Synopsis
The uEXPLORER total-body PET/CT system provides a very high level of detection sensitivity and simultaneous coverage of the entire body for dynamic imaging for quantification of tracer kinetics. This paper describes the fundamentals and potential benefits of total-body kinetic modeling and parametric imaging focusing on the noninvasive derivation of blood input function, multiparametric imaging, and high-temporal resolution kinetic modeling. Along with its attractive properties, total-body kinetic modeling also brings significant challenges, such as the large scale of total-body dynamic PET data, the need for organ and tissue appropriate input functions and kinetic models, and total-body motion correction. These challenges, and the opportunities using deep learning, are discussed.
Keywords: Total-body PET, dynamic imaging, kinetic modeling, parametric imaging, deep learning
Technical Terms: Total-body PET, dynamic imaging, kinetic modeling, parametric imaging, image-derived input function, high temporal resolution, dual-tracer, time-delay correction, model selection, deep learning, convolutional neural network
1. Introduction
Positron emission tomography (PET) is a sensitive molecular imaging method that uses radiolabeled tracers to monitor the biological and physiological function of the scanned subject in vivo1. PET is commonly used in the clinic to acquire static images of radioactivity distribution at a specific time interval (e.g., 60 minutes) post radiotracer injection. The standardized uptake value (SUV) is provided as a semi-quantitative measure of tracer uptake which, however, is affected by many factors, including body habitus, dietary preparation, and scan timing. Dynamic PET imaging can also be performed by taking multiple consecutive frames, typically starting at the time of injection. The acquired four-dimensional (4D: 3D space and 1D time) data from dynamic PET may reflect a broad spectrum of physiological and metabolic information, including blood flow, tracer delivery, transport, and metabolism. Dynamic PET data are commonly analyzed using compartmental modeling to quantify physiological parameters in a region of interest (ROI) or at the image voxel level (i.e., parametric imaging)2,3.
Although dynamic PET with tracer kinetic modeling has many potential advantages over static PET imaging, its widespread use and clinical implementation have been hampered by several factors such as high noise, the need of blood input function, long scan time, etc4. Dynamic PET also has been largely restricted to single-organ imaging, e.g., for brain and heart, due to the short axial field-of-view (AFOV) of conventional PET scanners (usually ~20 cm)5. Implementation of whole-body dynamic imaging has become feasible on conventional PET scanners6-8, but it is challenging to achieve high temporal resolution, and the data is sparse in the temporal domain due to the need to move the subject relative to the scanner to image different parts of the body. The recent advent of the uEXPLORER total-body PET system (and other long AFOV scanners that cover all or most of the vital organs9,10) provides unprecedented levels of detection sensitivity and simultaneous coverage of the entire body for dynamic imaging5,11,12. The total-body kinetic modeling and parametric imaging enabled by this system may have potential in both molecular imaging research and clinical applications13-16. Meanwhile, challenging technical problems co-exist with the opportunities given the large scale of total-body dynamic data sizes and the need for consideration of physiological heterogeneities in different organs and of the presence of motion that occurs throughout the dynamic series of images. This paper provides a brief introduction of total-body dynamic PET imaging and kinetic modeling (section 2.1-2.3), describes its potential benefits and limitations (section 2.4-2.6), and discusses potential directions to address the challenging problems using deep learning (section 3). Image examples shown in this paper are mainly from the widely used tracer 18F-fluorodeoxyglucose (FDG), but the concept is also applicable to most other radiotracers.
2. Total-body dynamic PET and its potential for kinetic modeling
2.1. Basis of dynamic PET and kinetic modeling
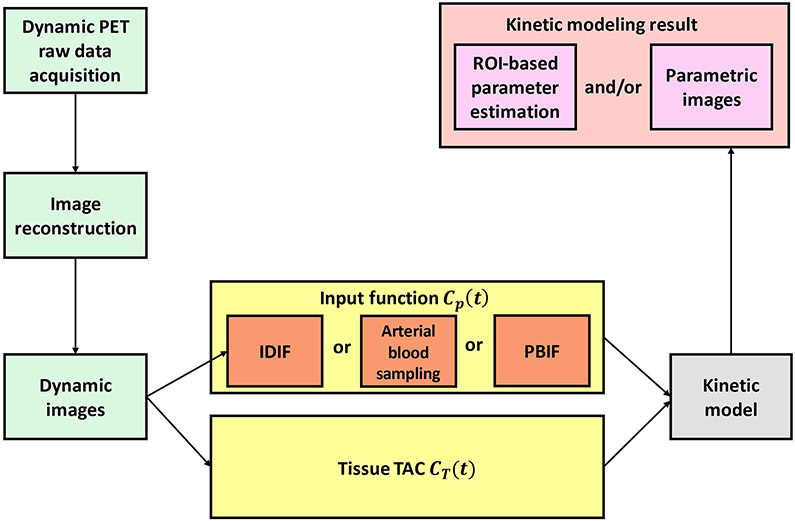
Figure 1 shows the flowchart for dynamic PET imaging and kinetic modeling. An example of tracer kinetic modeling with compartmental models17 for metabolic imaging is given in Figure 2 for 18F-FDG. The FDG net influx rate is directly proportional to the metabolic rate of glucose and is a macro parameter of interest 2,3. Compartmental modeling usually needs nonlinear curve fitting to estimate the kinetic parameters, which is computationally expensive and also sensitive to noise. As an alternative to compartmental modeling, the Patlak model18 is a linear graphical method that can approximate Ki using the slope of a graphical plot of the blood input function and tissue time-activity curve (TAC). It has the advantages of computational efficiency and noise robustness for parametric imaging. Examples of compartmental models and graphical methods for modeling ligand-receptor kinetics are reviewed by Watabe et al.19. The potential of kinetic modeling and parametric imaging has been demonstrated in both research studies and clinical diagnosis20,21.
Figure 1.
Flowchart of PET kinetic modeling and parametric imaging. Raw PET projection data in the format of sinogram or list-mode are acquired and reconstructed into dynamic images. For each image voxel or a region of interest (ROI), a time activity curve (TAC) CT(t)is extracted from the dynamic sequence and fitted using a kinetic model and a blood input function to estimate kinetic parameters. The input function can be either noninvasively derived from the dynamic images or invasively measured by arterial blood sampling or is a population-based input function (PBIF). Kinetic modeling can be ROI-based or voxel-based (i.e., parametric imaging).
