Abstract
The genotypes of 107 strains of Cryptococcus isolated from the environment or from patients from various geographical areas were determined by multilocus enzyme electrophoresis (MLEE). We analyzed the relationships between genotype structure and serotype and between genotype structure and strain origin. Twelve of the 14 enzyme-encoding loci studied were polymorphic, giving rise to 48 electrophoretic types. The genotypes of C. neoformans and C. laurentii were very similar. MLEE could not distinguish between these two pathogenic species. A correlation between the genetic multilocus structure and the origin of the sample (from the environment or patients) existed. A second analysis detected a correlation between genotype distribution and serotype. The second analysis considered three serotype groups (B, C, and A plus D plus A/D), proving that serotypes A, D, and A/D are closely related. MLEE is a useful epidemiological tool for improving our understanding of the biology of this fungus.
Cryptococcus neoformans is a yeast-like fungus. This species has two varieties (14): C. neoformans var. neoformans, which is ubiquitous and which groups together serotypes A and D and the very controversial serotype A/D (2, 4, 28), and C. neoformans var. gattii (serotypes B and C), which is found mainly in tropical and subtropical regions.
This opportunistic pathogen causes cryptococcosis, a deep mycosis with a specific tropism for the lungs and brain. It affects immunocompromised individuals and patients suffering from severe hematological malignancies (10). Its incidence has increased over the last 10 years because of the increase in the number of patients with AIDS and those undergoing transplantation (10).
Epidemiological studies of cryptococcosis are essential if we are to understand the biology of this fungus so as to obtain improved treatments for this infection (9, 22). Serotyping was used as an epidemiological tool for many years (1, 9). The development of DNA typing methods increases the power of discrimination of strains. Random amplified polymorphic DNA analysis (5, 12, 22), karyotyping (11, 17), and fingerprinting (12, 22) are routinely used to demonstrate intraspecific diversity in the genus Cryptococcus. Multilocus enzyme electrophoresis (MLEE), first used for the discrimination of Candida strains (18, 19), has provided good results for Aspergillus (20, 21) and Cryptococcus (5, 6). It is a very effective typing method (15, 21) and provides information about both the phenotype and the genotype (5, 6, 18, 19, 20).
We typed 107 strains of Cryptococcus of very different origins (patients and environment) and from different geographical areas by MLEE (5). Cryptococcus laurentii is slightly pathogenic for immunocompromised individuals, and some strains were included in this study to explore the genetic diversity of the genus Cryptococcus.
This study was performed to obtain taxonomic information about the two species C. neoformans and C. laurentii and to identify any relationships between genotype information and the serotype or origin of the strains. This should improve our understanding of the biology of this fungus.
MATERIALS AND METHODS
The 107 strains tested in the present study were kindly donated by the Institute of Tropical Medicine, Antwerp, Belgium. We used 48 strains that originated from the environment (e.g., soil, fruits, and eucalyptus), 57 strains that originated from patients, and 2 reference strains (Table 1). The isolates were grown on Sabouraud agar (Biomérieux, Marcy l’Etoile, France) at 33°C.
TABLE 1.
