Abstract
Mitochondria are double-membrane organelles that contain their own genome, the mitochondrial DNA (mtDNA), and reminiscent of its endosymbiotic origin. Mitochondria are responsible for cellular respiration via the function of the electron oxidative phosphorylation system (OXPHOS), located in the mitochondrial inner membrane and composed of the four electron transport chain (ETC) enzymes (complexes I-IV), and the ATP synthase (complex V). Even though the mtDNA encodes essential OXPHOS components, the large majority of the structural subunits and additional biogenetical factors (more than seventy proteins) are encoded in the nucleus and translated in the cytoplasm. To incorporate these proteins and the rest of the mitochondrial proteome, mitochondria have evolved varied, and sophisticated import machineries that specifically target proteins to the different compartments defined by the two membranes. The intermembrane space (IMS) contains a high number of cysteine-rich proteins, which are mostly imported via the MIA40 oxidative folding system, dependent on the reduction, and oxidation of key Cys residues. Several of these proteins are structural components or assembly factors necessary for the correct maturation and function of the ETC complexes. Interestingly, many of these proteins are involved in the metalation of the active redox centers of complex IV, the terminal oxidase of the mitochondrial ETC. Due to their function in oxygen reduction, mitochondria are the main generators of reactive oxygen species (ROS), on both sides of the inner membrane, i.e., in the matrix and the IMS. ROS generation is important due to their role as signaling molecules, but an excessive production is detrimental due to unwanted oxidation reactions that impact on the function of different types of biomolecules contained in mitochondria. Therefore, the maintenance of the redox balance in the IMS is essential for mitochondrial function. In this review, we will discuss the role that redox regulation plays in the maintenance of IMS homeostasis as well as how mitochondrial ROS generation may be a key regulatory factor for ETC biogenesis, especially for complex IV.
Keywords: mitochondria, biogenesis, protein import, redox signaling, ROS, respiratory chain assembly, MIA
Introduction
Mitochondrial biogenesis is essential for cell fitness and viability. Mitochondria are double membrane bound organelles composed of an outer membrane (OM), intermembrane space (IMS), inner membrane (IM), and matrix that harbors its own mitochondrial genome, the mtDNA. This mtDNA encodes only 13 polypeptides in humans or 8 polypeptides in yeast cells that are translated into key components of the respiratory chain complexes embedded in the IM. Assembly of the respiratory chain complexes relies on the protein import process through which most of these subunits find their way into the organelle.
A large number of proteins imported into the intermembrane space (IMS) are cysteine rich proteins that are imported via the mitochondrial import and assembly (MIA) pathway which involves oxidation of specific cysteine residues and subsequent folding of proteins into their mature conformation thereby trapping them in this compartment. As many of the substrates of this pathway include a variety of chaperones, proteases, mitochondrial dynamics factors and assembly factors for the respiratory chain complexes, and redox regulation of the IMS plays a crucial role in a wide variety of processes within the mitochondria. In this review we give a brief overview of the protein import pathways moving on to a more detailed presentation of the redox-regulated MIA pathway with a focus on the yeast and human systems. We then discuss the redox regulation features of mitochondria dynamics and proteases in the IMS following on with an analysis of the redox regulation mechanisms that are pertinent for the assembly process of the electron transport chain (ETC) complexes. Finally, we examine the possible role of reactive oxygen species (ROS) on maintaining the homeostasis of the respiratory chain.
Overview of Mitochondrial Protein Import Pathways
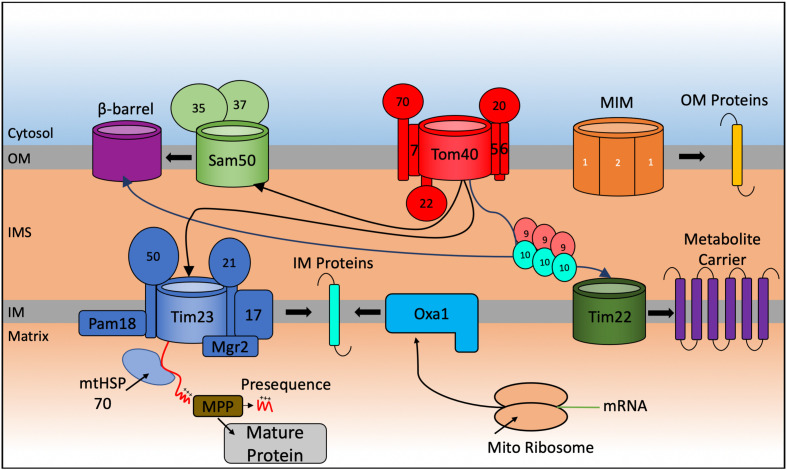
The mitochondrial proteome consists of 1,000–1,500 proteins (Sickmann et al., 2003; Reinders et al., 2006; Pagliarini et al., 2008; Rath et al., 2021) but only eight in the yeast S. cerevisiae and thirteen in humans, and other mammalian species, are encoded in the mitochondrial genome and translated inside the organelle. This implies that ∼99% of all mitochondrial proteins (1163 polypeptides in human mitochondria according to the last compendium of MitoCarta 3.0 (Rath et al., 2021) are nuclear encoded, translated on cytosolic ribosomes and guided to the mitochondria by a variety of cytosolic chaperones. From here they can be sorted within the organelle (Figure 1).
FIGURE 1.
Schematic representation of the general import pathways for proteins destined for localization to the mitochondria. Incoming nuclear-encoded proteins destined for import into the mitochondria follow a number of particular import pathways depending on the sub compartment where they become localized. Matrix targeted proteins follow the presequence pathway, passing through the TOM40, and TIM23 translocases where they become cleaved by matrix processing proteins (MPP) in the matrix to form the mature protein. Preproteins targeted to the outer membrane (OM) are inserted by the Sam complex if they are β-barrel proteins and via the mitochondrial import machinery (MIM) if they are α-helical containing proteins. Integration of preproteins into the inner membrane (IM) involves interaction with the Tim9-Tim10 chaperone complex that targets proteins to the TIM22 translocase for membrane insertion or via a variation of the TIM23 complex that promotes integration of the protein into the IM. Mitochondrial encoded proteins are synthesized on mito ribosomes located in the matrix and inserted into the IM via the Oxa1 complex.
Protein Entry Into Mitochondria
The translocase of the outer membrane (TOM) complex forms the entry gate for most imported mitochondrial proteins. The TOM complex is composed of seven different functional and receptor subunits. Precursors with a cleavable N-terminal presequence are recognized on the surface of mitochondria by the Tom20 receptor, while the Tom70 receptor recognizes precursor proteins that lack such a presequence but contain an internal targeting signal instead (Brix et al., 1997; Yamamoto et al., 2009; Backes et al., 2018). Tom22 is the central receptor that helps recruit both Tom20 and Tom70 to the TOM core complex and is involved in transferring preproteins from the receptor subunits to the translocation pore (Van Wilpe et al., 1999; Yamano et al., 2008). The structure of the S. cerevisiae TOM complex was recently solved using cryo-electron microscopy at a 3.8Å resolution (Araiso et al., 2019). This detailed structure revealed the architecture of the TOM entry gate providing a framework how the additional subunits Tom5, Tom6, Tom7 and Tom22 are organized within the complex. We will not provide a detailed analysis here as the TOM structural and functional features have been reviewed extensively (Shiota et al., 2015; Araiso et al., 2020).
Protein Sorting in the Inner Mitochondrial Compartments
Following translocation through the TOM complex in the OM, proteins are sorted through different import pathways into the matrix, the IM and the IMS (Jensen and Johnson, 2001; Grevel et al., 2019; Hansen and Herrmann, 2019). Most of the precursor proteins targeted to the matrix are synthesized with an N-terminal positively charged cleavable presequence (Vögtle et al., 2009) and follow the matrix targeting pathway, which accounts for almost two thirds of all mitochondrial proteins. The presequence translocase of the inner membrane (TIM23 complex) is the key translocon that allows passage of these matrix-targeted precursor proteins through the inner membrane. The positively charged region of the presequence is recognized by the TOM receptors Tom20 and Tom22 that facilitate the translocation across the OM. The Tim50 subunit feeds the positively charged region into the Tim23 channel (Dayan et al., 2019). The translocation of these precursors through the pore of the TIM23 complex is dependent on the membrane potential (Δψ) across the inner membrane generated from the activity of the respiratory chain complexes. Additionally, the TIM23 complex is also involved in releasing proteins, containing a strongly hydrophobic transmembrane segment adjacent to the matrix-targeting presequence, laterally into the inner membrane. These proteins start to engage with the Tim23 channel guided by their presequence but become stalled because of their strong hydrophobic “stop-transfer” signal in the translocation pore of the Tim23 channel, preventing their complete import into the matrix. A specific cleavage of the ‘stop-transfer’ signal by the IMS localized IMP protease releases the mature protein into the IMS. The translocation-arrest mechanism of such bipartite presequence containing preproteins requires a specific conformation of the TIM23 complex that contains Tim17, Tim21, and Tim23, termed the TIM23SORT translocase (Chacinska et al., 2010). By contrast, the TIM23 complex associated with the matrix-localized Pam18 (TIM23PAM) blocks the lateral release into the lipid bilayer and instead promotes import into the matrix (Schendzielorz et al., 2018). Protein transport into the matrix requires the ATP-dependent activity of the mitochondrial heat shock protein 70 (mtHsp70) (Pais et al., 2011) in addition to the membrane potential. Once in the matrix, the presequence is cleaved off from the precursor by the matrix processing peptidase (MPP), giving rise to the mature mitochondrial protein (Yang et al., 1991).
The mitochondrial metabolite inner membrane carrier proteins (at least 35 proteins in S. cerevisiae and more than 50 in mammalian cells) are critical for the trafficking of small metabolites and are synthesized without a cleavable presequence. They interact with the Tom70 receptor, engage with the TOM channel and are guided to the inner membrane TIM22 complex, by a number of small TIM chaperones in the IMS that escort them in the aqueous IMS preventing their aggregation. This import and insertion process constitutes the carrier pathway, and it is membrane potential-dependent but independent of ATP hydrolysis in the matrix (Sirrenberg et al., 1996).
Protein Insertion Into the Mitochondrial Outer Membrane
The mitochondrial outer membrane contains either β-barrel membrane proteins (which are absent from the inner membrane) or α-helical transmembrane proteins. Precursors of the β-barrel proteins are first recognized by the TOM complex and then transferred to the sorting and assembly (SAM) complex from the IMS side of the OM. A recent cryo-electron microscopy structure of the SAM complex revealed that it is made up of two copies of the central subunit Sam50, forming a β-barrel channel (Takeda et al., 2021) that allows the lateral release of the precursor proteins into the outer membrane (Paschen et al., 2003; Klein et al., 2012). The SAM complex also contains the Sam35 and Sam37 proteins found on the cytosolic side of the outer membrane. Sam35 is thought to stabilize the precursor-Sam50 interaction, while Sam37 is involved in the release of substrate proteins from the SAM complex (Chan and Lithgow, 2008; Kutik et al., 2008). Transfer from the IMS to the OM is mediated by the IMS small TIM chaperones that transiently bind to exposed hydrophobic regions preventing their aggregation (Wenz et al., 2015; Weinhäupl et al., 2018). α-helical membrane proteins in the OM follow a dedicated pathway via the mitochondrial import machinery (MIM) complex (Becker et al., 2011; Dimmer et al., 2012).
Import of Proteins Into the Intermembrane Space – The MIA Pathway
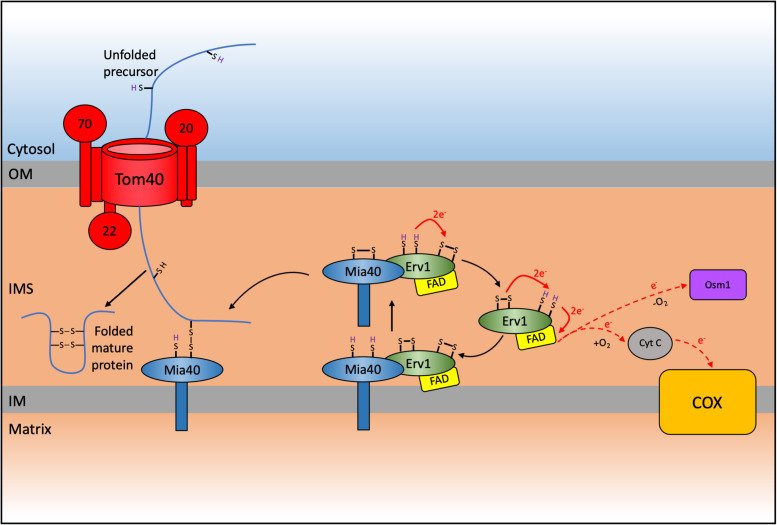
A detailed analysis of the proteome of the mitochondrial IMS for the yeast S. cerevisiae revealed that around 50 proteins were localized to this sub-compartment (Vögtle et al., 2012). In humans, the IMS proteome accounts for about 5% of the total mitochondrial proteome (53 proteins) (Rath et al., 2021). The majority of these IMS-located proteins lack a typical mitochondrial targeting sequence and are instead characterized by conserved cysteine residues organized into twin CXnC (typically either CX3C or CX9C) motifs that are necessary for their import, correct folding and maturation. The oxidative folding or mitochondrial protein import and assembly (MIA) pathway relies on the function of Mia40, in yeast, and CHCHD4 (also known as MIA40), in humans, as the key protein that facilitates the disulfide bonds in these proteins (Mesecke et al., 2005; Figure 2).
FIGURE 2.
Schematic representation of the mitochondrial import and assembly (MIA) pathway in yeast. A subset of proteins destined for the intermembrane space (IMS) are imported via the MIA pathway which results in the oxidative folding of these substrates thereby trapping them in the IMS. Nuclear encoded CXnC motif containing proteins are first imported through the TOM complex. These proteins contain an IMS-Targeting signal (ITS) that interacts with Mia40 resulting in the oxidation and subsequent folding of the substrate protein, thereby trapping it in the IMS. This interaction leaves Mia40 in a reduced state that must be reoxidized to allow it to further oxidize incoming substrates. This reoxidation is facilitated by interaction with Erv1 with subsequent electron transfer to cytochrome c and complex IV (COX) of the respiratory chain in aerobic conditions, or onto Osm1 in anaerobic conditions. The system varies in humans, where CHCHD4/MIA40 is a soluble IMS protein that interacts with membrane bound AIFM1 and with ALR/GFER/ERV1 (Reinhardt et al., 2020).
Many of these classical MIA substrates are vital for the biogenesis and function of mitochondria as, for example, the members of the small Tim protein family, including Tim 8, Tim 9, Tim 10, and Tim13 (Sideris and Tokatlidis, 2007). These proteins form the Tim9-Tim10 and Tim8-Tim13 chaperone complexes that are essential to deliver hydrophobic proteins to the TIM22 complex for insertion into the inner membrane. Other examples of MIA substrates are proteins necessary for the biogenesis of the ETC complexes. These will be discussed in detail in subsequent sections and include proteins with a dual CX9C motif that are involved mostly in the assembly and maintenance of cytochrome c oxidase (COX), also known as complex IV (CIV) (Gabriel et al., 2007; Koch and Schmid, 2014a; Habich et al., 2019a; Gladyck et al., 2021). It is, however, clear that Mia40 can recognize proteins that do not have CXnC motifs, like the yeast Erv1 protein (with a CX15C motif) (Terziyska et al., 2007; Kallergi et al., 2012), yeast Atp23 (with 10 cysteine residues not organized in any specific cysteine motif (Weckbecker et al., 2012), yeast Mix23 (with an unusual twin cysteine CX13C/CX14C motif (Vögtle et al., 2012; Zöller et al., 2020), or human COA7 containing thirteen Cys residues (Mohanraj et al., 2019). Other proteins without classical CXnC motifs that have been described as substrates of CHCHD4/MIA40 also include p53, AK2 and MICU1 (Zhuang et al., 2013; Petrungaro et al., 2015). It is also of note that Ccs1, the copper chaperone for Cu,Zn superoxide dismutase (Sod1), is a Mia40 substrate in yeast but the IMS import of the human ortholog (CCS1) does not depend on CHCHD4/MIA40 (Groß et al., 2011; Suzuki et al., 2013).
The other main component of the yeast MIA pathway is the FAD-dependant sulfhydryl oxidase Erv1 (Chacinska et al., 2004; Mesecke et al., 2005). The human ortholog is known as ERV1, ALR, or GFER (Lange et al., 2001; Di Fonzo et al., 2009). Mia40 and Erv1 function as a disulfide relay system to catalyze the import of proteins into the IMS through an oxidative folding process. This results in introduction of disulfide bonds into the imported proteins, promoting their folding into their native conformation and thereby trapping them in the IMS. Substrates of the MIA pathway are translated cytosolically and pass through the TOM complex in a reduced and unfolded state (Lu et al., 2004; Sideris and Tokatlidis, 2007). During translocation these substrates interact with Mia40, which acts as a trans site receptor to drive protein import into the IMS (Milenkovic et al., 2007; Sideris and Tokatlidis, 2007; Peleh et al., 2016).