Figure. 2.
Two-tissue compartment model for 18F-FDG. FDG is transported from plasma to tissue cells with the delivery rate K1 and from tissue to plasma with the rate k2. FDG is phosphorylated in cells into FDG 6-phosphate with the rate k3 and the process can be reversed at the rate k4. The total activity measured by PET is CT(t) that is a sum of different compartments. vb denoting the fractional blood volume and Cwb(t) is FDG activity in the whole-blood.
One challenge of kinetic modeling is to obtain the input function Cp(t) . Conventionally it would be obtained using arterial blood sampling which, however, is invasive. To reduce invasiveness, population-based input functions (PBIF) can be used in combination with one or two blood samples22. As the spatial resolution of PET scanners has improved over time, it has become feasible to non-invasively derive a blood input function from dynamic PET images if a large blood region (e.g., left ventricle or aorta) is available in the scanner FOV. This type of input function is usually referred to as the image-derived input function (IDIF).
2.2. Limitations of dynamic whole-body imaging on conventional PET scanners
Conventional PET scanners have a short AFOV, typically 15-30 cm, as Figure 3(A) shows. To acquire a set of whole-body dynamic images in sequence, the scanner must use multiple bed positions and multiple passes8,23, as shown in Figure 3(B). As a result, early-phase data that has unique information linked to blood flow and blood volume is only available for the organs imaged in the first bed position for conventional scanners but is missing for most of the body. There are also large temporal gaps in dynamic frames at any given scanned location. Furthermore, due to the isotropic nature of the annihilation photon emission, the detection sensitivity within the short AFOV is low11, leading to a high noise level in dynamic images.
Figure 3.
Illustration of dynamic whole-body imaging on a conventional PET scanner (A) with a multi-bed multi-pass strategy (B).
The acquisition of an input function Cp(t) for whole-body kinetic modeling with short AFOV PET scanners is another challenge. If measuring of the IDIF is desired, the location of the first bed position may need to be shifted from the main organ of interest (e.g. the brain) to a location covering the aorta, losing valuable early information in the organ of interest. This means some (complex) kinetic model models (e.g. two-tissue compartment model) can no longer be used for analyzing the tissue of interest.
2.3. Total-body dynamic imaging on uEXPLORER
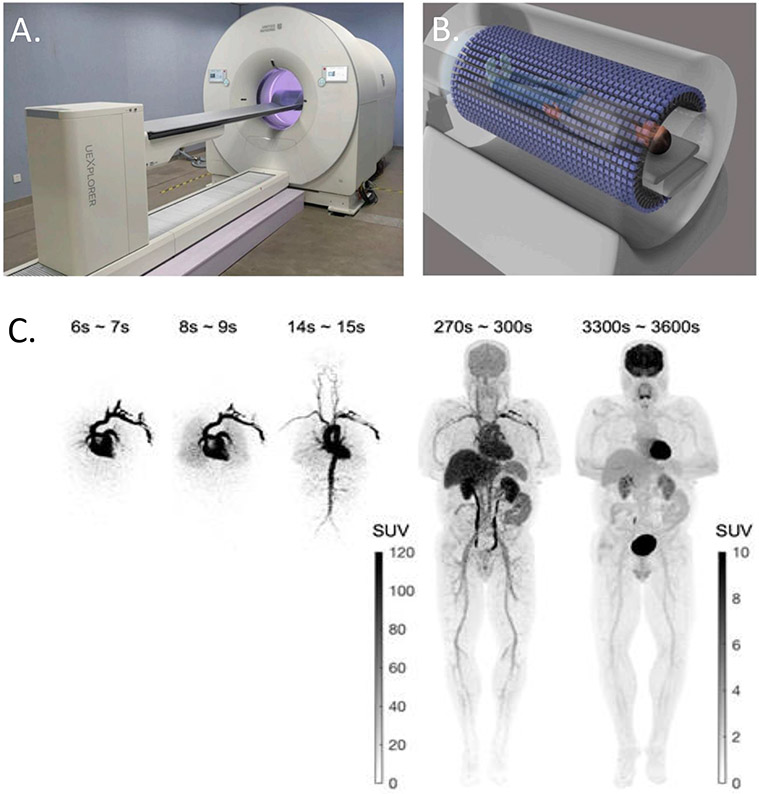
The development of the uEXPLORER total-body PET/CT system12, with a 194 cm AFOV, is an important step in addressing some limitations of conventional PET scanners, as shown in Figure 4. It allows simultaneous dynamic imaging of the entire body. This eliminates the large temporal gaps in conventional dynamic whole-body imaging. The total-body axial coverage also increases the scanner detection sensitivity by 20-40 fold and the image signal-to-noise ratio by 5-6 fold5 for imaging the entire body. In addition, the high sensitivity allows dynamic imaging with much higher temporal resolution (HTR), such as 1 s per time frame13 or even 0.1 s per frame24 compared to 10-40 s per frame in traditional protocols, which may be used to explore novel clinical applications.
Figure 4. Total-body dynamic PET imaging using uEXPLORER.
(A) The uEXPLORER total-body PET system. (B) Total-body imaging. (C) Total-body dynamic 18F-FDGimages of a healthy subject. The images were reconstructed with no point spread function modeling and no post-reconstruction smoothing.
2.4. Potential benefits of total-body dynamic PET for kinetic modeling: examples
2.4.1. Noninvasive IDIF
Total-body dynamic images acquired from the uEXPLORER system bring several potential benefits for non-invasive IDIF determination. As compared to dynamic whole-body imaging on conventional PET scanners, the total-body coverage of uEXPLORER allows an IDIF to be extracted from major blood pools in the body without compromising the temporal resolution of imaging any other organs. This is advantageous even for single-organ dynamic PET imaging. For example, for dynamic brain imaging with conventional PET, the carotid arteries are the largest available blood pool present in the FOV from which to derive an IDIF; with a diameter of 5-6 mm, the carotids have little signal and suffer from severe partial volume effects. Total-body dynamic PET overcomes this problem by providing a low-noise IDIF from the large blood pool present in the FOV such as the left ventricle or ascending/descending aorta. Figure 5 shows an example of brain parametric imaging from a healthy subject scan on the uEXPLORER, where carotid partial volume effects result in increased Ki estimates.
