Abstract
Kazakhstan covers a vast territory, and it has always been a land of nomadic pastoralism, where domesticated horses and sheep were moved by nomadic people across the steppe. Previous reports suggest that sheep breeds from Kazakhstan have an intermediate genetic composition between Asian and European breeds; however, this data appears to be limited. Therefore, we studied the genetic diversity of ancient domestic sheep from two Late Bronze Age settlements, Toksanbai and Kent, located in the Pre-Caspian region of Kazakhstan and central Kazakhstan, respectively. We have applied ZooMS analysis for taxonomic identification of small ruminant remains to select ancient specimens of domestic sheep (Ovis aries). To assign sheep mitochondrial DNA (mtDNA) haplogroups, the single nucleotide polymorphisms (SNPs) from the control region were analyzed by real-time PCR and direct sequencing. Identical distribution of mtDNA haplogroups A (8/14; 57%), B (5/14; 36%), and C (1/14; 7%) was observed in the specimens from Toksanbai (n = 14) and Kent (n = 14). Ovine haplogroup A was predominant in both settlements. Both archeological sites had similar patterns of haplogroup distribution, indicating early sheep introduction into the region. These results are important to gain a better understanding of sheep migrations in the Eurasian steppe and highlight the importance of genomic analysis of earlier local lineages.
Keywords: Haplotyping, aDNA, Mitochondrial DNA, ZooMS, Bronze age, Kazakhstan, Sheep, Ovis aries
Haplotyping; aDNA; Mitochondrial DNA; ZooMS; Bronze age; Kazakhstan; Sheep; Ovis aries.
1. Introduction
Sheep are known to be one of the first domesticated animals. The history of domestic sheep began in the Fertile Crescent around 11,000 BP [1]. Subsequently, sheep were introduced to Eurasia through several routes that bypass the Caspian Sea [2, 3, 4]. During this period, 5,000–7,000 years ago, Central Asia played a significant role in the “transport interchange” of migratory flows [2, 3].
Over 80% of the total area of Kazakhstan is classified as agricultural land. From ancient times, animal husbandry in this region has mostly been represented by two domestic species: horses and sheep. Currently, sheep breeding is an important traditional sector of agricultural production. Domestication of these animals in large numbers has been a characteristic feature of nomads in Eurasian steppes until the 19th century because of the appropriate climatic conditions and vast areas of these natural pastures. At present, sheep are one of the most widespread animals in Kazakhstan, and their number exceeds 18 million heads represented by well-known domestic breeds.
Several genetic markers are widely used in domestic animal studies [5, 6, 7]. One of the effective methods of studying genetic history of animals is the analysis of highly polymorphic genetic sites inherited along one parent line without recombination. Many such studies have focused on mitochondrial DNA [4, 8, 9, 10, 11], including those analyzing the genetic diversity of modern domestic sheep [12]. These analyses have revealed that there are five distinct lineages (haplogroups A, B, C, D, and E) of domestic sheep. Limited data indicated that sheep breeds from Kazakhstan have an intermediate genetic composition between that of Asian and European breeds [2, 4, 13]. Therefore, studying archeological sites located in this region may shed light on the migration routes and genetic structure of ancient sheep [14].
In Kazakhstan, there are archeological excavation sites with confirmed dominance of sheep and horses, as these animals are well adapted to nomadic lifestyles [15, 16]. However, sheep remains have not been reported in many Bronze Age settlements in Kazakhstan. For example, Botai and other sites in the north-central steppe dating from approximately 3500–2500 BC were found to be extremely rich in animal remains, mostly those of horses, which often comprised more than 99% of all fauna [17]. It is known that both local environmental variables and cultural factors influence herd composition [18]. In the present study, we investigated the archeological specimens of sheep from two Late Bronze settlements, namely Kent and Toksanbai, with respect to their mtDNA haplogroup composition. The selected settlements are located at 1,500 km from each other and differed in terms of size and culture. However, both have a high proportion of herded livestock specific to nomadic herding practices.
The proto-city Kent is a large Late Bronze Age settlement in the Karaganda region, located in the central Kazakhstan highlands within Karkaralinsk National Park. Kent belongs to the Begazy-Dandybaev material culture group [19]. Domestic animals in Kent are mostly represented by small ruminants, cattle, horses, and dogs. The bones of wild animals constitute only 1% of the remains in this area. Kent is dominated by animals adapted to roaming, such as sheep and horses. It has been reported that small ruminants are the most common domestic animals in Kent (48.8%), and their abundance is almost double that of cattle (26.5%) [20]. The proportion of horses in the settlement of Kent was reported to be 21.2%, suggesting that the proportion of cattle and horses among the bone remains was almost the same.
Toksanbai is a Late Bronze Age settlement in west Kazakhstan, located on the edge of a chink (“chink” refers to a cliff up to 350 m high formed as a result of marine abrasion during the retreat of the Caspian Sea on the Ustyurt plateau). The settlement probably existed in the middle Holocene Subboreal period [21]. A great diversity of mammalian bones was discovered in the Toksanbai settlement with more than 35,000 bones, of which, 19% have been attributed to domestic animals (sheep, goats, cattle, horses, camels, and dogs) and 81% to wild mammals (tolai hares, rodents, wolves, red foxes, corsac foxes, steppe polecats, Asiatic wild asses, saigas, goitered gazelles, and mouflons). In Toksanbai, small ruminants prevailed along with domestic animals.