Origins and ETs of Cryptococcus strains
| Strain, serotype, and strain no. | Origina | ET | Strain, serotype, and strain no. | Origina | ET | |
|---|---|---|---|---|---|---|
| C. neoformans | ||||||
| Serotype A | ||||||
| 4091 | Europe, patient | ET 1 | ||||
| 4092 | Africa, patient | ET 2 | ||||
| 4093 | Europe, patient | ET 2 | ||||
| 4094 | Africa, patient | ET 2 | ||||
| 4095 | Europe, environment | ET 2 | ||||
| 4096 | Africa, environment | ET 2 | ||||
| 4097 | Africa, environment | ET 2 | ||||
| 4098 | Africa, patient | ET 2 | ||||
| 4099 | Africa, patient | ET 3 | ||||
| 4100 | Europe, environment | ET 2 | ||||
| 4101 | Europe, environment | ET 2 | ||||
| 4102 | Europe, environment | ET 3 | ||||
| 4103 | Europe, environment | ET 2 | ||||
| 4104 | Africa, patient | ET 32 | ||||
| 4124 | Europe, patient | ET 2 | ||||
| 4125 | Europe, patient | ET 35 | ||||
| 4127 | Europe, environment | ET 10 | ||||
| 4128 | Africa, environment | ET 10 | ||||
| 4129 | Africa, environment | ET 36 | ||||
| 4141 | Africa, patient | ET 13 | ||||
| 4142 | Europe, environment | ET 40 | ||||
| 4143 | South America, patient | ET 41 | ||||
| 4186 | South America, environment | ET 14 | ||||
| 4187 | Europe, environment | ET 15 | ||||
| 4188 | Europe, environment | ET 13 | ||||
| 4189 | Europe, environment | ET 14 | ||||
| 4190 | Europe, environment | ET 13 | ||||
| 4191 | Europe, environment | ET 16 | ||||
| 4192 | Europe, environment | ET 15 | ||||
| 4193 | Europe, environment | ET 16 | ||||
| 4194 | Europe, environment | ET 17 | ||||
| 4195 | Europe, environment | ET 18 | ||||
| 4196 | Europe, environment | ET 18 | ||||
| 4197 | Europe, environment | ET 14 | ||||
| 4198 | Africa, patient | ET 18 | ||||
| 4199 | Africa, environment | ET 18 | ||||
| 4200 | Africa, environment | ET 14 | ||||
| 4201 | Africa, environment | ET 18 | ||||
| 4202 | Africa, environment | ET 18 | ||||
| 4203 | Africa, environment | ET 14 | ||||
| 4204 | Africa, environment | ET 18 | ||||
| 4205 | Africa, environment | ET 18 | ||||
| 4206 | Africa, environment | ET 14 | ||||
| 4207 | Europe, environment | ET 19 | ||||
| 4208 | Europe, environment | ET 14 | ||||
| 4209 | Europe, environment | ET 19 | ||||
| 4210 | Europe, environment | ET 14 | ||||
| 4211 | Europe, environment | ET 20 | ||||
| 4212 | Europe, environment | ET 20 | ||||
| 4213 | Europe, environment | ET 20 | ||||
| Serotype B | ||||||
| 4105 | Africa, patient | ET 4 | ||||
| 4106 | Asia, patient | ET 5 | ||||
| 4107 | Europe, patient | ET 33 | ||||
| 4108 | Africa, patient | ET 6 | ||||
| 4109 | Africa, patient | ET 42 | ||||
| 4112 | South America, patient | ET 8 | ||||
| 4137 | Africa, patient | ET 12 | ||||
| 4138 | United States, patient | ET 38 | ||||
| 4139 | Asia, patient | ET 39 | ||||
| 4140 | South America, patient | ET 48 | ||||
| 4214 | Asia, patient | ET 21 | ||||
| 4215 | Asia, patient | ET 20 | ||||
| 4216 | Asia, patient | ET 20 | ||||
| 4217 | Asia, patient | ET 21 | ||||
| 4218 | Asia, patient | ET 21 | ||||
| 4220 | South America, patient | ET 22 | ||||
| 4221 | South America, patient | ET 22 | ||||
| 4222 | Australia, environment | ET 12 | ||||
| 4223 | Australia, environment | ET 23 | ||||
| 4224 | United States, patient | ET 24 | ||||
| Serotype C | ||||||
| 4110 | Africa, patient | ET 43 | ||||
| 4119 | United States, patient | ET 34 | ||||
| 4120 | Africa, patient | ET 44 | ||||
| 4121 | Africa, patient | ET 28 | ||||
| 4225 | Africa, patient | ET 23 | ||||
| 4226 | Africa, patient | ET 25 | ||||
| 4227 | Africa, patient | ET 26 | ||||
| 4228 | Africa, patient | ET 25 | ||||
| 4229 | Africa, patient | ET 25 | ||||
