Abstract
In the budding yeast, Saccharomyces cerevisiae, replicators can function outside the chromosome as autonomously replicating sequence (ARS) elements; however, within chromosome III, certain ARSs near the transcriptionally silent HML locus show no replication origin activity. Two of these ARSs comprise the transcriptional silencers E (ARS301) and I (ARS302). Another, ARS303, resides between HML and the CHA1 gene, and its function is not known. Here we further localized and characterized ARS303 and in the process discovered a new ARS, ARS320. Both ARS303 and ARS320 are competent as chromosomal replication origins since origin activity was seen when they were inserted at a different position in chromosome III. However, at their native locations, where the two ARSs are in a cluster with ARS302, the I silencer, no replication origin activity was detected regardless of yeast mating type, special growth conditions that induce the transcriptionally repressed CHA1 gene, trans-acting mutations that abrogate transcriptional silencing at HML (sir3, orc5), or cis-acting mutations that delete the E and I silencers containing ARS elements. These results suggest that, for the HML ARS cluster (ARS303, ARS320, and ARS302), inactivity of origins is independent of local transcriptional silencing, even though origins and silencers share key cis- and trans-acting components. Surprisingly, deletion of active replication origins located 25 kb (ORI305) and 59 kb (ORI306) away led to detection of replication origin function at the HML ARS cluster, as well as at ARS301, the E silencer. Thus, replication origin silencing at HML ARSs is mediated by active replication origins residing at long distances from HML in the chromosome. The distal active origins are known to fire early in S phase, and we propose that their inactivation delays replication fork arrival at HML, providing additional time for HML ARSs to fire as origins.
In eukaryotic chromosomes, duplication of genetic information occurs during the S phase of the cell cycle and is coordinately regulated with the separation of the sister chromatids in mitosis (45). A chromosome initiates duplication at multiple DNA replication origins, and each origin is regulated to fire only once per S phase (16, 26, 47). Timing of initiation is also regulated within S phase, and DNA replication origins fire in a characteristic order (22). The temporal firing order often correlates with transcriptional activity: early-replicating regions of chromosomes are associated with active genes, and late-replicating regions are associated with silent genes (27, 28). Genetic elements that activate transcription are sometimes closely associated with active replication origins (15). Conversely, genetic elements that silence transcription are in some cases intimately associated with silent replication origins. It is not understood how DNA replication origins are silenced in chromosomes.
Genetic elements that function in cis to activate a DNA replication origin comprise the replicator. In the budding yeast, Saccharomyces cerevisiae, replicators can be isolated as genomic fragments that function as autonomous replicating sequence (ARS) elements in plasmids (64). ARS elements share a number of essential and important cis-acting determinants with the chromosomal replicators from which they were derived (18, 31, 32, 40, 57, 66). However, it remains puzzling why certain ARSs that mediate DNA replication in a plasmid are not detectably active as replication origins within the chromosome.
A number of ARS elements have been suggested to be inactive as replication origins in S. cerevisiae chromosome III (20, 54). Several such ARSs map near the HML locus, a transcriptionally silent mating-type locus on the left arm of the chromosome. ARS301 and ARS302 function in silencing transcription at HML (see below). ARS303 maps near ARS302 but is not essential for transcriptional silencing. Additional ARSs that are not detectably active as replication origins in the chromosome map at other locations, including near the transcriptionally active mating-type locus, MAT (48). The nature of the determinants responsible for the inactivity of replication origin function at HML ARSs in the chromosome is presently unknown.
Certain HML ARS elements are intimately associated with cis-acting elements called transcriptional silencers (7, 8). HML silencers E and I correspond to ARS301 and ARS302, respectively (39, 48). An ARS consensus sequence which interacts with the six-subunit origin recognition complex (ORC) and is essential for the initiation of DNA replication at active origins also has a role in maintenance of transcriptional repression via ORC binding at the silencer elements (2, 24, 42). In addition, binding sites for Rap1p and Abf1p, which stimulate replication initiation at some ARSs (41), contribute to ARS-mediated transcriptional silencing (11, 61). Four silent information regulator proteins, Sir1p to Sir4p, are essential for transcriptional repression (55), and they mediate interactions among ORC, Rap1p, and histones H3 and H4 to form a heterochromatin-like structure (29, 67, 68). Transcriptional silencers that flank the HMR locus on the right arm of chromosome III also contain ARSs but, unlike the ARSs at HML, HMR ARSs are active as replication origins in the chromosome (56, 57). At HML, all of the ARSs have been suggested to be inactive as origins (20). A pre-replicative complex (pre-RC) involving ORC can form at ARS301, the HML E silencer (58). This presents a paradox since the existence of a pre-RC is thought to reflect a potentially active replication origin, yet ARS301 is inactive as an origin.
The relationship between transcriptional silencing and inactive replication origins at HML ARSs has not been extensively studied. Strains with sir1 or sir4 mutations that relieved transcriptional repression showed no detectable initiation of replication from ARS301 and ARS302, the E and I silencers (20). The large number of genes involved in the transcriptional silencing process and the fact that the process shares key cis- and trans-acting components involved in the activation of replication origins indicates that further investigation is needed before firm conclusions can be drawn about why replication origins are inactive at HML. Mutations in SIR3 have not been examined but may be relevant since Sir3p has some unique features, including structural similarities with Orc1p and Cdc6p which are required for replication initiation (3) and an enhanced ability to propagate silenced chromatin (12, 53). Importantly, no cis-acting mutations that eliminate the transcriptional silencers (38) and no trans-acting mutations that selectively disrupt the transcriptional silencing function of ORC (19, 25) have been examined for their effects on replication origin activity at HML ARSs.
Other hypotheses have been considered to account for the inactivity of HML ARSs as chromosomal replication origins. One hypothesis is that some aspect of chromosome context, such as proximity to telomeres, is important. Telomeres are determinants of late replication timing at certain origins (23), and the HML ARSs are within 10 to 15 kb of the left telomere in chromosome III. However, no activity of HML ARSs was detected in a circular chromosome III derivative that lacked telomeres (20). Also, a strain containing an insertion of HML ARS301 near the MAT locus far from the telomere showed no replication origin activity at that ARS. Another hypothesis is that HML ARSs are specialized replication origins that are active in some special growth conditions or stage in the yeast life cycle (21). There is as yet no support for this hypothesis, but only one study has been performed. In meiosis, HML ARSs tested which have no origin activity in mitotic S phase are also not detectably active in premeiotic S phase (14).
Yeast mating type has marked effects on the left arm of chromosome III containing HML in terms of DNA recombination competence and chromatin structure (69, 70). MATa cells activate the entire left arm of chromosome III, while MATα cells inactivate the left arm, including HML. The effect of mating type in isogenic strains on replication origin activity at HML ARSs has not been previously examined. Certain HML ARSs reside near CHA1 (catabolism of hydroxy amino acids), whose expression is highly inducible in special growth conditions (5, 52, 65). CHA1 is normally transcriptionally repressed, but in special growth conditions that induce gene expression, a repressive chromatin structure present over the gene promoter region is disrupted (44). The hypothesis that certain HML ARSs are specialized replication origins that function only in growth conditions that induce CHA1 gene expression and open chromatin structure has not been tested.
ARS303 resides in a 1.4-kb region between HML and the CHA1 gene, and its function is not known. Sequences containing ARS303 are not required for HML transcriptional silencing. Unambiguous assessment of replication origin activity associated with ARS303 was previously not possible since the precise location of the ARS in the 1.4-kb region was unknown. Here we further localized and characterized ARS303 and found that it is near ARS302, the I transcriptional silencer, and in the process we discovered that it is closely associated with a new ARS, ARS320. We show that both ARS303 and ARS320 are competent as replication origins when inserted at a different location in chromosome III. At their native location, however, where both ARSs are closely clustered with ARS302, no replication origin activity is detected independent of yeast mating type, special growth conditions that induce the transcriptionally repressed CHA1 gene, trans-acting mutations that abrogate transcriptional silencing at HML (sir3, orc5), or cis-acting mutations that delete the E and I silencers (ARS301 and ARS302). Surprisingly, deletion of active replication origins at remote locations in the chromosome results in replication origin activity at the HML ARS cluster. Also, origin activity is seen at HML ARS301, previously known to form a pre-RC (58), but thought to be inactive as a replication origin. Our results show that replication origin silencing at HML ARSs is mediated by active replication origins located at long distances from HML. The distal active origins are known to fire early in S phase (54), and we propose that their deletion delays fork arrival at HML, providing additional time for HML ARSs to fire as origins.