2. Materials and methods
2.1. Samples and sampling
The sheep (Ovis aries) bone samples used in this study were obtained from two archeological sites, namely Kent settlement in the Karaganda region, central Kazakhstan (49°12′N 75°56′E) and the ancient settlement of Toksanbai in west Kazakhstan (45°51′N 56°33′E) (Figure 1). The samples from Kent were excavated in 2015 and supplied by the staff of the Saryarka Archaeological Institute, Karaganda University named after E.A. Buketov (Karaganda). The samples from Toksanbai were excavated in 2016 and supplied by the staff of the Institute of Archaeology named after A.Kh. Margulan (Almaty). The samples were stored at ambient temperature in a dark place in separate zipper storage bags. Morphology-based species identification was carried out by Dr. Pavel Kosintsev from the Institute of Plant and Animal Ecology (Yekaterinburg, Russia). The morphological inspection was performed only for taxa identification. The selected 66 bone samples of domestic sheep from spatially separated sites within the settlements were transferred to the National Center for Biotechnology (Nur-Sultan) for further study.
Figure 1.
Geographic locations of archeological sample sites on the map of Kazakhstan. Geographic locations of archeological sample sites are indicated in black. TKS, Toksanbai settlement; KNT, Kent settlement.
The specimens analyzed in the present study were not radiocarbon-dated directly, but dating was estimated from an archeological context and corresponding historical periods. According to archeological data, Toksanbai was dated to the Late Bronze Age. Radiocarbon dating of the cultural layer was 3780 ± 80 BP (GIN-7979) and 3240 ± 40 BP (GIN-8093) [21]. Kent is dated by ceramic typology to the Late/Final Bronze Age (3250-2850 BP) and belongs to the Begazy-Dandybaev material culture group presenting a single cultural layer [18].
2.2. Protein isolation
Bone specimens were sampled from Kent (n = 25) and Toksanbai (n = 41), and approximately 100 mg of bone powder was used for protein isolation and mass spectrometry analysis [22]. The samples were suspended in 1 mL 0.5 M EDTA (pH 8.0) and incubated overnight at 4 °C with gentle agitation. After centrifugation for 15 min at maximum speed at 4 °C, the supernatant was discarded. The pellets were washed twice with Milli-Q water (0.5 mL), re-suspended in 800 μL of 50 mM ammonium bicarbonate (pH 7.40), and incubated for 24 h at 75 °C with gentle agitation. The supernatant fraction was collected and stored at –20 °C (fraction A) following centrifugation for 15 min at maximum speed and 4 °C. The remaining pellets were re-suspended in 800 μL of 50 mM ammonium bicarbonate (pH 7.40) and further incubated for 24 h at 75 °C. After centrifugation, the supernatant was combined with fraction A and stored at –20 °C. Protein concentrations were estimated using a Qubit 2.0 fluorometer (Thermo, MA, USA) with Qubit Protein Assay (Thermo, MA, USA) following the manufacturer's instructions. A volume of solution containing approximately 50 μg of proteins was transferred to an Eppendorf tube for in-solution protein digestion.
2.3. Mass spectrometry analysis
After reduction and alkylation, the samples were digested by trypsin (20 ng/uL) at 37 °C overnight, and the obtained peptide mixtures were purified and concentrated using ZipTip-C18 (Millipore, Cork, Ireland). The eluted peptides were then dried using a centrifugal evaporator (Eppendorf, Hamburg, Germany) and re-suspended in 10 μL of 0.1% trifluoroacetic acid. The sample solution (1 μL) was mixed with 1 μL of α-cyano-4-hydroxycinnamic acid matrix solution [1% in ACN/H2O 1:1 (v/v)] and allowed to dry. Each collagen digest mixture was analyzed in triplicates by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) using a Microflex LT mass spectrometer (Bruker Daltonics, Leipzig, Germany).
In addition, peptide mixtures were analyzed using online nanoflow reversed-phase C18 liquid chromatography tandem mass spectrometry (LC-MS/MS). A trapping column (Acclaim PepMap 100 C18 pre-column) and a Dionex HPLC pump were used for chromatography. Peptides were separated on an Acclaim Pep-Map RSLC column (Thermo, MA, USA) using a 75 min multistep acetonitrile gradient at a flow rate of 0.3 mL/min. An unmodified captive spray ion source (capillary 1300 V, dry gas 3.0 L/min, dry temperature 150 °C) was used to interface the LC system to the Impact II mass spectrometer (Bruker Daltonics, Bremen, Germany). Full scan MS spectra were acquired at a spectral rate of 2.0 Hz, followed by the acquisition of one MS/MS spectrum. The MS/MS peak lists in Mascot generic format were searched on a local server using the Mascot 2.6.1 software (Matrix Science, London, UK) against the NCBI-nr protein database taxonomically restricted to Mammalia.