The yeast Mia40 has an N-terminal transmembrane domain that inserts the protein to the inner mitochondrial membrane and exposes its C-terminal domain to the IMS (Chacinska et al., 2004; Naoé et al., 2004; Terziyska et al., 2005). The human MIA40 (or CHCHD4) lacks this N-terminal domain and instead is found as a soluble protein of around 15 kDa in the mitochondrial IMS (Hofmann et al., 2005) where it interacts with the membrane anchored AIFM1 and this interaction facilitates the import of MIA40 into the IMS (Hangen et al., 2015). The C-terminal region was recently demonstrated to be important in maintaining the stability of the protein in the cytosol where it can often reside for extended periods as a consequence of slow import kinetics during AIFM1-mediated import (Murschall et al., 2020). However, all Mia40 homologs contain a highly conserved core domain of around 8 kDa that harbors six conserved cysteine residues that are folded into a coiled-helix-coiled-helix (CHCH) domain (Banci et al., 2009). The structure of the human and yeast homologs was solved by NMR and X-ray crystallography, respectively (Banci et al., 2009; Kawano et al., 2009). The invariant cysteines in this domain form a redox-active CPC motif that can readily switch between oxidized and reduced states, followed by a twin CX9C motif that forms two structural disulfide bonds. The CHCH domain folds to form a characteristic hydrophobic cleft where substrates can bind and interact with Mia40 (Banci et al., 2009; Sideris et al., 2009). Most Mia40 substrates contain a hydrophobic sequence, also known as an IMS-targeting signal (ITS, Sideris et al., 2009) or alternatively termed the mitochondrial IMS-sorting signal (MISS, Milenkovic et al., 2009). This is a 9 amino acid internal peptide sequence that is necessary and sufficient for IMS targeting of proteins to Mia40. Deletion of the ITS results in a complete loss of import (Sideris et al., 2009). The sequence aids the first interaction with Mia40 via hydrophobic stacking (Milenkovic et al., 2009; Sideris et al., 2009). This initial interaction orientates the substrate toward Mia40 to allow the formation of a transient intermolecular disulfide bond between the second cysteine of the redox active CPC motif of Mia40 and the docking cysteine of the ITS in the imported substrate (Banci et al., 2009; Koch and Schmid, 2014b). This transient disulfide bond is subject to a nucleophilic attack by another cysteine in the substrate to form an intramolecular disulfide bond. This promotes the release of Mia40 and the correct folding of the protein, thereby trapping it in the IMS and completing its import process.
The introduction of a disulfide into a substrate by Mia40 leaves it in a reduced state and unable to oxidize any further imported proteins. Thus, to maintain a functional MIA pathway, Mia40 needs to continually be re-oxidized by interaction with Erv1, which contains three cysteine pairs that are highly conserved (Mesecke et al., 2005). The FAD-binding catalytic core domain has a CXXC redox-active disulfide (Cys130 – Cys133) and a C-terminal CX16C structural disulfide (Cys156 and 179). A third disulfide bond (Cys30 – Cys33) is present within the flexible N-terminal of the protein, which is natively disordered and is involved in the interaction with reduced Mia40 and the transfer of electrons toward the redox active-site disulfide (Lionaki et al., 2010; Banci et al., 2011a, Banci et al., 2013). From here electrons are transferred to a final electron acceptor via the FAD cofactor (Banci et al., 2012). Under aerobic conditions Erv1 can either transfer electrons directly onto molecular oxygen which results in the production of hydrogen peroxide within the IMS, or onto cytochrome c, the mobile electron transfer protein that donates electrons to CIV (Allen et al., 2005; Dabir et al., 2007; Daithankar et al., 2009; Banci et al., 2012; Peker et al., 2021). Alternatively, under anaerobic conditions the fumarate reductase Osm1 can be used as an electron acceptor (Neal et al., 2017).
Intriguingly, Mia40 itself is a substrate of the MIA pathway requiring interactions with endogenous Mia40 during its own import, which occurs in three steps. First, Mia40 is inserted through the Tim23 translocon in the inner mitochondrial membrane. Next, the core domain of the protein is folded through interactions with endogenous Mia40. Lastly, Mia40 interacts with Erv1 to oxidize the CPC motif to produce functional Mia40 that can oxidize incoming substrates of the MIA pathway (Chatzi et al., 2013).
The Small Tim Chaperones
The small Tim proteins that function as chaperones in the IMS were the first discovered substrates of the MIA pathway, interacting with Mia40 to control their redox-regulated import (Chacinska et al., 2004). These small Tims possess non-cleavable internal targeting signals (ITS) that harbor conserved cysteine motifs to target them to the mitochondria (Milenkovic et al., 2009; Sideris et al., 2009). The cysteines are arranged in a CX3C motif, a classical substrate motif for Mia40 (Koehler, 2004). The ITS signal directs the small Tims to the IMS where they are imported by the MIA pathway through direct interaction with Mia40 (Chacinska et al., 2004; Sideris et al., 2009). This interaction results in the oxidation of the small Tims, leading to the formation of intramolecular disulfide bonds and trapping the proteins in the IMS.
The small Tims form hexameric protein complexes consisting of either Tim9 – Tim10 or Tim8 – Tim13 with three subunits of each Tim protein. Structural analysis of the complexes revealed the subunits take on the form of an α-propellor with two helical blades that radiate from a narrow central pore (Webb et al., 2006; Beverly et al., 2008). The subunits are stabilized by the formation of intramolecular and structural disulfide bonds. Within the IMS these complexes act as chaperones to transport incoming hydrophobic precursors across this compartment to the TIM22 complex for their insertion into the IMM. The helper of Tims 13 (Hot13p) was also shown to be involved in maintaining the small Tims in their active conformation (Curran et al., 2004).
The presence of the redox-regulated disulfide machinery in the IMS raises the question of a broader level of redox regulation in this sub-compartment that may affect not just the MIA pathway itself or import substrates for this pathway, but other important proteins and process linked to the IMS. In the next sub-sections, we will discuss in particular the redox regulation of mitochondrial dynamics and the effect of redox control on the IMS protease system. Both of these are critical for many functions of mitochondria and highlight the broader ramifications of mitochondria-specific redox control pathways.
Redox Regulation of Mitochondrial Dynamics
Mitochondria undergo coordinated processes of fission and fusion, known as mitochondrial dynamics, to regulate their size, shape, and integrity. There are several key proteins involved in the control of mitochondrial dynamics that have been demonstrated to be redox-regulated. Mitofusin proteins (Mfn1/Mfn2) are large GTPases that are implicated in mitochondrial fusion. This process has been shown to be triggered by oxidative stress, characterized by an accumulation of oxidized glutathione (GSSG), which in turn leads to the assembly of higher order Mfn oligomers mediated by the formation of disulfide bonds between conserved cysteine residues in the C-terminal region (Shutt et al., 2012). This region has been recently identified to be topologically located in the IMS and the conserved Cys demonstrated to be redox sensitive (Mattie et al., 2018). Redox regulation in this compartment mediates the formation of disulfide bonds in the Mfn proteins thereby driving oligomerization and triggering mitochondrial fusion. Therefore, the redox state of the IMS is likely to play a role in controlling mitochondrial dynamics.
ROS modulator 1 (ROMO1) was identified as a key regulator of mitochondrial fusion, with knockout cells displaying increased fragmented mitochondrial networks. ROMO1 is also involved in the generation of ROS by complex III (CIII) of the ETC and can form disulfide bridges to become incorporated into inactive high-molecular weight complexes (Chung et al., 2006; Norton et al., 2014). Disulfide formation occurs between the Cys15 and Cys79 residues in response to oxidative stress, inhibiting fusion and maintaining a fragmented mitochondrial network (Norton et al., 2014). More recently ROMO1 was found to be required for the import of the human inner membrane YME1L protease (Richter et al., 2019).
During mitochondrial fusion there is separate merging of the outer and inner mitochondrial membranes. Fusion of inner mitochondrial membranes involves heterotypic interactions between the dynamin-like GTPase optic atrophy 1 (OPA1) and cardiolipin (Ban et al., 2017; Liu and Chan, 2017). Mammalian OPA1 is an inner membrane associated protein that exists in either a long form (L-OPA1) or soluble short-form (S-OPA1) conformation (Del Dotto et al., 2018). Conversion of OPA1 between these two isoforms occurs as a result of proteolytic cleavage of L-OPA1 that is catalyzed by the OMA1 and YME1L proteases (Anand et al., 2014). A balance of both the long- and short- isoforms of OPA1 depends on these proteases and is required to maintain normal mitochondrial morphology.
Redox Regulation of Proteases in the IMS
YME1L, the human ortholog of yeast Yme1, is an integral membrane protein with its C-terminal domain exposed to the intermembrane (Stiburek et al., 2012). Yme1 is a subunit of the inner membrane anchored oligomeric complex known as the i-AAA protease that displays both ATP-dependent proteolytic and chaperone-like activities in the IMS (Leonhard et al., 1996, 1999; Schreiner et al., 2012). Yme1 is involved in the degradation of misfolded inner membrane proteins and more recently shown to be implicated in the clearance of the small Tim9 and Tim10 IMS chaperones (Leonhard et al., 2000; Baker et al., 2012). YME1L is rapidly degraded by OMA1, an ATP-independent protease, upon oxidative stress conditions (Rainbolt et al., 2015). This suggests that YME1L is a stress-sensitive protease and thus degradation of YME1L under oxidative stress conditions may provide a mechanism to segregate damaged mitochondria from the healthy pool, by restricting the ability of YME1L to regulate the fusion activity of OPA1 through its degradation. More recently it was demonstrated that YME1L adopts unique conformations depending on the ATP and ROS concentrations (Brambley et al., 2019). Under oxidative stress conditions, YME1L undergoes a conformational change, exposing OMA1 recognition sites that were previously unreachable in basal conditions, further supporting the mechanism of YME1L degradation by OMA1 during oxidative stress (Brambley et al., 2019).
Oma1, responsible for mediating OPA1 degradation, is an IMS facing ATP-independent metalloprotease embedded in the inner mitochondrial membrane displaying enhanced activity in response to stress conditions (Käser et al., 2003; Head et al., 2009). Oma1 is synthesized as a pre-pro-protein of 60 kDa that upon import into the mitochondria is proteolytically processed to a mature 40 kDa form (Head et al., 2009). The protein is normally dormant under normal physiological conditions but becomes rapidly activated upon changes in membrane potential, chronic hyperpolarization and oxidative stress (Bohovych et al., 2014). The 40 kDa Oma1 is proposed to be a stress-sensitive pro-protein that undergoes further autocatalytic cleavage in response to stress insults to produce the active form of the protein that is involved in proteolysis of substrates (Baker et al., 2014).
Oma1 exists in a semi-oxidative state in both yeast and mammalian cell types and the activity and stability of the homo-oligomeric complex was found to be altered in a redox sensitive manner. Two evolutionary conserved cysteine residues, Cys272 and Cys332, which are exposed to the IMS can form a disulfide bond that likely influences the stability of the oligomeric complex. It is proposed that the formation of this disulfide is controlled by the redox status of the cysteine residues which act as a redox sensor to influence the proteolytic activity of Oma1 in response to deviations from normal cell homeostasis (Bohovych et al., 2019).
A subset of IMS-located proteases has been shown to be involved in the proteolytic control of ETC assembly and stability, including the m-AAA, i-AAA and Oma1. These enzymes control the proteolysis of unassembled, misfolded OXPHOS subunits to prevent the formation of dysfunctional ETC subunits (Bohovych et al., 2015). Oma1 in particular is involved in the degradation of non-hemylated cytochrome c oxidase subunit 1 (Cox1) (Bestwick et al., 2010b; Khalimonchuk et al., 2012). In human mitochondria, the proteolytic activity of YME1L was shown to be involved in preventing the accumulation of non-assembled respiratory chain subunits including COX4, MT-ND1 and NDUFB6 (Stiburek et al., 2012).
In S. Cerevisiae, Atp23 was identified as an IMS protease required for the processing of the mtDNA-encoded subunit 6 of the mitochondrial ATPase (Atp6) (Osman et al., 2007). Atp23 contains ten cysteine residues, that during the import and folding into mitochondria form five disulfide bonds. The reduced Atp23 precursor is translocated into the IMS where Mia40 is then involved in introducing the disulfide bonds to promote folding of Atp23 into its mature conformation (Weckbecker et al., 2012).
The morphology of the inner membrane is regulated by the mitochondrial contact site and cristae organization system (MICOS) complex (Harner et al., 2011). Mic19, a peripheral inner membrane component of this complex is involved in the maintenance of the architechture of the inner membrane. Human MIC19 has five cysteine residues, four of which are arranged in a twin CX9C motif in the coiled-coil helix coiled-coil helix fomain, a typical motif for Mia40 substrates. This domain was shown to be essential for import of MIC19 into mitochondria and MIC19 demonstrated to form a transient disulfide-bond intermediate involving its Cys193 residue (Darshi et al., 2011, Darshi et al., 2012). Yeast Mic19, however, lacks the typical CX9C motif, and instead contains two conserved cys residues in a CX10C motif. Yeast Mic19 was identified in two different redox states in the mitochondria. The oxidized form displays an intermolecular disulfide with Mic60, which is not essential for its integration into the complex, but does play a role in the stability of the MICOS complex (Sakowska et al., 2015). This suggests that Mic19 may be a redox-sensor regulator of MICOS function and, therefore, of maintenance of inner membrane morphology.
Redox-Regulation of OXPHOS Components and Biogenesis Factors
Many of the import, dynamics and redox-regulated processes mentioned so far culminate in the biogenesis of the oxidative phosphorylation (OXPHOS) system. This is physically located in the mitochondrial inner membrane (therefore in contact to the IMS on the one side) and it is composed of the four complexes (complexes I-IV) of the respiratory or ETC, which transfer reducing equivalents from NADH or FADH2 to oxygen, reducing it to water, using the two mobile electron carriers: coenzyme Q (CoQ) and cytochrome c. Electron transfer through complexes I, III and IV is coupled with proton pumping from the matrix to the IMS. This electrochemical gradient generates the protonmotive force (pmf) employed by the ATP synthase (or complex V) for the synthesis of the majority of the cellular ATP in aerobic eukaryotes (Wikström et al., 2015). Complexes I, III, IV, and V are large multimeric enzymes whose structures span the IMM, with subunits that protrude either in the matrix, the IMS or both (Letts and Sazanov, 2017). The processes of assembly and maturation of these complexes are intricate and require the action of a significant number of proteins, generically called assembly factors, that are not part of the mature complex but are necessary for ETC biogenesis. The structure and assembly of the respiratory complexes seem to be conserved for the most part between yeast and humans, except for the absence of a multimeric, and proton pumping complex I in S. cerevisiae (Signes and Fernandez-Vizarra, 2018). There is an additional level of complexity in the organization of the ETC, which is the formation of supercomplex structures. In yeast, all of CIV is associated with the invariantly dimeric CIII (CIII2) in III2IV1–2 stoichiometries (Hartley et al., 2019; Rathore et al., 2019). In mammalian mitochondria, CI, CIII2 and CIV associate with different stoichiometries into supercomplexes, in which CIII2 is always present, that coexist with the individual non-associated complexes (Letts and Sazanov, 2017; Lobo-Jarne and Ugalde, 2018).
Several structural subunits and assembly factors directly involved in OXPHOS biogenesis are Cys-containing proteins located in the IMS and, therefore, potentially subject to redox regulation (Habich et al., 2019b; Reinhardt et al., 2020). In addition, many of these are MIA40/CHCHD4 substrates (Table 1).
TABLE 1.
IMS-located Cys-rich structural subunits of the ETC complexes and assembly factors.
*Two isoforms of COX6B exist: the ubiquitously expressed COX6B1 and the testis specific COX6B2 (Sinkler et al., 2017).
The Cys residues in these proteins are important for their biogenesis (i.e., their import and stability in the IMS), but in some cases also for their function (Gladyck et al., 2021). Most members of this group of proteins are also interesting from a medical point of view, as pathological variants in several genes encoding them have been found associated with respiratory chain deficiency and mitochondrial disease (Fernandez-Vizarra and Zeviani, 2021). Interestingly, the majority of IMS-located assembly factors appear to be involved in the maturation of CIV (Khalimonchuk and Winge, 2008; Longen et al., 2009; Bourens et al., 2013; Timón-Gómez et al., 2018; Gladyck et al., 2021). In addition to these, COA8 is a particular case of a CIV assembly factor, located in the IM facing the matrix, containing Cys residues within and immediately after its MTS and whose import and/or stability appears to be redox regulated (Signes et al., 2019).
Role of IMS Proteins in Complex I Biogenesis
The MIA pathway has been thoroughly studied using the yeast S. cerevisiae as a reference model (Chatzi and Tokatlidis, 2013). However, there is no multimeric complex I (CI) present in this unicellular organism, having instead three monomeric NADH dehydrogenases denominated Ndi1, Nde1, and Nde2 (Velázquez and Pardo, 2001). Nonetheless, the strong dependence of the mammalian CI biogenesis on the MIA pathway was made evident by the fact that Apoptosis Inducing Factor Mitochondria Associated 1 (AIFM1) deficiency, which was discovered to be an important interactor of CHCHD4 in the IMS (Hangen et al., 2015). AIFM1 was originally discovered as a caspase-independent effector of cell death when migrating from the mitochondrion to the nucleus (Susin et al., 1999). However, it soon became clear that AIFM1 defects were related with a strong mitochondrial CI deficiency (Vahsen et al., 2004; Bénit et al., 2008). Subsequent studies revealed that loss of AIFM1 function produced a more generalized combined respiratory chain deficiency in mouse and human cells (Ghezzi et al., 2010; Delavallée et al., 2020), but the full ramifications of the AIFM1-CHCHD4 interaction remain to be resolved (Reinhardt et al., 2020). More recently and using Drosophila melanogaster models, it was suggested that the dependency of CI biogenesis on AIFM1 was not direct but a consequence of decreased Mic19 levels instead (Murari et al., 2020).