MATERIALS AND METHODS
Reagents.
Restriction enzymes, T4 DNA ligase, Klenow fragment, and T4 DNA polymerase were obtained from New England BioLabs, Inc. [α-32P]dATP was purchased from Amersham International. Medium reagents were from Difco Laboratories and American Biorganics, Inc. 5-Fluoro-orotic acid (FOA) was from Toronto Research Chemicals, Inc. All other chemicals were purchased from Sigma Chemical Company.
Bacteria, plasmids, and yeast.
The Escherichia coli strain used for transformation and plasmid propagation was DH5α (BRL). The haploid Saccharomyces cerevisiae strain used for integrative transformation and the plasmid loss assay was YPH98 (MATa ade2-101 lys2-801 ura3-52 trp1-1 leu2-1) and was obtained from Philip Hieter (Johns Hopkins University). The following S. cerevisiae strains, obtained from James Broach (Princeton University), were used for two-dimensional (2-D) gel analysis: DMY1 (HMLα MATa ura3-52 leu2-3,112 ade2-1 lys1-1 his5-2 can1-100), DMY2 (DMY1; sir3::LEU2), DMY94 (DMY1; E+::URA3 I−Δ242), and DMY95 (DMY1; E−Δ79-113::URA3 I−Δ242) (38). S. cerevisiae JRY4554 (W303-1A; MATα orc5-1,2 HMR-SS) and JRY4556 (W303-1A; MATα orc5-1,3 HMR-SS) (W303-1A = MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100) (24), obtained from Jasper Rine (University of California, Berkeley), were also used in 2-D gel analysis. CHA1 gene induction experiments were done in strains X2180-A (MATa SUC2 mal mel gal2 CUP1) and X2180-B (MATα SUC2 mal mel gal2 CUP1), which were purchased from the Yeast Genetic Stock Center (University of California, Berkeley). YIp5 plasmids containing DNA fragments of yeast chromosome III (49) were gifts from Carol Newlon (UMD-New Jersey Medical School). The yeast strain with the ORI305 replicator deleted, Δ305, is a derivative of the YPH98 strain (YRH80/433) and was constructed as previously described (31). The yeast strain with both ORI305 and ORI306 replicators deleted, Δ305/Δ306, was obtained by transplacement of pARSΔ306 into homologous sequences near ARS306 in the Δ305 strain by using methods previously described (31). Plasmid pARSΔ306 was constructed by M. L. He by deleting a 220-bp HindIII/BglII fragment containing ARS306 (18) from pARS306. The latter plasmid is a YIp5 derivative (URA3 selectable marker) which contains ARS306 in a 1.2-kb EcoRI-BsrGI fragment of the chromosome III C1G region (49). Yeast transformation was performed with XhoI-linearized pARSΔ306, and integrants were selected after growth on synthetic medium in the absence of uracil (see below). Homologous recombinants that evicted the integrated plasmid sequences were obtained after selection against URA3 by growth on medium containing FOA (31). The correct homologous recombinants harboring a deletion at the ORI306 replicator in the chromosome were identified by restriction enzyme mapping after Southern blotting. The absence of origin activity at the Δ306 locus was confirmed by 2-D gel electrophoresis of replication intermediates (see below).
Bacteria were grown at 37°C, in Luria-Bertani medium supplemented with ampicillin (50 μg/ml). Plasmid DNA was obtained from E. coli DH5α cells by the boiling method, and minipreps of yeast genomic DNA were carried out as previously described (31). Yeast cells were grown at 30°C either in complete medium, yeast-peptone-dextrose (YPD), or in synthetic dextrose minimal medium (SD) containing supplements (see below) as described by Sherman et al. (59). SD was supplemented with adenine sulfate (20 mg/liter), lysine (30 mg/liter), leucine (30 mg/liter), and tryptophan (20 mg/liter), and this was called supplemented minimal medium (SMM). For the FOA selection process, SMM-agar plates were further supplemented with uracil (50 mg/liter) and FOA (1 g/liter).
Linker substitution mutations by PCR.
Linker substitution mutations in ARS303 and ARS320 were generated by using two mutant primers for each consensus sequence, one on the top and one on the bottom strand. Mutant primers contained a KpnI sequence (lower cased below) positioned so that it would substitute most of the potential ARS consensus sequence whose function was being tested. The sequences of mutant primers were as follows: 303KL1, 5′-TCGGCAgggtacccATACCTTAGATGTTACCAGCTGGGAA-3′; 303KL1op, 5′-AGGTATgggtacccTGCCGACATTTGCAGCTCTTTCATCA-3′; 320KL2, 5′-CCTATAgggtacccTATGTTATGTTATGTTTGCTATGACT-3′; and 320KL2op, 5′-AACATAgggtacccTATAGGTTGTACTTTGTCAATAGAAA-3′. Two outside primers were also used that correspond to the HindIII and PstI restriction sites of the HindIII/BamHI restriction fragment (see Fig. 1). In each mutagenesis, mutant primers were used with corresponding outside primer to generate two DNA fragments, each with a newly introduced KpnI restriction site on one end. Each PCR reaction was performed by using 8 U of AmpliTaq polymerase (Perkin-Elmer), a deoxynucleoside triphosphate mix in a final concentration of 0.25 mM, 10 ng of template DNA, and 10 pmol of each primer. YIp5 vector with HindIII/BamHI fragment containing ARS303/ARS320 (see Fig. 1, map) was used as template DNA. After PCR reaction, products were cut with KpnI and the appropriate restriction enzyme for the other end (HindIII or PstI), ligated together (via the KpnI site), and cloned back into YIp5 or pVHA vectors. Correct plasmid constructs were confirmed by restriction enzyme mapping of the DNA. The DNA sequence of the insert was determined to confirm the mutations and to verify that no undesired mutations were introduced by PCR.
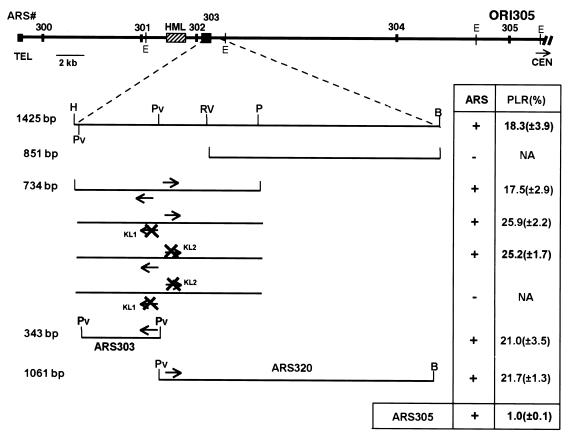
FIG. 1.
Localization of ARS303 and discovery of ARS320. Subcloning and mutational analysis were used in localization of ARS303. The top diagram shows a map of the first 40 kb of the left arm of chromosome III drawn to scale by using DNA sequence information (50). On the map are previously known ARS elements (numbered 300 to 305), the silent mating-type locus HML, the left telomere, and some relevant restriction sites. The HindIII/BamHI fragment (1,425 bp) is blown up below the chromosome III map, with relevant restriction sites and potential ARS consensus sequences (arrows) shown. Introduced KpnI linkers (KL1 and KL2) that disrupt two potential ACSs are shown as a crossed arrows. The ARS function of each subclone is indicated as “+” for active ARS derivatives or “−” for inactive ARS derivatives. Percent PLR determinations are average values from at least three different experiments. E, EcoRI; H, HindIII; B, BamHI; Pv, PvuII; RV, EcoRV; P, PstI.
High-frequency transformation (HFT) assay.
For the purpose of determining whether a particular fragment has ARS activity, several different fragments (see Fig. 1) were subcloned into the YIp5 vector in appropriate restriction sites. The resulting plasmids were transformed into yeast strain YPH98 by using a lithium-acetate procedure and plated on selective medium (SMM with no uracil added) as described earlier (31). The resulting transformants were scored as Ars+ if they could be passaged after being streaked on additional selective plates and grown in selective liquid medium.
