeeding
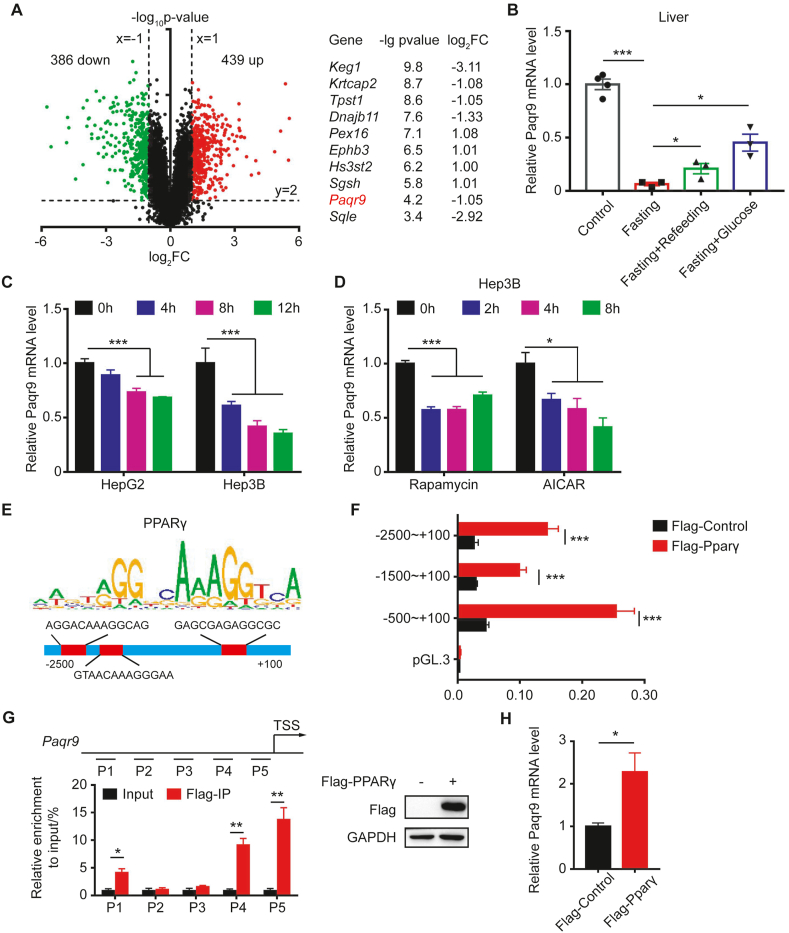
The liver is one of the most important organs that participate in homeostatic regulation during fasting. To identify how calorie restriction alters gene transcription in the liver, we mined an existing RNA-seq dataset of mouse liver (GSE92502). We compared the data of 24-hour fasting to the control and found that a total of 825 genes were significantly changed (439 genes upregulated and 386 genes downregulated), with P-values < 0.01 and estimated absolute log2–fold change >1 (Figure 1A). To characterize the functional changes associated with fasting, we performed KEGG pathway and GO enrichment analysis with DAVID 6.8 [17,18]. As expected, we found that nearly half of the 825 genes were associated with lipid metabolism, glucose homeostasis, the or oxidation-reduction process (data not shown).
Figure 1.
Paqr9 expression in the liver is regulated by fasting.A. A volcano plot showing RNA-Seq results of liver fasted for 24 hours from GEO dataset GSE92502. Downregulated or upregulated genes were divided by log2–fold change >1 or < −1 with P-value < 0.01. Red dots for upregulated genes and green dots for downregulated genes. The right panel depicts the top 10 most significantly changed genes (except for those involved in lipid metabolism and oxidation pathways). B.Paqr9 mRNA level in the liver after 48 hours fasting and refeeding or peritoneal injection with 10% glucose solution for 3 hours (n = 3 for control and n = 4 for other groups). C.Paqr9 mRNA level in Hep3B and HepG2 cells under starvation condition for different times. D.Paqr9 mRNA level in Hep3B cells treated with 50 nM rapamycin or 1 mM AICAR for different times. E. PPARγ motif analysis and potential binding sites at the Paqr9 promoter. F. Dual-luciferase reporter assay. Different lengths of putative Paqr9 promoter were cloned into luciferase-containing plasmid and transiently expressed in the Hep3B cells along with PPARγ-expressing plasmid as indicated. Relative luciferase activity was calculated by dividing firefly luciferase activity by renilla activity. G. ChIP-qPCR with different regions of Paqr9 promoter. P1 to P5 were five different segments covering the region of −2,500 bp to the transcription start site (TSS) region. Both input and immunoprecipitation results are shown. The expression of Flag-tagged PPARγ is confirmed by Western blotting (right panel). H.Paqr9 mRNA level in Hep3B with overexpression of PPARγ. The mRNA expression results were relative to β-actin in B and relative to α-tubulin in C, D, and H. All the quantitative data are shown as mean ± S.E.M., ∗ for P < 0.05 and ∗∗∗ for P < 0.001.
In order to identify new genes and pathways involved in metabolic regulation during fasting, we focused on the genes that have not been described as modifying glucose and lipid metabolism in the past. Ranking these genes by P-value, we found that PAQR9 was one of the most significantly changed genes (Figure 1A). To confirm that Paqr9 is regulated by fasting, we examined the change in Paqr9 gene expression in the mouse liver during fasting and refeeding. As shown in Figure 1B, Paqr9 expression in the liver was robustly downregulated after 48 h fasting. Refeeding or peritoneal injection with 10% glucose solution following fasting could increase expression of Paqr9 in the liver (Figure 1B). Consistently, the protein level of PAQR9 in the liver was significantly reduced by fasting and partially elevated by refeeding (Figure S1). We also analyzed changes in Paqr9 expression in vitro. In two liver cell lines, HepG2 and Hep3B, glucose deprivation significantly reduced Paqr9 expression (Figure 1C), consistent with the in vivo findings. As AMPK and mTOR signaling pathways are the two most important modules involved in nutrient sensing [[19], [20], [21]], we analyzed the changes in Paqr9 expression upon treatment with rapamycin, an mTOR signal inhibitor, or AICAR, an AMPK phosphorylation activator. The expression of Paqr9 was downregulated by treatment with rapamycin or AICAR (Figure 1D), indicating that these two nutrient sensing pathways likely mediate the fasting-induced changes of Paqr9 expression.