2.4. DNA extraction
Ancient DNA was extracted as described previously [23] in a laboratory specialized for aDNA analysis, thus limiting DNA contamination. All equipment and PCR cabinets were UV-irradiated before use. Pre- and post-PCR procedures were performed in separate rooms. A single extraction of every sample was carried out except for samples TKS-07 and KNT-25 that were extracted twice. Negative controls were used throughout the procedure (extraction control without any bone material and amplification control). The efficacy of sheep aDNA extraction was evaluated by real-time PCR targeting Ccr-2 (C–C chemokine receptor type 2) single nucleotide polymorphisms (SNPs) specific to Ovis aries (LOC101117706). Primer pairs (mm-U3 and mm-R2), TaqMan probes (mm Probe) were used to perform real-time PCR using CFX96 (Bio-Rad, CA, USA), as described elsewhere [14]. TaqMan Universal PCR Master Mix (Thermo, CA, USA) supplied at 2X concentration and containing Uracil-DNA Glycosylase was used as a master mix in the PCR mixture (20 μL). DNA damage expected for aDNA was not assessed in this study.
2.5. Assignment of mitochondrial DNA haplogroups
Five custom TaqMan probes (Life Technologies, USA) were used to target sheep mtDNA polymorphisms 15459, 15476, 15484, 15509, and 15512 (Table 1). The custom TaqMan probes were designed by Custom TaqMan Assay Design Tool (Thermo, CA, USA) and were described earlier [13]. Haplogroups were assigned based on differences in the specific sites of mtDNA [24] (Table 2). Fluorescence accumulation was analyzed for VIC and FAM channels. The analysis was carried out using CFX Manager 3.1 (Bio-Rad, CA, USA). Mitochondrial DNA haplogroups were assigned to 28 aDNA samples (Table 3).
Table 1.
Mitochondrial DNA control region primers and probes.
| mtDNA position (AF010406) | Sequence of primers, 5′-3′ | Sequence of probes, 5′-3′ |
|---|---|---|
| 15459 | F-ACACCCAAAGCTGAAGTTCTACTTAAA R- TTTTAGCAAGTTTAATACTGGAGAGGTTCTT |
VIC-CAACGATACTTATCAATATAT FAM-TCAACGATACTTATTAATATAT |
| 15476 | F-CTTAAACTATTCCCTGAATCATTATCAACGATACT R-TCCGTGTTGTATGTTTGGGAGTTTT |
VIC-AGGCTCTTTATATTTTTG FAM-AGGCTCTTTATGTTTTTG |
| 15484 | F-CTATTCCCTGAATCATTATCAACGATACTTATCA R-TCCGTGTTGTATGTTTGGGAGTTTT |
VIC-ACTGGAGAGGCTCTTTA FAM-ACTGGAGAGGTTCTTTA |
| 15509 | F-CCAAAAATATAAAGAGCCTCTCCAGTATTAAACT R-GAGTGGGAAGTCCGTGTTGT |
VIC-ATGTTTGGGAGTTTTAGCA FAM-ATGTTTGGGAGTCTTAGCA |
| 15512 | F-AATATATTTCCAAAAATATAAAGAGCCTCTCCAGT R-GGGTTGTTATGTGGGCTTGTG |
VIC-AAACTTGCTAAAACTTCCCAC FAM-AACTTGCTAAAACCTCCCAC |
Table 2.
Mitochondrial DNA control region positions.
| Haplogroup | mtDNA position (AF010406) |
||||
|---|---|---|---|---|---|
| 15459 | 15476 | 15484 | 15509 | 15512 | |
| A | T | T | A | A | T |
| B | C | T | G | A | T |
| C | C | T | G | G | T |
| D | C | T | G | A | C |
| E | C | C | G | G | T |
Table 3.
Ancient DNA sample codes and their corresponding archaeological description.