Mitotic stability and plasmid loss rate assays.
For these assays, different ARS derivatives were cloned in a centromere-containing vector, pVHA, and transformed into the YPH98 strain. Assays were performed as described previously (31) but with some modifications. Single transformants were inoculated in 3 ml of selective, liquid minimal medium (SMM with no uracil) and grown for 22 h on 30°C. Cultures were plated on nonselective (YPD) and selective plates (SMM with no uracil) at dilutions that gave rise to ∼200 colonies on nonselective plates, and the initial percentage of plasmid-containing cells (I) under selection was determined. The same cultures were also used to inoculate 3 ml of nonselective, YPD medium, at 104 cells/ml. Cultures were grown for 12 generations at 30°C and plated on nonselective and selective plates as described above to determine the final percentage of plasmid-containing cells (F) after growth in the absence of selection for a specified number of generations (N). For transformants with extremely slow growth rates, twice and 200 times the amount of cells were plated on the selective plates to determine the I and F values, respectively. Mitotic stabilities (the percentage of plasmid-containing cells) before release of selection and after growth in nonselective medium correspond to I and F, respectively. The rate of plasmid loss per generation is expressed as 1 − (F/I)1/N.
Transplacement of HML ARS sequences into a different location of chromosome III.
For the transplacement of ARS303, ARS320, or both to the chromosomal location of ORI305, a chimeric plasmid was made by inserting an appropriate fragment (343-bp PvuII fragment for ARS303, 1,061-bp PvuII/BamHI fragment for ARS320, and 1,425-bp HindIII/BamHI fragment for ARS303/ARS320) in place of the ORI305 replicator between the SacI and ClaI sites of plasmid p305BP (31). After the SacI/ClaI restriction digest of p305BP, the larger fragment containing the vector was gel separated from the 335-bp fragment containing the ARS element and, after treatment with T4-DNA polymerase (producing blunt ends), used in a ligation reaction with each of three fragments described above. After identification with the appropriate restriction enzymes, correct chimeric plasmids were linearized within sequences flanking the ORI305 replicator to transplace HML ARSs into the Δ305 locus by integrative transformation and homologous recombination as previously described (31). The desired homologous recombinants were identified by the correct size restriction fragments seen after Southern blotting and hybridization with a 32P-labeled DNA probe specific for the region of interest.
2-D gel electrophoresis of DNA replication intermediates.
Replication intermediates from yeast cells were isolated as previously described (33). Total genomic DNA was isolated from exponentially growing cultures containing approximately 2 × 107 cells/ml and, after purification on by equilibrium sedimentation in a CsCl density gradient, were digested with appropriate restriction enzyme(s). The replication intermediates were enriched by binding them to a benzoylated, naphthoylated DEAE-cellulose column and elution with 1.8% solution of caffeine. The replication intermediates in the caffeine eluant were analyzed by 2-D gel electrophoresis (10) with minor modifications. In the first-dimension gel, electrophoresis was carried out for 22 h at 1 V/cm in 0.4% agarose in Tris-acetate-EDTA buffer containing 0.1 μg of ethidium bromide per ml. Approximately 8- to 10-cm-long lanes were cut from the first-dimension gel and transferred into the second-dimension gel, which consisted of 1% agarose in Tris-borate-EDTA buffer with 0.5 μg of ethidium bromide per ml. Second-dimension gels were run for 7 to 9 h at 4.5 V/cm at 4°C. The DNA was transferred from the gel to a nylon membrane (GeneScreen Plus; DuPont) by using a pressure blotter (Stratagene), and hybridizations with a 32P-labeled DNA probe specific for the region of interest were carried out as described earlier (39a). Radioactive signals were detected by using a PhosphorImager (Molecular Dynamics).
RESULTS
The map at the top of Fig. 1 shows part of the left arm of chromosome III with all of the ARS elements known prior to this work, as well as some landmarks, including a transcriptionally silent mating-type locus, HML, and the left telomere. Of the six ARS elements shown (numbered 300 to 305), only one, ARS305, maps physically with origin activity in the chromosome (14, 34, 49). ARS305 contains functional components of a chromosomal replication origin, ORI305 (31, 32). Other replication origins in the left arm of chromosome III, such as ORI306 and ORI307, also map physically with ARS elements and require ARS components for chromosomal origin function (18, 66, 71). To understand why ARSs localized near HML are inactive as chromosomal replication origins, we first localized ARS303 and then examined its replication activity in the chromosome under different experimental conditions and chromosomal contexts.
Localization of ARS303 and discovery of a new ARS element near HML.
ARS303 was originally defined as a 1.4-kb HindIII-BamHI chromosomal fragment, which is Ars+ in the HFT assay and which maps near the HML locus (49). Since the size of some ARS elements in S. cerevisiae can be reduced to ∼0.1 to 0.2 kb with retention of full function (43), we performed a series of subcloning and mutational analyses to further localize cis-acting genetic elements responsible for ARS activity within the original clone.
Three different, overlapping fragments, HindIII-BamHI (H-B; 1,425 bp), EcoRV-BamHI (RV-B; 851 bp), and HindIII-PstI (H-P; 734 bp) were subcloned into the plasmid pVHA (Fig. 1, maps). All three plasmids were tested for the ability to provide HFT of a Ura− yeast strain (YPH98) under selection for the URA3 marker in the plasmid. As shown in Fig. 1 in the column labeled “ARS,” the fragments H-B and H-P are Ars+ in pVHA and the RV-B fragment is Ars−. These data further localize ARS activity within the 734-bp H-P fragment.
Computer analysis of the DNA sequence of the H-P fragment revealed many matches to the ARS consensus sequence (ACS) (11/11, 10/11, or 9/11). The occurrence of a broad easily unwound region, which is 3′ to the T-rich strand of the ACS, has been demonstrated for several ARS elements in S. cerevisiae (46). A combined search for both features (ACS match and a low free energy of unwinding 3′ to the ACS T-rich strand [45a]) suggested two matches to the ACS to be candidates for the essential ACS: one on the bottom strand, starting from position 346, and one on the top strand, starting from position 448 of the H-P fragment. These two potential ACSs were individually mutated by replacing them with a KpnI linker (KL1 and KL2; Fig. 1). When introduced separately, none of the mutations abolished the ARS activity of the H-P fragment. Surprisingly, only simultaneous linker substitution of both consensus sequences completely abolished ARS activity (Fig. 1). This result, together with the fact that two consensus sequences are in a tail-to-tail orientation, strongly suggested the presence of two independent ARS elements within the analyzed fragment.
Two ARS elements were identified by additional subcloning. Nonoverlapping fragments, PvuII-PvuII (Pv-Pv; 343 bp) and PvuII-BamHI (Pv-B; 1,061 bp) inserted individually into the pVHA vector each provided HFT of yeast cells, confirming the existence of two independent ARS elements within the originally cloned H-B fragment. We assigned the previously existing name, ARS303, to the ARS element with the ACS 5′-ATTTATATTTT-3′ in the bottom (minus) strand. We designated the new ARS element with the ACS 5′-TTTTATGTTAT-3′ in the top (plus) strand as ARS320 (Fig. 1). The ARS303 essential sequence resides in a perfect match to the ACS, and in ARS320 the essential sequence it resides in a 10/11 match with overlapping 9/11 matches. No ARS activity was detected when the same linker substitutions of these ACS matches were tested in the separated ARS elements (data not shown), confirming an essential role for the substituted sequences in plasmid replication.
A quantitative plasmid loss rate (PLR) assay was performed on the ARS+ derivatives to assess their replication efficiency. The assay is independent of possible effects of the ARS element on plasmid segregation or on selectable marker expression since the pVHA vector contains a centromere to promote proper segregation and the assay is done in the absence of selection for the URA3 marker. The lower the PLR, the higher the replication efficiency of the ARS. As seen in Fig. 1 in the column labeled PLR, all of the ARS derivatives tested have a high PLR compared to an efficient replicator, ARS305. Thus, ARS function is inefficient for the separated ARS303 and ARS320 elements, as well as for the various restriction fragments or mutant derivatives of the two ARS elements tested (Fig. 1). Attempts to further localize ARS320 indicated that the PvuII-PstI fragment was Ars+, but the PLR was not determined.