3.2. PPARγ is involved in the transcriptional regulation of Paqr9 expression
We next explored how Paqr9 expression was regulated during nutrient deficiency. We analyzed the putative promoter region of the Paqr9 gene with two online tools (http://alggen.lsi.upc.es and http://cistrome.org) to predict the potential transcription factors (TFs) regulating Paqr9 expression, and then compared them with the known TFs affected by both mTOR and AMPK pathways. One of the predicted TFs was PPARγ, which has been shown to be associated in mTOR regulation on adipogenesis [22,23]. Motif analysis revealed 3 potential binding sites of PPARγ in the promoter region of the Paqr9 gene (Figure 1E). To confirm whether PPARγ was involved in the regulation of Paqr9 gene expression, we used a dual-luciferase reporter assay to analyze the promoter activity of the Paqr9 gene. We co-transfected HEK293T cells with a Flag-tagged PPARγ, a plasmid expressing renilla-luciferase, and a plasmid of the fly-luciferase reporter gene under the control of different lengths of the putative promoter region of the Paqr9 gene. The luciferase activity of the Paqr9 promoter was elevated by PPARγ overexpression (Figure 1F). On the other hand, PPARα overexpression had a very minor effect on the promoter of Paqr9 (Figure S2). Consistently, neither PPARα deletion nor PPARα overexpression changed the expression of Paqr9 under normal and low-glucose starvation conditions (Figure S3). Furthermore, −500/+100 bp had the highest responsiveness to PPARγ overexpression (Figure 1F & Figure S2), indicating that the PPARγ binding site within this segment is involved in regulation. We next performed a ChIP-qPCR analysis and confirmed that PPARγ could directly bind to the Paqr9 promoter (Figure 1G). Consistently, overexpression of PPARγ increased the expression of Paqr9 in Hep3B cells (Figure 1H), further confirming that PPARγ is involved in the transcriptional regulation of Paqr9 gene expression.
3.3. Deletion of Paqr9 has no effect on metabolic phenotype under feeding state, but attenuates ketogenesis and fatty acids β-oxidation under fasting condition
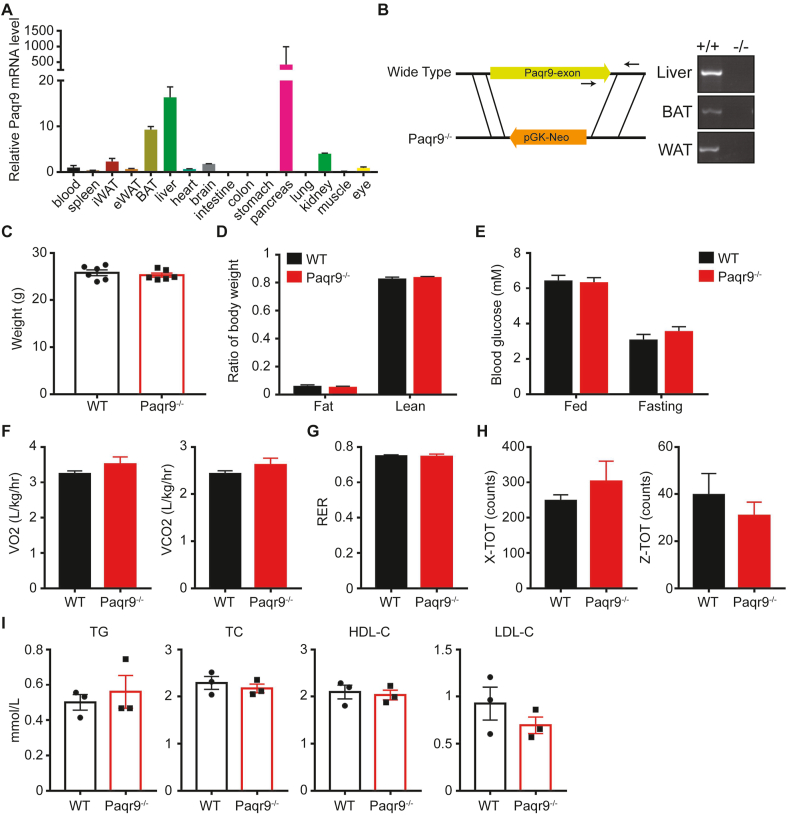
We next analyzed the in vivo function of Paqr9. First, we detected the tissue expression pattern of Paqr9 in mice by RT-PCR. Paqr9 showed the highest expression in pancreas, followed by liver and brown adipose tissue (BAT) (Figure 2A). We generated a systemic Paqr9 knockout mouse model through a targeted disruption of the Paqr9 gene by homologous recombination (Figure 2B). Deletion of Paqr9 was confirmed by RT-PCR using RNA isolated from both the wild type and Paqr9-deleted mouse tissues, including liver and adipose tissues (Figure 2B).
Figure 2.
Deletion of Paqr9 does not alter metabolic phenotype under normal chow feeding.A. The mRNA level of Paqr9 in different tissues of C57BL/6 mice relative to β-actin (n = 4 for each tissue). B. Generation and identification of the Paqr9−/− mouse. The left panel depicts the scheme of the Paqr9−/− mouse. Note that Paqr9 gene only has one exon. The right panel shows genotyping results using primers denoted by arrows in the left panel. C. Body weight of male wild type (WT) and Paqr9−/− mice at 10 weeks old (n = 6 for each group). D. Body composition of the mice. E. Blood glucose of the mice at feeding or fasting for 16 hours. F. Oxygen consumption and carbon dioxide production of the mice during a 24-hour period. G. Respiratory exchange ratio (RER) of the mice. H. Movement of the mice on the X and Z axes. I. Blood levels of triglyceride (TG), total cholesterol (TC), HDL cholesterol, and LDL cholesterol of the mice. All data were analyzed with Student's t-test. All the quantitative data are shown as mean ± S.E.M.