Figure 5. Total-body IDIF extraction.
(A) Maximum intensity projection (MIP) image of the dynamic, uEXPLORER dataset of a healthy volunteer. (B) Example IDIFs derived from different blood pool ROIs. (C) Parametric images of FDG net influx rate Ki using different ROIs for IDIF extraction. The carotid-extracted IDIF results in much higher Ki estimates than the LV-extracted IDIF.
The ability to acquire low-noise input functions, non-invasively, and measured using the same device as the tissue TACs, aids in unbiased kinetic parameter estimation. While most organs are supplied by a single blood input from the arterial system, some organs have dual blood supplies. The liver, for example, is supplied by both the hepatic artery and the portal vein. However, IDIF extraction from the portal vein is difficult with conventional dynamic PET imaging, due to the combined effect of limited spatial resolution, the small size of the portal vein (about 10 mm), and high noise levels present in a small ROI. This challenge can be reduced by total-body dynamic PET imaging on uEXPLORER which with much higher sensitivity and higher spatial resolution will allow better estimation of the IDIF from such vessels.
2.4.2. Multi-parametric imaging
Limited by the short AFOV, conventional PET scanners cannot simultaneously capture total-body kinetic signals, especially in the early phase of radiotracer bolus distribution. Thus, parametric imaging has usually been limited to the linear Patlak plot method in dynamic whole-body PET studies with conventional scanners. This method provides a slope image for the macro kinetic parameter Ki and an intercept image related to a combination of fractional blood volume vb and volume of distribution. However, the full potential of compartmental modeling that allows quantification of micro kinetic parameters (e.g., the tracer delivery rate K1 and fractional blood volume vb) are difficult to explore with such systems. For example, a whole-body Ki image can be obtained with conventional scanners, whereas a whole-body K1 image cannot.
Total-body dynamic imaging with the uEXPLORER system has the potential to address this shortcoming and enable high-quality total-body kinetic modeling and parametric imaging of micro kinetic parameters. Figure 6 shows an example for parametric imaging of FDG uptake rate Ki fractional blood volume vb, and FDG delivery rate K1 from a uEXPLORER scan16.
Figure 6.

Total-body parametric images estimated from a 60-minute dynamic 18F-FDG scan of a patient with metastatic cancer on the uEXPLORER: (A) SUV, (B) FDG net influx rate Ki (C) fractional blood volume vb, and (D) FDG delivery rate K1.
Among the different kinetic parameters measured, the tracer delivery rate K1 is of particular clinical interest due to its connection to blood flow25. This parameter is different from the influx rate Ki and hence can provide complementary information, e.g., potentially for simultaneous evaluation of perfusion-metabolism mismatch using FDG for myocardial viability26. Parametric imaging of the relative delivery rate R1 (ratio of K1 between a tissue region and a reference region) of tau tracers and beta amyloid tracers is also being explored as a surrogate of cerebral blood flow to simultaneously assess both blood flow and misfolded protein changes in neurodegenerative disease using a single tracer. Readers are referred to the section III.D of a recent review article on parametric imaging for more details4.
Total-body parametric imaging of the fractional blood volume vb may also add useful physiological and pathological information. For example, chronic obstructive pulmonary disease (COPD) changes lung blood volume27. vb may also reveal the local blood supply and microenvironment of a tumor, thus may be helpful to improve tumor diagnosis and characterization28.
2.4.3. High-temporal resolution kinetic modeling
The ability of uEXPLORER for HTR imaging (e.g., 1 s per frame) allows not only better temporal sampling of the blood input function, but also more accurate modeling of fast tracer kinetics. For example, after bolus injection, radiotracer signal in the lungs is supplied by the pulmonary artery and bronchial artery. However, differentiation of the contributions from the two blood supplies is challenging if the temporal resolution of dynamic imaging is insufficient. With total-body dynamic PET, it becomes feasible to measure lung TACs with high temporal resolution and derive the bronchial arterial input function and pulmonary arterial input function from the left ventricle and right ventricle, respectively. Our preliminary results have demonstrated HTR imaging has a significant effect on the quantification of 18F-FDG K1 and vb in the lungs29.
The second example is the multiphase Patlak plot. Patlak plots derived with standard temporal resolution commonly show a single late-time linear phase. With HTR imaging, preliminary results from Zuo et al.30 demonstrated two additional approximately linear phases: one at around 20-30s (first-pass) and the other at around 1-2 minutes (early-time) (Figure 7). Total-body parametric images of the slopes of the first-pass, early-time and standard late-time linear phases also demonstrate different spatial patterns. It is worth noting that Patlak plot has been used with high temporal resolution in dynamic contrast-enhanced magnetic resonance imaging for assessing blood brain barrier permeability31. Thus, we postulated that the first-pass and early-time Patlak slopes may be related to blood flow and tracer permeability, though the precise meaning and physiological basis for these earlier linear phases remain to be determined.
Figure 7.
High temporal resolution Patlak plot and parametric imaging. (A) Patlak plot of one-hour dynamic FDG data; (B) zoom in of the first 2-minute data; (C) parametric images of the slope at three different phases – standard Ki (30-60 minutes), first-pass Ki (20-30s) and early-time Ki (1-2 minutes). Shown are the maximum intensity projection images. The unit of Ki is mL/min/mL.
Another example of HTR kinetic modeling is for the separation of blood flow and tracer-specific transport from the overall tracer delivery rate K1, e.g., through time-varying kinetic modeling that is currently under development32,33. For 18F-FDG, HTR kinetic modeling may make it possible to derive blood flow, glucose transport, and glucose metabolism simultaneously from a single dynamic scan.
2.4.4. Dual-tracer dynamic PET imaging
Single-scan dual-tracer (or multi-tracer) PET imaging has attracted a lot of interest over the past decades (see the review paper from Kadrmas and Hoffman34). To recover separate images of each tracer from the same scan, dynamic imaging and kinetic modeling can be used to separate the two tracer signals from each other. The robustness of single-scan dual-tracer methods has typically been limited by data noise. The increased sensitivity of total-body PET scanners is offering new opportunities to make this framework more robust and feasible for clinical investigation. The total-body coverage will also allow simultaneous dual-tracer dynamic imaging of the entire body.