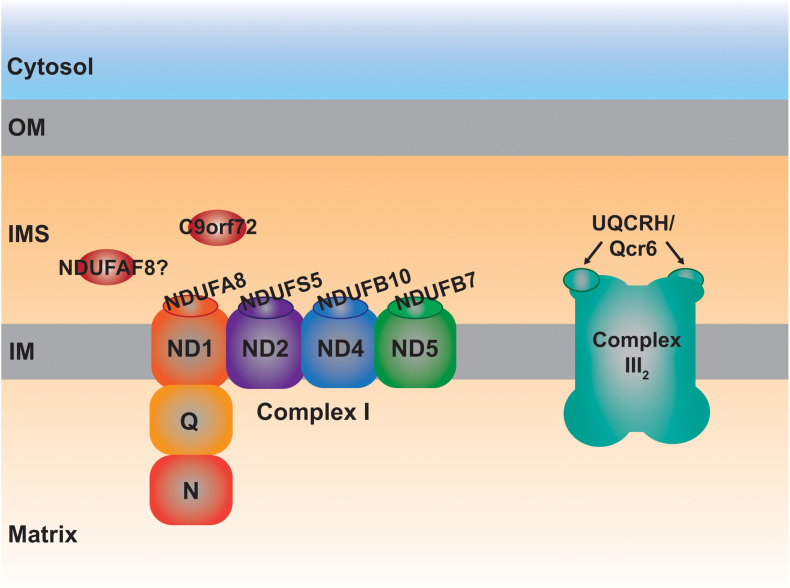
More specifically, four CI supernumerary subunits, i.e., outside the fourteen evolutionarily conserved core subunits, are located in the IMS (Zhu et al., 2016). These are NDUFA8, NDUFB7, NDUFS5 and NDUFB10 (Table 1 and Figure 3), the first three subunits are CHCHD4 substrates containing typical twin Cx9C motifs (Szklarczyk D. et al., 2011; Fischer et al., 2013). NDUFB10 is also imported into the IMS via CHCHD4 (Friederich et al., 2017), although it appears to contain two extra disulfide bonds, all of which most probably contribute to the stability of the complex (Zhu et al., 2016). In fact, lack of any of these four subunits is detrimental to the assembly of CI (Stroud et al., 2016). In accordance, a pathological variant in a conserved Cys residue in NDUFB10 and those in NDUFA8 predicting changes in conserved Arg residues located at the vicinity of the CX9C motifs result in the structural loss of CI (Friederich et al., 2017; Tort et al., 2020; Yatsuka et al., 2020), reinforcing the importance of these domains for the import and/or stability of these IMS subunits and, consequently, that of the whole complex.
FIGURE 3.
Schematic representation of the MIA40 protein substrates located in the IMS involved in the assembly of complexes I and III2. Each one of the IMS CI structural subunits, NDUFA8, NDUFS5, NDUFB10, and NDUFB7 belongs to a different module (ND1, ND2, ND4, and ND5, respectively) of the membrane arm, responsible for proton pumping. The peripheral arm of CI is composed of the quinone binding (Q) and NADH dehydrogenase (N) functional modules. UQCRH, Qcr6 in yeast, is an IMS-located supernumerary subunit.
NDUFAF8 is a CI assembly factor that was identified through proteomic studies as an interactor of other CI-related proteins and then determined to be encoded by a mitochondrial disease gene (Floyd et al., 2016; Alston et al., 2020). Sequence analysis predicted its IMS localization due to the presence of twin CX9C motifs. However, even though the exact submitochondrial localization has not been proven experimentally, it is likely that NDUFAF8 is located in the matrix due to its strong interaction with NDUFAF5, a well-known matrix protein (Floyd et al., 2016; Alston et al., 2020).
Defects in C9orf72 have been associated with the development of amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD) but the molecular functions of this protein are still unclear (Braems et al., 2020). Very recently it was determined that a proportion of C9orf72 is imported into the mitochondrial IMS via AIFM1/CHCHD4 where it binds to the prohibitin complex and the m-AAA protease (Merkwirth and Langer, 2009) regulating the proteolysis of the CI assembly factor TIMMDC1 (Guarani et al., 2014) and, consequently, the amounts of fully assembled CI (Wang et al., 2021).
Role of IMS Proteins in Complex III Biogenesis
Qcr6 in yeast and UQCRH in mammals, is the only structural component of the ten subunits that constitute the cytochrome bc1 complex (or complex III; CIII2) localized completely in the IMS and containing Cx9C motifs (Iwata et al., 1998; Hunte et al., 2000; Szklarczyk R. et al., 2011; Vögtle et al., 2012). Qcr6/UQCRH is a supernumerary subunit, not involved in the catalytic Q-cycle, whose function is still unclear. The first observations in Δqcr6 strains showed normal growth in non-fermentable carbon sources and CIII2 activity. However, further investigations revealed a temperature-sensitive petite phenotype associated with a defect in the assembly of the bc1 complex and lack of maturation of cytochrome c1 (Cyt1), one of the three catalytical subunits (Yang and Trumpowers, 1994). Using different deletion strains, including that of Qcr6, it was determined that the subunit was incorporated on its own in the middle states of the assembly pathway (Zara et al., 2009). However, proteomic studies using a human cell line with a strong defect in CIII2 assembly, revealed the formation of intermediates containing UQCR10, cytochrome c1 (CYC1) and most probably UQCRH as well (Protasoni et al., 2020). One can speculate that Qcr6/UQCRH would somehow contribute to the maturation of CYC1 before it is incorporated into nascent CIII2 also in human mitochondria. This possible function and the involvement of a redox regulatory mechanism through the Cys residues present in UQCRH, remains an interesting aspect to investigate.
Role of IMS Proteins in Complex IV Biogenesis
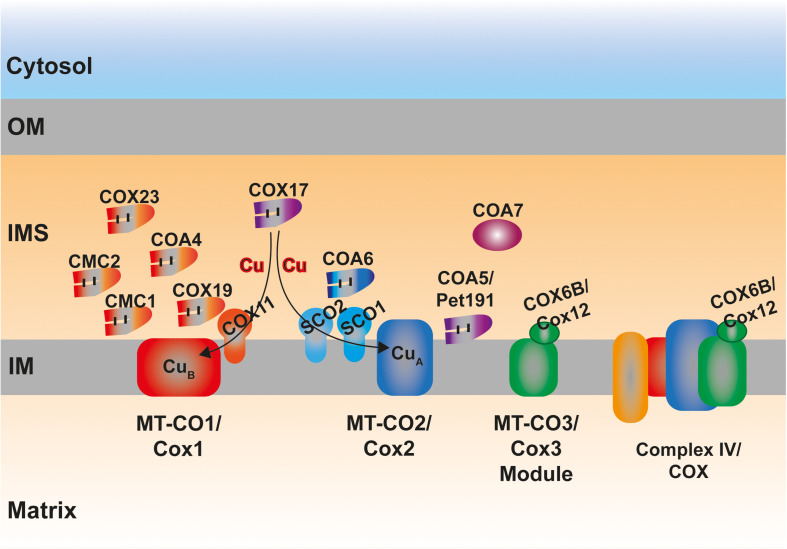
Of all the ETC complexes, cytochrome c oxidase (COX) or complex IV (CIV) is probably the most heavily dependent on Cys-rich redox regulated IMS proteins for its biogenesis and function. COX catalysis, oxidizing cytochrome c, and reducing molecular oxygen, involves the presence of two heme a moieties and one copper (CuB center) ion in Cox1/MT-CO1, and two Cu ions (CuA center) in Cox2/MT-CO2 (Wikström et al., 2018). The processes of metal cofactor insertion, i.e. hemylation and Cu delivery, occur in the IMS and therefore, a significant number of Cys-rich proteins located in this compartment seem to be involved in these processes, especially in the formation of the CuA site, as well as in the stabilization of these two COX subunits in early assembly stages (Khalimonchuk and Winge, 2008; Jett and Leary, 2018; Timón-Gómez et al., 2018; Gladyck et al., 2021; Table 1 and Figure 4). In addition to all the assembly factors that will be described below, there is one structural subunit exclusively localized in the IMS, which is yeast Cox12 or human COX6B1 (ubiquitous isoform) or COX6B2 (testis-specific isoform), containing one CX9C and one CX10C motif (Tsukihara et al., 1996). The first mammalian CIV structure was in dimeric form, appearing COX6B1 to serve as a bridge and stabilize the COX dimer, as well as modulating the interaction with cytochrome c (Kadenbach and Hüttemann, 2015). However, it was recently determined that the monomer harboring COX6B1, could also be isolated and that it displayed high activity (Shinzawa-Itoh et al., 2019). Apart from a stabilization role, a function in Cu delivery to the active center of Cox2/MT-CO2 was also proposed for Cox12/COX6B1 (Ghosh et al., 2016). In any case, this structural subunit is clearly important for completion of the assembly process and for the activity of the enzyme, as mutations destabilizing COX6B1 or Cox12 result in assembly defects and COX deficiency in human and yeast (Massa et al., 2008; Abdulhag et al., 2015).
FIGURE 4.
Schematic representation of the MIA40 protein substrates located in the IMS involved in the assembly of complex IV (COX). The proteins involved in the stabilization and metalation of MT-CO1 and MT-CO2 are indicated in red and blue color, respectively. COX17 is the archetypal twin CX9C protein, mediating copper (Cu) delivery to the active centers of both MT-CO1 and MT-CO2. The exact molecular role of COA5/Pet191 and COA7 is still unknown. COX6B, Cox12 in yeast, is the only COX subunit fully localized in the IMS.
Cmc1 was firstly identified in yeast as an IMS-located protein containing twin CX9C motifs and imported by the MIA pathway that was necessary for COX expression (Horn et al., 2008; Bourens et al., 2012; Fischer et al., 2013). Later, it was determined that human CMC1 was necessary for the stabilization of MT-CO1, being released from the first intermediate of the MT-CO1 module once this subunit joins the early nuclear-encoded subunits COX4 and COX5A (Bourens and Barrientos, 2017a). The exact function of CMC1 is still not clear but it was proposed to either bind Cu itself or to somehow assist the process of MT-CO1 metalation (Horn et al., 2008; Bourens and Barrientos, 2017a).
Cmc2 was identified in yeast via a screening of all the genes encoding for CX9C motif-containing proteins (Longen et al., 2009). Deletion of Cmc2 produced a respiratory-deficient phenotype associated with low COX activity (Longen et al., 2009; Horn et al., 2010). Cmc2 is homologous to and interacts with Cmc1, cooperating in COX assembly with a different non-overlapping molecular role. The human and C. elegans CMC2 orthologs contain the conserved Cys residues, are localized in mitochondria and appear to be involved in CIV biogenesis as well (Horn et al., 2010).
Cmc3 is another member the group of Cx9C proteins identified in yeast, also related with COX biogenesis (Longen et al., 2009). This protein was later renamed Coa4 (cytochrome c oxidase assembly number 4) and its involvement in CIV assembly was confirmed being identified as a genetic suppressor of mutations in Shy1 modeling those found in the human ortholog SURF1 and associated with Leigh syndrome (Petruzzella et al., 1998; Zhu et al., 1998; Bestwick et al., 2010b). The twin CX9C motives in Coa4/Cmc3 are highly conserved in the human protein, which is known also as CHCHD8, being also imported into the IMS using the oxidative folding pathway (Fischer et al., 2013), although its direct relationship with human CIV biogenesis has not been proven experimentally yet.
Pet191 was also determined to be important for COX activity in a screen for respiration deficient yeast strains (McEwen et al., 1993). It contains a variation of the twin Cx9C motifs (i.e., two Cx9C and one Cx10C) and there are still conflicting evidences to whether its import in the IMS is mediated by Mia40 and Erv1 or not (Khalimonchuk and Winge, 2008; Bragoszewski et al., 2013). Although its involvement in COX biogenesis was clearly proven, Pet191 is not a Cu-binding protein (Khalimonchuk and Winge, 2008). C2orf64, also known as COA5, is the human ortholog of S. cerevisiae Pet191 (Szklarczyk et al., 2012). A pathological variant in C2orf64 was associated with COX activity and assembly defects. Although the exact molecular role of this protein remains unknown, its levels are increased in cells where the MT-CO2 copper chaperones SCO1 and SCO2 are defective (Leary et al., 2013). In addition, the pattern of CIV subassemblies accumulated in the patient-derived cultured cells indicated a function in the early steps of assembly (Huigsloot et al., 2011).
Coa6 in yeast is a COX assembly factor as well as an IMS-localized protein and a substrate of the MIA import pathway (Vögtle et al., 2012). The human and zebrafish orthologs also contain the conserved CX9CXnCX10C motifs and have been proven to be important for the correct biogenesis of CIV (Szklarczyk et al., 2012; Ghosh et al., 2014; Baertling et al., 2015; Pacheu-Grau et al., 2015; Stroud et al., 2015). The fact that Cu supplementation restored the growth of defective strains in fermentative substrates, the fact that COA6 binds Cu in vitro, together with the connection of the protein with SCO1 and SCO2 and with MT-CO2 maturation, led to the idea that COA6 was a Cu-binding protein involved in the process of delivery of this metal to the CuA site (Ghosh et al., 2014, 2016; Baertling et al., 2015; Pacheu-Grau et al., 2015; Stroud et al., 2015; Bourens and Barrientos, 2017b; Aich et al., 2018). However, more recent studies have suggested that COA6 is a thiol-reductase binding SCO1 and/or SCO2 and catalyzing the reduction of critical Cys residues in SCO1 and/or SCO2 as well as MT-CO2 allowing the transfer and binding of Cu to MT-CO2 (Soma et al., 2019; Pacheu-Grau et al., 2020). The exact COA6 binding partners and mechanisms are still not completely clear as the interpretation of the results varied in different laboratories depending on the experimental approach used (Maghool et al., 2020).
COA7/C1orf63/SELRC1 is a protein only present in metazoa that was originally studied as a Mitofilin interactor whose decreased levels were associated with defects in the respiratory chain (Kozjak-Pavlovic et al., 2014). Although one report localized this protein within the mitochondrial matrix (Lyons et al., 2016), more detailed biochemical analyses determined its IMS localization (Kozjak-Pavlovic et al., 2014; Mohanraj et al., 2019). Although it does not contain the typical Cx3C or Cx9C motifs, it is a Cys-rich protein that it is imported inside the mitochondria via CHCHD4 (Mohanraj et al., 2019). The first indication that COA7 is involved in COX biogenesis came from the fact that pathological variants in the COA7 gene are associated with mitochondrial CIV deficiency in humans (Lyons et al., 2016; Higuchi et al., 2018; Ban et al., 2021). Recent data suggest that it is a haem-binding protein involved in the early stages of COX assembly (Formosa et al., 2021). The right Cys composition in COA7 was shown to be crucial for IMS import efficiency and for its stability (Mohanraj et al., 2019).
Cox11 deficiency in yeast results in decreased COX levels (Tzagoloff et al., 1990). Originally proposed to be involved in heme a synthesis (Tzagoloff et al., 1993) it is clear now that it is a Cu binding protein involved in the formation of the CuB center in Cox1, through conserved Cys residues in its Cu-binding motif (Hiser et al., 2000; Carr et al., 2002; Banci et al., 2004). A gene encoding a protein showing high homology to yeast Cox11 is present in the human genome (Petruzzella et al., 1998) and although the direct involvement of COX11 in metalation human CIV assembly has not yet been proven, it was shown to interact with CIV subunits and assembly factors (Vidoni et al., 2017).
Cox17 is the archetypal twin CX9C IMS protein involved in Cu binding in mitochondria, and it has been extensively studied at the functional and structural levels both in yeast and human systems (Gladyck et al., 2021). Cox17 plays a fundamental role in COX biogenesis by donating Cu to Sco1 for metalation of Cox2 on one side, and to Cox11 for the formation of the CuB center, on the other (Horng et al., 2004; Cobine et al., 2006, 2021). The highly conserved residues contained in the Cx9C motifs and in the Cu binding motif are necessary for import using the MIA system, binding of Cu, oligomerization and promoting CIV assembly (Heaton et al., 2000, 2001; Palumaa et al., 2004; Mesecke et al., 2005; Banci et al., 2008a, b, 2011b). Although Cox17 is found both in the IMS and the cytosol and for that reason it was proposed to act as a Cu shuttle to the inside of mitochondria (Beers et al., 1997), other investigations point out to the fact that the only active partially oxidized form of Cox17 is the one present in the IMS (Maxfield et al., 2004; Palumaa et al., 2004; Banci et al., 2008b). COX17 is also an essential protein in mammals as knocking-out the mouse gene leads to embryonic lethality (Takahashi et al., 2002). Human cell lines with knock-down expression of COX17 display decreased amounts of MT-CO1 and MT-CO2 together with the accumulation of assembly intermediates containing MT-CO1 but lacking MT-CO2 (Oswald et al., 2009; Vanišová et al., 2019). Curiously, the proportion of CIV associated with complexes I and III within the supercomplexes was more sensitive to the loss of COX17 that the “free” complex (Oswald et al., 2009). In addition, intramitochondrial Cu amounts were reduced in human cells where COX17 expression was stably knocked down (Vanišová et al., 2019). All in all, these observations point out to a completely conserved function of COX17 between yeast and humans.
The amino acid sequence of Cox19 is homologous to that of Cox17, containing the twin CX9C motifs but lacking the additional Cys residues of the Cu binding domain (Gladyck et al., 2021). As indicated by its name it is also a necessary factor for COX biogenesis (Nobrega et al., 2002). Similarly, the yeast and human proteins also localize both in the cytosol and mitochondria, being the steady-state levels and distribution of COX19 Cu-dependent (Nobrega et al., 2002; Leary et al., 2013). The import into the IMS is mediated by Mia40/CHCHD4 (Fischer et al., 2013) and Cox19 also shows Cu binding properties in vitro (Rigby et al., 2007), whereas, in vivo Cu binding by Cox19 appears to be less likely. COX19 interacts with COX11 in a redox regulated manner, most likely to keep the Cu-binding Cys residues of COX11 reduced and, therefore, in an active form (Bode et al., 2015; Cobine et al., 2021).