Our subcloning and mutational analyses of ARS303 served to localize the ARS element and to define the essential ACS. The analyses also revealed the existence of a new, independent ARS element, ARS320, within the originally isolated 1.4-kb Ars+ fragment. While certain other ARSs are known to be closely spaced (35, 60), ARS303 and ARS320 are the most closely spaced ARS elements identified so far in S. cerevisiae chromosomes. Their essential ACS elements are only 102 bp apart. On average, ARSs are spaced every 14 kb in chromosome III (48). ARS303 and ARS320, together with ARS302 (Fig. 1, map at top), form a dense cluster of three ARSs in ∼1 kb of DNA. We refer to this region as the HML ARS cluster. The possible functional significance of this unusual clustering of ARS elements is presently unclear.
HML ARS elements ARS303 and ARS320 are inactive as replication origins in the chromosome independent of mating type.
Precise localization of essential cis-acting elements of ARS303 and ARS320 enabled us to optimize detection of potential replication origin activity associated with these ARSs in the chromosome. Origin firing creates replication bubbles whose detection by 2-D gel electrophoretic analysis is favored by positioning the essential core of the ARS in the central region of the restriction fragment. Therefore, we examined the origin activity at the HML locus by using a restriction enzyme digest that would appropriately center the essential core of ARS303 and the newly discovered ARS320 within the fragment used in 2-D gel analysis. Yeast mating type has profound effects on the left arm of chromosome III in terms of DNA recombination competence and chromatin structure (69, 70). MATα cells inhibit recombination along the entire left arm, while MATa cells activate recombination in the same region (70). The effect of mating type in isogenic strains has not been examined previously. Thus, we examined the two yeast mating types in isogenic strains for replication origin activity near HML.
Replication bubbles resulting from initiation of DNA replication within the restriction fragment analyzed are detected as a high rising arc (bubble arc) after neutral-neutral 2-D gel electrophoresis (10) and probing with appropriate DNA probe (Fig. 2A, Ori+). Passive replication originating outside of the restriction fragment analyzed results in fork-shaped molecules with a single replication fork, and these are detected as a distinct Y-arc (Fig. 2A, Ori−).
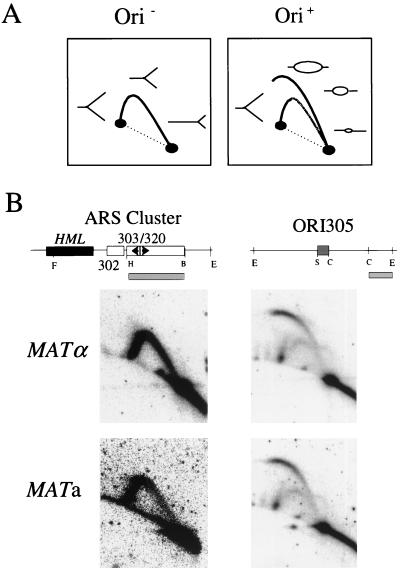
FIG. 2.
Replication origin activity is not detectable at the HML ARS cluster in MATα or MATa cells. The HML ARS cluster containing ARS302, ARS303, and ARS320 was analyzed by 2-D gel electrophoresis in both mating types of S. cerevisiae X2180, MATα and MATa. (A) Schematic drawing of replication intermediates that can be detected when analyzed restriction fragment is passively replicated by forks coming from the outside (Ori−) or when an active replication origin is centrally located within the analyzed fragment (Ori+). The arc in the Ori− example is called a Y arc. The right half of the arc is the early Y arc, and the left half is the late Y arc. The high rising arc in the Ori+ example is called a bubble arc. (B) 2-D gel analysis of replication intermediates from the HML region of chromosome III (left). The map above shows a portion of the left end of chromosome III (ca. 4.5 kb) containing the HML locus and the ARS302/ARS303/ARS320 region, with relevant restriction sites. The probe used in this analysis is shown as a shaded box below the map. FspI/EcoRI genomic digest produces a 3.9-kb fragment with the three ARS elements positioned in a central region to favor detection of replication bubbles. As a positive control, the same blot was reprobed for ORI305. The map above shows the EcoRI fragment of chromosome III containing ORI305 with relevant restriction sites and the probe used in this analysis (shaded box).
FspI and EcoRI digestions produce a 3.9-kb fragment at the HML locus, with the ACS elements of ARS303 and ARS320 within the central region of the fragment (Fig. 2B, arrows in left map). This fragment overlaps the HML ARS cluster, and so it includes ARS302 which has been suggested to be nonfunctional as a chromosomal origin (20). Even in a restriction fragment chosen to optimize detection of potential replication bubbles, no bubble arc was seen after 2-D gel analysis (Fig. 2, MATα, left panel). In order to test whether this result is due to artifactual loss of replication bubbles caused by breakage, the same blot was reprobed to detect ORI305, which is an efficient replication origin (31). An FspI/EcoRI genomic digest produces 5.8-kb EcoRI/EcoRI fragment containing ORI305 in a central position (Fig. 2B, right map). Abundant replication bubbles are detected at ORI305 (Fig. 2, MATα, right panel), indicating that the absence of detectable replication bubbles at the HML locus in the MATα strain is not due to extensive losses caused by breakage. The results extend the findings of Dubey et al. (20) for ARS302 to a different MATα strain. We conclude that in the MATα strain analyzed no ARSs in the HML ARS cluster (ARS303, ARS320, and ARS302) are detectably active as chromosomal replication origins.
In addition, the same 2-D gel analysis was performed in an isogenic strain with the opposite mating type, MATa. No replication initiation could be detected at the HML ARS cluster (Fig. 2, MATa, left panel), while efficient initiation was detected at ORI305 (Fig. 2, MATa, right panel). These results indicate that ARS303, ARS320, and ARS302 are not detectably active as chromosomal replication origins in the isogenic MATa strain. Each mating type of another pair of isogenic yeast strains (YPH98 and YPH102) was also analyzed, with the same results (data not shown). The persistence of origin inactivity independent of the mating type in isogenic strains suggests that inactivity of these ARSs as chromosomal replication origins does not depend on properties of the left arm of chromosome III, such as differences in DNA recombination competence or chromatin structure known to be specified by the two different mating types of yeast cells (69, 70).
HML ARS elements ARS303 and ARS320 are active as replication origins when inserted at a different location in the chromosome.
It is not known whether any HML ARS is competent to function as a chromosomal origin outside their native location near HML. In order to test this, we analyzed the replication origin activity of copies of ARS303 and ARS320 transplaced into a different location in chromosome III. ARS303 and ARS320 were inserted in place of the ORI305 replicator (Fig. 3A). A variety of mutations that affect ORI305 activity at this chromosomal locus have been previously characterized (31, 32). A DNA region larger than ones known to be sufficient for ORI305 activity in the chromosome was deleted (Fig. 3A, Δ305). This chromosomal locus was chosen for the transplacement of these HML ARSs for two primary reasons. First, deletion of DNA regions smaller than that in Δ305, and even point mutations in the essential ACS of the replicator, have been shown to completely abolish ORI305 activity in the chromosome (31). Thus, the Δ305 locus has no cryptic origin function. Second, the locus is permissive for origin function, not only for ORI305 but also for active origins from other yeast chromosomes (42a). Therefore, insertion of potential replicators into the Δ305 locus avoids any negative effects on origin function that might be encountered after transplacement into an uncharacterized chromosomal location.
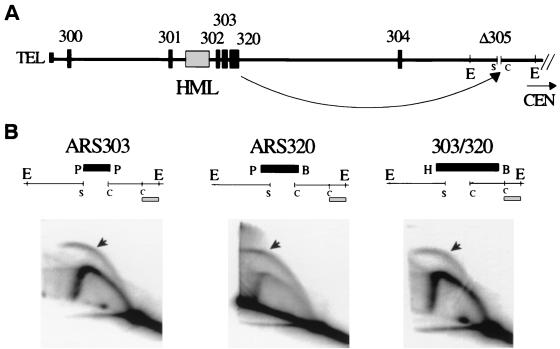
FIG. 3.