| 4230 | Africa, patient | ET 25 | ||||
| 4231 | United States, patient | ET 27 | ||||
| 4232 | United States, patient | ET 27 | ||||
| 4233 | United States, patient | ET 20 | ||||
| Serotype D | ||||||
| 4113 | United States, patient | ET 28 | ||||
| 4114 | Europe, patient | ET 9 | ||||
| 4115 | Europe, patient | ET 31 | ||||
| 4234 | Europe, patient | ET 28 | ||||
| 4235 | Europe, patient | ET 27 | ||||
| 4236 | Europe, patient | ET 28 | ||||
| 4237 | Europe, patient | ET 20 | ||||
| 4238 | Europe, patient | ET 27 | ||||
| 4239 | Europe, patient | ET 20 | ||||
| 4240 | Europe, environment | ET 29 | ||||
| 4241 | Europe, environment | ET 20 | ||||
| 4242 | Europe, environment | ET 27 | ||||
| 4243 | Europe, environment | ET 23 | ||||
| 4244 | Europe, environment | ET 30 | ||||
| 4245 | United States, reference strain | ET 31 | ||||
| 4246 | United States, reference strain | ET 28 | ||||
| Serotype A/D | ||||||
| 4116 | Europe, patient | ET 34 | ||||
| 4117 | Africa, patient | ET 2 | ||||
| 4118 | Africa, patient | ET 32 | ||||
| C. laurentii | ||||||
| 4111 | South America, patient | ET 7 | ||||
| 4130 | Europe, environment | ET 45 | ||||
| 4131 | Europe, environment | ET 10 | ||||
| 4132 | Europe, environment | ET 37 | ||||
| 4133 | Africa, patient | ET 10 | ||||
| 4134 | Europe, environment | ET 46 | ||||
| 4135 | Europe, environment | ET 11 | ||||
| 4136 | South America, patient | ET 47 | ||||
| 4137 | South America, patient | ET 12 | ||||
| 4138 | South America, environment | ET 38 |
Origin indicates the geographical origin of the strain and the origin of the sample.
Preparation of culture lysates.
The isolates were grown in 500-ml flasks containing 180 ml of Sabouraud agar medium (pH 8) at 33°C for 72 h. Yeasts were collected from the surface with glass beads (diameter, 8 mm) in 12 ml of sterile deionized water.
The resulting suspension was centrifuged at 4,000 × g for 30 min, and the yeasts were suspended in 8 ml of distilled water. The cells were mechanically disrupted in the cold by the glass bead method (bead diameter, 0.25 mm) for 2 min in a Science tech MSK homogenizer (B. Braun, San Francisco, Calif.). The cellular debris was removed by centrifugation at 15,000 × g for 15 min at 0°C. The supernatant of each lysate was aliquoted and was stored at −20°C before use in MLEE.
Enzyme electrophoresis.
Starch gel electrophoresis and specific enzyme staining were performed as described previously (16, 18, 19, 20). In this study, the following 14 enzyme systems were tested: peptidase A (PEP A; EC 3.4.11; substrate, Val-Leu), peptidase B (PEP B; EC 3.4.11; substrate, Leu-Gly-Gly), peptidase C (PEP C; EC 3.4.11; substrate, Leu-Ala), glucose phosphate isomerase (GPI; EC 5.3.1.9), malate dehydrogenase (MDH; EC 1.1.1.37), glucose-6-phosphate dehydrogenase (G6PD; EC 1.1.1.27), phosphoglucomutase (PGM; EC 2.7.5.1), alcohol dehydrogenase (ADH; EC 1.1.1.1), fructokinase (FK; EC 2.7.1.4), fumarase (FUM, EC 4.2.1.2), isocitrate dehydrogenase (IDH; EC 1.1.1.42), 6-phosphogluconate dehydrogenase (PGD; EC 1.1.1.43), pyruvate kinase (PK; EC 2.7.1.40), and sorbitol dehydrogenase (SDH; EC 1.1.1.14).
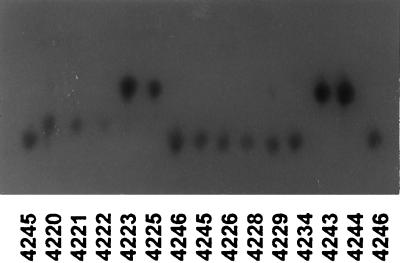
Twelve loci were polymorphic in the 107 strains examined, and each polymorphic locus had three to six alleles (see Fig. 1). We identified 58 alleles that gave 48 different electrophoretic types (ETs) (see Table 2). Alleles were numbered in decreasing order of mobility. The various polymorphic enzyme-encoding loci were characterized for each isolate. Distinct multilocus variants were designated ETs.