Cox23 was again identified in S. cerevisiae as a protein necessary for COX biogenesis showing high homology to Cox17 (Barros et al., 2004) with a potential function in Cox1 maturation (Dela Cruz et al., 2016). Structural analysis of the human homolog CHCHD7/COX23, predicted the presence of an ITS and the dependence on MIA40 for its folding creating the typical disulfide bridges between the twin CX9C motifs (Banci et al., 2012). Interestingly, the levels of human COX23 decrease when the Cu binding proteins SCO1 and SCO2 are defective or in the presence of a Cu chelating agent (Leary et al., 2013).
SCO1 and SCO2 are not members of the CHCHD family, but they are IM tethered proteins facing the IMS containing Cys residues important for Cu binding, essential for their function in the metalation of MT-CO2 (Nittis et al., 2001; Leary et al., 2004). As pointed out above, there is clearly an interplay between SCO1 and SCO2 and the twin CX9C motif-containing chaperones involved in Cu delivery to the COX active sites, and this is illustrated by the fact that yeast Sco1 and Sco2 were originally identified as suppressors of Cox17 deficiency (Moira Glerum et al., 1996b). However, contrary to Sco1, the lack of Sco2 did not affect COX (Moira Glerum et al., 1996b). This differs from human mitochondria, where defects in both SCO1 and SCO2 result in CIV deficiency and pathological mutations in both these genes are associated with manifestations of mitochondrial diseases (Papadopoulou et al., 1999; Jaksch et al., 2000; Valnot et al., 2000; Leary et al., 2004; Fernandez-Vizarra and Zeviani, 2021). In human cells, SCO1 and SCO2 have non-overlapping cooperative roles in MT-CO2 maturation, where SCO2 acts upstream of SCO1 oxidizing its Cu-binding Cys residues during the process of Cu delivery to MT-CO2 (Leary et al., 2004, Leary et al., 2009). The importance of these processes not only for COX biogenesis but for other aspects of cellular health, is pointed out by the fact that functional SCO1 and SCO2 are required to maintain Cu homeostasis (Leary et al., 2007; Cobine et al., 2021).
Reactive Oxygen Species-Regulated Assembly and Turnover of the ETC Complexes
The production of ROS is inherent to the aerobic metabolic routes located in mitochondria (Murphy, 2009; Brand, 2016). These ROS are mainly superoxide (O2–), the majority of which is produced at the levels of complexes I and III (Brand, 2010, 2016), which is subsequently dismutated to hydrogen peroxide (H2O2), and the highly reactive and unstable hydroxyl radical (OH), produced through the Fenton reaction (Lambert and Brand, 2009). Mitochondria also produce other redox active compounds such as reactive nitrogen species (RNS), mainly nitric oxide (NO) (Ghafourifar and Cadenas, 2005), or H2S (Kimura et al., 2010; Paul et al., 2021), which induce modifications of Cys in proteins, modulating their activities (Habich et al., 2019b). However, discussion of these species is beyond the scope of this review, which focuses on ROS directly produced by the ETC (Figure 5), due to electrons slipping off of the system and escaping from the redox active centers to partially reduce oxygen. The idea that ROS are only harmful molecules leading to detrimental effects for the cell and causing different diseases and aging, is now changing (Scialò et al., 2017; Ji and Yeo, 2021). Nowadays, ROS are recognized as signaling molecules mediating a myriad of cellular responses (Holmström and Finkel, 2014; Reczek and Chandel, 2015). However, in parallel, cells have had to develop antioxidant defense systems to protect themselves from ROS overaccumulation and oxidative damage, whilst these antioxidant systems themselves might be involved in the ROS-mediated signaling as well (Holmström and Finkel, 2014; Reczek and Chandel, 2015). Consequently, a correct balance between ROS production and scavenging is important to ensure the appropriate cellular responses while preventing oxidative stress (Ji and Yeo, 2021). In fact, a moderate production of ROS is the basis of the phenomenon called “mitohormesis” leading to a stress adaptive response and ultimately to improved mitochondrial function and life extension (Schulz et al., 2007; Palmeira et al., 2019). In addition, it is becoming evident that ROS produced in the mitochondria play an important role in mediating mitochondrial biogenesis to compensate for ETC dysfunction (Dogan et al., 2018).
FIGURE 5.
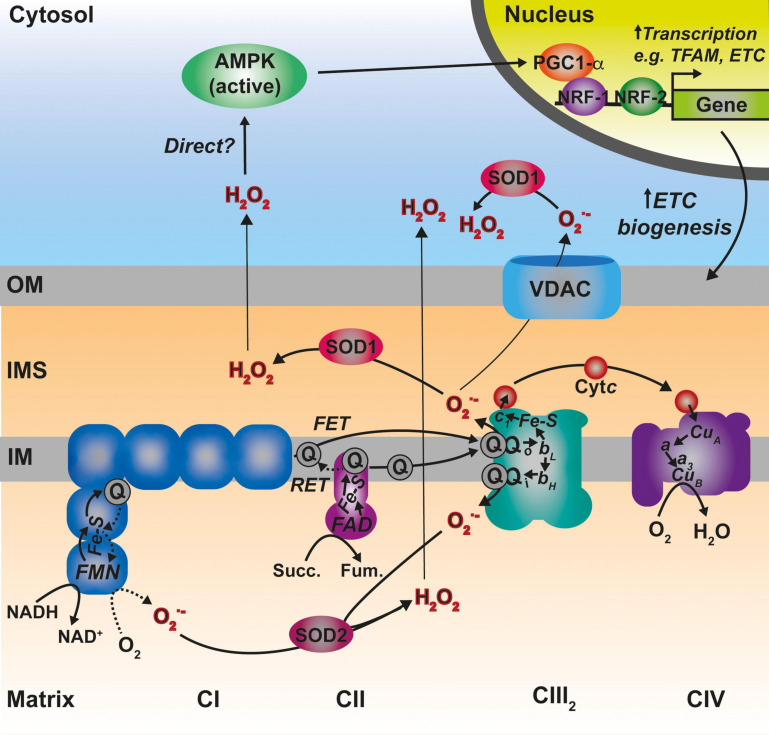
Main sites of ROS generation by the ETC and possible pathways of ROS-mediated regulation of mitochondrial biogenesis. Complex I (CI) represented in blue, generates superoxide (O2–) both via forward electron transfer (FET) or reverse electron transfer (RET) from a highly reduced coenzyme Q (Q) pool and high membrane potential. Two other prominent sites of O2– production are the two Q binding sites in CIII2 (Qo and Qi). O2– is dismutated to hydrogen peroxide (H2O2) by the two differently localized superoxide dismutases SOD1 and SOD2. See main text for details.
Superoxide radicals produced by CIII2 and glycerol-3-phosphate dehydrogenase (GPDH) are generated in both sides of the IM (the IMS and the matrix), whereas, all the other sites produce ROS in the matrix (Brand, 2010; Figure 5). In addition, ALR (Erv1) seems to be a significant contributor of O2– radicals within the IMS (Daithankar et al., 2012). O2– produced in the matrix side is dismutated to H2O2 by the mitochondrial Mn superoxide dismutase (SOD2) and can then diffuse to the cytosol (Boveris et al., 2006; Murphy, 2009), whereas, that released into the IMS can be transported through voltage-dependent ion channels into the cytosol (Han et al., 2003; Muller et al., 2004). Once in there, the Cu,Zn superoxide dismutase (SOD1) converts it to H2O2. SOD1 is dually localized in the cytosol and the IMS in mammals and yeast, and the IMS pool appears to be inactive in basal conditions and activated by modulating its redox state (Iñarrea et al., 2005, 2007). H2O2 in the cytosol is then able to modify the redox state of key Cys residues in target proteins, modulating the function of factors such as signaling regulatory kinases and phosphatases or transcription factors (Holmström and Finkel, 2014; Reczek and Chandel, 2015).
There are three aspects in which ROS production in mitochondria may influence directly the biogenesis and function of the ETC complexes: (1) by generating oxidation in the IMS and inducing changes in the redox state of key Cys residues and thus, in the structure and function of the redox-sensitive biogenesis factors; (2) the role of ROS produced inside mitochondria as signaling molecules to regulate ETC biogenesis; and (3) ROS-mediated protease activation inducing the degradation of components of the ETC complexes.
Redox Homeostasis in the IMS and Regulation of ETC Biogenesis
Given the essentiality of the Cys-containing subunits and assembly factors of the ETC complexes, a correct redox homeostasis in the IMS is crucial for the biogenesis of the respiratory complexes. Thus, it is important to maintain the redox sensitive residues in the correct oxidation state for import and retention in the IMS, as well as for protein function (Habich et al., 2019a; Dickson-Murray et al., 2021). Considering the number of redox regulated IMS-located factors involved, this is particularly relevant for the assembly and metalation of CIV (Khalimonchuk and Winge, 2008; Jett and Leary, 2018). Although there are still many open questions as to how the IMS maintains its redox homeostasis (Dickson-Murray et al., 2021), it is clear that it contains several systems that ensure it, having glutathione as a central component (Calabrese et al., 2017). One such mechanism for maintaining homeostasis in this compartment is the retro-translocation of reduced proteins back into the cytosol through the Tom40 import pore protein. This provides a means to regulate the abundance of IMS proteins either in response to various changes in cellular physiology or for the removal of misfolded proteins by presenting them to protein quality control machineries located in the cytosol (Bragoszewski et al., 2015). Surprisingly, the IMS is not as oxidizing as one would expect, being the glutathione pools as highly reduced in yeast and human mitochondria as in the cytosol (Kojer et al., 2012; Fischer et al., 2013). In such a reducing environment, the essential oxidative folding pathway is maintained by limiting glutaredoxin (Grx) activity in the IMS (Kojer et al., 2015). As mentioned before, the IMS, where most of the Cys-rich redox sensitive proteins reside, receives significant amounts of ROS. H2O2 oxidizes target thiols of the Cys residues to form disulfide bonds, but also to higher oxidation states such as sulfenic, sulfinic and sulfonic acids, changing the functional properties of the Cys-containing proteins (Habich et al., 2019b). Sulfonylation is irreversible and sulfinylation was thought to be as well until an enzyme called sulfiredoxin (Srx) was identified in yeast and humans, catalyzing the reduction of Cys sulfinic acids in vivo (Jacob et al., 2004). Enzymes aimed to detoxify H2O2 and/or remove thiol modifications have been found in the IMS of both yeast and human mitochondria (Vögtle et al., 2012; Hung et al., 2014). Peroxiredoxins 3 and 4 (PRDX3 and 4) react directly with H2O2, reducing it to water, resulting in the oxidation of Cys residues that can be reduced again by thioredoxin 1 (TRX1), TRX1 is then recycled by Thioredoxin reductase (TRR) (Cardenas-Rodriguez and Tokatlidis, 2017; Habich et al., 2019b). In addition, glutathione peroxidase 3 (Gpx3) is found in the IMS, where it becomes actively targeted to through the addition of an N-terminal extension of 18 amino acids (N18) and is thought to play a role in H2O2 detoxification (Kritsiligkou et al., 2017). Translation of this long form of Gpx3 with the N18 extension can be modulated by H2O2 stress (Gerashchenko et al., 2012). The cytoplasmic isoform of GPX4, the human homolog of Gpx3, has also been found in mitochondria (Liang et al., 2009; Tadokoro et al., 2020) but the role it plays there is still unclear.
ROS-Mediated Signaling Promoting ETC Biogenesis
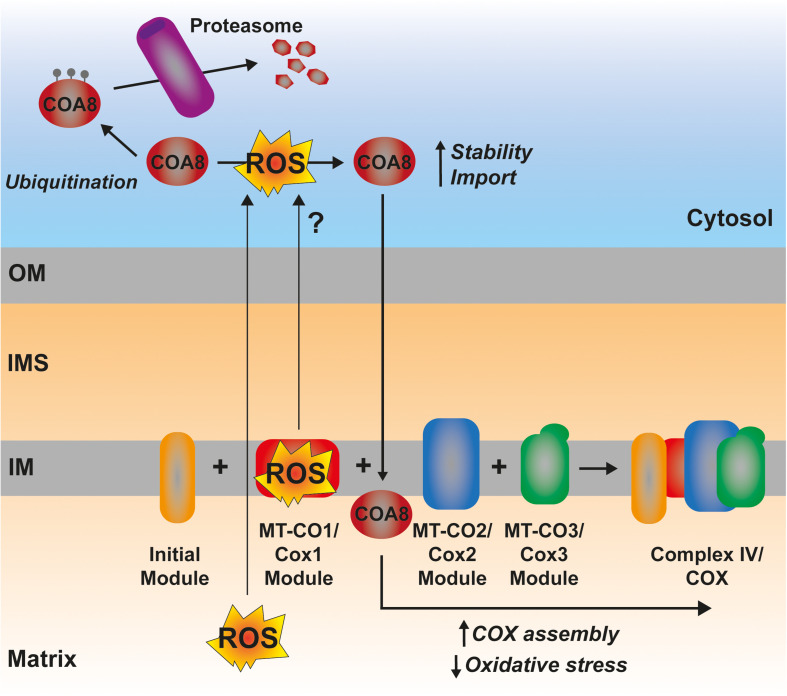
In certain situations, dysfunction or inhibition of CI, CIII, or CIV leads to increased ROS production (Verkaart et al., 2007; Kowaltowski et al., 2009; Brand, 2016; Dogan et al., 2018). However, as briefly discussed above, the nature and topology of these ROS is different depending on what complex is affected. In addition, the physio-pathological consequences can also diverge. CI can generate ROS both by “forward electron transfer (FET)” or by “reverse electron transfer (RET),” i.e., by transferring electrons from a highly reduced CoQ pool back to CI, in conditions where the membrane potential is high (Robb et al., 2018; Figure 5). This is relevant because ROS generated at the level of CI, but only if its via RET, are able to induce an adaptive program leading to improved mitochondrial activity and increased life span in D. melanogaster (Scialò et al., 2016). O2– generated at the level of the first CoQ binding site of cIII2 (Qo), closest to the IMS, has been described to be the origin of signaling pathways such as the hypoxic responses (Guzy et al., 2005; Bell et al., 2007). At the mitochondrial level, the ROS-mediated hypoxic transcriptional program leads to the expression of specific COX isoforms, regulating the activity of the complex under these conditions (Bourens et al., 2013). Also, ROS produced in the mitochondria activate AMPK, which activates PGC-1α, a transcriptional master regulator of mitochondrial biogenesis (Jäer et al., 2007; Rabinovitch et al., 2017; Figure 5). This ROS-mediated signaling axis is necessary to induce an increase in mitochondrial mass to counteract CIV deficiency in muscle (Dogan et al., 2018). Apart from these mitochondrial remodeling effects mediated by transcriptional changes, an attractive possibility is that ROS produced in the mitochondria can directly modulate the activity of biogenetical factors such as COA8 (Figure 6). The absence of COA8 (formerly known as APOPT1) leads to CIV deficiency and the accumulation of MT-CO1-containing partially assembled species (Melchionda et al., 2014; Signes and Fernandez-Vizarra, 2018). COA8 was shown to be readily degraded by the proteasome in the cytosol but stabilized inside mitochondria when cells were stressed with both exogenous H2O2 or with mitochondrially targeted paraquat (mitoPQ), which induces the production of O2– in the matrix (Signes and Fernandez-Vizarra, 2018). The COA8 sequence contains Cys residues within the mitochondrial targeting sequence that could potentially be oxidized by H2O2 in the cytosol, inducing its stabilization and/or mitochondrial import (Signes and Fernandez-Vizarra, 2018). Yeast strains deleted of Sco1 or Cox11 show increased sensitivity to acute hydrogen peroxide stress (Khalimonchuk et al., 2007; Veniamin et al., 2011). This effect was linked to impaired COX assembly leading to the accumulation of hemylated Cox1 intermediates acting as pro-oxidant species (Khalimonchuk et al., 2007). Interestingly, COA8-null patient derived fibroblasts produce more ROS when they are oxidatively challenged (Melchionda et al., 2014) and dCoa8 knock-down flies are more sensitive to oxidative stress (Brischigliaro et al., 2019). A hypothesis that can be derived from these observations is that ROS produced because of the accumulation of pro-oxidant MT-CO1 subcomplexes serve as a signal to induce the assembly of CIV mediated by COA8, which also appeared to have a role in protecting MT-CO1 from oxidative stress induced degradation (Signes and Fernandez-Vizarra, 2018). More experimental data must be collected to confirm or disprove these ideas and to determine whether there are more redox sensitive assembly factors for CIV and the rest of the complexes, which may be regulated in this way to promote a rapid biogenetical response to regulate ETC function.
FIGURE 6.
Possible mechanism of ROS-mediated CIV assembly regulation by COA8. The ROS produced by a hemylated partially assembled MT-CO1 could represent a signal triggering the increased import and/or stabilization of COA8, promoting CIV assembly and, thereby, reducing oxidative stress.