We first analyzed the metabolic phenotype of Paqr9−/− mice fed with normal chow. Body weight and body composition were not changed by Paqr9 deletion (Figure 2C,D). The level of blood glucose was not altered by Paqr9 deletion, either, under both feeding and fasting conditions (Figure 2E). Furthermore, analysis with a metabolic cage revealed that oxygen consumption, carbon dioxide production, respiratory exchange ratio (RER), and physical activities were not changed by Paqr9 deletion (Figure 2F–H). In addition, the lipid profile in the blood of the mice was not altered by Paqr9 deletion (Figure 2I). Collectively, these data indicate that Paqr9 deletion does not give rise to obvious changes in glucose and lipid metabolism under the condition of normal chow feeding.
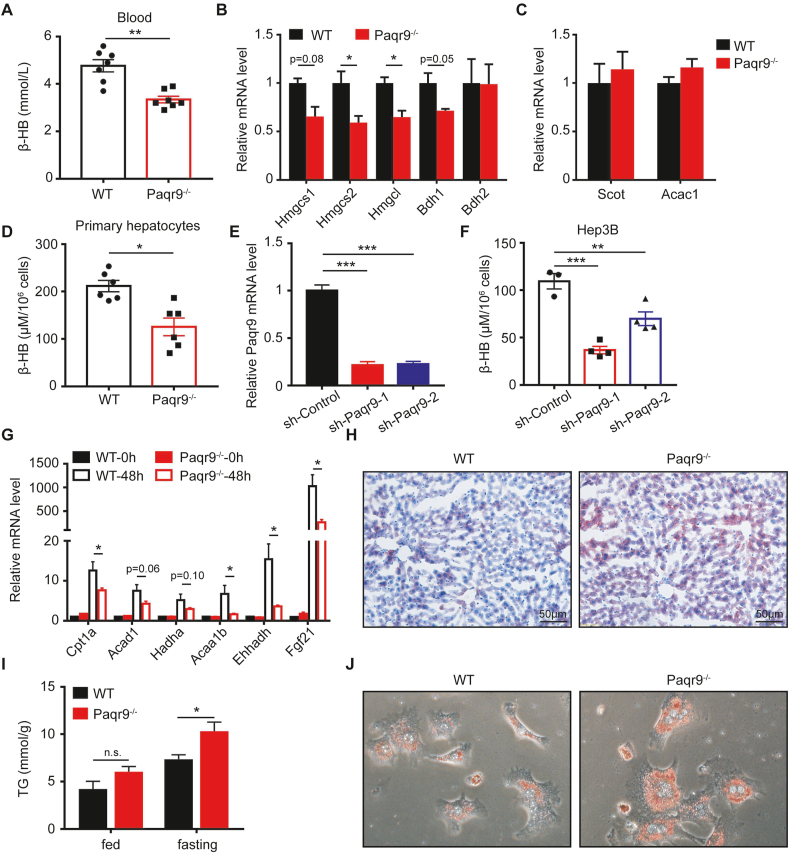
Having originally found that Paqr9 expression was reduced by fasting (Figure 1), we investigated whether deletion of Paqr9 affected the metabolism during fasting. After 24 h fasting, the blood level of ketone bodies in the Paqr9−/− mice was significantly lower than that of the wild type mice (Figure 3A). To elucidate whether the observed change in ketone bodies was partially caused by changes in ketogenesis or ketolysis, we analyzed the expression of genes involved in the ketogenesis pathway in the liver and ketolysis pathway in the skeletal muscle (Figure 3B–C). We found that the expression of Hmgcs2 and Hmgcl, two key genes in ketogenesis, were significantly downregulated in the liver of Paqr9−/− mice (Figure 3B). On the other hand, the expression of two key genes for ketolysis was not affected by Paqr9 deletion in the skeletal muscle (Figure 3C), indicating that deletion of Paqr9 mainly affects ketogenesis but not ketolysis during fasting.
Figure 3.
Deletion of Paqr9 reduces hepatic ketogenesis and fatty acid oxidation in vivo and in vitro.A. Concentration of β-hydroxybutyrate (β-HB) in serum of the male wild type (WT) and Paqr9−/− mice after fasting for 24 hours (n = 6 for WT and n = 7 for Paqr9−/− mice). B. The mRNA levels of ketogenic genes in the liver of the mice, as in A. C. The mRNA levels of ketone degradation genes in the skeletal muscle of the mice, as in A. D. Concentration of β-HB in culture medium of primary hepatocytes isolated from the mice. The cells were treated with 500 μM sodium caprylate for 24 hours before measurement. E. Efficiency of Paqr9 knockdown in Hep3B cells transfected with shRNA-containing plasmids. F. Concentration of β-HB in culture medium of the cells, as in E. G. The mRNA levels of fatty acid β-oxidation genes in the livers of the mice after fasting for 48 hours. H. Representative images of Oil Red O staining of the livers from the mice after fasting for 24-hour fasting. I. Triglyceride level in the livers of the mice under feeding or fasting for 24 hours. J. Representative images of Oil Red O staining of primary hepatocytes isolated from the mice. The hepatocytes were treated with 1 g/L glucose and 500 μM sodium caprylate for 36 hours. All data were analyzed with Student's t-test. All the quantitative data are shown as mean ± S.E.M., ∗ for P < 0.05, ∗∗∗ for P < 0.001, and n. s. for non-significant.