2.5. Potential clinical impact
Compared to the semi-quantitative SUV used routinely in the clinic today, total-body kinetic modeling on the uEXPLORER opens up the window for studying systemic diseases quantitatively and in a multi-parametric fashion using a single radiotracer. One obvious example is studying the heterogeneity of metastatic tumor characteristics, both before and after treatment. Total-body PET allows for kinetic data to be acquired simultaneously for all metastatic lesions in the body, and multi-parametric imaging may provide a sensitive assay for assessing the response or likely response of each lesion to treatment. For example, dysregulated cellular metabolism and angiogenesis are both hallmarks of cancer35. High-quality multi-parametric imaging of FDG Ki (reflecting metabolism) and K1 (reflecting perfusion and glucose transport) has the potential to provide complementary information of tumor metabolism and angiogenesis for more accurate tumor characterization for response assessment.
Similar approaches could be used to quantify inflammatory or infection burden across the entire body with appropriate radiotracers. For example, in arthritis, total-body dynamic imaging allows all the joints of the body to be assessed simultaneously with kinetic modeling approaches. Finally, absolute quantification of perfusion and blood volume across the entire body would likely have a broad clinical impact, especially if scans can be performed quickly and at relatively low radiation dose. Clinical applications would include a range of cardiovascular diseases (for example, peripheral arterial disease) where the vascular health of the entire body can be assessed and quantified. Whole-body parametric imaging of rapid physiological processes and tracer distributions is particularly challenging on conventional short axial field of scanners as there simply is insufficient time to move the bed and collect data across the entire body.
2.6. Limitations of total-body dynamic PET
While total-body dynamic PET brings several potential benefits, some limitations of conventional dynamic PET imaging still remain. For example, parametric imaging of FDG Ki usually requires a one-hour long scan, which is one of the main hurdles that limit the widespread adoption of dynamic FDG-PET in clinics. This hurdle remains with total-body dynamic PET imaging. Many radiotracers have the issue of being metabolized during the imaging time. Metabolite correction is needed for determining the true parent plasma input function36, which remains a challenge for total-body kinetic modeling, though new noninvasive methods are being explored. Another limitation of total-body dynamic PET imaging is the limited availability of total-body PET systems. At the time of writing there are approximately 10 installations of total-body or long (> 1 meter) AFOV scanner worldwide. This implies that initially total-body dynamic PET studies will be only conducted in a small number of institutions, although the number of these scanners is expected to increase quite quickly in the coming years.
Total-body kinetic modeling also brings new significant challenges, including the large scale of total-body dynamic datasets and wide physiological heterogeneity in different organs that should be accounted for by using organ and tissue appropriate models. Total-body parametric imaging is further complicated by non-rigid subject motion that occurs throughout the dynamic acquisition. Kinetic parameter estimation with compartmental modeling is also highly nonlinear, suffers from local minima, and is sensitive to noise, resulting in a challenging task for conventional ROI-based modeling that is exacerbated when performing parametric imaging where models are applied on a voxel-by-voxel basis. In addition, the computational efficiency of nonlinear parametric imaging needs to be further improved for practical use because up to 10 million image voxels per acquisition are typically processed in total-body parametric imaging.
3. Opportunities using deep learning for total-body kinetic modeling
Deep learning has attracted broad attention for its huge potential in almost every field, including PET37,38. One major advantage of deep learning is its ability for end-to-end training of a mapping from problem inputs to answers. Once the model is trained, the prediction is fast. In this section, we discuss a few examples for potential deep learning-based solutions to address the specific challenges in total-body kinetic modeling.
3.1. Voxel-wise corrections for blood input function
In total-body dynamic PET, the IDIF is usually extracted from an ROI selected within a large central blood pool. For a peripheral tissue region that is far from the blood ROI, the arrival time of the radiotracer is delayed compared to the start time of the extracted blood input function. Thus, this time delay of the input function needs to be corrected to achieve accurate kinetic quantification. Time delay correction has been considered impactful only for high-temporal resolution imaging in conventional dynamic PET but is also essential in total-body imaging because the time delay can be 30-40 s in peripheral tissues. In addition, a dispersion correction to account for the mixing of the injected radiotracer in the blood may also be critical to model the input function for a specific organ14,39.
Corrections for these factors can be pursued by joint estimation which estimates the parameters simultaneously with other kinetic parameters during the fitting of TACs. A recent example is given by Feng et al.4 in total-body imaging of early FDG kinetics. However, the joint estimation method can be computationally expensive. Alternatively, time delay can be corrected voxel-wise before parametric imaging40. Such a pre-correction method has the advantage of reducing the computational burden. While analytical approaches such as the leading-edge method may not achieve the best accuracy, one of our ongoing efforts is to explore deep learning for regression to train a model that can predict the time delay and/or dispersion of the blood input from the tissue TAC data in combination with the blood input function. The labeling data for training can be built from patient data using the joint estimation method.
3.2. Learning-based input function
For tracer kinetic modeling, a plasma input function is what is actually needed, while an IDIF represents the tracer activity in the whole blood. The tracer fraction in plasma may be pre-determined based on population data, but it is challenging to adapt this to individual patients41. In addition, an IDIF may suffer from partial volume and spill-over effects, which leads to a potential discrepancy between the IDIF and true blood input function.
Machine learning has the potential to provide a robust and fast prediction of the input function from tissue TACs. For example, Kuttner et al. estimated the IDIF from multiple blood pool TACs or multiple tissue TACs using a Gaussian Process model and long short-term memory network42. Instead of requiring manual ROI placements, machine learning can also estimate the IDIF directly from dynamic images43.
Metabolite correction is needed for some radiotracers36 and can be done by arterial blood sampling (or if the metabolism is slow and the arterial-venous difference is small, venous blood sampling may be sufficient) followed by high-performance liquid chromatography analysis, which is complex and invasive, and therefore is often mainly used to derive a population-based model. Mathematical metabolite correction44-46 is an alternative solution that jointly estimates the parent plasma input function during TAC fitting47. However, this method requires a complex optimization due to the high nonlinearity of the model and can easily get stuck at a local minimum and has a high computational cost. One possible solution is to use deep learning to learn an end-to-end solution from the TAC data by using the global optimization or arterial sampling result as the training labels. Once trained in this manner, the model can be efficient to use.