HML ARS303 and ARS320 are active as replication origins when inserted at a different location in the chromosome. (A) The map shows first 40 kb of the left arm of chromosome III with native locations of HML ARSs, as well as the position of their transplacement at the location of inactive ORI305 (Δ305). (B) Maps above each 2-D gel show the details of the transplacement in each strain: ARS303-containing fragment (PvuII/PvuII, 343 bp) (left), ARS320-containing fragment (PvuII/BamHI, 1,061 bp) (middle), or ARS303/ARS320-containing fragment (HindIII/BamHI, 1,425 bp) (right) were transplaced into the Δ305 locus between SacI and ClaI restriction sites. Replication origin activity of ARS303, ARS320, and ARS303/ARS320 were analyzed by 2-D gel electrophoresis. EcoRI genomic digest (producing 5.8-kb fragment for ARS303 transplacement, 6.5-kb fragment for ARS320 transplacement, and 6.9-kb for ARS303/ARS320 transplacement) was used for the 2-D gel analysis. The ClaI/EcoRI fragment was used as a probe (indicated as hatched box below each map).
Three new strains with ARS303, ARS320, or ARS303/ARS320 in place of the ORI305 replicator in the chromosome were confirmed by Southern blot analysis (Fig. 3B, maps, and data not shown). Chromosomal origin activity at the Δ305 region in these three strains was analyzed by 2-D gel electrophoresis. As seen in Fig. 3B (2-D gel panels), bubble arcs, indicative of active replication origins, were detected in all three strains. The data show that ARS303 and ARS320, when present individually or together in their natural closely spaced configuration, have the ability to initiate DNA replication in the chromosomal context of the Δ305 locus.
Our data show for the first time that two ARSs, ARS303 and ARS320, which are inactive as replication origins at their native loci near HML, can function as replication origins when transplaced into a different location of the same chromosome. These findings suggest, as one possibility, that replication origin activity of ARS303 and ARS320 at the HML locus is somehow silenced.
Inactivity of the HML ARS cluster is independent of cis elements and trans-acting factors that silence transcription.
The existence of common functional elements between transcriptional silencing and replication initiation suggests a functional interdependence between the two processes. We examined the effects of mutations in certain cis elements and trans-acting factors that participate in transcriptional silencing for their effects on replication origin activity at the HML ARS cluster.
Deletions that removed the cis-acting silencer elements E (ARS301) and I (ARS302) were tested. These elements bind several proteins, including ORC, Rap1p, and Abf1p, that contribute to transcriptional silencing at HML and to replication initiation in other chromosomal contexts. The silencer elements also contribute to establishing a heterochromatin-like structure at and near HML. Deletion of both the E and I elements abrogates transcriptional silencing at HML (38). A disruption of the SIR3 gene, sir3::LEU2, that abrogates silencing of HML transcription was also examined (38). Sir3p functions in transcriptional silencing via binding Sir4p, Rap1p, and histones to condense chromatin structure (13, 30, 68). We also examined two ORC mutants, orc5-1,3 and orc5-1,2, which are competent for the initiation of DNA replication but defective in transcriptional silencing (25).
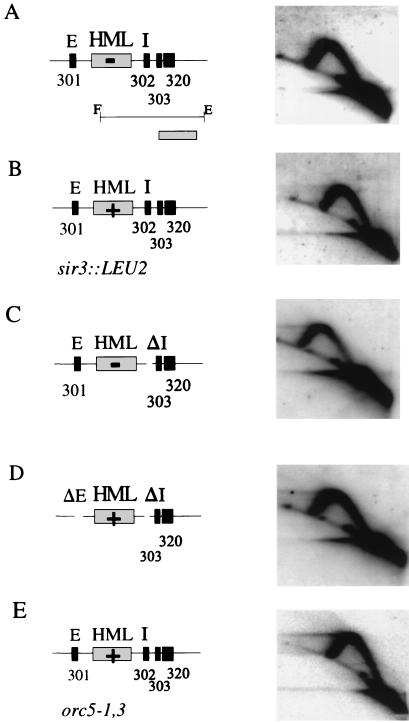
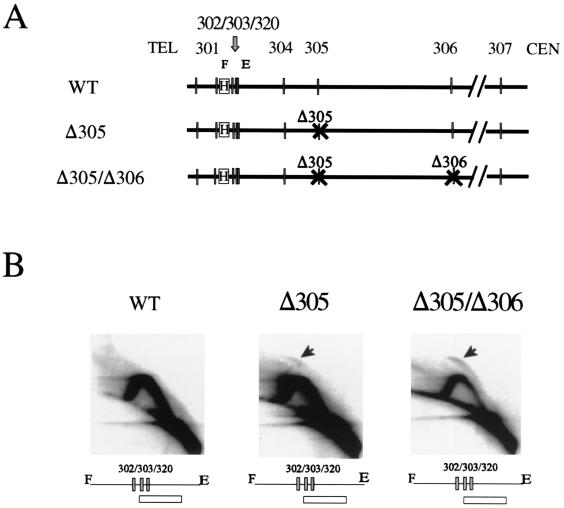
The parental strain used for construction of the E and I silencer mutations and the sir3 mutation was used as a control. In the wild-type parental strain, the HML ARS cluster containing ARS302, ARS303, and ARS320 shows a Y arc indicative of passive replication, but no bubble arc (Fig. 4A). Thus, no replication origin activity is detected at the HML ARS cluster in the parental strain, a finding consistent with the results obtained above in other wild-type strains. Also, Y arcs, but no bubble arcs, were seen in strains with trans-acting mutations in sir3 (Fig. 4B), orc5-1,2 (not shown), orc5-1,3 (Fig. 4E), or cis-acting mutations that delete either the I silencer alone (Fig. 4C) or both the E and I silencers (Fig. 4D). Thus, in all of the strains tested, ARS303 and ARS320 remain inactive as replication origins. In addition, when present, ARS302 also remains inactive (Fig. 4A, B, and E). Several of these mutations are known to abrogate transcriptional silencing at HML (Fig. 4, HML+). Also, mutations in the SIR1 or SIR4 genes that abrogate transcriptional silencing at HML showed no replication origin activity at ARS302 in the chromosome (20). Thus, in a variety of strains with mutations that derepress transcription at HML, the ARSs ARS303, ARS320 and, when present, ARS302 are silent as replication origins.
FIG. 4.
Origin inactivity at the HML ARS cluster is independent of mutations that abrogate transcriptional silencing. Five different strains were used to analyze chromosomal origin activity at the HML locus. The maps in all panels show the position of the HML locus, the E and I silencer elements, the ARS elements, and the transcriptional status of HML (−, silent; +, active). (A) DMY1, the parental strain. (B) DMY2, strain with a mutation disrupting the SIR3 gene (shown as sir3::LEU2). (C) DMY94, strain with the I silencer deleted (ΔI). (D) DMY95, strain with E and I silencers deleted (ΔEΔI). (E) JRY4556, strain with a mutation in ORC5 (orc5-1,3) which is defective in transcriptional silencing but competent in replication. Replication intermediates within the 3.9-kb FspI/EcoRI fragment (see map in panel A) were detected by using the HindIII/BamHI fragment as a probe (shown as hatched box in panel A). 2-D gels show no replication bubble at HML ARS303, ARS320, and ARS302 (where present) in all five strains. HML, silent mating-type locus; E and I, silencer elements; 303, ARS303; 320, ARS320; 302, ARS302; 301, ARS301.
Replication origin silencing at the HML ARS cluster is unaffected by special growth conditions that induce expression of the neighboring CHA1 gene.
Replication origins are sometimes located near genes that are transcriptionally active or whose transcription is induced by special growth conditions or during development (4, 36). One possibility is that the normally silent origins at the HML ARSs are specialized replication origins that are activated in the growth conditions that result in the induction of expression at the neighboring gene, CHA1 (catabolism of hydroxy amino acids). CHA1 encodes the enzyme l-serine (l-threonine) dehydratase, which permits growth of S. cerevisiae in conditions where the hydroxy amino acids are the only source of nitrogen (5). In such a growth condition, CHA1 mRNA levels are among the most abundant of all chromosome III transcripts (65), while in standard growth conditions the mRNA is not detectable (52). The CHA1 gene is normally transcriptionally repressed, and the special growth conditions induce CHA1 expression by disrupting a positioned nucleosome which occludes the promoter region for gene transcription (44).