FIG. 1.
Polymorphism observed in C. neoformans with GPI as the enzymatic system.
TABLE 2.
Allelic composition of each ET
| ET | No. of alleles at the following locusa:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FUM | G6PD | SDH | ADH | PEP A | PEP B | PEP D | IDH | PGD | PGM | GPI | FK | |
| ET 1 | 2 | 5 | 3 | 2 | 3 | 1 | 3 | 5 | 1 | 4 | 4 | 2 |
| ET 2 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 1 | 4 | 4 | 2 |
| ET 3 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 3 | 3 | 4 | 2 |
| ET 4 | 2 | 5 | 3 | 2 | 5 | 3 | 5 | 4 | 2 | 3 | 5 | 2 |
| ET 5 | 2 | 5 | 3 | 2 | 5 | 3 | 5 | 4 | 2 | 3 | 4 | 2 |
| ET 6 | 2 | 5 | 3 | 2 | 5 | 3 | 5 | 4 | 2 | 4 | 5 | 2 |
| ET 7 | 3 | 2 | 1 | 5 | 4 | 2 | 4 | 4 | 4 | 5 | 2 | 2 |
| ET 8 | 2 | 3 | 3 | 2 | 5 | 3 | 5 | 4 | 2 | 3 | 5 | 2 |
| ET 9 | 2 | 5 | 3 | 2 | 3 | 1 | 3 | 5 | 2 | 4 | 5 | 2 |
| ET 10 | 2 | 4 | 3 | 2 | 2 | 1 | 2 | 5 | 1 | 4 | 4 | 2 |
| ET 11 | 2 | 1 | 5 | 1 | 1 | 2 | 1 | 5 | 2 | 3 | 3 | 2 |
| ET 12 | 2 | 5 | 3 | 2 | 4 | 3 | 4 | 5 | 2 | 3 | 4 | 2 |
| ET 13 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 2 | 3 | 4 | 2 |
| ET 14 | 1 | 3 | 2 | 4 | 2 | 3 | 2 | 1 | 3 | 2 | 3 | 1 |
| ET 15 | 2 | 5 | 3 | 2 | 1 | 1 | 1 | 5 | 2 | 4 | 3 | 2 |
| ET 16 | 1 | 3 | 2 | 4 | 3 | 3 | 2 | 1 | 3 | 2 | 3 | 1 |
| ET 17 | 2 | 5 | 3 | 2 | 3 | 1 | 3 | 5 | 3 | 3 | 4 | 2 |
| ET 18 | 3 | 4 | 6 | 6 | 1 | 1 | 1 | 6 | 2 | 4 | 1 | 1 |
| ET 19 | 1 | 3 | 2 | 4 | 2 | 3 | 2 | 1 | 3 | 2 | 3 | 2 |
| ET 20 | 1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 1 |
| ET 21 | 1 | 2 | 3 | 4 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 |
| ET 22 | 2 | 3 | 3 | 2 | 4 | 2 | 4 | 3 | 2 | 3 | 4 | 2 |
| ET 23 | 1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 1 |
| ET 24 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 2 | 2 |
| ET 25 | 3 | 3 | 4 | 3 | 3 | 2 | 3 | 3 | 3 | 4 | 5 | 3 |
| ET 26 | 3 | 3 | 4 | 3 | 2 | 2 | 2 | 3 | 3 | 4 | 5 | 3 |
| ET 27 | 1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 |
| ET 28 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 3 | 3 | 5 | 2 |
| ET 29 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 1 | 3 | 5 | 2 |
| ET 30 | 1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 |
| ET 31 | 2 | 5 | 3 | 2 | 3 | 1 | 3 | 5 | 3 | 3 | 5 | 2 |
| ET 32 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 1 | 3 | 4 | 2 |
| ET 33 | 3 | 5 | 4 | 2 | 2 | 2 | 2 | 5 | 1 | 4 | 4 | 3 |
| ET 34 | 2 | 5 | 3 | 2 | 2 | 1 | 2 | 5 | 3 | 3 | 4 | 2 |
| ET 35 | 2 | 3 | 3 | 2 | 2 | 1 | 2 | 5 | 1 | 4 | 4 | 2 |
| ET 36 | 2 | 3 | 3 | 2 | 2 | 1 | 2 | 3 | 1 | 4 | 4 | 2 |
| ET 37 | 3 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 2 |
| ET 38 | 3 | 2 | 4 | 2 | 3 | 3 | 3 | 5 | 3 | 4 | 4 | 3 |
| ET 39 | 2 | 2 | 4 | 2 | 5 | 3 | 5 | 3 | 2 | 3 | 4 | 2 |
| ET 40 | 3 | 2 | 3 | 2 | 4 | 2 | 4 | 3 | 3 | 3 | 3 | 2 |
| ET 41 | 3 | 5 | 4 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 2 |
| ET 42 | 2 | 5 | 3 | 2 | 2 | 2 | 2 | 5 | 1 | 3 | 4 | 2 |
| ET 43 | 4 | 5 | 4 | 3 | 2 | 2 | 2 | 5 | 1 | 6 | 5 | 3 |
| ET 44 | 3 | 3 | 4 | 3 | 3 | 2 | 3 | 3 | 1 | 6 | 5 | 3 |
| ET 45 | 2 | 3 | 3 | 2 | 2 | 3 | 2 | 3 | 1 | 4 | 3 | 2 |
| ET 46 | 2 | 2 | 3 | 2 | 2 | 3 | 2 | 3 | 1 | 4 | 3 | 2 |
| ET 47 | 3 | 2 | 4 | 2 | 3 | 2 | 3 | 3 | 1 | 3 | 3 | 2 |
| ET 48 | 2 | 2 | 4 | 2 | 2 | 3 | —b | 3 | — | 3 | 4 | 2 |