ROS-Mediated Degradation of ETC Components
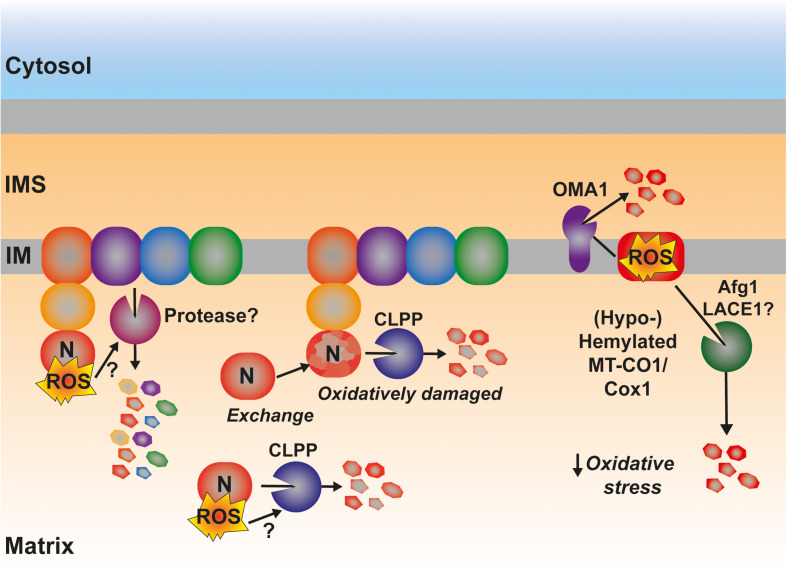
A much less explored yet interesting component in the control of the amounts and function of the ETC is the turnover, i.e., balance between synthesis and degradation, of its components (Szczepanowska and Trifunovic, 2021). In yeast, the accumulation of pro-oxidant COX subcomplexes is attenuated by the activity of the AAA-ATPase Afg1, facilitating the degradation of Cox1, Cox2, and Cox3 (Khalimonchuk et al., 2007; Figure 7). Afg1 human ortholog is LACE1, and it was shown to be involved in the degradation of nuclear-encoded CIV subunits (Cesnekova et al., 2016). As mentioned earlier, Cox1 accumulated due to the absence of the assembly factor Coa2 is degraded by Oma1 (Khalimonchuk et al., 2012) and Oma1 is redox regulated both in yeast and human mitochondria (Bohovych et al., 2019). Also in human mitochondria, in the case of defective CIV assembly at different stages with the accumulation of MT-CO1 module subassemblies, is associated with an increased turnover of the mtDNA-encoded CIV subunits (Leary et al., 2009; Bourens et al., 2014; Bourens and Barrientos, 2017a, b; Signes and Fernandez-Vizarra, 2018). Another ETC component in mammals, whose turnover is potentially regulated by ROS is CI (Figure 7). The assembly of this huge complex happens in a modular fashion and the N-module containing the redox active Fe-S clusters and flavin (FMN) is preassembled and then added in the last step of assembly (Guerrero-Castillo et al., 2017). CLPP is responsible for the turnover of both the pre-assembled and assembled N-module (Szczepanowska et al., 2020). In this case, one could think that this might be a way of preventing the accumulation of a highly reactive pro-oxidant species and to remove oxidatively damaged protein components in proximity to the N-module FMN center, which is one of the main sites of ROS production in mitochondria (Brand, 2016; Hirst and Roessler, 2016). Then again, mammalian CI shows a high interdependence with the other components of the ETC. Strong CIII2 and CIV assembly defects produce a secondary decline in CI amounts (Diaz et al., 2006; Protasoni et al., 2020; Čunátová et al., 2021). This was explained by the existence of an active degradation of fully assembled CI in response to RET-mediated ROS production and oxidative damage (Guarás et al., 2016). However, the exact mechanism and the protease/s mediating this accelerated turnover have not been defined yet, and other levels of regulation, i.e., repression of mitochondrial translation and attenuated assembly of the N-module, have been shown (Protasoni et al., 2020; Čunátová et al., 2021). In the case of the CIII2 defects originated by mutations in the central subunit MT-CYB, mitochondrial translation was not affected (Protasoni et al., 2020; Tropeano et al., 2020). However, the steady-state levels of CI subunits and specially those of the N-module were severely reduced (Protasoni et al., 2020; Páleníková et al., 2021). One can imagine that if the N-module is being preassembled but not incorporated into fully assembled CI, it will need to be degraded before its assembly, as previously shown (Szczepanowska et al., 2020). One can hypothesize that this might be ROS mediated and induced by a pro-oxidant intermediate, because xeno-expression of an alternative oxidase (AOX), which decreases ROS production by the ETC (Dogan et al., 2018; Robb et al., 2018; Szibor et al., 2020), resulted in increased amounts of N-module subunits and fully assembled CI in MT-CYB mutated human cells (Protasoni et al., 2020).
FIGURE 7.
Possible involvement of ROS in the regulation of the degradation by active proteolysis of some of the catalytical components of the ETC. ROS produced by RET at the level of CI have been proposed to trigger the degradation of the full complex. The mechanistic details are still unknown. The matrix protease CLPP is responsible for the degradation of the catalytic N-module of CI, preventing its accumulation as an assembly intermediate in the mitochondrial matrix as well as promoting the interchange of an oxidatively damaged N-module for a newly synthesized one. Some Cox1 (MT-CO1) intermediates are actively degraded by the inner membrane-bound protease OMA1, which is redox activated. Other pro-oxidant Cox1 intermediates are degraded through a mechanism mediated by the AAA-ATPase Afg1 in yeast being LACE1 its human ortholog. See main text for details.
Whether the enhanced proteolysis of the redox active, and potentially damaging, CI and CIV subassemblies is directly modulated by ROS, and which are quality control proteases that are involved and regulated in this way, constitute extremely interesting topics to explore in the future.
Conclusion and Perspectives
Mitochondria contain dedicated import and sorting machineries to direct mitochondrial proteins that are nuclear encoded and synthesized in the cytoplasm. The MIA pathway makes use of reduction and oxidation reactions involving particular Cys residues so that the correctly folded and functional proteins are retained in the IMS. Therefore, redox homeostasis in the IMS plays a crucial role in the regulation of the ETC complexes because essential factors involved in their maturation are proteins containing twin Cx9C motifs and substrates of Mia40/CHCHD4. In order to guarantee a correct ETC biogenesis, redox homeostasis of the IMS needs to be maintained in spite of its proximity to the ETC. The ETC contains major sites of ROS production, which were traditionally envisioned as merely harmful molecules contributing to aging and disease. It is now becoming clearer that an exquisite balance between ROS production and scavenging is essential to maintain mitochondrial activity. Another emerging concept is that they constitute signaling molecules important for a correct mitochondrial biogenesis, mediating feedback mechanisms regulating the biogenesis and degradation of ETC components. The mechanisms and players underlying these phenomena are just starting to be unraveled. In this respect, an interesting hypothesis to explore is the fact that ROS might be a sensor of unbalanced ETC activity, triggering mechanisms aimed to compensate it, through biogenetical factors and proteases whose activity is directly redox regulated.
Author Contributions
SG and EF-V wrote the first draft and produced the figures and table. EF-V and KT coordinated the writing. KT obtained funding. All authors edited the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Paul A. Cobine (Auburn University, Auburn, AL, United States), Scot C. Leary (University of Saskatchewan, Saskatoon, SK, Canada), Oleh Khalimonchuk (University of Nebraska, Lincoln, NE, United States), David Pacheu-Grau (University of Zaragoza, Spain), and the members of our lab for helpful discussions and comments on the manuscript.
Funding
Work in our laboratory was supported by UKRI-BBSRC (grants BB/R009031/1 and BB/T003804/1) and a BBSRC-EPSRC Impact Accelerator grant and a Wellcome Trust early concept development grant (University of Glasgow). Structural biology work in our laboratory benefited from access to the Solution NMR, CERM/CIRMMP, Florence, Italy, an Instruct-ERIC center. Financial support was provided by Instruct-ERIC (PID: 12148).
References
- Abdulhag U. N., Soiferman D., Schueler-Furman O., Miller C., Shaag A., Elpeleg O., et al. (2015). Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet. 23 159–164. 10.1038/ejhg.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aich A., Wang C., Chowdhury A., Ronsör C., Pacheu-Grau D., Richter-Dennerlein R., et al. (2018). COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis. Elife 7:32572. 10.7554/eLife.32572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S., Balabanidou V., Sideris D. P., Lisowsky T., Tokatlidis K. (2005). Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353 937–944. 10.1016/j.jmb.2005.08.049 [DOI] [PubMed] [Google Scholar]
- Alston C. L., Veling M. T., Heidler J., Taylor L. S., Alaimo J. T., Sung A. Y., et al. (2020). Pathogenic Bi-allelic Mutations in NDUFAF8 Cause Leigh Syndrome with an Isolated Complex I Deficiency. Am. J. Hum. Genet. 106 92–101. 10.1016/j.ajhg.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R., Glerum D. M., Tzagoloff A. (1997). Isolation of a cDNA encoding the human homolog of COX17, a yeast gene essential for mitochondrial copper recruitment. Hum. Genet. 99 329–333. 10.1007/s004390050367 [DOI] [PubMed] [Google Scholar]
- Anand R., Wai T., Baker M. J., Kladt N., Schauss A. C., Rugarli E., et al. (2014). The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204 919–929. 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araiso Y., Imai K., Endo T. (2020). Structural snapshot of the mitochondrial protein import gate. FEBS J. 10.1111/febs.15661 [DOI] [PubMed] [Google Scholar]
- Araiso Y., Tsutsumi A., Qiu J., Imai K., Shiota T., Song J., et al. (2019). Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575 395–401. 10.1038/s41586-019-1680-7 [DOI] [PubMed] [Google Scholar]
- Backes S., Hess S., Boos F., Woellhaf M. W., Gödel S., Jung M., et al. (2018). Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 217 1369–1382. 10.1083/jcb.201708044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertling F., van den A. M., Brand M., Hertecant J. L., Al-Shamsi A., van den P., et al. (2015). Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 36 34–38. 10.1002/humu.22715 [DOI] [PubMed] [Google Scholar]
- Baker M. J., Lampe P. A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., et al. (2014). Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33 578–593. 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. J., Mooga V. P., Guiard B., Langer T., Ryan M. T., Stojanovski D. (2012). Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J. Mol. Biol. 424 227–239. 10.1016/j.jmb.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Ban R., Liu Z., Shimura M., Tong X., Wang J., Yang L., et al. (2021). Biallelic COA7-Variants Leading to Developmental Regression With Progressive Spasticity and Brain Atrophy in a Chinese Patient. Front. Genet. 12 1–8. 10.3389/fgene.2021.685035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K., et al. (2017). Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 19 856–863. 10.1038/ncb3560 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Calderone V., Cefaro C., Ciofi-Baffoni S., Gallo A., et al. (2011a). Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc. Natl. Acad. Sci. U. S. A. 108 4811–4816. 10.1073/pnas.1014542108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L., Bertini I., Calderone V., Cefaro C., Ciofi-Baffoni S., Gallo A., et al. (2012). An electron-transfer path through an extended disulfide relay system: The case of the redox protein ALR. J. Am. Chem. Soc. 134 1442–1445. 10.1021/ja209881f [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cantini F., Ciofi-Baffoni S., Gonnelli L., Mangani S. (2004). Solution structure of Cox11, a novel type of β-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J. Biol. Chem. 279 34833–34839. 10.1074/jbc.M403655200 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gajda K., Felli I. C., et al. (2013). An intrinsically disordered domain has a dual function coupled to compartment-dependent redox control. J. Mol. Biol. 425 594–608. 10.1016/j.jmb.2012.11.032 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A. (2011b). Functional role of two interhelical disulfide bonds in human Cox17 protein from a structural perspective. J. Biol. Chem. 286 34382–34390. 10.1074/jbc.M111.246223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L., Bertini I., Cefaro C., Ciofi-Baffoni S., Gallo A., Martinelli M., et al. (2009). MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16 198–206. 10.1038/nsmb.1553 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Hadjiloi T., Martinelli M., Palumaa P. (2008a). Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105 6803–6808. 10.1073/pnas.0800019105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Janicka A., Martinelli M., Kozlowski H., et al. (2008b). A structural-dynamical characterization of human Cox17. J. Biol. Chem. 283 7912–7920. 10.1074/jbc.M708016200 [DOI] [PubMed] [Google Scholar]
- Barros M. H., Johnson A., Tzagoloff A. (2004). COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 279 31943–31947. 10.1074/jbc.M405014200 [DOI] [PubMed] [Google Scholar]
- Becker T., Wenz L. S., Krüger V., Lehmann W., Müller J. M., Goroncy L., et al. (2011). The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194 387–395. 10.1083/jcb.201102044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers J., Glerum D. M., Tzagoloff A. (1997). Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 272 33191–33196. 10.1074/jbc.272.52.33191 [DOI] [PubMed] [Google Scholar]
- Bell E. L., Klimova T. A., Eisenbart J., Moraes C. T., Murphy M. P., Budinger G. R. S., et al. (2007). The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 177 1029–1036. 10.1083/jcb.200609074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénit P., Goncalves S., Dassa E. P., Brière J. J., Rustin P. (2008). The variability of the Harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS One 3:3208. 10.1371/journal.pone.0003208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick M., Jeong M.-Y., Khalimonchuk O., Kim H., Winge D. R. (2010a). Analysis of Leigh Syndrome Mutations in the Yeast SURF1 Homolog Reveals a New Member of the Cytochrome Oxidase Assembly Factor Family. Mol. Cell. Biol. 30 4480–4491. 10.1128/mcb.00228-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick M., Khalimonchuk O., Pierrel F., Winge D. R. (2010b). The Role of Coa2 in Hemylation of Yeast Cox1 Revealed by Its Genetic Interaction with Cox10. Mol. Cell. Biol. 30 172–185. 10.1128/mcb.00869-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly K. N., Sawaya M. R., Schmid E., Koehler C. M. (2008). The Tim8-Tim13 Complex Has Multiple Substrate Binding Sites and Binds Cooperatively to Tim23. J. Mol. Biol. 382 1144–1156. 10.1016/j.jmb.2008.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode M., Longen S., Morgan B., Peleh V., Dick T. P., Bihlmaier K., et al. (2013). Inaccurately assembled cytochrome c oxidase can lead to oxidative stress-induced growth arrest. Antioxidants Redox Signal. 18 1597–1612. 10.1089/ars.2012.4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode M., Woellhaf M. W., Bohnert M., Van Der Laan M., Sommer F., Jung M., et al. (2015). Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol. Biol. Cell 26 2385–2401. 10.1091/mbc.E14-11-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I., Dietz J. V., Swenson S., Zahayko N., Khalimonchuk O. (2019). Redox Regulation of the Mitochondrial Quality Control Protease Oma1. Antioxidants Redox Signal. 31 429–443. 10.1089/ars.2018.7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I., Donaldson G., Christianson S., Zahayko N., Khalimonchuk O. (2014). Stress-triggered activation of the metalloprotease Oma1 involves its C-terminal region and is important for mitochondrial stress protection in yeast. J. Biol. Chem. 289 13259–13272. 10.1074/jbc.M113.542910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I., Fernandez M. R., Rahn J. J., Stackley K. D., Bestman J. E., Anandhan A., et al. (2015). Metalloprotease OMA1 Fine-tunes Mitochondrial Bioenergetic Function and Respiratory Supercomplex Stability. Sci. Rep. 5:13989. 10.1038/srep13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M., Barrientos A. (2017a). A CMC 1 -knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. EMBO Rep. 18 477–494. 10.15252/embr.201643103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M., Barrientos A. (2017b). Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 292 7774–7783. 10.1074/jbc.M117.778514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M., Boulet A., Leary S. C., Barrientos A. (2014). Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum. Mol. Genet. 23 2901–2913. 10.1093/hmg/ddu003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M., Dabir D. V., Tienson H. L., Sorokina I., Koehler C. M., Barrientos A. (2012). Role of twin Cys-Xaa9-Cys motif cysteines in mitochondrial import of the cytochrome c oxidase biogenesis factor Cmc1. J. Biol. Chem. 287 31258–31269. 10.1074/jbc.M112.383562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M., Fontanesi F., Soto I. C., Liu J., Barrientos A. (2013). Redox and reactive oxygen species regulation of mitochondrial cytochrome C oxidase biogenesis. Antioxidants Redox Signal. 19 1940–1952. 10.1089/ars.2012.4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A., Valdez L. B., Zaobornyj T., Bustamante J. (2006). Mitochondrial metabolic states regulate nitric oxide and hydrogen peroxide diffusion to the cytosol. Biochim. Biophys. Acta - Bioenerg. 1757 535–542. 10.1016/j.bbabio.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Braems E., Swinnen B., Van Den Bosch L. (2020). C9orf72 loss-of-function: a trivial, stand-alone or additive mechanism in C9 ALS/FTD? Acta Neuropathol. 140 625–643. 10.1007/s00401-020-02214-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragoszewski P., Gornicka A., Sztolsztener M. E., Chacinska A. (2013). The Ubiquitin-Proteasome System Regulates Mitochondrial Intermembrane Space Proteins. Mol. Cell. Biol. 33 2136–2148. 10.1128/mcb.01579-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragoszewski P., Wasilewski M., Sakowska P., Gornicka A., Böttinger L., Qiu J., et al. (2015). Retro-translocation of mitochondrial intermembrane space proteins. Proc. Natl. Acad. Sci. U. S. A. 112 7713–7718. 10.1073/pnas.1504615112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambley C. A., Marsee J. D., Halper N., Miller J. M. (2019). Characterization of Mitochondrial YME1L Protease Oxidative Stress-Induced Conformational State. J. Mol. Biol. 431 1250–1266. 10.1016/j.jmb.2019.01.039 [DOI] [PubMed] [Google Scholar]