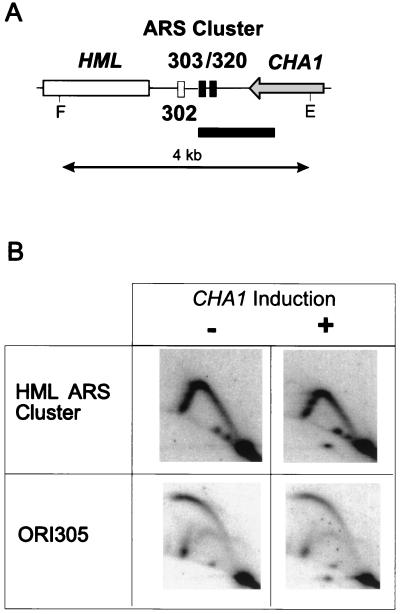
To test for possible replication origin activation at HML ARS cluster in this special growth condition, replication intermediates were isolated from S. cerevisiae X2180-B (MATα) grown in synthetic minimal media without or with l-serine as the sole nitrogen source. Northern blot analysis confirmed the induction of CHA1 mRNA (data not shown). 2-D gel electrophoresis analysis of DNA replication intermediates showed a Y arc but no bubble arc at the HML ARS cluster region containing ARS302, ARS303, and ARS320, indicating that this region remains inactive as a replication origin in this special growth condition (Fig. 5B, HML ARS cluster). Similar results were seen for an isogenic MATa strain (X2180-A; data not shown). The absence of detectable replication bubbles at the HML ARS cluster is not due to their artifactual breakage under the special growth conditions of CHA1 induction, since the same membrane reprobed to detect replication intermediates at ORI305 shows a strong bubble-arc signal (Fig. 5B, ORI305). We conclude that the HML ARS cluster is not used as a replication origin under the special growth conditions required for induction of the neighboring CHA1 gene.
FIG. 5.
Effect of CHA1 induction on chromosomal replication activity of the HML ARS cluster. Replication activity at the HML ARS cluster region was analyzed in special growth conditions where expression of the neighboring CHA1 gene was induced by more than 100 fold. (A) Map of relevant locus. HindIII/BamHI fragment used as a probe in 2-D gel analysis is shown as black box below the map. (B) 2-D gel analysis of replication intermediates at the HML locus and at ORI305 (positive control for detection of replication bubble). F, FspI; E, EcoRI.
Activation of silent origins in the HML ARS cluster after deletion of replication origins at remote locations in the chromosome.
The HML region is duplicated by replication forks that originate from early firing origins at remote locations in chromosome III (54). ORI305 is the origin closest to HML (Fig. 6A, WT) and is one of the earliest firing origins on chromosome III. ORI305 is a highly efficient replication origin, and 2-D gel analysis shows that the origin fires in practically every S phase in a variety of yeast strains (31, 34; the present study). The next closest origin in the chromosome is ORI306 (Fig. 6A, WT), which is also early firing and highly efficient (54, 71). Inactivation of the closest origins is expected to move the source of replication forks farther away from HML, increasing the time it takes for replication forks to reach the HML ARS cluster. We wondered whether inactivating ORI305 and ORI306 would affect replication origin silencing at HML.
FIG. 6.
2-D gel analysis of the HML ARS cluster in S. cerevisiae strains with or without deletion of the closest replication origins. ARS303, ARS320, and ARS302 were analyzed by 2-D electrophoresis in a wild-type (WT) strain and in two mutant strains deleted for ORI305 (Δ305) or both ORI305 and ORI306 (Δ305/Δ306). (A) Map with the details of the left portion of chromosome III in each strain. The box marked “H” represents the HML locus. (B) 2-D gel analysis of all three strains. The same restriction digest, FspI/EcoRI, was used as in previous experiments (see the maps below each column), producing a 3.9-kb fragment at the HML locus. The HindIII/BamHI fragment (1.4 kb) was used as the probe (shown as an open box below the maps). Arrows in 2-D gels point to a bubble arc resulting from replication origin activity at the HML ARS cluster.
Deletion of ORI305 or ORI306, resulting in the strains Δ305 or Δ306, respectively, is known to inactivate replication origin function at the respective loci (18, 31). We first tested whether inactivating ORI305 and ORI306 in this way affected origin silencing at the HML ARS cluster. Replication intermediates were isolated from the parental strain (Fig. 6A, WT) and two mutant strains (Fig. 6A, Δ305 and Δ305/Δ306), and then the DNA was digested with FspI and EcoRI and subsequently analyzed by 2-D gel electrophoresis. In mutant strains where one or two active origins were deleted, replication initiation occurred at the HML locus, as evidenced by the detection of bubble arcs (Fig. 6B, Δ305 and Δ305/Δ306). Upon close inspection, a bubble arc is just barely detectable in the strain Δ305 with ORI305 deleted. The frequency of initiation was greater in the strain deleted for both ORI305 and ORI306 (Δ305/Δ306) compared to the strain deleted for ORI305 alone, as evidenced by a stronger bubble arc signal accompanied by a weaker Y-arc signal. The origin at the HML ARS cluster functions at a low level, as indicated by the low ratio of bubble arc to early-Y-arc signals. The results show that replication origin function is clearly detectable at the HML ARS cluster after the deletion of ORI305 and ORI306, two known early-firing origins that are the closest to HML in the chromosome.
A silent origin at ARS301, but not at ARS304, is activated after deletion of active origins at remote locations in the chromosome.
We also tested whether ARS301 and ARS304 on the left arm of chromosome III (see map in Fig. 1) would be active replication origins at their native locations after deletion of ORI305 and ORI306. Both ARS301 and ARS304 are active on a plasmid but are inactive as replication origins at their native locations in the chromosome (14, 20). Paradoxically, ARS301 is known to form a prereplication complex in the chromosome (58), despite its inactivity as an origin. Genomic DNA from all three strains of yeast (WT, Δ305, and Δ305/Δ306) was isolated, digested with FspI (for ARS301) and FspI/BglI (for ARS304), and analyzed by 2-D gel electrophoresis.
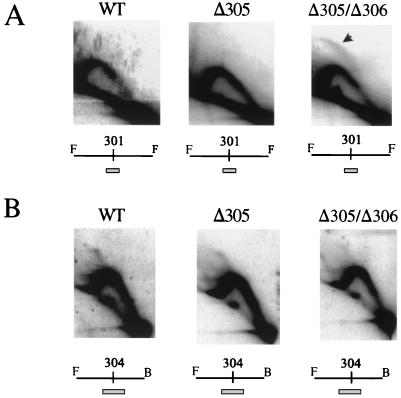
ARS301, the HML E silencer, showed no detectable bubble arc in the strain where only ORI305 was inactivated by deletion (Fig. 7A, Δ305 strain), in contrast to a barely detectable bubble arc seen at the HML ARS cluster (Fig. 6B). However, when both ORI305 and ORI306 were deleted, replication origin activity was detected within the genomic DNA fragment containing ARS301, as evidenced by the appearance of a bubble arc (Fig. 7A, Δ305/Δ306 strain, arrow). The origin functions at low levels, as indicated by the low ratio of bubble arc to early-Y-arc signals.
FIG. 7.
2-D gel analysis of ARS301 and ARS304 regions in S. cerevisiae strains with or without deletion of the closest replication origins. Replication intermediates at silent origins at ARS301 and ARS304 in chromosome III were analyzed by 2-D gel electrophoresis. (A) 2-D gel analysis of the ARS301 region in three different strains (columns marked WT, Δ305, and Δ305/Δ306). Genomic DNA was digested with FspI producing a 3.9-kb genomic fragment containing ARS301 (shown on the maps below each 2-D gel picture). The SacI/EcoRI fragment (463 bp) was used as a probe (shown as a box below the maps). The arrow indicates the bubble arc resulting from the initiation of replication within the fragment. (B) 2-D gel analysis of ARS304 region in same three strains of yeast. FspI and BglI restriction enzymes were used to generate the 3-kb genomic fragment containing ARS304 in a central position (maps below each 2D gel). The 0.8-kb XbaI/PacI fragment was used as a probe (shown as a box below the maps). No bubble arcs could be detected in this analysis, indicating that ARS304 does not become detectably active under these experimental conditions.