We isolated primary hepatocytes from wile type and Paqr9−/− mice to further analyze ketogenesis in vitro. With the treatment of sodium caprylate, a short-chain fatty acid, for 36 h, the concentration of β-hydroxybutyric acid (β-HB) was reduced in the medium of hepatocytes isolated from Paqr9−/− mice (Figure 3D), consistent with the in vivo finding. To provide further evidence, we constructed Paqr9-downregulated Hep3B cell lines with two different shRNAs (Figure 3E). The secretion of β-HB was decreased by Paqr9 knockdown in these cells (Figure 3F). These results thus indicate that PAQR9 has a positive effect on ketogenesis both in vivo and in vitro.
Because ketogenesis is tightly coupled to β-oxidation of fatty acids [24], we investigated whether fatty acid oxidation (FAO) was affected by Paqr9 deletion. As shown in Figure 3G, fasting for 48 h robustly induced the expression of all the genes involved in FAO in the liver. However, the fasting-induced expression of these genes was significantly reduced by Paqr9 deletion (Figure 3G). Accordingly, lipid accumulation in the liver was increased by Paqr9 deletion under fasting conditions, as analyzed by Oil Red O staining (Figure 3H). The triglyceride level in the liver was also elevated by Paqr9 deletion under fasting conditions (Figure 3I). Consistently, lipid accumulation was increased by Paqr9 deletion in isolated hepatocytes when treated with fatty acids and low concentrations of glucose (Figure 3J). In addition, we found that Paqr9 knockdown in Hep3B cells or Paqr9 deletion in mouse primary hepatocytes also led to decreases in the expression levels of numerous genes involved in ketogenesis and FAO (Figure S4). Overall, these results indicate that Paqr9 deletion leads to decreases in ketogenesis and FAO in the liver under fasting conditions, accompanied by an increase in lipid accumulation.
3.4. PAQR9 regulates protein stability of PPARα
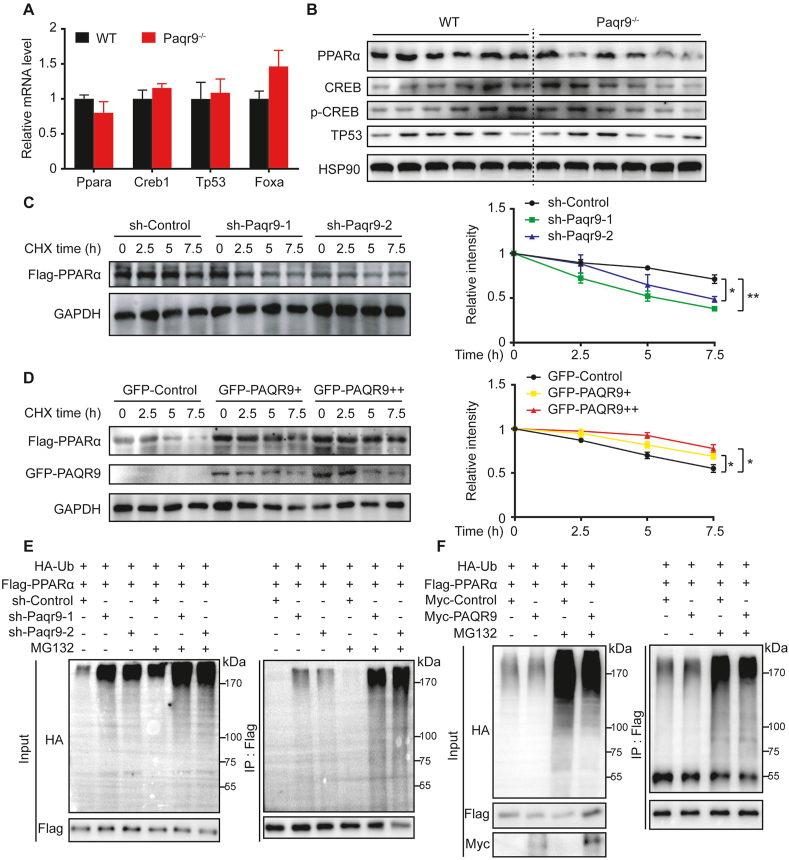
It has been reported that ketogenesis and FAO during fasting are regulated by a series of TFs [2,3]. We first analyzed the expression of a set of TFs over the course of 24 h fasting. At the mRNA level, none of the major TFs involved in ketogenesis and FAO was altered by Paqr9 deletion (Figure 4A). However, Western blotting revealed a significant reduction of PPARα protein levels in the livers of Paqr9−/− mice, while CREB and TP53 were not altered (Figure 4B). Analysis of more mouse samples also showed a lower PPARα protein level in Paqr9-deleted liver than in the wild type control (Figure S5).
Figure 4.
PAQR9 protects PPARα from degradation by reducing PPARα ubiquitination.A. The mRNA levels of representative transcription factors in the liver of the mice after fasting for 24 hours (n = 6 for each group). B. Western blotting to detect transcription factors in the livers of the mice as in A. Quantitation of the data is shown in the right panel. C. Analysis of protein stability of PPARα with PPARα knockdown. Hep3B cells with or without PPARα knockdown were transiently transfected with Flag-tagged PPARα and then treated with CHX (100 μg/mL) for various times, followed by immunoblotting with the antibodies as indicated. Quantitation of the immunoblotting results from three independent experiments is shown in the right panel. D. Analysis of protein stability of PPARα with Paqr9 overexpression. Hep3B cells were transiently transfected with Flag-tagged PPARα and GFP-tagged PAQR9 as indicated and treated with CHX (100 μg/mL) for various times. Quantitation of the immunoblotting from three independent experiments is shown in the right panel. E. Poly-ubiquitination of PPARα in Hep3B cells with Paqr9 knockdown. The cells were pretreated with MG132 (10 μM) for 6 hours, followed by immunoprecipitation and immunoblotting with the antibodies as indicated. F. Poly-ubiquitination of PPARα in Hep3B cells with Paqr9 overexpression. The cells were transiently transfected with Myc-tagged Paqr9 and treated with MG132 (10 μM) for 6 hours, followed by immunoprecipitation (IP) and immunoblotting with the antibodies as indicated. All the quantitative data are shown as mean ± S.E.M., ∗ for P < 0.05 and ∗∗ for P < 0.01.