3.3. Voxel-wise kinetic model selection
In conventional parametric imaging with short AFOV scanners, one kinetic model is commonly used for the kinetic parameter estimation for all image voxels. However, the physiology varies among organs in the body and is beyond the descriptive ability of a single kinetic model. An oversimplified model causes biased quantification, while a too complex model leads to unstable parameter estimation. This issue can be addressed by voxel-wise model selection which chooses the best of two or more candidate models. As shown in Figure 8(B), the parametric image generated by a standard two-tissue irreversible compartment model produces artifacts in the ventricular blood pool and blood vessels (pointed out by arrows). These artifacts can be suppressed by the appropriate model selection, as shown in Figure 8(C).
Figure 8.
Total-body kinetic model selection. (A) The model selection map. (B) Total-body Ki image using the two-tissue irreversible model without model selection. (C) Total-body Ki image with model selection.
Conventional methods for model selection use statistical metrics to balance the trade-off between the curve-fitting error and the model complexity. Golla et al. compared several model selection metrics, including Akaike Information Criterion, Model Selection Criterion, and Schwartz Criterion, and found strong agreement across them48. These methods can be implemented in a voxel-wise manner but are time-consuming for total-body imaging.
Deep learning may bring an efficient solution to kinetic model selection by formulating model selection as a binary or multi-class classification problem. The spatial correlation of neighboring voxels can also be incorporated into the learning model. Such an approach has been recently explored by Klyuzhin et al.49 and Fuller el al.50 for detecting transient responses in neurotransmitter PET.
3.4. Fast and robust prediction of kinetic parameters
Machine learning has also been applied for directly predicting kinetic parameters from the TAC data. A trained model has the potential to generate parametric images very efficiently and be robust to noise. Early attempts were demonstrated in 2001 using a shallow neural network for 13N-ammonia perfusion parametric imaging by Golish et al.51, in 2011 using a support vector machine for kinetic prediction with a shortened dynamic FDG-PET protocol52,53, and recently using a deep convolutional neural network for predicting the parametric images of macro parameters54.
While learning a direct mapping from the TAC space to kinetic parameter space remains a big challenge, it is more feasible to predict high-quality parametric images from low-quality parametric images generated by conventional kinetic modeling. For example, parametric imaging commonly uses an indirect approach that reconstructs the dynamic images first and then performs kinetic modeling voxel by voxel to generate parametric images, as discussed in the sections above. In comparison, the ‘direct method’ combines the kinetic model into the forward projection model of PET image reconstruction and estimates the parametric images directly from the projection data. Direct parametric image reconstruction has been demonstrated to outperform the indirect method55. However, direct parametric image reconstruction requires a more complex optimization and is time-consuming. Feng et al.56 and Xie et al.57 used convolutional neural network learning to recover high-quality Patlak parametric images (estimated by the direct method) from the Patlak images estimated by the indirect method.
3.5. Single-Subject Deep Learning
One general challenge with deep learning is the requirement of collecting a large number of patient scans for building the training dataset. This is particularly challenging for deep learning for kinetic modeling because each patient has only one blood input function, though many tissue TACs are available for building a database. With a limited number of blood input functions, the generalization capability of a trained model remains a concern.
Single-subject deep learning may be a feasible alternative solution and is being pursued in our group. In this method, a deep learning model is trained on the fly with a low computational cost by using a small fraction of voxels labeled with conventional kinetic modeling. The remaining voxels are unlabeled but will be predicted efficiently using the trained model. One advantage of this learning method is all the training and testing samples share the same blood input function. The labeled and unlabeled data can be further combined through a semi-supervised learning framework58.
3.6. Voxel-wise motion correction
As the image data size and FOV increase with total-body PET, various types of subject motion, involuntary (e.g. respiratory, cardiac, bladder filling) and voluntary (e.g. head tilting) are always present in the FOV. For the purposes of total-body parametric imaging, positioning changes in the tissue will be reflected in the individual voxel TACs, which in turn may affect kinetic parameter estimation. Motion tracking hardware59,60 and data-driven methods 61 such as the centroid of distribution method62-64 can be applied to total-body dynamic imaging. Note that total-body motion is generally non-rigid and requires time-consuming algorithms for motion correction. Deep-learning based solutions may have the advantage to efficiently and non-rigidly register respiratory-gated PET images65, and may provide improved results with high-quality images generated from total-body PET.
4. Summary
Total-body kinetic modeling on the uEXPLORER system enables quantitative multi-parametric imaging of the entire body simultaneously. With recent developments in deep learning, the remaining challenges of total-body kinetic modeling and parametric imaging may be addressed. Many opportunities are emerging for exploiting total-body kinetic modeling in various research and clinical applications.
Clinics Care Points.
Reading List
Must Read
Carson RE. Tracer Kinetic Modeling in PET. In: Positron Emission Tomography. Springer-Verlag; 2005:127-159.
Rahmim A, Lodge MA, Karakatsanis NA, et al. Dynamic whole-body PET imaging: principles, potentials and applications. Eur J Nucl Med Mol Imaging. 2019;46(2):501-518. doi:10.1007/s00259-018-4153-6
Badawi RD, Shi H, Hu P, et al. First Human Imaging Studies with the EXPLORER Total-Body PET Scanner. J Nucl Med. 2019;60(3):299-303. doi:10.2967/jnumed.119.226498.
Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-Body PET: Maximizing Sensitivity to Create New Opportunities for Clinical Research and Patient Care. J Nucl Med. 2018;59(1):3-12. doi:10.2967/jnumed.116.184028
Pantel AR, Viswanath V, Daube-Witherspoon ME, et al. PennPET Explorer: Human Imaging on a Whole-Body Imager. Journal of Nuclear Medicine. 2020;61:144-151.
Nice to Read
Golish SR, Hove JD, Schelbert HR, Gambhir SS. A Fast Nonlinear Method for Parametric Imaging of Myocardial Perfusion by Dynamic 13N-Ammonia PET J Nucl Med. 2001;42(6):924-931.