| Sample code | Archaeological site | Location | Bone type | Haplogroup |
|---|---|---|---|---|
| TKS-02 | Toksanbai | Western Kazakhstan | Tooth | B |
| TKS-04 | Toksanbai | Western Kazakhstan | Metacarpal | B |
| TKS-07 | Toksanbai | Western Kazakhstan | Tooth | C |
| TKS-08 | Toksanbai | Western Kazakhstan | Radius | B |
| TKS-09 | Toksanbai | Western Kazakhstan | Tooth | A |
| TKS-10 | Toksanbai | Western Kazakhstan | Metatarsal | A |
| TKS-17 TKS-20 TKS-23 |
Toksanbai Toksanbai Toksanbai |
Western Kazakhstan Western Kazakhstan Western Kazakhstan |
Tibia Talus Tooth |
B A A |
| TKS-27 TKS-31 |
Toksanbai Toksanbai |
Western Kazakhstan Western Kazakhstan |
Tooth Talus |
A A |
| TKS-34 | Toksanbai | Western Kazakhstan | Talus | A |
| TKS-39 | Toksanbai | Western Kazakhstan | Tooth | B |
| TKS-41 KNT-1 KNT-3 KNT-4 KNT-7 KNT-10 KNT-11 KNT-12 KNT-14 KNT-16 KNT-17 KNT-19 KNT-20 KNT-24 KNT-25 |
Toksanbai Kent Kent Kent Kent Kent Kent Kent Kent Kent Kent Kent Kent Kent Kent |
Western Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan Central Kazakhstan |
Fragment of talus Tooth Tooth Tooth Tooth Tooth Tooth Tooth Tooth Tooth Tooth Tooth Tooth Vertebra Radius |
A B B A A A A A A A B A B B C |
2.6. Primers, sequencing, and data analysis
A set of primers (L15391 5#-CCACTATCAACACCCAAAG-3# and H15534 5#-AAGTCCGTGTTGTATGTTTG-3#) was used to sequence the 144 bp mitochondrial control region of the ovine reference sequence (GenBank AF010406) [25]. Two independent amplifications were carried out for every sample. Uracil-DNA glycosylase (New England Biolabs, UK) was used to remove uracil residues from DNA. Polymerase chain reaction products were sequenced in both forward and reverse directions using a 3130xl DNA Analyzer (Applied Biosystems, Tokyo, Japan) and BigDye Terminator v.1.1 Cycle Sequencing Kit (Thermo, MA, USA). The sequences were aligned to the reference sequence (GenBank AF010406) using SeqScape software v. 2.6 (Applied Biosystems, CA, USA). The sequences were trimmed at sequencing starts to remove poor quality 5′ regions and submitted to GenBank with accession numbers MZ848101-MZ848128.
3. Results
3.1. Mass spectrometry analysis of sheep-specific peptides
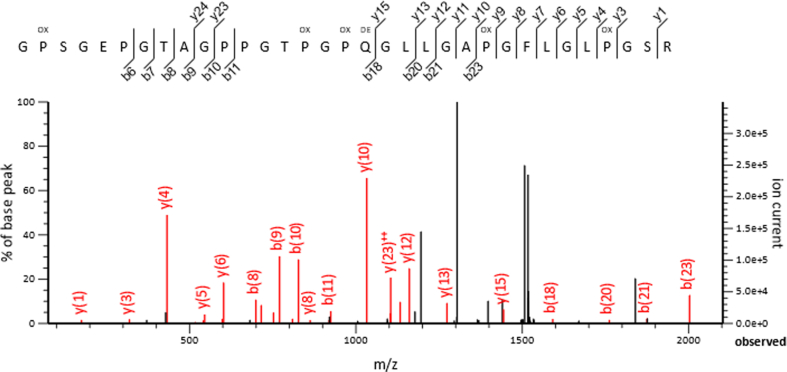
The method of zooarcheology by mass spectrometry (ZooMS) was used to discriminate sheep from goats [26]. MS/MS fragmentation spectra of tryptic digestion of collagen from our archeological samples were obtained after protein isolation. A typical tandem mass spectrum of a precursor ion with m/z 1517.7413 and charge +2 was attributed to the marker peptide GPSGEPGTAGPPGTPGPQGLLGAPGFLGLPGSR (Figure 2). The differentiation marker peptide belonged to the collagen alpha-2(I) chain protein XP_004007775 [Ovis aries] and was observed to contain post-translational modifications, including five hydroxylated P (Ox) residues and one deamidated Q (De) residue.
Figure 2.
Tandem mass spectrometry (MS/MS) presenting sheep-specific marker peptide. The MS/MS fragmentation spectrum of a precursor ion with m/z 1517.7413 and charge +2 was attributed to the differentiation marker peptide GPSGEPGTAGPPGTPGPQGLLGAPGFLGLPGSR presenting collagen alpha-2(I) chain protein (Ovis aries). This peptide has post-translational modifications, including five hydroxylated P (Ox) residues and one deamidated Q (De) residue characteristic for archeological specimens.
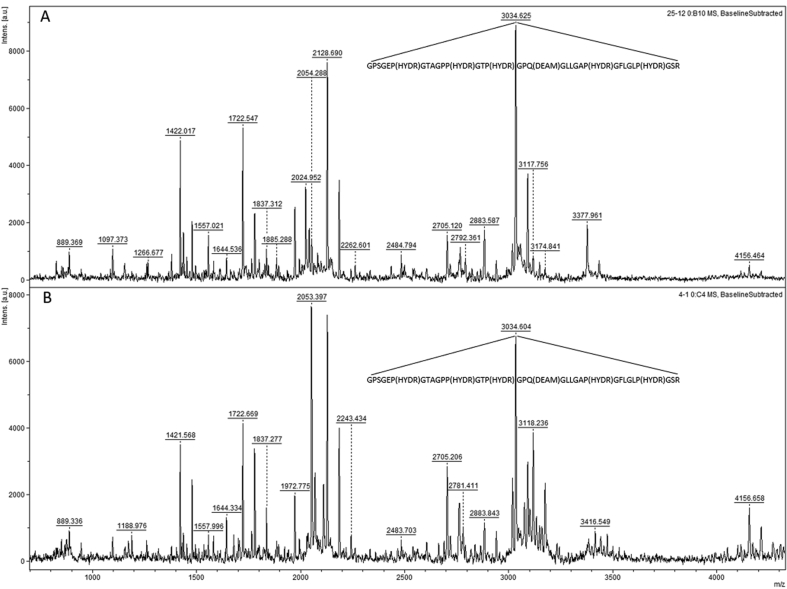
In addition to tandem LC-MS/MS, MALDI-MS mass spectra of tryptic digestion of collagen from the archeological samples were searched for the same marker peptide GPSGEP(Ox)GTAGPP(Ox)GTP(Ox)GPQ(De)GLLGAP(Ox)GFLGLP(Ox)GSR (m/z 3034.6, charge +1) specific to sheep (Figure 3). As a result, 66 ancient sheep specimens were identified by ZooMS and selected for aDNA analysis.