As the protein but not mRNA of PPARα was affected by Paqr9 deletion, we next analyzed whether PAQR9 affected the protein degradation rate of PPARα. In Hep3B cells, knockdown of Paqr9 significantly accelerated the protein degradation rate of PPARα (Figure 4C). Conversely, overexpression of PAQR9 extended the half-life of PPARα protein (Figure 4D).
It was previously reported that PPARα is degraded by a proteasome-mediated pathway through poly-ubiquitination by HUWE1 [12]. We thus analyzed the poly-ubiquitination level of PPARα protein. Knockdown of Paqr9 in Hep3B cells enhanced the level of poly-ubiquitination of PPARα protein in the presence of MG132 (Figure 4E). In contrast, PAQR9 overexpression inhibited poly-ubiquitination of PPARα protein in the presence of MG132 (Figure 4F). Therefore, these results indicate that PAQR9 inhibits the degradation of PPARα protein via reduction of poly-ubiquitination of PPARα protein.
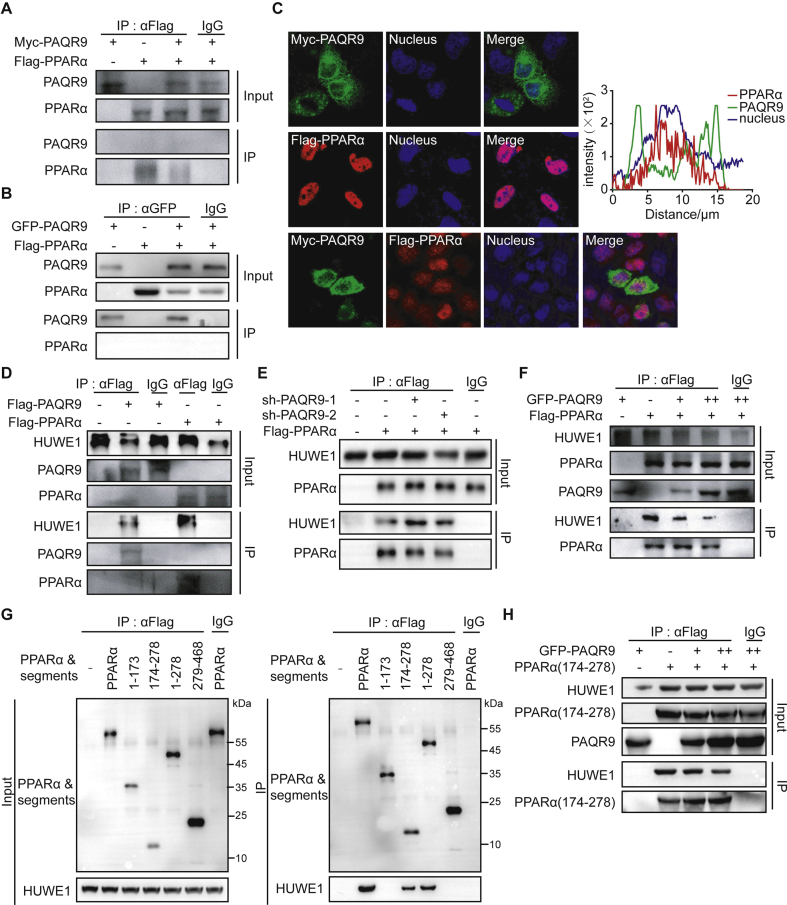
3.5. PAQR9 does not interact with PPARα but competes with PPARα for interaction with HUWE1
As our results revealed that PAQR9 protects PPARα from ubiquitination-mediated degradation, we investigated the underlying mechanism. Based on a previous study on post-translational regulation of PPARα [12], we performed co-immunoprecipitation assays to detect the potential interaction between PAQR9 and PPARα. However, no interaction was found between these two proteins through co-immunoprecipitation assays (Figure 5A–B). Furthermore, when expressed in HeLa cells, the GFP-fused PAQR9 was localized in the ER as previously reported [15], and PPARα was localized in the nucleus (Figure 5C, upper and middle panels). When both PAQR9 and PPARα were co-expressed, though PAQR9 was seen close to the nucleus, they were scarcely co-localized (Figure 5C, lower panel), further indicating that there is no interaction between PAQR9 and PPARα.
Figure 5.
PAQR9 reduces PPARα ubiquitination by competing with its interaction with HUWE1.A-B. Co-immunoprecipitation assay to analyze interaction of PPARα with PAQR9. HEK293T cells were transfected with the plasmids as indicated, followed by immunoprecipitation (IP) and immunoblotting with the antibodies as indicated. C. Localization of PAQR9 and PPARα in HeLa cells. The cells were transiently transfected with the plasmids as indicated and used in immunofluorescent staining. The nucleus was stained with Hoechst 33,342. The right panel shows plots of signal intensity (y axis) against distance in mm (x axis) to indicate occurrence of overlaps between the red and green channels for PAQR9/PPARα co-expressing cells. D. Co-immunoprecipitation assay to analyze interaction of PPARα and PAQR9 with HUWE1. Hep3B cells were transfected with the plasmids as indicated, followed by IP and immunoblotting. E. The interaction of PPARα with HUWE1 is enhanced by Paqr9 knockdown. Hep3B cells with or without Paqr9 knockdown were transiently transfected with GFP-fused PAQR9, followed by IP and immunoblotting. F. The interaction of PPARα with HUWE1 is reduced by Paqr9 overexpression. Hep3B cells were transiently transfected with different amount of GFP-fused PAQR9, followed by IP and immunoblotting. G. Identification of the region of PPARα that interacts with HUWE1. Different segments of PPARα were overexpressed in Hep3B cells, followed by IP and immunoblotting. H. The interaction of the 174–278aa segment of PPARα with HUWE1 is reduced by Paqr9 overexpression. Hep3B cells were transiently transfected with GFP-fused PAQR9 and Flag-tagged PPARα segment, followed by IP and immunoblotting.