Pan L, Cheng C, Haberkorn U, Dimitrakopoulou-Strauss A. Machine learning-based kinetic modeling: a robust and reproducible solution for quantitative analysis of dynamic PET data. Phys Med Biol. 2017;62(9):3566.
Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging.
Gallezot J-D, Lu Y, Naganawa M, Carson RE. Parametric Imaging With PET and SPECT IEEE Trans Radiat Plasma Med Sci. 2020;4(1):1-23. doi:10.1109/TRPMS.2019.2908633
Wang G, Rahmim A, Gunn RN. PET Parametric Imaging: Past, Present, and Future. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):663-675. doi:10.1109/TRPMS.2020.3025086.
Key Points.
Total-body PET kinetic modeling on the uEXPLORER PET/CT system addresses several factors that serve as barriers to implementation of dynamic whole-body PET imaging on conventional PET scanners.
Total-body kinetic modeling with increased detection sensitivity supports multi-parametric imaging which has clinical potential, but also brings several technical challenges.
Deep learning provides multiple opportunities in total-body kinetic modeling, including non-invasive input function estimation, kinetic model selection, and kinetic parameter estimation. Applications of deep learning can help with further improvement of accuracy, robustness, and efficiency.
Acknowledgement
The authors acknowledge the contribution of all EXPLORER team members. We also thank the two anonymous reviewers for helpful comments that led to the improvement of this paper.
Disclosure statement
The authors have a research agreement with United Imaging Healthcare. UC Davis also has a revenue-sharing agreement with United Imaging Healthcare. This work was funded in part by NIH grant R01 CA206187.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- 1.Jones T, Townsend D. History and future technical innovation in positron emission tomography. J Med Imag. 2017;4(1):011013. doi: 10.1117/1.JMI.4.1.011013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris ED, Endres CJ, Schmidt KC, Christian BT, Jr RFM, Fisher RE. Kinetic Modeling in Positron Emission Tomography. Emission tomography: The fundamentals of PET and SPECT. 2004;46(1):499–540. [Google Scholar]
- 3.Carson RE. Tracer Kinetic Modeling in PET. In: Positron Emission Tomography. Springer-Verlag; 2005:127–159. [Google Scholar]
- 4.Wang G, Rahmim A, Gunn RN. PET Parametric Imaging: Past, Present, and Future. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):663–675. doi: 10.1109/TRPMS.2020.3025086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-Body PET: Maximizing Sensitivity to Create New Opportunities for Clinical Research and Patient Care. J Nucl Med. 2018;59(1):3–12. doi: 10.2967/jnumed.116.184028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, Rahmim A. Dynamic whole-body PET parametric imaging: I. Concept, acquisition protocol optimization and clinical application. Phys Med Biol. 2013;58(20):7391–7418. doi: 10.1088/0031-9155/58/20/7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Panin V, Smith AM, et al. Design and Implementation of Automated Clinical Whole Body Parametric PET With Continuous Bed Motion. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):696–707. doi: 10.1109/TRPMS.2020.2994316 [DOI] [Google Scholar]
- 8.Rahmim A, Lodge MA, Karakatsanis NA, et al. Dynamic whole-body PET imaging: principles, potentials and applications. Eur J Nucl Med Mol Imaging. 2019;46(2):501–518. doi: 10.1007/s00259-018-4153-6 [DOI] [PubMed] [Google Scholar]
- 9.Karp JS, Viswanath V, Geagan MJ, et al. PennPET Explorer: Design and Preliminary Performance of a Whole-Body Imager. J Nucl Med. 2020;61(1):136–143. doi: 10.2967/jnumed.119.229997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberts I, Hünermund J-N, Prenosil G, et al. Clinical performance of long axial field of view PET/CT: a head-to-head intra-individual comparison of the Biograph Vision Quadra with the Biograph Vision PET/CT. Eur J Nucl Med Mol Imaging. Published online April2, 2021:1–10. doi: 10.1007/s00259-021-05282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherry SR, Badawi RD, Karp JS, Moses WW, Price P, Jones T. Total-body imaging: Transforming the role of positron emission tomography. Sci Transl Med. 2017;9(381):1. doi: 10.1126/scitranslmed.aaf6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer BA, Berg E, Schmall JP, et al. Performance evaluation of the uEXPLORER Total-body PET/CT scanner based on NEMA NU 2-2018 with additional tests to characterize long axial field-of-view PET scanners. J Nucl Med. Published online 2020:jnumed.120.250597. doi: 10.2967/jnumed.120.250597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badawi RD, Shi H, Hu P, et al. First Human Imaging Studies with the EXPLORER Total-Body PET Scanner*. J Nucl Med. 2019;60(3):299–303. doi: 10.2967/jnumed.119.226498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng T, Zhao Y, Shi H, et al. Total-Body Quantitative Parametric Imaging of Early Kinetics of FDG. J Nucl Med. Published online September18, 2020:jnumed.119.238113. doi: 10.2967/jnumed.119.238113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vera DB, Schulte B, Henrich T, et al. First-in-human total-body PET imaging of HIV with 89Zr- VRC01 on the EXPLORER. J Nucl Med. 2020;61(supplement 1):545. [Google Scholar]
- 16.Wang G, Parikh M, Nardo L, et al. Total-Body Dynamic PET of Metastatic Cancer: First Patient Results. J Nucl Med. 2020;61(supplement 1):208. [Google Scholar]
- 17.Gunn RN, Gunn SR, Cunningham VJ. Positron Emission Tomography Compartmental Models. J Cereb Blood Flow Metab. 2001;21(6):635–652. doi: 10.1097/00004647-200106000-00002 [DOI] [PubMed] [Google Scholar]