Figure 3.
Matrix-assisted laser desorption/ionization-time of flight (MALDI-ToF) mass spectra of tryptic digestion of collagen from archaeological samples KNT-25 (A) and TKS-4 (B) with the marker peptide GPSGEP(Ox)GTAGPP(Ox)GTP(Ox)GPQ(De)GLLGAP(Ox)GFLGLP(Ox)GSR (m/z, 3034.6 and charge +1 specific to sheep. TKS, Toksanbai settlement; KNT, Kent settlement.
3.2. Mitochondrial DNA analysis
The effectiveness of aDNA isolation was verified by amplification of the Ccr-2 gene [14]. Samples with the threshold cycle number (CT) values of less than 35 cycles were considered positive. A total of 28 PCR reactions gave a positive result at this threshold line, while 38 PCR reactions were negative. Out of 66 ancient sheep samples confirmed by the collagen-peptide marker, aDNA was successfully isolated from 28 samples (42% success rate). Mitochondrial DNA haplogroups were assigned using five custom TaqMan probes targeting sheep mtDNA control region positions 15459, 15476, 15484, 15509, and 15512 of the reference genome (accession number AF010406) [24]. Negative extraction controls were amplified to detect possible contamination. In addition, the negative amplification controls were used in PCR experiments. No contamination was observed with exception of preliminary experiments that were not included in this study. The genotyping results were validated by direct sequencing of the specified region of mtDNA (15391–15534) with at least two sequences per sample (Table 4). There was no difference in the sequences derived from the independent amplifications. Identical distribution of haplogroups A (8/14; 57%), B (5/14; 36%), and C (1/14; 7%) was demonstrated in the investigated ancient samples from Toksanbai and Kent, with the predominance of haplogroup A. Haplogroups D and E were not discovered in the studied specimens.
Table 4.
Mitochondrial haplotype sequences of control region fragment in relation to reference sequence AF010406.
| Sample | 15459 | 15467 | 15476 | 15484 | 15509 | 15512 |
|---|---|---|---|---|---|---|
| GenBank AF010406.1 | C | T | T | G | A | T |
| TKS-02 | . | . | . | . | . | . |
| TKS-04 | . | . | . | . | . | . |
| TKS-07 | . | . | . | . | G | . |
| TKS-08 | . | . | . | . | . | . |
| TKS-09 | T | . | . | A | . | . |
| TKS-10 | T | . | . | A | . | . |
| TKS-17 | . | . | . | . | . | . |
| TKS-20 | T | . | . | A | . | . |
| TKS-23 | T | . | . | A | . | . |
| TKS-27 | T | . | . | A | . | . |
| TKS-31 | T | . | . | A | . | . |
| TKS-34 | T | . | . | A | . | . |
| TKS-39 | . | . | . | . | . | . |
| TKS-41 | T | . | . | A | . | . |
| KNT-1 | . | . | . | . | . | . |
| KNT-3 | . | . | . | . | . | . |
| KNT-4 | T | . | . | A | . | . |
| KNT-7 | T | . | . | A | . | . |
| KNT-10 | T | . | . | A | . | . |
| KNT-11 | T | C | . | A | . | . |
| KNT-12 | T | C | . | A | . | . |
| KNT-14 | T | . | . | A | . | . |
| KNT-16 | T | . | . | A | . | . |
| KNT-17 | . | . | . | . | . | . |
| KNT-19 | T | C | . | A | . | . |
| KNT-20 | . | . | . | . | . | . |
| KNT-24 | . | . | . | . | . | . |
| KNT-25 | . | . | . | . | G | . |
4. Discussion
4.1. Identification of taxa
We used paleoproteomic methods to select sheep archeological biosamples. These methods are routinely used to identify fragmented skeletal elements or specimens with degraded aDNA [26]. The use of morphological features for the identification of taxa may be misleading in the case of sheep/goat remains, when many bones of sheep and goats are difficult to distinguish because of their similar characteristics [27]. This is particularly problematic in the case of young sheep/goat individuals, when morphological markers are not fully developed yet [26]. In the present study, ancient proteins were digested into peptides, separated, and analyzed by mass spectrometry for taxonomic identification. Marker peptides with known m/z values were used to differentiate closely related species (the ZooMS approach). ZooMS was used to verify sheep identification prior to aDNA analysis by examining the differentiation marker peptide GPSGEPGTAGPPGTPGPQGLLGAPGFLGLPGSR by LC-MS/MS and/or MALDI-ToF mass spectrometry. A total of 66 ancient sheep samples were confirmed by the collagen-peptide marker and subjected to aDNA analysis.