It was previously reported that HUWE1 was the one of most abundant proteins pulled down by PAQR9, discovered through mass spectrometry analysis [15]. We thus employed immunoprecipitation assays to analyze the potential interaction between PAQR9 and HUWE1. When Flag-tagged PAQR9 was expressed in Hep3B cells, the anti-Flag antibody was able to pull down HUWE1 (Figure 5D). On the other hand, Flag-tagged PPARα could also pull down HUWE1 (Figure 5D). To investigate the relationship among HUWE1, PAQR9, and PPARα, we performed immunoprecipitation assays in Paqr9-deleted Hep3B cells. As a result, deletion of PAQR9 enhanced the interaction of HUWE1 with PPARα (Figure 5E), while overexpression of PAQR9 reduced the interaction between HUWE1 and PPARα in a dose-dependent manner (Figure 5F). These results, therefore, indicate that PAQR9 competitively inhibited the interaction between PPARα and HUWE1 to regulate the degradation of PPARα.
We next investigated which region of PPARα was involved in the interaction with HUWE1. We cloned different segments of PPARα and analyzed their interaction with HUWE1 through co-immunoprecipitation (Figure 5G). We found that the region covering 174–278 amino acid residues of PPARα interacted with HUWE1 (Figure 5G). Furthermore, overexpression of PAQR9 dose-dependently reduced the interaction of the PPARα segment (174–278aa) with HUWE1 (Figure 5H), consistent with the result with full-length PPARα (Figure 5F). We also analyzed the subcellular compartment in which PPARα interacts with HUWE1. We found that PPARα was mainly localized in the nucleus while HUWE1 was mainly localized in the cytoplasm (Figure S6). However, the interaction of PPARα with HUWE1 was able to occur in both the cytoplasm and nucleus (Figure S6).
We also analyzed whether or not the degradation of PAQR9 itself was affected by PPARα. The half-life of PAQR9 was extended by PPARα overexpression (Figure S7), while poly-ubiquitination of PAQR9 was attenuated by PPARα overexpression (Figure. S7). Overall, these results indicate that PAQR9 and PPARα associate with HUWE1 in a mutually exclusive manner, and that therefore the degree of poly-ubiquitination/degradation of PPARα is affected by the relative amount of PAQR9 and vice versa.
3.6. PAQR9 regulation on ketogenesis and fatty acid oxidation is mediated by PPARα
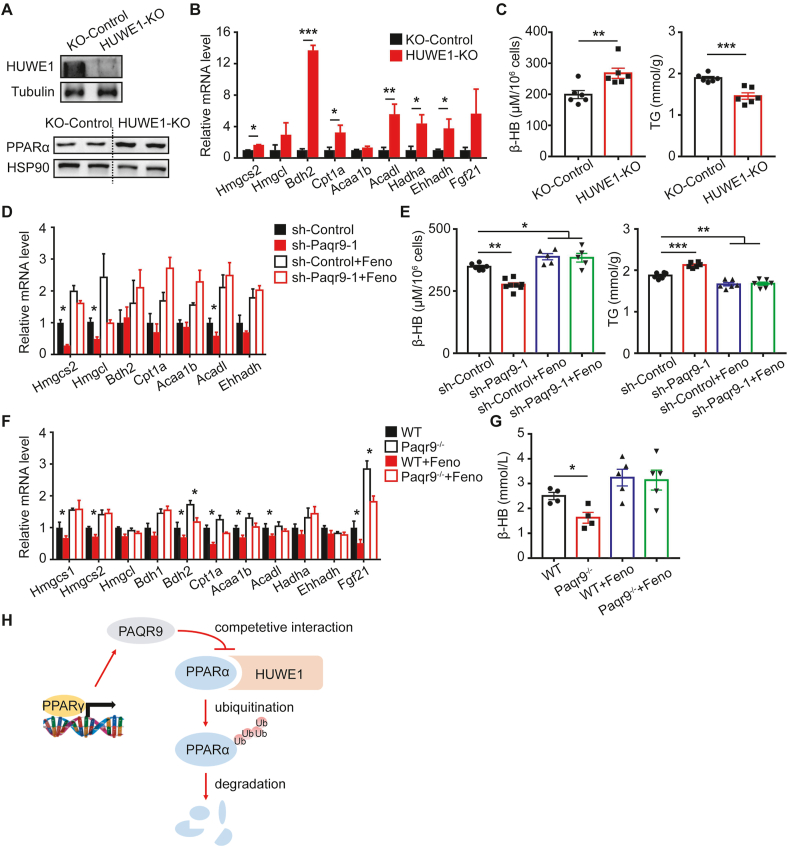
Having identified that PAQR9 and PPARα associate with HUWE1 in a mutually exclusive way, we further investigated whether HUWE1 is involved in fasting-induced ketogenesis and FAO in hepatocytes. We established a HUWE1-deleted Hep3B cell line using Cas9/CRISPR technology (Figure 6A). We found that knockout of HUWE1 elevated the protein level of PPARα in Hep3B cells (Figure 6A). Upon sodium caprylate treatment, HUWE1 deletion led to increases in the expression of genes involved in ketogenesis and FAO (Figure 6B). Consistently, HUWE1 knockout significantly elevated the production of β-HB and decreased lipid accumulation in Hep3B cells (Figure 6C). These results further confirmed the involvement of HUWE1 in regulating the function of PPARα.
Figure 6.