- 18.Patlak CS, Blasberg RG, Fenstermacher JD. Graphical Evaluation of Blood-to-Brain Transfer Constants from Multiple-Time Uptake Data. J Cereb Blood Flow Metab. 1983;3(1):1–7. doi: 10.1038/jcbfm.1983.1 [DOI] [PubMed] [Google Scholar]
- 19.Watabe H, Ikoma Y, Kimura Y, Naganawa M, Shidahara M. PET kinetic analysis—compartmental model. Ann Nncl Med. 2006;20(9):583–588. doi: 10.1007/BF02984655 [DOI] [PubMed] [Google Scholar]
- 20.Hooker JM, Carson RE. Human Positron Emission Tomography Neuroimaging. Annu Rev Biomed Eng. 2019;21:551–581. [DOI] [PubMed] [Google Scholar]
- 21.Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging. Published online January2021:1–19. doi: 10.1007/s00259-020-04843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberl S, Anayat AR, Fulton RR. Evaluation of two population-based input functions for quantitative neurological FDG PET studies. Eur J Nucl Med. 1997;24(3):299–304. [DOI] [PubMed] [Google Scholar]
- 23.Gallezot J-D, Lu Y, Naganawa M, Carson RE. Parametric Imaging With PET and SPECT. IEEE Trans Radiat Plasma Med Sci. 2020;4(1):1–23. doi: 10.1109/TRPMS.2019.2908633 [DOI] [Google Scholar]
- 24.Zhang X, Cherry SR, Xie Z, Shi H, Badawi RD, Qi J. Subsecond total-body imaging using ultrasensitive positron emission tomography. Proc Natl Acad Sci USA. 2020;117(5):2265–2267. doi: 10.1073/pnas.1917379117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullani NA, Herbst RS, O’Neil RG, Gould KL, Barron BJ, Abbruzzese JL. Tumor Blood Flow Measured by PET Dynamic Imaging of First-Pass 18F-FDG Uptake: A Comparison with 15O-Labeled Water-Measured Blood Flow. J Nucl Med. 2008;49(4):517–523. doi: 10.2967/jnumed.107.048504 [DOI] [PubMed] [Google Scholar]
- 26.Abraham A, Nichol G, Williams KA, et al. 18 F-FDG PET Imaging of Myocardial Viability in an Experienced Center with Access to 18 F-FDG and Integration with Clinical Management Teams: The Ottawa-FIVE Substudy of the PARR 2 Trial. J Nucl Med. 2010;51(4):567–574. doi: 10.2967/jnumed.109.065938 [DOI] [PubMed] [Google Scholar]
- 27.Chen DL, Cheriyan J, Chilvers ER, et al. Quantification of Lung PET Images: Challenges and Opportunities. J Nucl Med. 2017;58(2):201–207. doi: 10.2967/jnumed.116.184796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrakopoulou-Strauss A, Strauss LG, Schwarzbach M, et al. Dynamic PET 18F-FDG Studies in Patients with Primary and Recurrent Soft-Tissue Sarcomas: Impact on Diagnosis and Correlation with Grading. J Nucl Med. 2001;42(5):713–720. [PubMed] [Google Scholar]
- 29.Wang Y, Cherry S, Badawi R, Wang G. Effect of dual-input function and dispersion on lung FDG-PET kinetic quantification using the EXPLORER total-body PET/CT scanner. J Nucl Med. 2020;61(supplement 1):13.31712326 [Google Scholar]
- 30.Zuo Y, Cherry S, Badawi R, Wang G. Multiphase Patlak Plot Enabled by High Temporal Resolution Total-body Dynamic PET. J Nucl Med. 2020;61 (supplement 1):207. [Google Scholar]
- 31.Cramer SP, Larsson HB. Accurate Determination of Blood-Brain Barrier Permeability Using Dynamic Contrast-Enhanced T1-Weighted MRI: A Simulation and in vivo Study on Healthy Subjects and Multiple Sclerosis Patients. J Cereb Blood Flow Metab. 2014;34(10):1655–1665. doi: 10.1038/jcbfm.2014.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Sarkar S, Kim E, Badawi R. Time-Varying Kinetic Modeling of High Temporal- Resolution Dynamic 18F-FDG PET Data for Multiparametric Imaging. J Nucl Med. 2018;59(supplement 1):503. [Google Scholar]
- 33.Wang G, Spencer B, Sarkar S, et al. Quantification of Glucose Transport Using High Temporal Resolution Dynamic PET Imaging. J Nucl Med. 2019;60(supplement 1):521. [Google Scholar]
- 34.Kadrmas DJ, Hoffman JM. Methodology for Quantitative Rapid Multi-Tracer PET Tumor Characterizations. Theranostics. 2013;3(10):757–773. doi: 10.7150/thno.5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 36.Gunn RN, Sargent PA, Bench CJ, et al. Tracer Kinetic Modeling of the 5-HT1AReceptor Ligand [carbonyl-11C]WAY-100635 for PET. NeuroImage. 1998;8(4):426–440. doi: 10.1006/nimg.1998.0379 [DOI] [PubMed] [Google Scholar]
- 37.Domingues I, Pereira G, Martins P, Duarte H, Santos J, Abreu PH. Using deep learning techniques in medical imaging: a systematic review of applications on CT and PET. Artif Intell Rev. 2020;53(6):4093–4160. doi: 10.1007/s10462-019-09788-3 [DOI] [Google Scholar]
- 38.Gong K, Berg E, Cherry SR, Qi J. Machine Learning in PET: From Photon Detection to Quantitative Image Reconstruction. Proc IEEE. 2020;108(1):51–68. doi: 10.1109/JPROC.2019.2936809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, Corwin MT, Olson KA, Badawi RD, Sarkar S. Dynamic PET of human liver inflammation: impact of kinetic modeling with optimization-derived dual-blood input function. Phys Med Biol. 2018;63(15):155004. doi: 10.1088/1361-6560/aac8cb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li E, Spencer BA, Schmall JP, Wang G, Cherry SR. Pulse-timing methods for time delay estimation in dynamic total-body PET kinetic modeling. In: IEEE NSS&MIC; 2020. [Google Scholar]
- 41.Keiding S Bringing Physiology into PET of the Liver. Journal of Nuclear Medicine. 2012;53(3):425–433. doi: 10.2967/jnumed.111.100214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuttner S, Wickstrøm KK, Lubberink M, et al. Cerebral blood flow measurements with 15 O-water PET using a non-invasive machine-learning-derived arterial input function. J Cereb Blood Flow Metab. Published online February8, 2021:0271678X2199139. doi: 10.1177/0271678X21991393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Ma T, Yao S, et al. Direct Estimation of Input Function Based on Fine-tuned Deep Learning Method in Dynamic PET Imaging. J Nucl Med. 2020;61(supplement 1):1394–1394. [Google Scholar]