In addition to taxonomic identification, MS/MS fragmentation data were also used to detect post-translational modifications such as deamidation of glutamine (Q), which is a characteristic modification of archeological specimens [28], as well as oxidation of proline (P) residues (Figure 2).
4.2. Ancient sheep lineage
Mitochondrial DNA was successfully analyzed in ancient sheep remains from two Late Bronze Age settlements located in the central and western parts of Kazakhstan. Mitochondrial haplogroups A, B, and C were detected in ancient domestic sheep. The distribution of haplogroups was identical in the studied settlements, with the predominance of ovine haplogroup A (57%), followed by haplogroup B (36%) and haplogroup C (7%) in both areas.
Similar patterns of haplogroup distribution in the two investigated settlements, which are located 1,500 km away from each other, may indicate early sheep introduction into the region. Both localities were inhabited by cattle breeders and hunters [29]. The Toksanbai settlement belongs to the Northeast Pre-Caspian region, and Kent is situated in the Kent mountains, 250 km southeast of Karaganda City in the central part of Kazakhstan [30]. Kent is known to be the biggest settlement from the Bronze Age in Kazakhstan, whereas Toksanbai is a relatively compact settlement. Animals adapted to a nomadic lifestyle were prevalent at both sites. The high proportion of sheep and horses in these two settlements brings the herd composition closer to the species structure of modern nomadic herds in Eurasian steppes. The transition to mobile nomadic cattle breeding may have started in Eurasian steppes, not at the beginning of the Early Iron Age, but in the Late Bronze Age.
4.3. Ancient sheep and modern Kazakh sheep breeds
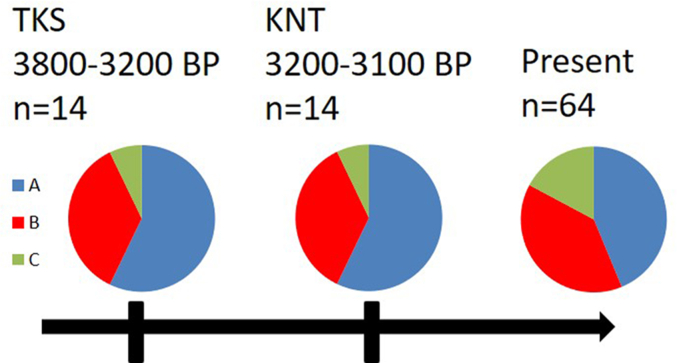
In our previous study, a sample of 64 unrelated animals from six local sheep breeds was studied [13]. A similar distribution of haplogroups in modern local sheep breeds was reported. Genotyping indicated that 28 tested animals belonged to haplogroup A (44%), 25 – to haplogroup B (39%), and 11 – to haplogroup C (17%). Haplogroups A and B were the most frequent and were found in all studied sheep breeds. Haplogroup A is currently prevalent in Asian countries, indicating that most of the Kazakh sheep breeds are genetically closer to the Asian genotype than to European one. In contrast, haplogroup B occurs at a high frequency in European breeds and countries of the Eastern Mediterranean. Haplogroup C has been simultaneously identified by two research groups, among a sample of domestic breeds of sheep in China and Turkey [31, 32]. Interestingly, the frequency of haplogroup C is higher in the local breeds than in the studied ancient ones (Figure 4). In the modern breeds haplogroup C was found in the South Kazakh merino sheep (40%), Kazakh mutton-semifine-wool sheep (20%), and Kalchengel type of Kazakh mutton-semifine-wool sheep (29%), and virtually absent in the other breeds. In general, haplogroup C was reported mainly in Central Asia, India, China, and the Caucasus.
Figure 4.
Mitochondrial haplogroup distribution in Late Bronze Age archaeological sites Toksanbai (TKS) and Kent (KNT), along with present data on the modern breeds from Kazakhstan (present data from Mukhametzharova et al.).
Dymova et al. carried out archeological mitochondrial DNA D-loop fragment analysis based on about 4,000–1,000 years old sheep bone remains in Altai (Russia) on the border with Kazakhstan [14]. There were all five haplogroups (A, B, C, D, and E lineages) present. This diversity led to the conclusion that the Altai region might be a migratory area for many sheep and peoples in the past. It is not certain if these sheep populations migrated to Kazakhstan since Bronze Age settlements from north-central and north-eastern Kazakhstan were exclusively rich in animal remains of horses [17].