PAQR9 affects ketogenesis and fatty acid oxidation through PPARα.A. HUWE1 deletion increases the protein level of PPARα in Hep3B cells. Upper panel: Western blotting to detect the efficiency of HUWE1 knockout. Lower panel: Western blotting to analyze PPARα protein level. B. Hep3B cells with or without HUWE1 knockout were treated with 1 g/L glucose and 500 μM sodium caprylate for 48 hours, followed by quantitative RT-PCR to analyze expression of genes involved in ketogenesis and FAO (n = 6 for each group). C. The cells, as in B, were used to measure β-HB level in culture medium and triglyceride concentration in the cells. D. HepB3 cells with or without Paqr9 knockdown were treated with 500 μM sodium caprylate for 24 hours (n = 6 for each group), followed by quantitative RT-PCR (n = 6 for each group). The cells were treated with 5 μM of fenofibrate (Feno) as indicated. E. The cells, as in D, were used to measure β-HB level in culture medium and triglyceride concentration in the cells. F. Wild type and Paqr9-deleted mice were gavaged with corn oil or fenofibrate (10 μg/g body weight) for 5 days and then fasted for 24 hours (n = 4 for mice with corn oil and n = 5 for mice with fenofibrate). The livers of the mice were used in quantitative RT-PCR. G. The mice, as in F, were used to determine β-HB concentration in the serum. H. A graphic summary to depict the inhibitory effect of PAQR9 on HUWE1-mediated poly-ubiquitination and degradation of PPARα protein. Note that PAQR9 competes with PPARα for HUWE1 binding.
We next investigated whether the regulation of ketogenesis and FAO by PAQR9 is mediated by PPARα. As expected, treatment of Hep3B cells with fenofibrate, a clinically used PPARα agonist, was able to elevate the expression of genes involved in ketogenesis and FAO (Figure 6D). Fenofibrate also increased β-HB production and reduced triglyceride accumulation in Hep3B cells (Figure 6E). The reduced expression of ketogenesis and FAO genes by Paqr9 knockdown was abrogated by fenofibrate treatment (Figure 6D). The alteration of β-HB production and triglyceride accumulation by Paqr9 knockdown was also abrogated by fenofibrate treatment (Figure 6E). Similarly, the reduced expression of ketogenesis/FAO genes in the liver and decreased β-HB level in the blood by Paqr9 deletion were abrogated by fenofibrate treatment (Figure 6F and 6G). Collectively, these results indicate that PAQR9 regulation of ketogenesis and FAO is mediated by PPARα.
4. Discussion
Our study reveals for the first time that PAQR9 has a functional role in the regulation of energy homeostasis during starvation. The expression of the Paqr9 gene itself is altered during fasting and refeeding. Deletion of the Paqr9 gene attenuated starvation-induced ketogenesis and FAO in the liver. Consistently, fasting-induced lipid accumulation in the liver was elevated by Paqr9 deletion. Mechanistically, PAQR9 regulates the protein stability of PPARα, a master regulator of ketogenesis and FAO, by competitive interaction with HUWE1 (Figure 6H). In addition, we found that Paqr9 deletion had no effect on blood insulin level, but increased blood glucagon level under fasting conditions (Figure S8). As glucagon mainly regulates gluconeogenesis during starvation, it is worth investigating in the future whether or not PAQR9 affects this process. However, the fasting blood glucose level was not affected by Paqr9 deletion (Figure 2E). It is worth noting that glucagon can also regulate FAO and ketogenesis in the liver [25,26], and that investigation of the functional link between PAQR9 and glucagon will be a pursuit of interest in the future. In addition, knowing that FAO also occurs in brown adipose tissue (BAT), we analyzed the effect of Paqr9 deletion on FAO-related genes in BAT. We found that numerous genes involved in FAO in BAT were decreased by Paqr9 deletion (Figure S9), similar to the results in the liver. These results indicated that PAQR9 has a general effect on FAO in different tissues.
Our work also indicated a regulatory role of the PPARγ-PAQR9-PPARα pathway in modulating energy homeostasis during fasting. We found that PPARγ upregulates PAQR9 expression, which in turn could reduce PPARα degradation through competition for HUWE1 binding. Under this theory, activation of PPARγ signaling can enhance the functions of PPARα in the liver via mediation of PAQR9. Interestingly, a recent study demonstrated that PPARα and PPARγ agonists have a potentially synergistic effect in inhibiting the formation of non-alcoholic steatohepatitis (NASH), shown as reduction of lipid accumulation and inflammatory response in the liver [27]. The combined use of PPARα and PPARγ agonists also synergistically ameliorates type 2 diabetes in mice [28]. However, another study revealed a contradictory result, showing that a combination of PPARα and PPARγ agonists increased triglyceride levels in mouse liver [29]. Based on our study, we postulate that a combination of PPARα and PPARγ agonists can boost ketogenesis and FAO during starvation. This is an interesting question that is worth further investigation in the future.
A paradoxical observation in our study is that Paqr9 expression is reduced by fasting, and such reduction would decrease the interaction of Paqr9 with HUWE1, leading to an increase in PPARα ubiquitination/degradation and consequent reduction in PPARα function. In this scenario, fasting-induced PAQR9 reduction would serve as a negative feedback loop to keep PPARα function in check. We found that starvation could promote poly-ubiquitination of PPARα protein (Figure S10). We thus hypothesize that ubiquitination-mediated PPARα degradation may comprise a negative feedback during starvation to avoid over-activating the PPARα pathway. Decreased expression of Paqr9 during starvation may aid in such a negative feedback by enhancing PPARα ubiquitination/degradation via HUWE1. If this were the case, overexpression of PAQR9 in the liver would augment the function of PPARα under starvation. This question can be addressed in the future using mouse models with ectopic overexpression of Paqr9 in the liver.
In addition to PAQR9, post-translational regulation of PPARα can be modulated by another member of the PAQR family: PAQR3. As reported before, PAQR3 negatively regulates the function of PPARα by promoting its interaction with HUWE1, thus enhancing protein degradation of PPARα [12]. On the other hand, PAQR9 positively regulates the function of PPARα by preventing its interaction with HUWE1, as demonstrated in this study. Therefore, PAQR3 and PAQR9 have opposite effects on PPARα regulation. Our preliminary experiments also revealed that these two proteins had an antagonistic effect on the degradation of PPARα protein (data not shown), although the molecular details still need further characterization. Nevertheless, our studies have pinpointed the importance of PAQR family members in the regulation of metabolic processes during starvation via post-translational modulation of PPARα protein.