ARS304 is located more than 10 kb from HML, in between HML and ORI305 (Fig. 6A, maps). No detectable replication origin activity was associated with ARS304 in the parental strain (Fig. 7B, WT) or when ORI305 was deleted (Fig. 7B, Δ305). In contrast to the HML ARS cluster and to ARS301, no origin activity was seen at ARS304 in the mutant strains even when both ORI305 and ORI306 were deleted (Fig. 7B, Δ305/Δ306). In all three strains, only Y arcs are seen, indicating that ARS304 is passively replicated. No traces of bubble arcs are detected even after gross overexposure of the Y-arc signals (data not shown). ARS304 may be inactive as an origin in a chromosome, or it may be silenced by a mechanism that differs from the mechanism that silences the HML ARS cluster and ARS301.
Overall, our results show that silent origins at the HML locus, including the HML ARS cluster and ARS301, function as active replication origins after deletion of two known early-firing origins from remote locations in the chromosome. In contrast, no origin activity was detectable at ARS304 in the chromosome.
DISCUSSION
The inactivity of various HML ARSs as DNA replication origins in the chromosome has long been puzzling since ARS elements are active replication origins in plasmids. Here we localized two HML ARSs, ARS303 and the newly discovered ARS320 (Fig. 1). These ARSs exhibited no detectable replication origin activity at their native location between the HML locus and the CHA1 gene (Fig. 2). However, we showed that these ARSs are in fact competent as replication origins in the chromosome when inserted at a position remote from their native location (Fig. 3). These are the first HML ARSs shown to exhibit replication origin activity in a chromosome. The results suggested that the replication origin activity of these ARSs is somehow silenced at the HML locus.
The locations of the ARSs near the transcriptionally silent HML locus, coupled with the fact that transcriptional silencers and replication origins share common functional components, including ARSs and ORC, kept open the possibility that the normal inactivity of these replication origins was linked to the complex pathway for transcriptional silencing of the mating-type locus HML. However, we found no replication origin activity at the HML ARS cluster regardless of the trans-acting mutations in the SIR3 or ORC5 genes that abrogate the silencing of transcription (Fig. 4). These findings on trans-acting mutations extend those of Dubey et al. (20), who showed that ARS301 and ARS302 are not detectably active as origins in sir1 or sir4 mutants which abrogate transcriptional silencing. Sir3p was of particular interest here because it exhibits structural similarities with two proteins involved in replication initiation, Cdc6p, as well as with the largest subunit of ORC, Orc1p (3). One hypothesis was that ORC is involved in a mutually exclusive interaction with either Sir3p or Cdc6p and the particular interaction determines whether HML ARSs function in transcriptional silencing or in replication initiation. Along this same line of reasoning, the orc5-1,3 mutant was also of interest because it is competent for initiation of DNA replication but defective in transcriptional silencing (19, 25). However, the absence of detectable replication origin activity at the HML ARS cluster in either sir3 or orc5-1,3 mutants renders such a hypothesis less likely (Fig. 4). Furthermore, cis-acting mutations which delete both the E and I transcriptional silencer elements had no effect on origin silencing at HML ARS303 and ARS320. Deletion of the E and I silencers removes ARS301 and ARS302 along with the DNA binding sites for the silencer proteins ORC, Rap1p, and Abf1p, abrogating transcriptional silencing at HML (38). In addition to ARSs and ORC, Rap1p and Abf1p can be components of active replication origins in other contexts (41). Our findings suggest that origin silencing at ARS303, ARS320 and, where present, ARS302 is independent of transcriptional silencing, despite the fact that replication origins and transcriptional silencers share key cis- and trans-acting components.
Other plausible hypotheses for silencing of replication origins near HML include an unfavorable nuclear localization or inaccessibility of the DNA in a heterochromatin-like structure. HML is located near the left telomere of chromosome III which is tethered to the nuclear periphery along with Sir3p (51), a component of the heterochromatin-like structure at and near HML (30). DNA in HML is highly inaccessible to exogenous factors such as enzymes (63). In mutant strains disrupted in SIR3, telomeres lose their perinuclear localization, and DNA in HML becomes accessible in a more open chromatin structure. Despite these marked changes in HML nuclear localization and accessibility known to accompany abrogation of transcriptional silencing, we found that SIR3 disruption and other mutations that abrogate transcriptional silencing, including orc5-1,3 and deletion of E and I silencers, did not lead to loss of replication origin silencing at ARS303, ARS320 and, where present, ARS302.
Another hypothesis is that silent origins are specialized replication origins that function only in some stage in the yeast life cycle or under special growth conditions (21). With respect to meiosis, HML ARSs tested which have no origin activity in the chromosome during mitotic S phase are also not detectably active in premeiotic S phase (14). We examined yeast mating type since it is known to have profound effects on DNA recombination in the entire left arm of chromosome III (70). However, no detectable replication origin activity was seen at the HML ARS cluster in either MATα or MATa cell types (Fig. 2). We also examined special cell growth conditions that induce expression of the nearby CHA1 gene (52, 65) by disrupting a transcriptionally repressive chromatin structure (44). However, we found that the HML ARS cluster showed no detectable replication origin activity under the special growth conditions required for CHA1 induction (Fig. 5). Thus, the hypothesis that silent origins in yeast are specialized replication origins that function only in some stage in the yeast life cycle or in special growth conditions is currently without support.
Surprisingly, the HML ARS cluster and HML ARS301 exhibited replication origin activity at their natural locations when two replication origins, located 25 kb (ORI305) and 59 kb (ORI306) away, were inactivated by deletion from the chromosome (Fig. 6 and 7A). These data represent the first demonstration of replication origin activity at the HML ARS cluster and at HML ARS301 in their normal chromosomal locations. A preliminary report of the effects of deleting active chromosomal replicators to the left of MAT suggested no apparent origin activation at HML in a strain carrying a wild-type S. cerevisiae chromosome III as a balancer chromosome (48); however, consistent with our findings in a strain with one copy of S. cerevisiae chromosome III, low levels of origin activity were detected upon examination of HML ARSs in a strain carrying a brewing yeast balancer chromosome (16a). Our findings show that origin silencing at the HML locus is mediated by active replication origins located in the chromosome at long distances from HML. In animal cells, distal genetic elements can positively affect replication origin activity (1, 37). Our findings show that in yeast cells, genetic elements that comprise chromosomal replication origins can negatively affect detection of replication origin activity at distal replicators.
The ability of the HML ARS cluster to function as a chromosomal origin is consistent with our findings that two of the ARSs, ARS303 and ARS320, are competent as replication origins in the chromosome when inserted at a position remote from the HML locus, even when the ARSs are clustered together in their natural configuration (Fig. 3). In contrast, the two ARSs are silent as origins at the HML locus independent of whether ARS302, the I silencer, is present (Fig. 4A) or has been deleted (Fig. 4C and D). These observations, combined with the fact that in the double origin deletion mutant (Δ305/Δ306) origin activation was seen not only at the HML ARS cluster but also at the solitary ARS301, suggest that clustering of multiple ARS elements is neither necessary nor sufficient for origin silencing. It is interesting in this connection that an HMR silencer region, which contains three independent subregions with ARS activity in a narrow region of DNA, is normally active as a chromosomal origin (35).
Replication origins are known to interfere with or dominate over one another when closely spaced, at distances of less than ∼7 kb, resulting in the reduction in activity of both origins or the elimination of activity of one of the two origins (9, 40). The mechanisms for interference and dominance between closely spaced origins are unknown. It is possible that the close spacing of the HML ARS cluster with respect to ARS301 in the chromosome (Fig. 1, chromosome map) may contribute to the reduced origin activity seen at both loci in the Δ305/Δ306 strain, where these ARSs are associated with active replication origins. Other possible explanations include a low intrinsic activity of some of the replicators as seen in ARS assays (Fig. 1) or inhibition of origin activity by flanking sequences in the chromosome (62).