- Brand M. D. (2010). The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 45 466–472. 10.1016/j.exger.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M. D. (2016). Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 100 14–31. 10.1016/j.freeradbiomed.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Brischigliaro M., Corrà S., Tregnago C., Fernandez-Vizarra E., Zeviani M., Costa R., et al. (2019). Knockdown of APOPT1/COA8 Causes Cytochrome c Oxidase Deficiency, Neuromuscular Impairment, and Reduced Resistance to Oxidative Stress in Drosophila melanogaster. Front. Physiol. 10:1143. 10.3389/fphys.2019.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J., Dietmeier K., Pfanner N. (1997). Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272 20730–20735. 10.1074/jbc.272.33.20730 [DOI] [PubMed] [Google Scholar]
- Calabrese G., Morgan B., Riemer J. (2017). Mitochondrial Glutathione: Regulation and Functions. Antioxidants Redox Signal. 27 1162–1177. 10.1089/ars.2017.7121 [DOI] [PubMed] [Google Scholar]
- Cardenas-Rodriguez M., Tokatlidis K. (2017). Cytosolic redox components regulate protein homeostasis via additional localisation in the mitochondrial intermembrane space. FEBS Lett. 591 2661–2670. 10.1002/1873-3468.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H. S., George G. N., Winge D. R. (2002). Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 277 31237–31242. 10.1074/jbc.M204854200 [DOI] [PubMed] [Google Scholar]
- Cesnekova J., Rodinova M., Hansikova H., Houstek J., Zeman J., Stiburek L. (2016). The mammalian homologue of yeast Afg1 ATPase (lactation elevated 1) mediates degradation of nuclear-encoded complex IV subunits. Biochem. J. 473 797–804. 10.1042/BJ20151029 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., et al. (2004). Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23 3735–3746. 10.1038/sj.emboj.7600389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., van der Laan M., Mehnert C. S., Guiard B., Mick D. U., Hutu D. P., et al. (2010). Distinct Forms of Mitochondrial TOM-TIM Supercomplexes Define Signal-Dependent States of Preprotein Sorting. Mol. Cell. Biol. 30 307–318. 10.1128/mcb.00749-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. C., Lithgow T. (2008). The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 19 126–136. 10.1091/mbc.E07-08-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi A., Sideris D. P., Katrakili N., Pozidis C., Tokatlidis K. (2013). Biogenesis of yeast Mia40 - Uncoupling folding from import and atypical recognition features. FEBS J. 280 4960–4969. 10.1111/febs.12482 [DOI] [PubMed] [Google Scholar]
- Chatzi A., Tokatlidis K. (2013). The mitochondrial intermembrane space: A hub for oxidative folding linked to protein biogenesis. Antioxidants Redox Signal. 19 54–62. 10.1089/ars.2012.4855 [DOI] [PubMed] [Google Scholar]
- Chung Y. M., Kim J. S., Yoo Y., Do. (2006). A novel protein, Romo1, induces ROS production in the mitochondria. Biochem. Biophys. Res. Commun. 347649–655. 10.1016/j.bbrc.2006.06.140 [DOI] [PubMed] [Google Scholar]
- Cobine P. A., Moore S. A., Leary S. C. (2021). Getting out what you put in: Copper in mitochondria and its impacts on human disease. Biochim. Biophys. Acta - Mol. Cell Res. 1868:118867. 10.1016/j.bbamcr.2020.118867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P. A., Pierrel F., Winge D. R. (2006). Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta - Mol. Cell Res. 1763 759–772. 10.1016/j.bbamcr.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Čunátová K., Reguera D. P., Vrbacký M., Fernández-Vizarra E., Ding S., Fearnley I. M., et al. (2021). Loss of COX4I1 Leads to Combined Respiratory Chain Deficiency and Impaired Mitochondrial Protein Synthesis. Cells 10:10020369. 10.3390/cells10020369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Leuenberger D., Leverich E. P., Hwang D. K., Beverly K. N., Koehler C. M. (2004). The role of Hot13p and redox chemistry in the mitochondrial TIM22 import pathway. J. Biol. Chem. 279 43744–43751. 10.1074/jbc.M404878200 [DOI] [PubMed] [Google Scholar]
- Dabir D. V., Leverich E. P., Kim S. K., Tsai F. D., Hirasawa M., Knaff D. B., et al. (2007). A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 26 4801–4811. 10.1038/sj.emboj.7601909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daithankar V. N., Farrell S. R., Thorpe C. (2009). Augmenter of liver regeneration: Substrate specificity of a flavin-dependent oxidoreductase from the mitochondrial intermembrane space. Biochemistry 48 4828–4837. 10.1021/bi900347v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daithankar V. N., Wang W., Trujillo J. R., Thorpe C. (2012). Flavin-linked erv-family sulfhydryl oxidases release superoxide anion during catalytic turnover. Biochemistry 51 265–272. 10.1021/bi201672h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M., Mendiola V. L., Mackey M. R., Murphy A. N., Koller A., Perkins G. A., et al. (2011). ChChd3, an inner mitochondrial membrane protein, is essential for maintaining Crista integrity and mitochondrial function. J. Biol. Chem. 286 2918–2932. 10.1074/jbc.M110.171975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M., Trinh K. N., Murphy A. N., Taylor S. S. (2012). Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space. J. Biol. Chem. 287 39480–39491. 10.1074/jbc.M112.387696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan D., Bandel M., Günsel U., Nussbaum I., Prag G., Mokranjac D., et al. (2019). A mutagenesis analysis of Tim50, the major receptor of the TIM23 complex, identifies regions that affect its interaction with Tim23. Sci. Rep. 9:38353. 10.1038/s41598-018-38353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Dotto V., Fogazza M., Carelli V., Rugolo M., Zanna C. (2018). Eight human OPA1 isoforms, long and short: What are they for? Biochim. Biophys. Acta - Bioenerg. 1859 263–269. 10.1016/j.bbabio.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Dela Cruz R., Jeong M. Y., Winge D. R. (2016). Cox1 mutation abrogates need for Cox23 in cytochrome c oxidase biogenesis. Microb. Cell 3 275–284. 10.15698/mic2016.07.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavallée L., Mathiah N., Cabon L., Mazeraud A., Brunelle-Navas M. N., Lerner L. K., et al. (2020). Mitochondrial AIF loss causes metabolic reprogramming, caspase-independent cell death blockade, embryonic lethality, and perinatal hydrocephalus. Mol. Metab. 40:2020. 10.1016/j.molmet.2020.101027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fonzo A., Ronchi D., Lodi T., Fassone E., Tigano M., Lamperti C., et al. (2009). The Mitochondrial Disulfide Relay System Protein GFER Is Mutated in Autosomal-Recessive Myopathy with Cataract and Combined Respiratory-Chain Deficiency. Am. J. Hum. Genet. 84 594–604. 10.1016/j.ajhg.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F., Fukui H., Garcia S., Moraes C. T. (2006). Cytochrome c Oxidase Is Required for the Assembly/Stability of Respiratory Complex I in Mouse Fibroblasts. Mol. Cell. Biol. 26 4872–4881. 10.1128/mcb.01767-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson-Murray E., Nedara K., Modjtahedi N., Tokatlidis K. (2021). The mia40/chchd4 oxidative folding system: Redox regulation and signaling in the mitochondrial intermembrane space. Antioxidants 10:10040592. 10.3390/antiox10040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer K. S., Papić D., Schumann B., Sperl D., Krumpe K., Walther D. M., et al. (2012). A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J. Cell Sci. 125 3464–3473. 10.1242/jcs.103804 [DOI] [PubMed] [Google Scholar]
- Dogan S. A., Cerutti R., Benincá C., Brea-Calvo G., Jacobs H. T., Zeviani M., et al. (2018). Perturbed Redox Signaling Exacerbates a Mitochondrial Myopathy. Cell Metab. 28 764.e–775.e. 10.1016/j.cmet.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra E., Zeviani M. (2021). Mitochondrial disorders of the OXPHOS system. FEBS Lett. 595 1062–1106. 10.1002/1873-3468.13995 [DOI] [PubMed] [Google Scholar]
- Fischer M., Horn S., Belkacemi A., Kojer K., Petrungaro C., Habich M., et al. (2013). Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell 24 2160–2170. 10.1091/mbc.E12-12-0862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd B. J., Wilkerson E. M., Veling M. T., Minogue C. E., Xia C., Beebe E. T., et al. (2016). Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol. Cell 63 621–632. 10.1016/j.molcel.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa L. E., Maghool S., Sharpe A. J., Reljic B., Wong L., Stroud D. A., et al. (2021). Mitochondrial COA7 is a heme-binding protein involved in the early stages of complex 1 IV assembly. 2 3. bioRxiv 2021:447992. 10.1101/2021.06.10.447992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich M. W., Erdogan A. J., Coughlin C. R., Elos M. T., Jiang H., O’Rourke C. P., et al. (2017). Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Hum. Mol. Genet. 26 702–716. 10.1093/hmg/ddw431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K., Milenkovic D., Chacinska A., Müller J., Guiard B., Pfanner N., et al. (2007). Novel Mitochondrial Intermembrane Space Proteins as Substrates of the MIA Import Pathway. J. Mol. Biol. 365 612–620. 10.1016/j.jmb.2006.10.038 [DOI] [PubMed] [Google Scholar]
- Gerashchenko M. V., Lobanov A. V., Gladyshev V. N. (2012). Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 109 17394–17399. 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafourifar P., Cadenas E. (2005). Mitochondrial nitric oxide synthase. Trends Pharmacol. Sci. 26 190–195. 10.1016/j.tips.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D’Adamo P., et al. (2010). Severe X-Linked Mitochondrial Encephalomyopathy Associated with a Mutation in Apoptosis-Inducing Factor. Am. J. Hum. Genet. 86 639–649. 10.1016/j.ajhg.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Pratt A. T., Soma S., Theriault S. G., Griffin A. T., Trivedi P. P., et al. (2016). Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis. Hum. Mol. Genet. 25 660–671. 10.1093/hmg/ddv503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Trivedi P. P., Timbalia S. A., Griffin A. T., Rahn J. J., Chan S. S. L., et al. (2014). Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 23 3596–3606. 10.1093/hmg/ddu069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyck S., Aras S., Hüttemann M., Grossman L. I. (2021). Regulation of COX Assembly and Function by Twin CX9C Proteins-Implications for Human Disease. Cells 10:10020197. 10.3390/cells10020197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevel A., Pfanner N., Becker T. (2019). Coupling of import and assembly pathways in mitochondrial protein biogenesis. Biol. Chem. 2019:310. 10.1515/hsz-2019-0310 [DOI] [PubMed] [Google Scholar]
- Groß D. P., Burgard C. A., Reddehase S., Leitch J. M., Culotta V. C., Hell K. (2011). Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system. Mol. Biol. Cell 22 3758–3767. 10.1091/mbc.E11-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V., Paulo J., Zhai B., Huttlin E. L., Gygi S. P., Harper J. W. (2014). TIMMDC1/C3orf1 Functions as a Membrane-Embedded Mitochondrial Complex I Assembly Factor through Association with the MCIA Complex. Mol. Cell. Biol. 34 847–861. 10.1128/mcb.01551-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarás A., Perales-Clemente E., Calvo E., Acín-Pérez R., Loureiro-Lopez M., Pujol C., et al. (2016). The CoQH2/CoQ Ratio Serves as a Sensor of Respiratory Chain Efficiency. Cell Rep. 15 197–209. 10.1016/j.celrep.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Guerrero-Castillo S., Baertling F., Kownatzki D., Wessels H. J., Arnold S., Brandt U., et al. (2017). The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 25 128–139. 10.1016/j.cmet.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., et al. (2005). Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1 401–408. 10.1016/j.cmet.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Habich M., Salscheider S. L., Murschall L. M., Hoehne M. N., Fischer M., Schorn F., et al. (2019a). Vectorial Import via a Metastable Disulfide-Linked Complex Allows for a Quality Control Step and Import by the Mitochondrial Disulfide Relay. Cell Rep. 26 759.e–774.e. 10.1016/j.celrep.2018.12.092 [DOI] [PubMed] [Google Scholar]
- Habich M., Salscheider S. L., Riemer J. (2019b). Cysteine residues in mitochondrial intermembrane space proteins: more than just import. Br. J. Pharmacol. 176 514–531. 10.1111/bph.14480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Antunes F., Canali R., Rettori D., Cadenas E. (2003). Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 278 5557–5563. 10.1074/jbc.M210269200 [DOI] [PubMed] [Google Scholar]
- Hangen E., Féraud O., Lachkar S., Mou H., Doti N., Fimia G. M., et al. (2015). Interaction between AIF and CHCHD4 Regulates Respiratory Chain Biogenesis. Mol. Cell 58 1001–1014. 10.1016/j.molcel.2015.04.020 [DOI] [PubMed] [Google Scholar]
- Hansen K. G., Herrmann J. M. (2019). Transport of Proteins into Mitochondria. Protein J. 2019:6. 10.1007/s10930-019-09819-6 [DOI] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., et al. (2011). The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30 4356–4370. 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley A. M., Lukoyanova N., Zhang Y., Cabrera-Orefice A., Arnold S., Meunier B., et al. (2019). Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc 1. Nat. Struct. Mol. Biol. 26 78–83. 10.1038/s41594-018-0172-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B., Griparic L., Amiri M., Gandre-Babbe S., Van Der Bliek A. M. (2009). Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187 959–966. 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton D., Nittis T., Srinivasan C., Winge D. R. (2000). Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 275 37582–37587. 10.1074/jbc.M006639200 [DOI] [PubMed] [Google Scholar]
- Heaton D. N., George G. N., Garrison G., Winge D. R. (2001). The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 40 743–751. 10.1021/bi002315x [DOI] [PubMed] [Google Scholar]
- Helman G., Compton A. G., Hock D. H., Walkiewicz M., Brett G. R., Pais L., et al. (2021). Multiomic analysis elucidates Complex I deficiency caused by a deep intronic variant in NDUFB10. Hum. Mutat. 42 19–24. 10.1002/humu.24135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Okunushi R., Hara T., Hashiguchi A., Yuan J., Yoshimura A., et al. (2018). Mutations in COA7 cause spinocerebellar ataxia with axonal neuropathy. Brain 141 1622–1636. 10.1093/brain/awy104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Roessler M. M. (2016). Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex i. Biochim. Biophys. Acta - Bioenerg. 1857 872–883. 10.1016/j.bbabio.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser L., Di Valentin M., Hamer A. G., Hosler J. P. (2000). Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 275 619–623. 10.1074/jbc.275.1.619 [DOI] [PubMed] [Google Scholar]
- Hofmann S., Rothbauer U., Mühlenbein N., Baiker K., Hell K., Bauer M. F. (2005). Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J. Mol. Biol. 353 517–528. 10.1016/j.jmb.2005.08.064 [DOI] [PubMed] [Google Scholar]
- Holmström K. M., Finkel T. (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15 411–421. 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- Horn D., Al-Ali H., Barrientos A. (2008). Cmc1p Is a Conserved Mitochondrial Twin CX 9 C Protein Involved in Cytochrome c Oxidase Biogenesis. Mol. Cell. Biol. 28 4354–4364. 10.1128/mcb.01920-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., Zhou W., Trevisson E., Al-Ali H., Harris T. K., Salviati L., et al. (2010). The conserved mitochondrial twin Cx9C Protein Cmc2 is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J. Biol. Chem. 285 15088–15099. 10.1074/jbc.M110.104786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng Y. C., Cobine P. A., Maxfield A. B., Carr H. S., Winge D. R. (2004). Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J. Biol. Chem. 279 35334–35340. 10.1074/jbc.M404747200 [DOI] [PubMed] [Google Scholar]
- Huigsloot M., Nijtmans L. G., Szklarczyk R., Baars M. J. H., Van Den Brand M. A. M., Hendriksfranssen M. G. M., et al. (2011). A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 88 488–493. 10.1016/j.ajhg.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H. W., Udeshi N. D., Cracan V., Svinkina T., et al. (2014). Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol. Cell 55 332–341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C., Koepke J., Lange C., Roßmanith T., Michel H. (2000). Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure 8 669–684. 10.1016/S0969-2126(00)00152-0 [DOI] [PubMed] [Google Scholar]
- Iñarrea P., Moini H., Han D., Rettori D., Aguiló I., Alava M. A., et al. (2007). Mitochondrial respiratory chain and thioredoxin reductase regulate intermembrane Cu,Zn-superoxide dismutase activity: Implications for mitochondrial energy metabolism and apoptosis. Biochem. J. 405 173–179. 10.1042/BJ20061809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñarrea P., Moini H., Rettori D., Han D., Martínez J., García I., et al. (2005). Redox activation of mitochondrial intermembrane space Cu,Zn-superoxide dismutase. Biochem. J. 387 203–209. 10.1042/BJ20041683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S., Lee J. W., Okada K., Lee J. K., Iwata M., Rasmussen B., et al. (1998). Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281 64–71. 10.1126/science.281.5373.64 [DOI] [PubMed] [Google Scholar]
- Jacob C., Holme A. L., Fry F. H. (2004). The sulfinic acid switch in proteins. Org. Biomol. Chem. 2 1953–1956. 10.1039/b406180b [DOI] [PubMed] [Google Scholar]