For definitions of the abbreviations, see the text. Alleles are encoded in decreasing order of their anodal mobility.
—, no enzyme activity was detected for this locus.
Statistical analysis. (i) Factorial correspondence analysis.
We analyzed the results by factorial correspondence analysis (FCA) using the PRAXIS-p.c., version 2.0, software package (Praxème, R&D, Biométrie, Centre National de la Recherche Scientifique, Montpellier, France).
FCA involves the construction of a contingency table (samples versus alleles), in which each isolate is represented in terms of its allelic composition. This method of analysis simultaneously characterizes the ET according to all the genetic variables (alleles) and determines the contribution of each allele to the overall variability of the sample. The calculation of variance is based on the χ2 test, which measures the extent to which a sample distribution deviates from a theoretical distribution and is an integral part of the correspondence analysis (3, 7). FCA was presented as a plane projection of the two most informative axes accounting for the genetic structure of the ETs.
(ii) Canonical correspondence analysis.
We used canonical correlation analysis (CCA) to compare and correlate genetic information and the origin or serotype of strains of various ETs using the PRAXIS-p.c., version 2.0, software package. This CCA was performed with two contingency tables: one (X) in which each ET was represented in terms of its allelic composition and a second one (Y) in which the strains were organized into various groups (sample origin and serotype) (23–25). The independence of X and Y was testing by the approximation-to-permutation test.
RESULTS
MLEE gave clearly reproducible results. Identical results were obtained when isolates were subcultured many times on Sabouraud’s medium and when electrophoresis was performed with samples from every fourth isolate passage.
Genetic diversity and taxonomy.
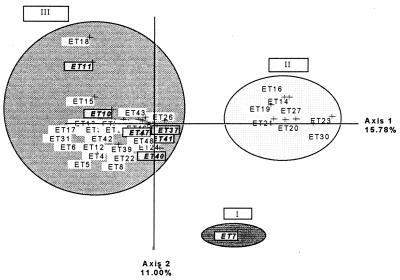
Twelve loci were polymorphic in the 107 strains examined, and each polymorphic locus had three to six alleles (Fig. 1). We identified 58 alleles giving 48 different ETs (Table 2). In the first FCA, strains formed three clusters (Fig. 2) in the most informative plane (accounting for 26.78% of the total genetic variability). Cluster I consisted of one strain of C. laurentii, and the GPI 1 allele was responsible for this structure (Table 2). Cluster II consisted of C. neoformans strains only, and cluster III contained both C. neoformans and C. laurentii strains. This projection shows the extensive diversity of the C. neoformans species. The C. neoformans strains of cluster II are separated from those of cluster III by axis 1, which is the most informative (15.78% of the total variability). This shows that the strains from the various clusters have very different genotypes. The inclusion of C. laurentii strains in a C. neoformans cluster suggests that these species are genetically related and that there is greater genotype diversity in the C. neoformans sample than in the C. laurentii sample. The alleles responsible for this structure are indicated in Table 2 and are confirmed in Table 3 because the least frequent alleles are the most discriminatory.