Despite the absence of detectable origin activity at HML ARS301 in wild-type strains containing ORI305 and ORI306, a pre-RC that includes ORC is known to form in the chromosome (58). Prior to our work, pre-RC formation at the silent ARS301 origin in a chromosome presented a paradox. The existence of a pre-RC usually reflects a potentially active origin, but the available evidence suggested that ARS301 was not active as a chromosomal replication origin (20). Our findings here help to resolve this apparent paradox by showing that ARS301 at its native chromosomal location can be active as a DNA replication origin after deletion of ORI305 and ORI306, suggesting that the ARS301 pre-RC is competent for origin firing in this condition. In this regard, the properties of ARS301, the E silencer at HML, begin to resemble more closely the E and I silencers at HMR which are competent for origin firing (56, 57).
What is the mechanism for origin silencing at HML? The HML region is duplicated by replication forks that originate from early-firing origins in chromosome III (54). If the HML region was passively replicated before the silent origins could fire, the controls that restrict origin firing to once per S phase would then prevent the HML ARSs from functioning as origins during completion of that S phase. The early-firing active origins include ORI305, ORI306, and ORI307 (see maps in Fig. 6A). Deletion of ORI305 and ORI306 makes ORI307 the closest origin to HML. The possibility that chromosome III sequences that do not have ARS function in a plasmid become active as replication origins and function efficiently is unlikely since deletions of known origins associated with ARSs have failed to reveal any cryptic origins in the left arm of chromosome III (17, 18, 31, 48). ARS304, the only remaining ARS between ORI307 and the HML ARS cluster, is not active as an origin in wild-type cells (Fig. 7B, WT) (14). Importantly, we found that ARS304 is not activated as an origin as a result of deleting ORI305 and ORI306 (Fig. 7B, Δ305/Δ306). Thus, deletion of ORI305 and ORI306 likely moves the source of replication forks that duplicate HML from 25 kb (ORI305) to 93 kb (ORI307; distances are from HML ARS303). Such a large increase in distance necessitates a large delay in time that forks arrive at HML. Thus, based on our findings and on information regarding the positions of origins and ARSs on chromosome III, as well as the timing of origin firing (49, 54), we propose the following model to account for the activation of silent origins: a delay in the arrival of forks at HML caused by deletion of the closest, early-firing origins relieves origin silencing by permitting additional time for HML ARSs to fire as origins. A likely possibility is that HML ARSs require additional time because they are activated late in S phase (ARS301, see below). Another possibility is that certain HML ARSs require more time to fire because they are intrinsically inefficient as replicators (Fig. 1, ARS303 and ARS320). The latter possibility would appear not apply to ARS301, which is known to function efficiently as a replicator in a plasmid (58).
The fact that low-level origin activation was seen at the HML ARS cluster, as well as at HML ARS301, after deletion of ORI305 and ORI306 suggests that the basis for origin silencing at all of the HML ARSs may be similar and independent of the intrinsic replicator efficiency in plasmids. ARS301 is known to fire late in S phase in a plasmid (6). Late-S-phase activation of the ARS301 origin in the chromosome would be consistent with the model for activation of silent origins proposed above. Also consistent is the fact that every ARS functions as a replication origin in a plasmid. Plasmids, unlike the chromosome, have no additional yeast replication origins and thus no replication forks to duplicate the ARS before it fires as an origin. Finally, the proposed model is consistent with our finding that ARS303 and ARS320 can be active replication origins in the chromosome when inserted in place of ORI305, which is located in an early-firing, chromosomal context (23, 54). Our discovery that silent origins can be active as origins under certain conditions opens the possibilities of testing the proposal that the HML ARSs are late-activated origins at their natural locations and of identifying the molecular mechanism for origin silencing in a eukaryotic chromosome.
ACKNOWLEDGMENTS
We thank Martha Eddy for technical assistance; Carol Newlon, Jasper Rine, James Broach, and Phil Hieter for yeast strains and plasmids; and Yangzhou Wang and Bill Held for helpful comments on the manuscript.
This work was supported in part by shared resources of the Roswell Park Cancer Center Support Grant (P30 CA16056) and by a grant (GM-30614) from the National Institutes of Health.
REFERENCES
- 1.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 2.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 4.Benard M, Lagnel C, Pallotta D, Pierron G. Mapping of a replication origin within the promoter region of two unlinked, abundantly transcribed actin genes of Physarum polycephalum. Mol Cell Biol. 1996;16:968–976. doi: 10.1128/mcb.16.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornaes C, Ignjatovic M W, Schjerling P, Kielland-Brandt M C, Holmberg S. A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol Cell Biol. 1993;13:7604–7611. doi: 10.1128/mcb.13.12.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousset K, Diffley J F. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand A H, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 8.Brand A H, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- 9.Brewer B J, Fangman W L. Initiation at closely spaced replication origins in a yeast chromosome. Science. 1993;262:1728–1731. doi: 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- 10.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 11.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 13.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins I, Newlon C S. Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol Cell Biol. 1994;14:3524–3534. doi: 10.1128/mcb.14.5.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePamphilis M L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993;3:161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 16.DePamphilis M L. Initiation of DNA replication in eukaryotic chromosomes. J Cell Biochem Suppl. 1998;31:8–17. [PubMed] [Google Scholar]
- 16a.Dershowitz, A., and C. S. Newlon. Personal communication.
- 17.Dershowitz A, Newlon C S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993;13:391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshpande A M, Newlon C S. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillin A, Rine J. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics. 1997;147:1053–1062. doi: 10.1093/genetics/147.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J G, Broach J R, Newlon C S, Huberman J A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fangman W L, Brewer B J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- 22.Fangman W L, Brewer B J. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson B M, Fangman W L. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 24.Foss M, McNally F J, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 25.Fox C A, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert D M. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Curr Opin Genet Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- 27.Goldman M A, Holmquist G P, Gray M C, Caston L A, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- 28.Hatton K S, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 30.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 31.Huang R Y, Kowalski D. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 1993;12:4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang R Y, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huberman J A, Spotila L D, Nawotka K A, el-Assouli S M, Davis L R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 34.Huberman J A, Zhu J G, Davis L R, Newlon C S. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurst S T, Rivier D H. Identification of a compound origin of replication at the HMR-E locus in Saccharomyces cerevisiae. J Biol Chem. 1999;274:4155–4159. doi: 10.1074/jbc.274.7.4155. [DOI] [PubMed] [Google Scholar]
- 36.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 37.Kalejta R F, Li X, Mesner L D, Dijkwel P A, Lin H B, Hamlin J L. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 38.Mahoney D J, Broach J R. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989;9:4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahoney D J, Marquardt R, Shei G J, Rose A B, Broach J R. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 39a.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Marahrens Y, Stillman B. Replicator dominance in a eukaryotic chromosome. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 42.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley J F. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 42a.Miller, C. A., and D. Kowalski. Unpublished data.
- 43.Miller C A, Kowalski D. cis-Acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5360–5369. doi: 10.1128/mcb.13.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreira J M, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 45a.Natale, D., and D. Kowalski. Unpublished data.
- 46.Natale D A, Umek R M, Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 48.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 49.Newlon C S, Lipchitz L R, Collins I, Deshpande A, Devenish R J, Green R P, Klein H L, Palzkill T G, Ren R B, Synn S, et al. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta J P, Benit P, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 51.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 52.Petersen J G, Kielland-Brandt M C, Nilsson-Tillgren T, Bornaes C, Holmberg S. Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics. 1988;119:527–534. doi: 10.1093/genetics/119.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivier D H, Ekena J L, Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivier D H, Rine J. An origin of DNA replication and a transcription silencer require a common element. Science. 1992;256:659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- 58.Santocanale C, Diffley J F. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 59.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 60.Shirahige K, Iwasaki T, Rashid M B, Ogasawara N, Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 62.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 63.Singh J, Klar A J. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 64.Stinchcomb D T, Struhl K, Davis R W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka S, Isono K. Correlation between observed transcripts and sequenced ORFs of chromosome III of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1149–1153. doi: 10.1093/nar/21.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Theis J F, Newlon C S. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 68.Weiss K, Simpson R T. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu C, Weiss K, Yang C, Harris M A, Tye B K, Newlon C S, Simpson R T, Haber J E. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 1998;12:1726–1737. doi: 10.1101/gad.12.11.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X, Moore J K, Haber J E. Mechanism of MAT alpha donor preference during mating-type switching of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:657–668. doi: 10.1128/mcb.16.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu J, Newlon C S, Huberman J A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]