- Jäer S., Handschin C., St-Pierre J., Spiegelman B. M. (2007). AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. U. S. A. 104 12017–12022. 10.1073/pnas.0705070104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H. G., Gerbitz K. D., et al. (2000). Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 9 795–801. 10.1093/hmg/9.5.795 [DOI] [PubMed] [Google Scholar]
- Jensen R. E., Johnson A. E. (2001). Opening the door to mitochondrial protein import. Nat. Struct. Biol. 8 1008–1010. 10.1038/nsb1201-1008 [DOI] [PubMed] [Google Scholar]
- Jett K. A., Leary S. C. (2018). Building the CuA site of cytochrome c oxidase: A complicated, redox-dependent process driven by a surprisingly large complement of accessory proteins. J. Biol. Chem. 293 4644–4652. 10.1074/jbc.R117.816132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L. L., Yeo D. (2021). Oxidative stress: an evolving definition. Fac. Rev. 10:13. 10.12703/r/10-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B., Hüttemann M. (2015). The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion 24 64–76. 10.1016/j.mito.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Kallergi E., Andreadaki M., Kritsiligkou P., Katrakili N., Pozidis C., Tokatlidis K., et al. (2012). Targeting and maturation of Erv1/ALR in the mitochondrial intermembrane space. ACS Chem. Biol. 7 707–714. 10.1021/cb200485b [DOI] [PubMed] [Google Scholar]
- Käser M., Kambacheld M., Kisters-Woike B., Langer T. (2003). Oma1, a Novel Membrane-bound Metallopeptidase in Mitochondria with Activities Overlapping with the m-AAA Protease. J. Biol. Chem. 278 46414–46423. 10.1074/jbc.M305584200 [DOI] [PubMed] [Google Scholar]
- Kawano S., Yamano K., Naoé M., Momose T., Terao K., Nishikawa S. I., et al. (2009). Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc. Natl. Acad. Sci. U. S. A. 106 14403–14407. 10.1073/pnas.0901793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O., Bird A., Winge D. R. (2007). Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282 17442–17449. 10.1074/jbc.M702379200 [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., Winge D. R. (2012). Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287 7289–7300. 10.1074/jbc.M111.313148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O., Rigby K., Bestwick M., Pierrel F., Cobine P. A., Winge D. R. (2008). Pet191 is a cytochrome c oxidase assembly factor in Saccharomyces cerevisiae. Eukaryot. Cell 7 1427–1431. 10.1128/EC.00132-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O., Winge D. R. (2008). Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim. Biophys. Acta - Mol. Cell Res. 1783 618–628. 10.1016/j.bbamcr.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Goto Y. I., Kimura H. (2010). Hydrogen sulfide increases glutathione production and suppresses oxidative stress in mitochondria. Antioxidants Redox Signal. 12 1–13. 10.1089/ars.2008.2282 [DOI] [PubMed] [Google Scholar]
- Klein A., Israel L., Lackey S. W. K., Nargang F. E., Imhof A., Baumeister W., et al. (2012). Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. J. Cell Biol. 199 599–611. 10.1083/jcb.201207161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J. R., Schmid F. X. (2014a). Mia40 combines thiol oxidase and disulfide isomerase activity to efficiently catalyze oxidative folding in mitochondria. J. Mol. Biol. 426 4087–4098. 10.1016/j.jmb.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Koch J. R., Schmid F. X. (2014b). Mia40 targets cysteines in a hydrophobic environment to direct oxidative protein folding in the mitochondria. Nat. Commun. 5:4041. 10.1038/ncomms4041 [DOI] [PubMed] [Google Scholar]
- Koehler C. M. (2004). The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 29 1–4. 10.1016/j.tibs.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Kojer K., Bien M., Gangel H., Morgan B., Dick T. P., Riemer J. (2012). Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 31 3169–3182. 10.1038/emboj.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojer K., Peleh V., Calabrese G., Herrmann J. M., Riemer J. (2015). Kinetic control by limiting glutaredoxin amounts enables thiol oxidation in the reducing mitochondrial intermembrane space. Mol. Biol. Cell 26 195–204. 10.1091/mbc.E14-10-1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaltowski A. J., de Souza-Pinto N. C., Castilho R. F., Vercesi A. E. (2009). Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 47 333–343. 10.1016/j.freeradbiomed.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V., Prell F., Thiede B., Götz M., Wosiek D., Ott C., et al. (2014). C1orf163/RESA1 is a novel mitochondrial intermembrane space protein connected to respiratory chain assembly. J. Mol. Biol. 426 908–920. 10.1016/j.jmb.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Kritsiligkou P., Chatzi A., Charalampous G., Mironov A., Grant C. M., Tokatlidis K. (2017). Unconventional Targeting of a Thiol Peroxidase to the Mitochondrial Intermembrane Space Facilitates Oxidative Protein Folding. Cell Rep. 18 2729–2741. 10.1016/j.celrep.2017.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., et al. (2008). Dissecting Membrane Insertion of Mitochondrial β-Barrel Proteins. Cell 132 1011–1024. 10.1016/j.cell.2008.01.028 [DOI] [PubMed] [Google Scholar]
- Lambert A. J., Brand M. D. (2009). Reactive oxygen species production by mitochondria. Methods Mol. Biol. 554 165–181. 10.1007/978-1-59745-521-3_11 [DOI] [PubMed] [Google Scholar]
- Lange H., Lisowsky T., Gerber J., Mühlenhoff U., Kispal G., Lill R. (2001). An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2 715–720. 10.1093/embo-reports/kve161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S. C., Cobine P. A., Kaufman B. A., Guercin G. H., Mattman A., Palaty J., et al. (2007). The Human Cytochrome c Oxidase Assembly Factors SCO1 and SCO2 Have Regulatory Roles in the Maintenance of Cellular Copper Homeostasis. Cell Metab. 5 9–20. 10.1016/j.cmet.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Leary S. C., Cobine P. A., Nishimura T., Verdijk R. M., De Krijger R., De Coo R., et al. (2013). COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol. Biol. Cell 24 683–691. 10.1091/mbc.E12-09-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S. C., Kaufman B. A., Pellecchia G., Guercin G. H., Mattman A., Jaksch M., et al. (2004). Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 13 1839–1848. 10.1093/hmg/ddh197 [DOI] [PubMed] [Google Scholar]
- Leary S. C., Sasarman F., Nishimura T., Shoubridge E. A. (2009). Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 18 2230–2240. 10.1093/hmg/ddp158 [DOI] [PubMed] [Google Scholar]
- Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000). Membrane protein degradation by AAA proteases in mitochondria: Extraction of substrates from either membrane surface. Mol. Cell 5 629–638. 10.1016/S1097-2765(00)80242-7 [DOI] [PubMed] [Google Scholar]
- Leonhard K., Herrmann J. M., Stuart R. A., Mannhaupt G., Neupert W., Langer T. (1996). AAA proteases with catalytic sites on apposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15 4218–4229. 10.1002/j.1460-2075.1996.tb00796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard K., Stiegler A., Neupert W., Langer T. (1999). Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 398 348–351. 10.1038/18704 [DOI] [PubMed] [Google Scholar]
- Letts J. A., Sazanov L. A. (2017). Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 24 800–808. 10.1038/nsmb.3460 [DOI] [PubMed] [Google Scholar]
- Liang H., Yoo S. E., Na R., Walter C. A., Richardson A., Ran Q. (2009). Short form glutathione peroxidase 4 is the essential isoform required for survival and somatic mitochondrial functions. J. Biol. Chem. 284 30836–30844. 10.1074/jbc.M109.032839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionaki E., Aivaliotis M., Pozidis C., Tokatlidis K. (2010). The N-terminal shuttle domain of Erv1 determines the affinity for Mia40 and mediates electron transfer to the catalytic Erv1 core in yeast mitochondria. Antioxidants Redox Signal. 13 1327–1339. 10.1089/ars.2010.3200 [DOI] [PubMed] [Google Scholar]
- Liu R., Chan D. C. (2017). OPA1 and cardiolipin team up for mitochondrial fusion. Nat. Cell Biol. 19 760–762. 10.1038/ncb3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Jarne T., Ugalde C. (2018). Respiratory chain supercomplexes: Structures, function and biogenesis. Semin. Cell Dev. Biol. 76 179–190. 10.1016/j.semcdb.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., et al. (2009). Systematic Analysis of the Twin Cx9C Protein Family. J. Mol. Biol. 393 356–368. 10.1016/j.jmb.2009.08.041 [DOI] [PubMed] [Google Scholar]
- Lu H., Allen S., Wardleworth L., Savory P., Tokatlidis K. (2004). Functional TIM10 Chaperone Assembly Is Redox-regulated in Vivo. J. Biol. Chem. 279 18952–18958. 10.1074/jbc.M313045200 [DOI] [PubMed] [Google Scholar]
- Lyons A. M., Ardissone A., Reyes A., Robinson A. J., Moroni I., Ghezzi D., et al. (2016). COA7 (C1orf163/RESA1) mutations associated with mitochondrial leukoencephalopathy and cytochrome c oxidase deficiency. J. Med. Genet. 53 846–849. 10.1136/jmedgenet-2016-104194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghool S., Ryan M. T., Maher M. J. (2020). What role does coa6 play in cytochrome c oxidase biogenesis: A metallochaperone or thiol oxidoreductase, or both? Int. J. Mol. Sci. 21 1–13. 10.3390/ijms21196983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa V., Fernandez-Vizarra E., Alshahwan S., Bakhsh E., Goffrini P., Ferrero I., et al. (2008). Severe Infantile Encephalomyopathy Caused by a Mutation in COX6B1, a Nucleus-Encoded Subunit of Cytochrome C Oxidase. Am. J. Hum. Genet. 82 1281–1289. 10.1016/j.ajhg.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie S., Riemer J., Wideman J. G., McBride H. M. (2018). A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 217 507–515. 10.1083/jcb.201611194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield A. B., Heaton D. N., Winge D. R. (2004). Cox17 Is Functional When Tethered to the Mitochondrial Inner Membrane. J. Biol. Chem. 279 5072–5080. 10.1074/jbc.M311772200 [DOI] [PubMed] [Google Scholar]
- McEwen J. E., Hong K. H., Park S., Preciado G. T. (1993). Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 23 9–14. 10.1007/BF00336742 [DOI] [PubMed] [Google Scholar]
- Melchionda L., Haack T. B., Hardy S., Abbink T. E. M., Fernandez-Vizarra E., Lamantea E., et al. (2014). Mutations in APOPT1, encoding a mitochondrial protein, cause cavitating leukoencephalopathy with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 95 315–325. 10.1016/j.ajhg.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C., Langer T. (2009). Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta - Mol. Cell Res. 1793 27–32. 10.1016/j.bbamcr.2008.05.013 [DOI] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., et al. (2005). A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121 1059–1069. 10.1016/j.cell.2005.04.011 [DOI] [PubMed] [Google Scholar]
- Milenkovic D., Gabriel K., Guiard B., Schulze-Specking A., Pfanner N., Chacinska A. (2007). Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: Site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J. Biol. Chem. 282 22472–22480. 10.1074/jbc.M703294200 [DOI] [PubMed] [Google Scholar]
- Milenkovic D., Ramming T., Müller J. M., Wenz L. S., Gebert N., Schulze-Specking A., et al. (2009). Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 20 2530–2539. 10.1091/mbc.E08-11-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K., Wasilewski M., Benincá C., Cysewski D., Poznanski J., Sakowska P., et al. (2019). Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7. EMBO Mol. Med. 11:201809561. 10.15252/emmm.201809561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moira Glerum D., Shtanko A., Tzagoloff A. (1996a). Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271 14504–14509. 10.1074/jbc.271.24.14504 [DOI] [PubMed] [Google Scholar]
- Moira Glerum D., Shtanko A., Tzagoloff A. (1996b). SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271 20531–20535. 10.1074/jbc.271.34.20531 [DOI] [PubMed] [Google Scholar]
- Muller F. L., Liu Y., Van Remmen H. (2004). Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 279 49064–49073. 10.1074/jbc.M407715200 [DOI] [PubMed] [Google Scholar]
- Murari A., Rhooms S. K., Goparaju N. S., Villanueva M., Owusu-Ansah E. (2020). An antibody toolbox to track complex I assembly defines AIF’s mitochondrial function. J. Cell Biol. 219:202001071. 10.1083/JCB.202001071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417 1–13. 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murschall L. M., Gerhards A., MacVicar T., Peker E., Hasberg L., Wawra S., et al. (2020). The C-terminal region of the oxidoreductase MIA40 stabilizes its cytosolic precursor during mitochondrial import. BMC Biol. 18:824. 10.1186/s12915-020-00824-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoé M., Ohwa Y., Ishikawa D., Ohshima C., Nishikawa S. I., Yamamoto H., et al. (2004). Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 279 47815–47821. 10.1074/jbc.M410272200 [DOI] [PubMed] [Google Scholar]
- Neal S. E., Dabir D. V., Wijaya J., Boon C., Koehler C. M. (2017). Osm1 facilitates the transfer of electrons from Erv1 to fumarate in the redox-regulated import pathway in the mitochondrial intermembrane space. Mol. Biol. Cell 28 2773–2785. 10.1091/mbc.E16-10-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittis T., George G. N., Winge D. R. (2001). Yeast Sco1, a Protein Essential for Cytochrome c Oxidase Function Is a Cu(I)-binding Protein. J. Biol. Chem. 276 42520–42526. 10.1074/jbc.M107077200 [DOI] [PubMed] [Google Scholar]
- Nobrega M. P., Bandeira S. C. B., Beers J., Tzagoloff A. (2002). Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 277 40206–40211. 10.1074/jbc.M207348200 [DOI] [PubMed] [Google Scholar]
- Norton M., Ng A. C. H., Baird S., Dumoulin A., Shutt T., Mah N., et al. (2014). ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Sci. Signal. 7:2004374. 10.1126/scisignal.2004374 [DOI] [PubMed] [Google Scholar]
- Osman C., Wilmes C., Tatsuta T., Langer T. (2007). Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1FO-ATP synthase. Mol. Biol. Cell 18 627–635. 10.1091/mbc.E06-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald C., Krause-Buchholz U., Rödel G. (2009). Knockdown of Human COX17 Affects Assembly and Supramolecular Organization of Cytochrome c Oxidase. J. Mol. Biol. 389 470–479. 10.1016/j.jmb.2009.04.034 [DOI] [PubMed] [Google Scholar]
- Pacheu-Grau D., Bareth B., Dudek J., Juris L., Vögtle F. N., Wissel M., et al. (2015). Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 21 823–833. 10.1016/j.cmet.2015.04.012 [DOI] [PubMed] [Google Scholar]
- Pacheu-Grau D., Wasilewski M., Oeljeklaus S., Gibhardt C. S., Aich A., Chudenkova M., et al. (2020). COA6 Facilitates Cytochrome c Oxidase Biogenesis as Thiol-reductase for Copper Metallochaperones in Mitochondria. J. Mol. Biol. 432 2067–2079. 10.1016/j.jmb.2020.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., et al. (2008). A Mitochondrial Protein Compendium Elucidates Complex I Disease Biology. Cell 134 112–123. 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais J. E., Schilke B., Craig E. A. (2011). Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol. Biol. Cell 22 4740–4749. 10.1091/mbc.E11-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páleníková P., Harbour M. E., Prodi F., Minczuk M., Zeviani M., Ghelli A., et al. (2021). Duplexing complexome profiling with SILAC to study human respiratory chain assembly defects. Biochim. Biophys. Acta - Bioenerg. 2021:1862. 10.1016/j.bbabio.2021.148395 [DOI] [PubMed] [Google Scholar]
- Palmeira C. M., Teodoro J. S., Amorim J. A., Steegborn C., Sinclair D. A., Rolo A. P. (2019). Mitohormesis and metabolic health: The interplay between ROS, cAMP and sirtuins. Free Radic. Biol. Med. 141 483–491. 10.1016/j.freeradbiomed.2019.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumaa P., Kangur L., Voronova A., Sillard R. (2004). Metal-binding mechanism of Cox17, a copper chaperone for cytochrome c oxidase. Biochem. J. 382 307–314. 10.1042/BJ20040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou L. C., Sue M. C., Davidson M. M., Tanji K., Nishino I., Sadlock J. E., et al. (1999). Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 23 333–337. 10.1038/15513 [DOI] [PubMed] [Google Scholar]
- Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., et al. (2003). Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426 862–866. 10.1038/nature02208 [DOI] [PubMed] [Google Scholar]
- Paul B. D., Snyder S. H., Kashfi K. (2021). Effects of hydrogen sulfide on mitochondrial function and cellular bioenergetics. Redox Biol. 38:101772. 10.1016/j.redox.2020.101772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peker E., Erdogan A. J., Volkov A. N., Riemer J. (2021). Erv1 and Cytochrome c Mediate Rapid Electron Transfer via A Collision-Type Interaction. J. Mol. Biol. 433:167045. 10.1016/j.jmb.2021.167045 [DOI] [PubMed] [Google Scholar]
- Peleh V., Cordat E., Herrmann J. M. (2016). Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. Elife 5:16177. 10.7554/eLife.16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro C., Zimmermann K. M., Küttner V., Fischer M., Dengjel J., Bogeski I., et al. (2015). The Ca2+-dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2+ uptake. Cell Metab. 22 721–733. 10.1016/j.cmet.2015.08.019 [DOI] [PubMed] [Google Scholar]
- Petruzzella V., Tiranti V., Fernandez P., Ianna P., Carrozzo R., Zeviani M. (1998). Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 54 494–504. 10.1006/geno.1998.5580 [DOI] [PubMed] [Google Scholar]
- Protasoni M., Pérez-Pérez R., Lobo-Jarne T., Harbour M. E., Ding S., Peñas A., et al. (2020). Respiratory supercomplexes act as a platform for complex III -mediated maturation of human mitochondrial complexes I and IV. EMBO J. 39:2019102817. 10.15252/embj.2019102817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch R. C., Samborska B., Faubert B., Ma E. H., Gravel S. P., Andrzejewski S., et al. (2017). AMPK Maintains Cellular Metabolic Homeostasis through Regulation of Mitochondrial Reactive Oxygen Species. Cell Rep. 21 1–9. 10.1016/j.celrep.2017.09.026 [DOI] [PubMed] [Google Scholar]