FIG. 2.
First plane projection of FCA in two informative axes of all ETs observed. Axes 1 and 2 represent 15.78 and 11.00% of the overall variability, respectively. ETs represent the observed ET (Table 2). The ET group at the upper left (cluster III) includes C. laurentii strains.
TABLE 3.
Allele frequencies for each locus studied
| Locus | Frequency of the following allelea:
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| G6PD | 0.021 | 0.292 | 0.229 | 0.042 | 0.417 | |
| SDH | 0.021 | 0.146 | 0.583 | 0.208 | 0.021 | 0.021 |
| ADH | 0.021 | 0.688 | 0.083 | 0.167 | 0.021 | 0.021 |
| PEP A | 0.063 | 0.563 | 0.188 | 0.083 | 0.104 | |
| PEP B | 0.333 | 0.396 | 0.271 | |||
| PEP D | 0.64 | 0.574 | 0.170 | 0.085 | 0.106 | |
| IDH | 0.063 | 0.104 | 0.292 | 0.104 | 0.417 | 0.021 |
| PGD | 0.298 | 0.255 | 0.383 | 0.064 | ||
| PGM | 0.063 | 0.125 | 0.479 | 0.271 | 0.021 | 0.042 |
| GPI | 0.021 | 0.146 | 0.271 | 0.313 | 0.250 | |
| FK | 0.104 | 0.771 | 0.125 | |||
Alleles are encoded in decreasing order of their anodal mobility.
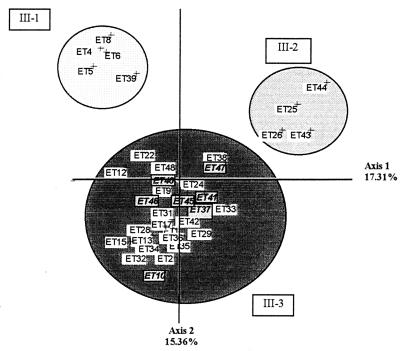
We investigated genotype differences between the ETs of cluster III using a second FCA. We obtained three clusters from the original group III (Fig. 3). Clusters III-1 and III-2 are separated by axis 1 (accounting for 17.31% of the total variability). Cluster III-1 contains exclusively human serotype B strains, and cluster III-2 contains only human serotype C strains. Cluster III-3 is separated from the other two clusters by axis 2 (accounting for 15.36% of the total variability) and consists of both C. neoformans (serotypes A, B, C, D, and A/D) and C. laurentii strains. PEP A5 (the numbers after the locus designation indicate allele numbers) was present only in isolates from cluster III-1, and ADH 3 was present only in those from cluster III-2 (Table 2).
FIG. 3.
First plane projection of FCA in two informative axes of ETs of cluster III (Fig. 2). Axes 1 and 2 represent 17.31 and 15.38% of the overall variability, respectively. ETs represent the observed ETs (Table 2). The ET group at the lower (cluster III-3) includes C. laurentii strains.
Genetic structure and serotype.
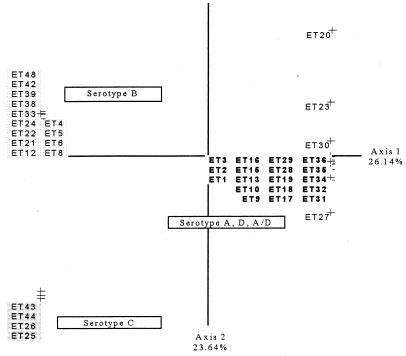
A CCA was carried out to identify a relationship between genetic structure and serotype. No link between serotype and genotype was found, and the permutation test revealed no significant relationship between genetic structure and serotype (P > 0.1). Serotypes A, D, and A/D are genetically similar, so we put them in the same cluster, in agreement with previous studies (22). We considered three serotyping groups (B, C, and A plus D plus A/D) (Fig. 4), and a correlation between the genotype structures of the strains and their serotypes was identified on the basis of a permutation test (P < 0.02).