The first possible source of domestic sheep introduction is the pre-Caspian Caucasus region, which is considered to be among the primary localities for sheep breeding after the Fertile Crescent [3]. An early introduction of sheep from the Pontic-Caspian steppe to western Kazakhstan by Afanasevo culture is likely [33]. The second and more recent source of introduction may be attributed to the route originating from eastern Central Asia, as recently described by Schroeder et al. [3, 34]. Using endogenous retroviruses as genetic markers, Chessa et al. discovered secondary population expansion of domestic sheep out of Southwest Asia, inhabited by pastoralist societies whose economy included sheep husbandry [5]. Both sources may have contributed to the genetic diversity of ancient sheep during the Late Bronze Age in Kazakhstan. A recent study of faunal remains of domestic sheep along the margins of Central Asia in the Ferghana Basin (Obishir V) and southern Turkmenistan (Djeitun) established the presence of domesticated sheep by ca. 6,000–6,500 BCE [27]. Thus, the limitation of this study is the absence of genomic analysis of earlier local lineages that is needed to gain a better understanding of sheep migrations in the Eurasian steppe. Another limitation is the absence of sequence-based comparisons to other ancient and/or modern sheep populations due to a very short fragment of ovine mitochondrial genome sequenced. These limitations highlight promising areas for future studies.
5. Conclusion
We investigated the genetic diversity of Late Bronze Age lineages of ancient sheep at two archeological sites in Kazakhstan. The diversity of mitochondrial DNA haplogroup distribution in ancient domestic sheep in two geographically distant settlements may indicate earlier sheep introduction to the region. The fact that three basal groups were detected in two distant settlements gives us a hint that the different genetic lineages of sheep were introduced to the Eurasian steppe long before the Late Bronze Age. Multiple introductory events may have been facilitated by suitable natural and climatic conditions and the availability of vast natural pastures in Kazakhstan. The predominance of mitochondrial haplogroups A and B in modern Kazakh sheep breeds resembles the distribution of haplogroups in ancient domestic sheep. The frequency of haplogroup C is higher in the local breeds than in the studied ancient ones. Further studies of ancient sheep at earlier time points and studies with additional genetic markers are needed to provide a closer look at early migratory routes and the evolutionary history of domestic sheep in Kazakhstan.
Declarations
Author contribution statement
Pavel Tarlykov: Conceived and designed the experiments; Wrote the paper.
Sabina Atavliyeva: Performed the experiments; Analyzed and interpreted the data.
Dana Auganova: Performed the experiments.
Ilyas Akhmetollayev, Tatyana Loshakova, Victor Varfolomeev, Yerlan Ramankulov: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant No. AP08955854).
Data availability statement
Data associated with this study has been deposited at GenBank under the accession numbers MZ848101-MZ848128.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Dr. Pavel Kosintsev from the Institute of Plant and Animal Ecology (Yekaterinburg, Russia) for the morphology-based species identification. We are grateful to Dr. Maksim Filipenko and Dr. Maya Dymova from the Institute of Chemical Biology and Fundamental Medicine (Novosibirsk, Russia) for methodological support.
References
- 1.Zeder M.A. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lv F.H., Peng W.F., Yang J. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 2015;32:2515–2533. doi: 10.1093/molbev/msv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder O., Benecke N., Frolich K. Endogenous retroviral insertions indicate a secondary introduction of domestic sheep lineages to the Caucasus and central Asia between the Bronze and Iron Age. Genes. 2017;8 doi: 10.3390/genes8060165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapio M., Marzanov N., Ozerov M. Sheep mitochondrial DNA variation in European, caucasian, and central Asian areas. Mol. Biol. Evol. 2006;23:1776–1783. doi: 10.1093/molbev/msl043. [DOI] [PubMed] [Google Scholar]
- 5.Chessa B., Pereira F., Arnaud F. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324:532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meadows J.R., Hanotte O., Drogemuller C. Globally dispersed Y chromosomal haplotypes in wild and domestic sheep. Anim. Genet. 2006;37:444–453. doi: 10.1111/j.1365-2052.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- 7.Wallner B., Vogl C., Shukla P. Identification of genetic variation on the horse Y chromosome and the tracing of male founder lineages in modern breeds. PloS One. 2013;8 doi: 10.1371/journal.pone.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez I., Capote J., Traore A. Mitochondrial analysis sheds light on the origin of hair sheep. Anim. Genet. 2013;44:344–347. doi: 10.1111/j.1365-2052.2012.02398.x. [DOI] [PubMed] [Google Scholar]