Funding sources
This study was funded by the National Natural Science Foundation of China (31630036 to YC) and the Ministry of Science and Technology of China (2016YFA0500103 to Y.C.).
Author contributions
Y.C. and Y.L. conceptualized and designed the study. Y.L. performed the experiment. L.C., X.Y., Z.L., and C.L. provided technical assistance. Y.L. and Y.C. wrote the manuscript and prepared the figures. All authors read and approved the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101331.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cahill G.F., Jr. Fuel metabolism in starvation. Annual Review of Nutrition. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein I., Hager G.L. Transcriptional and chromatin regulation during fasting - the genomic era. Trends in Endocrinology and Metabolism. 2015;26(12):699–710. doi: 10.1016/j.tem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein I., Baek S., Presman D.M., Paakinaho V., Swinstead E.E., Hager G.L. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Research. 2017;27(3):427–439. doi: 10.1101/gr.212175.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kersten S. Integrated physiology and systems biology of PPARalpha. Mol Metab. 2014;3(4):354–371. doi: 10.1016/j.molmet.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabacka M., Pierzchalska M., Dean M., Reiss K. Regulation of ketone body metabolism and the role of PPARalpha. International Journal of Molecular Sciences. 2016;17(12) doi: 10.3390/ijms17122093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metabolism. 2017;25(2):262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leone T.C., Weinheimer C.J., Kelly D.P. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S.S., Chan W.Y., Lo C.K., Wan D.C., Tsang D.S., Cheung W.T. Requirement of PPARalpha in maintaining phospholipid and triacylglycerol homeostasis during energy deprivation. The Journal of Lipid Research. 2004;45(11):2025–2037. doi: 10.1194/jlr.M400078-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez J.C., Gil-Gomez G., Hegardt F.G., Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. Journal of Biological Chemistry. 1994;269(29):18767–18772. [PubMed] [Google Scholar]
- 10.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura M.T., Yudell B.E., Loor J.J. Regulation of energy metabolism by long-chain fatty acids. Progress in Lipid Research. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Z., Xu D., Wang Z., Wang L., Han R., Wang Z. Hepatic PPARalpha function is controlled by polyubiquitination and proteasome-mediated degradation through the coordinated actions of PAQR3 and HUWE1. Hepatology. 2018;68(1):289–303. doi: 10.1002/hep.29786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y.T., Hu T., Arterburn M., Boyle B., Bright J.M., Emtage P.C. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. Journal of Molecular Evolution. 2005;61(3):372–380. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- 14.Pang Y., Dong J., Thomas P. Characterization, neurosteroid binding and brain distribution of human membrane progesterone receptors delta and {epsilon} (mPRdelta and mPR{epsilon}) and mPRdelta involvement in neurosteroid inhibition of apoptosis. Endocrinology. 2013;154(1):283–295. doi: 10.1210/en.2012-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You X., Lin Y., Hou Y., Xu L., Cao Q., Chen Y. PAQR9 Modulates BAG6-mediated protein quality control of mislocalized membrane proteins. Biochemical Journal. 2020;477(2):477–489. doi: 10.1042/BCJ20190620. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y., Huang M., Wang S., You X., Zhang L., Chen Y. PAQR11 modulates monocyte-to-macrophage differentiation and pathogenesis of rheumatoid arthritis. Immunology. 2021;163(1):60–73. doi: 10.1111/imm.13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S.C., Hardie D.G. AMPK: sensing glucose as well as cellular energy status. Cell Metabolism. 2018;27(2):299–313. doi: 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Inoki K., Kim J., Guan K.L. AMPK and mTOR in cellular energy homeostasis and drug targets. Annual Review of Pharmacology and Toxicology. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.E., Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53(11):2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 23.Le Bacquer O., Petroulakis E., Paglialunga S., Poulin F., Richard D., Cianflone K. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. Journal of Clinical Investigation. 2007;117(2):387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rui L. Energy metabolism in the liver. Comparative Physiology. 2014;4(1):177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habegger K.M., Heppner K.M., Geary N., Bartness T.J., DiMarchi R., Tschop M.H. The metabolic actions of glucagon revisited. Nature Reviews Endocrinology. 2010;6(12):689–697. doi: 10.1038/nrendo.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rix I., Nexoe-Larsen C., Bergmann N.C., Lund A., Knop F.K. In: Feingold K.R., Anawalt B., Boyce A., Chrousos G., de Herder W.W., Dhatariya K., editors. Endotext; South Dartmouth (MA): 2000. Glucagon physiology. [Google Scholar]
- 27.Zhang Y., Cui Y., Wang X.L., Shang X., Qi Z.G., Xue J. PPARalpha/gamma agonists and antagonists differently affect hepatic lipid metabolism, oxidative stress and inflammatory cytokine production in steatohepatitic rats. Cytokine. 2015;75(1):127–135. doi: 10.1016/j.cyto.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 28.Choi W.S., Lee J.J., Kim Y., Kim I.S., Zhang W.Y., Myung C.S. Synergistic improvement in insulin resistance with a combination of fenofibrate and rosiglitazone in obese type 2 diabetic mice. Archives of Pharmacal Research. 2011;34(4):615–624. doi: 10.1007/s12272-011-0412-9. [DOI] [PubMed] [Google Scholar]
- 29.Rull A., Geeraert B., Aragones G., Beltran-Debon R., Rodriguez-Gallego E., Garcia-Heredia A. Rosiglitazone and fenofibrate exacerbate liver steatosis in a mouse model of obesity and hyperlipidemia. A transcriptomic and metabolomic study. Journal of Proteome Research. 2014;13(3):1731–1743. doi: 10.1021/pr401230s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.