- 44.Burger C, Buck A. Tracer kinetic modelling of receptor data with mathematical metabolite correction. Eur J Nucl Med. 1996;23(5):539–545. doi: 10.1007/BF00833389 [DOI] [PubMed] [Google Scholar]
- 45.Sanabria-Bohórquez SM, Labar D, Levêque P, et al. [11C]Flumazenil metabolite measurement in plasma is not necessary for accurate brain benzodiazepine receptor quantification. Eur J Nucl Med. 2000;27(11):1674–1683. doi: 10.1007/s002590000336 [DOI] [PubMed] [Google Scholar]
- 46.Graham MM, Peterson LM, Hayward RM. Comparison of simplified quantitative analyses of FDG uptake. Nucl Med Biol. 2000;27(7):647–655. doi: 10.1016/S0969-8051(00)00143-8 [DOI] [PubMed] [Google Scholar]
- 47.Feng DD, Chen K, Wen L. Noninvasive Input Function Acquisition and Simultaneous Estimations With Physiological Parameters for PET Quantification: A Brief Review. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):676–683. doi: 10.1109/TRPMS.2020.3010844 [DOI] [Google Scholar]
- 48.Golla SSV, Adriaanse SM, Yaqub M, et al. Model selection criteria for dynamic brain PET studies. EJNMMI Phys. 2017;4(1):1–10. doi: 10.1186/s40658-017-0197-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klyuzhin IS, Bevington CWJ, Cheng J-C (Kevin), Sossi V. Detection of transient neurotransmitter response using personalized neural networks. Phys Med Biol. 2020;65(23):235004. doi: 10.1088/1361-6560/abc230 [DOI] [PubMed] [Google Scholar]
- 50.Fuller OK, Angelis GI, Meikle SR. Classification of Neurotransmitter Response in Dynamic PET Data Using Machine Learning Approaches. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):708–719. doi: 10.1109/TRPMS.2020.2984259 [DOI] [Google Scholar]
- 51.Golish SR, Hove JD, Schelbert HR, Gambhir SS. A Fast Nonlinear Method for Parametric Imaging of Myocardial Perfusion by Dynamic 13N-Ammonia PET. J Nucl Med. 2001;42(6):924–931. [PubMed] [Google Scholar]
- 52.Strauss LG, Pan L, Cheng C, Haberkorn U, Dimitrakopoulou-Strauss A. Shortened Acquisition Protocols for the Quantitative Assessment of the 2-Tissue-Compartment Model Using Dynamic PET/CT 18F-FDG Studies. J Nucl Med. 2011;52(3):379–385. doi: 10.2967/jnumed.110.079798 [DOI] [PubMed] [Google Scholar]
- 53.Pan L, Cheng C, Haberkorn U, Dimitrakopoulou-Strauss A. Machine learning-based kinetic modeling: a robust and reproducible solution for quantitative analysis of dynamic PET data. Phys Med Biol. 2017;62(9):3566. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, Ruan D, Liu H. Noninvasive Estimation of Macro-Parameters by Deep Learning. IEEE Trans Radiat Plasma Med Sci. 2020;4(6):684–695. doi: 10.1109/TRPMS.2020.2979017 [DOI] [Google Scholar]
- 55.Wang G, Qi J. An Optimization Transfer Algorithm for Nonlinear Parametric Image Reconstruction From Dynamic PET Data. IEEE Trans Med Imaging. 2012;31(10):1977–1988. doi: 10.1109/TMI.2012.2212203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng T, Zhao Y, Dong Y, Yao S. Acceleration of Whole-body Patlak Parametric Image Reconstruction using Convolutional Neural Network. J Nucl Med. 2019;60(supplement 1):518–518. [Google Scholar]
- 57.Xie N, Gong K, Guo N, et al. Clinically Translatable Direct Patlak Reconstruction from Dynamic PET with Motion Correction Using Convolutional Neural Network. In: Martel AL, Abolmaesumi P, Stoyanov D, et al. , eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. Vol 12267. Lecture Notes in Computer Science. Springer International Publishing; 2020:793–802. doi: 10.1007/978-3-030-59728-3_77 [DOI] [Google Scholar]
- 58.Zhou X, Belkin M. Semi-Supervised Learning. In: Academic Press Library in Signal Processing. Vol 1. Elsevier; 2014:1239–1269. doi: 10.1016/B978-0-12-396502-8.00022-X [DOI] [Google Scholar]
- 59.Jin X, Mulnix T, Gallezot J-D, Carson RE. Evaluation of motion correction methods in human brain PET imaging-A simulation study based on human motion data: Evaluation of human brain PET motion correction methods. Med Phys. 2013;40(10):102503. doi: 10.1118/1.4819820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bec J, Henry D, Kyme A, Fulton R, Badawi RD, Cherry SR. Optical motion tracking for use with the EXPLORER total-body PET scanner. J Nucl Med. 59(supplement 1):429. [Google Scholar]
- 61.Rahmim A, Olivier R, Habib Z. Strategies for motion tracking and correction in PET. PET Clin. 2007;2(2):251–266. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y, Naganawa M, Toyonaga T, et al. Data-Driven Motion Detection and Event-by-Event Correction for Brain PET: Comparison with Vicra. J Nucl Med. 2020;61(9):1397–1403. doi: 10.2967/jnumed.119.235515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren S, Jin X, Chan C, et al. Data-driven event-by-event respiratory motion correction using TOF PET list-mode centroid of distribution. Phys Med Biol. 2017;62(12):4741–4755. doi: 10.1088/1361-6560/aa700c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Y, Gallezot J-D, Naganawa M, et al. Data-driven voluntary body motion detection and non-rigid event-by-event correction for static and dynamic PET. Phys Med Biol. 2019;64(6):065002. doi: 10.1088/1361-6560/ab02c2 [DOI] [PubMed] [Google Scholar]
- 65.Li T, Zhang M, Qi W, Asma E, Qi J. Motion correction of respiratory-gated PET images using deep learning based image registration framework. Phys Med Biol. 2020;65(15):155003. doi: 10.1088/1361-6560/ab8688 [DOI] [PMC free article] [PubMed] [Google Scholar]