- 9.Hiendleder S., Kaupe B., Wassmuth R., Janke A. Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proc. Biol. Sci. 2002;269:893–904. doi: 10.1098/rspb.2002.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiendleder S., Mainz K., Plante Y., Lewalski H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: no evidence for contributions from urial and argali sheep. J. Hered. 1998;89:113–120. doi: 10.1093/jhered/89.2.113. [DOI] [PubMed] [Google Scholar]
- 11.Wood N.J., Phua S.H. Variation in the control region sequence of the sheep mitochondrial genome. Anim. Genet. 1996;27:25–33. doi: 10.1111/j.1365-2052.1996.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 12.Meadows J.R., Cemal I., Karaca O., Gootwine E., Kijas J.W. Five ovine mitochondrial lineages identified from sheep breeds of the near East. Genetics. 2007;175:1371–1379. doi: 10.1534/genetics.106.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukhametzharova I., Islamov Y., Shauyenov S. Genetic characterization of Kazakh native sheep breeds using mitochondrial DNA. Online J. Biol. Sci. 2018;18:341–348. [Google Scholar]
- 14.Dymova M.A., Zadorozhny A.V., Mishukova O.V. Mitochondrial DNA analysis of ancient sheep from Altai. Anim. Genet. 2017;48:615–618. doi: 10.1111/age.12569. [DOI] [PubMed] [Google Scholar]
- 15.Librado P., Gamba C., Gaunitz C. Ancient genomic changes associated with domestication of the horse. Science. 2017;356:442–445. doi: 10.1126/science.aam5298. [DOI] [PubMed] [Google Scholar]
- 16.Outram A.K., Stear N.A., Bendrey R. The earliest horse harnessing and milking. Science. 2009;323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 17.Gaunitz C., Fages A., Hanghøj K. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science. 2018;360:111. doi: 10.1126/science.aao3297. [DOI] [PubMed] [Google Scholar]
- 18.Miller V., Haruda A., Varfolomeev V., Goryachev A., Makarewicz C. Close management of sheep in ancient Central Asia: evidence for foddering, transhumance, and extended lambing seasons during the Bronze and Iron Ages. Sci. Technol. Archaeol. Res. 2020;6:41–60. [Google Scholar]
- 19.Yevdokimov V.V., Varfolomeev V.V. Karagandinskij Gosudarstvennyj Universitet; 2002. Ehpoxa Bronzy Central'nogo I Cevernogo Kazaxstana. [Google Scholar]
- 20.Outram A.K., Kasparov A.K. Vol. 1. 2007. Historical and Cultural Heritage of Saryarka; pp. 107–123. [Google Scholar]
- 21.Nekrasov A.E., Kosintsev P.A., Samashev Z. New data on avifauna of the Ustyurt plateau in the Holocene. Dokl. Biol. Sci. 2016;469:170–172. doi: 10.1134/S0012496616040025. [DOI] [PubMed] [Google Scholar]
- 22.Cappellini E., Jensen L.J., Szklarczyk D. Proteomic analysis of a pleistocene mammoth femur reveals more than one hundred ancient bone proteins. J. Proteome Res. 2012;11:917–926. doi: 10.1021/pr200721u. [DOI] [PubMed] [Google Scholar]
- 23.Rohland N., Hofreiter M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 24.Demirci S., Koban Bastanlar E., Dagtas N.D. Mitochondrial DNA diversity of modern, ancient and wild sheep(Ovis gmelinii anatolica) from Turkey: new insights on the evolutionary history of sheep. PloS One. 2013;8 doi: 10.1371/journal.pone.0081952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai D., Han L., Zhang X., Zhou H., Zhu H. DNA analysis of archaeological sheep remains from China. J. Archaeol. Sci. 2007;34:1347–1355. [Google Scholar]
- 26.Buckley M., Whitcher Kansa S., Howard S. Distinguishing between archaeological sheep and goat bones using a single collagen peptide. J. Archaeol. Sci. 2010;37:13–20. [Google Scholar]
- 27.Taylor W.T.T., Pruvost M., Posth C. Evidence for early dispersal of domestic sheep into Central Asia. Nature Human Behav. 2021 doi: 10.1038/s41562-021-01083-y. [DOI] [PubMed] [Google Scholar]
- 28.Smith C.I., Chamberlain A.T., Riley M.S., Stringer C., Collins M.J. The thermal history of human fossils and the likelihood of successful DNA amplification. J. Hum. Evol. 2003;45:203–217. doi: 10.1016/s0047-2484(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 29.Samashev Z.S., Ermolaeva A.S., Teplovodskaya T.M. [Settlement toksanbay in Ustyurt] article in Russian. Vestn. Akad. Guman. Nauk Resp. Kazakhstan. 1999;1:49–70. [Google Scholar]
- 30.Lightfoot E., Motuzaite-Matuzeviciute G., O'Connell T.C. How ‘pastoral’ is pastoralism? Dietary diversity in Bronze Age communities in the Central Kazakhstan steppes. Archaeometry. 2015;57:232–249. [Google Scholar]
- 31.Guo J., Du L.X., Ma Y.H. A novel maternal lineage revealed in sheep (Ovis aries) Anim. Genet. 2005;36:331–336. doi: 10.1111/j.1365-2052.2005.01310.x. [DOI] [PubMed] [Google Scholar]
- 32.Pedrosa S., Uzun M., Arranz J.J. Evidence of three maternal lineages in Near Eastern sheep supporting multiple domestication events. Proc. Biol. Sci. 2005;272:2211–2217. doi: 10.1098/rspb.2005.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anthony D.W. Princeton University Press; 2007. The horse, the wheel, and language. How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. [Google Scholar]
- 34.Frachetti M.D. Multiregional emergence of mobile pastoralism and nonuniform institutional complexity across Eurasia. Curr. Anthropol. 2012;53:2–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at GenBank under the accession numbers MZ848101-MZ848128.