- Rainbolt T. K., Saunders J. M., Wiseman R. L. (2015). YME 1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep. 16 97–106. 10.15252/embr.201438976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T. J., et al. (2021). MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49 D1541–D1547. 10.1093/nar/gkaa1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S., Berndtsson J., Marin-Buera L., Conrad J., Carroni M., Brzezinski P., et al. (2019). Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 26 50–57. 10.1038/s41594-018-0169-7 [DOI] [PubMed] [Google Scholar]
- Reczek C. R., Chandel N. S. (2015). ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33 8–13. 10.1016/j.ceb.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Zahedi R. P., Pfanner N., Meisinger C., Sickmann A. (2006). Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5 1543–1554. 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- Reinhardt C., Arena G., Nedara K., Edwards R., Brenner C., Tokatlidis K., et al. (2020). AIF meets the CHCHD4/Mia40-dependent mitochondrial import pathway. Biochim. Biophys. Acta - Mol. Basis Dis. 1866:165746. 10.1016/j.bbadis.2020.165746 [DOI] [PubMed] [Google Scholar]
- Richter F., Dennerlein S., Nikolov M., Jans D. C., Naumenko N., Aich A., et al. (2019). ROMO1 is a constituent of the human presequence translocase required for YME1L protease import. J. Cell Biol. 218 598–614. 10.1083/jcb.201806093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby K., Zhang L., Cobine P. A., George G. N., Winge D. R. (2007). Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 282 10233–10242. 10.1074/jbc.M610082200 [DOI] [PubMed] [Google Scholar]
- Robb E. L., Hall A. R., Prime T. A., Eaton S., Szibor M., Viscomi C., et al. (2018). Control of mitochondrial superoxide production by reverse electron transport at complex I. J. Biol. Chem. 293 9869–9879. 10.1074/jbc.RA118.003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowska P., Jans D. C., Mohanraj K., Riedel D., Jakobs S., Chacinska A. (2015). The Oxidation Status of Mic19 Regulates MICOS Assembly. Mol. Cell. Biol. 35 4222–4237. 10.1128/mcb.00578-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendzielorz A. B., Bragoszewski P., Naumenko N., Gomkale R., Schulz C., Guiard B., et al. (2018). Motor recruitment to the TIM23 channel’s lateral gate restricts polypeptide release into the inner membrane. Nat. Commun. 9:6492. 10.1038/s41467-018-06492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner B., Westerburg H., Forneb́ I., Imhof A., Neupert W., Mokranjac D. (2012). Role of the AAA protease Yme1 in folding of proteins in the intermembrane space of mitochondria. Mol. Biol. Cell 23 4335–4346. 10.1091/mbc.E12-05-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T. J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. (2007). Glucose Restriction Extends Caenorhabditis elegans Life Span by Inducing Mitochondrial Respiration and Increasing Oxidative Stress. Cell Metab. 6 280–293. 10.1016/j.cmet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Scialò F., Fernández-Ayala D. J., Sanz A. (2017). Role of mitochondrial reverse electron transport in ROS signaling: Potential roles in health and disease. Front. Physiol. 8:428. 10.3389/fphys.2017.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialò F., Sriram A., Fernández-Ayala D., Gubina N., Lõhmus M., Nelson G., et al. (2016). Mitochondrial ROS Produced via Reverse Electron Transport Extend Animal Lifespan. Cell Metab. 23 725–734. 10.1016/j.cmet.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K., Sugimura T., Misaki T., Tadehara Y., Yamamoto S., Hanada M., et al. (2019). Monomeric structure of an active form of bovine cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 116 19945–19951. 10.1073/pnas.1907183116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T., Imai K., Qiu J., Hewitt V. L., Tan K., Shen H. H., et al. (2015). Molecular architecture of the active mitochondrial protein gate. Science 349, 1544–1548. 10.1126/science.aac6428 [DOI] [PubMed] [Google Scholar]
- Shutt T., Geoffrion M., Milne R., McBride H. M. (2012). The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 13 909–915. 10.1038/embor.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., et al. (2003). The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 100 13207–13212. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., et al. (2009). A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187 1007–1022. 10.1083/jcb.200905134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris D. P., Tokatlidis K. (2007). Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol. Microbiol. 65 1360–1373. 10.1111/j.1365-2958.2007.05880.x [DOI] [PubMed] [Google Scholar]
- Signes A., Cerutti R., Dickson A. S., Benincá C., Hinchy E. C., Ghezzi D., et al. (2019). APOPT 1/COA 8 assists COX assembly and is oppositely regulated by UPS and ROS. EMBO Mol. Med. 11 201809582. 10.15252/emmm.201809582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signes A., Fernandez-Vizarra E. (2018). Assembly of mammalian oxidative phosphorylation complexes I–V and supercomplexes. Essays Biochem. 62 255–270. 10.1042/EBC20170098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkler C. A., Kalpage H., Shay J., Lee I., Malek M. H., Grossman L. I., et al. (2017). Tissue- and Condition-Specific Isoforms of Mammalian Cytochrome c Oxidase Subunits: From Function to Human Disease. Oxid. Med. Cell. Longev. 2017:153456. 10.1155/2017/1534056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C., Bauer M. F., Guiard B., Neupert W., Brunner M. (1996). Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384, 582–585. 10.1038/384582a0 [DOI] [PubMed] [Google Scholar]
- Soma S., Morgada M. N., Naik M. T., Boulet A., Roesler A. A., Dziuba N., et al. (2019). COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase. Cell Rep. 29 4114.e–4126.e. 10.1016/j.celrep.2019.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiburek L., Cesnekova J., Kostkova O., Fornuskova D., Vinsova K., Wenchich L., et al. (2012). YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 23 1010–1023. 10.1091/mbc.E11-08-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud D. A., Maher M. J., Lindau C., Vögtle F. N., Frazier A. E., Surgenor E., et al. (2015). COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet. 24 5404–5415. 10.1093/hmg/ddv265 [DOI] [PubMed] [Google Scholar]
- Stroud D. A., Surgenor E. E., Formosa L. E., Reljic B., Frazier A. E., Dibley M. G., et al. (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex i. Nature 538 123–126. 10.1038/nature19754 [DOI] [PubMed] [Google Scholar]
- Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., Brothers G. M., et al. (1999). Molecular characterization of mitochodrial apoptosis-inducing factor. Nature 397 441–446. 10.1038/17135 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Ali M., Fischer M., Riemer J. (2013). Human copper chaperone for superoxide dismutase 1 mediates its own oxidation-dependent import into mitochondria. Nat. Commun. 4:3430. 10.1038/ncomms3430 [DOI] [PubMed] [Google Scholar]
- Szczepanowska K., Senft K., Heidler J., Herholz M., Kukat A., Höhne M. N., et al. (2020). A salvage pathway maintains highly functional respiratory complex I. Nat. Commun. 11:15567. 10.1038/s41467-020-15467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K., Trifunovic A. (2021). Tune instead of destroy: How proteolysis keeps OXPHOS in shape. Biochim. Biophys. Acta - Bioenerg. 1862:18365. 10.1016/j.bbabio.2020.148365 [DOI] [PubMed] [Google Scholar]
- Szibor M., Gainutdinov T., Fernandez-Vizarra E., Dufour E., Gizatullina Z., Debska-Vielhaber G., et al. (2020). Bioenergetic consequences from xenotopic expression of a tunicate AOX in mouse mitochondria: Switch from RET and ROS to FET. Biochim. Biophys. Acta - Bioenerg. 1861:148137. 10.1016/j.bbabio.2019.148137 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., et al. (2011). The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:973. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R., Wanschers B. F. J., Cuypers T. D., Esseling J. J., Riemersma M., Van Den Brand M. A. M., et al. (2012). Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 13:12. 10.1186/gb-2012-13-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R., Wanschers B. F. J., Nabuurs S. B., Nouws J., Nijtmans L. G., Huynen M. A. (2011). NDUFB7 and NDUFA8 are located at the intermembrane surface of complex i. FEBS Lett. 585 737–743. 10.1016/j.febslet.2011.01.046 [DOI] [PubMed] [Google Scholar]
- Tadokoro T., Ikeda M., Ide T., Deguchi H., Ikeda S., Okabe K., et al. (2020). Mitochondria-dependent ferroptosis plays a pivotal role in doxorubicin cardiotoxicity. JCI Insight 5:132747. 10.1172/jci.insight.132747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Kako K., Kashiwabara S., Takehara A., Inada Y., Arai H., et al. (2002). Mammalian Copper Chaperone Cox17p Has an Essential Role in Activation of Cytochrome c Oxidase and Embryonic Development. Mol. Cell. Biol. 22 7614–7621. 10.1128/mcb.22.21.7614-7621.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., Tsutsumi A., Nishizawa T., Lindau C., Busto J. V., Wenz L. S., et al. (2021). Mitochondrial sorting and assembly machinery operates by β-barrel switching. Nature 590 163–169. 10.1038/s41586-020-03113-7 [DOI] [PubMed] [Google Scholar]
- Terziyska N., Grumbt B., Bien M., Neupert W., Herrmann J. M., Hell K. (2007). The sulfhydryl oxidase Erv1 is a substrate of the Mia40-dependent protein translocation pathway. FEBS Lett. 581 1098–1102. 10.1016/j.febslet.2007.02.014 [DOI] [PubMed] [Google Scholar]
- Terziyska N., Lutz T., Kozany C., Mokranjac D., Mesecke N., Neupert W., et al. (2005). Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 579 179–184. 10.1016/j.febslet.2004.11.072 [DOI] [PubMed] [Google Scholar]
- Timón-Gómez A., Nývltová E., Abriata L. A., Vila A. J., Hosler J., Barrientos A. (2018). Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 76 163–178. 10.1016/j.semcdb.2017.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort F., Barredo E., Parthasarathy R., Ugarteburu O., Ferrer-Cortès X., García-Villoria J., et al. (2020). Biallelic mutations in NDUFA8 cause complex I deficiency in two siblings with favorable clinical evolution. Mol. Genet. Metab. 131 349–357. 10.1016/j.ymgme.2020.10.005 [DOI] [PubMed] [Google Scholar]
- Tropeano C. V., Aleo S. J., Zanna C., Roberti M., Scandiffio L., Loguercio Polosa P., et al. (2020). Fine-tuning of the respiratory complexes stability and supercomplexes assembly in cells defective of complex III. Biochim. Biophys. Acta - Bioenerg. 1861:148133. 10.1016/j.bbabio.2019.148133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., et al. (1996). The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272 1136–1144. 10.1126/science.272.5265.1136 [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Capitanio N., Nobrega M. P., Gatti D. (1990). Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 9 2759–2764. 10.1002/j.1460-2075.1990.tb07463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Nobrega M., Gorman N., Sinclair P. (1993). On the functions of the yeast COX10 and COX11 gene products. Biochem. Mol. Biol. Int. 31 593–598. [PubMed] [Google Scholar]
- Vahsen N., Candé C., Brière J. J., Bénit P., Joza N., Larochette N., et al. (2004). AIF deficiency compromises oxidative phosphorylation. EMBO J. 23 4679–4689. 10.1038/sj.emboj.7600461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V., et al. (2000). Mutations of the SCO1 Gene in Mitochondrial Cytochrome c Oxidase Deficiency with Neonatal-Onset Hepatic Failure and Encephalopathy. Am. J. Hum. Genet. 67 1104–1109. 10.1016/s0002-9297(07)62940-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., et al. (1999). Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401 485–489. 10.1038/46802 [DOI] [PubMed] [Google Scholar]
- Vanišová M., Burská D., Křížová J., Daňhelovská T., Zeman J. (2019). Stable COX17 downregulation leads to alterations in mitochondrial ultrastructure, decreased copper content and impaired cytochrome c oxidase biogenesis in HEK293 cells. Folia Biol. 65 181–187. [DOI] [PubMed] [Google Scholar]
- Velázquez I., Pardo J. P. (2001). Kinetic characterization of the rotenone-insensitive internal NADH: Ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 389 7–14. 10.1006/abbi.2001.2293 [DOI] [PubMed] [Google Scholar]
- Veniamin S., Sawatzky L. G., Banting G. S., Glerum D. M. (2011). Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic. Biol. Med. 51 1589–1600. 10.1016/j.freeradbiomed.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Verkaart S., Koopman W. J. H., van Emst-de Vries S. E., Nijtmans L. G. J., van den Heuvel L. W. P. J., Smeitink J. A. M., et al. (2007). Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim. Biophys. Acta - Mol. Basis Dis. 1772 373–381. 10.1016/j.bbadis.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Vidoni S., Harbour M. E., Guerrero-Castillo S., Signes A., Ding S., Fearnley I. M., et al. (2017). MR-1S Interacts with PET100 and PET117 in Module-Based Assembly of Human Cytochrome c Oxidase. Cell Rep. 18 1727–1738. 10.1016/j.celrep.2017.01.044 [DOI] [PubMed] [Google Scholar]
- Vögtle F. N., Burkhart J. M., Rao S., Gerbeth C., Hinrichs J., Martinou J. C., et al. (2012). Intermembrane space proteome of yeast mitochondria. Mol. Cell. Proteomics 11 1840–1852. 10.1074/mcp.M112.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., et al. (2009). Global Analysis of the Mitochondrial N-Proteome Identifies a Processing Peptidase Critical for Protein Stability. Cell 139 428–439. 10.1016/j.cell.2009.07.045 [DOI] [PubMed] [Google Scholar]
- Wang T., Liu H., Itoh K., Oh S., Zhao L., Murata D., et al. (2021). C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab. 33 531.e–546.e. 10.1016/j.cmet.2021.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. T., Gorman M. A., Lazarou M., Ryan M. T., Gulbis J. M. (2006). Crystal structure of the mitochondrial chaperone TIM9∙10 reveals a six-bladed α-propeller. Mol. Cell 21 123–133. 10.1016/j.molcel.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Weckbecker D., Longen S., Riemer J., Herrmann J. M. (2012). Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 31 4348–4358. 10.1038/emboj.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhäupl K., Lindau C., Hessel A., Wang Y., Schütze C., Jores T., et al. (2018). Structural Basis of Membrane Protein Chaperoning through the Mitochondrial Intermembrane Space. Cell 175 1365.e–1379.e. 10.1016/j.cell.2018.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz L. S., Ellenrieder L., Qiu J., Bohnert M., Zufall N., van der Laan M., et al. (2015). Sam37 is crucial for formation of the mitochondrial TOM-SAM supercomplex, thereby promoting ß-barrel biogenesis. J. Cell Biol. 210 1047–1054. 10.1083/jcb.201504119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström M., Krab K., Sharma V. (2018). Oxygen Activation and Energy Conservation by Cytochrome c Oxidase. Chem. Rev. 118 2469–2490. 10.1021/acs.chemrev.7b00664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström M., Sharma V., Kaila V. R. I., Hosler J. P., Hummer G. (2015). New perspectives on proton pumping in cellular respiration. Chem. Rev. 115 2196–2221. 10.1021/cr500448t [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., et al. (2009). Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284 31635–31646. 10.1074/jbc.M109.041756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Yatsukawa Y. I., Esaki M., Aiken Hobbs A. E., Jensen R. E., Endo T. (2008). Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283 3799–3807. 10.1074/jbc.M708339200 [DOI] [PubMed] [Google Scholar]
- Yang M., Geli V., Oppliger W., Suda K., James P., Schatz G. (1991). The MAS-encoded processing protease of yeast mitochondria: interaction of the purified enzyme with signal peptides and a purified precursor protein. J. Biol. Chem. 266, 6416–6423. 10.1016/s0021-9258(18)38134-1 [DOI] [PubMed] [Google Scholar]
- Yang M., Trumpowers B. L. (1994). Deletion of QCR6, the gene encoding subunit six of the mitochondrial cytochrome bc1 complex, blocks maturation of cytochrome c1, and causes temperature-sensitive petite growth in Saccharomyces cerevisiae. J. Biol. Chem. 269 1270–1275. 10.1016/s0021-9258(17)42253-8 [DOI] [PubMed] [Google Scholar]
- Yatsuka Y., Kishita Y., Formosa L. E., Shimura M., Nozaki F., Fujii T., et al. (2020). A homozygous variant in NDUFA8 is associated with developmental delay, microcephaly, and epilepsy due to mitochondrial complex I deficiency. Clin. Genet. 98 155–165. 10.1111/cge.13773 [DOI] [PubMed] [Google Scholar]
- Zara V., Conte L., Trumpower B. L. (2009). Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta - Mol. Cell Res. 1793 89–96. 10.1016/j.bbamcr.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Zhu J., Vinothkumar K. R., Hirst J. (2016). Structure of mammalian respiratory complex i. Nature 536 354–358. 10.1038/nature19095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., et al. (1998). SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20 337–343. 10.1038/3804 [DOI] [PubMed] [Google Scholar]
- Zhuang J., Wang P. Y., Huang X., Chen X., Kang J. G., Hwang P. M. (2013). Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc. Natl. Acad. Sci. U. S. A. 110 17356–17361. 10.1073/pnas.1310908110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller E., Laborenz J., Krämer L., Boos F., Räschle M., Todd Alexander R., et al. (2020). The intermembrane space protein Mix23 is a novel stress-induced mitochondrial import factor. J. Biol. Chem. 295 14686–14697. 10.1074/jbc.RA120.014247 [DOI] [PMC free article] [PubMed] [Google Scholar]