FIG. 4.
First plane projection of the CCA in two informative axes of all observed ETs. Axes 1 and 2 represent 26.14 and 23.64% of the overall variability, respectively. ETs are grouped as a function of their serotypes. The ETs in boldface type are those of serotype A, D, and A/D strains, the ETs at the upper left are those of serotype B strains, and the ETs at the lower left are those of serotype C strains.
Genetic structure and origin.
A second CCA showed a clustering of the sample after the correlation between allelic composition and sample origin (patients and the environment) was tested. This was confirmed by a permutation test (P < 0.02). Isolates were grouped into three clusters on the projection (Fig. 5). One cluster contained strains isolated from both patients and the environment, the second contained strains from patients only, and the third contained strains from the environment only.
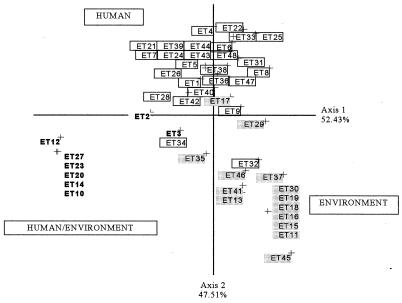
FIG. 5.
First plane projection of CCA in two informative axes of the observed ETs of C. neoformans. Axes 1 and 2 represent 52.43 and 47.51% of the overall variability, respectively. ETs are grouped as a function of the sample origin. Boxed ETs represent those of human strains, the shaded ETs at the lower right represent those of environmental strains, and the ETs in boldface type represent those of human or environmental strains.
DISCUSSION
We carried out a genetic analysis of 107 strains of Cryptococcus. The results obtained showed that there is extensive variation in this fungus, as reported in several other studies (1, 2, 8, 9, 11, 12, 17). We found 48 allele combinations among 107 isolates by studying 12 polymorphic loci. The level of diversity observed was similar to that reported in previous studies (4, 5) and was due to the large number of enzymes tested and the extensive heterogeneity of the strains studied. MLEE is among the numerous typing methods developed for the differentiation of C. neoformans isolates. It has a relatively high degree of discriminatory power but also allows assessment of the structure of the sample studied (4), including its genetic diversity. It was therefore used as the main technique for the present study.
Genetic diversity and taxonomy.
The first FCA (Fig. 2) produced three clusters. ET 7 consisted of one strain of C. laurentii. Cluster II contained only C. neoformans strains, whereas cluster III contained both C. neoformans and C. laurentii strains. There was more genetic divergence between some strains of C. neoformans than between C. laurentii and some strains of C. neoformans. The C. neoformans strains from clusters II and III differ more from each other than the C. neoformans and C. laurentii strains of cluster III do. Therefore, these fungi from different species are presumably closely related genetically.
A second FCA (Fig. 3) was used to identify differences between the ETs of cluster III. Most human strains of serotypes B and C were well separated on the projection. Serotypes A, D, and A/D were grouped together, and C. neoformans and C. laurentii were not always separated on this projection. This raises important taxonomic questions about the notion of species because C. neoformans and C. laurentii have very similar genotypes. The most informative enzyme systems (FUM, SDH, ADH, PGM, and PEP A) should be tested first to type an isolate and to determine to which cluster it belongs, but these enzymes cannot discriminate between some of the C. neoformans and C. laurentii strains used in this study.
Correlation between genotype and serotype.
Several studies have shown that multilocus genotypes correlate with variety (C. neoformans var. neoformans and C. neoformans var. gattii) and serotype (5, 6). However, the CCA, which tested for a possible link between serotype and genotype, detected no correlation when strains of the five serotypes (serotypes A, B, C, D, and A/D) were considered separately. This absence of a correlation was also confirmed by a nonsignificant value obtained in the approximation-to-permutation test (P > 0.1). If serotypes A, D, and A/D, which are clustered together in the FCA (Fig. 2), were placed in the same group, there was a correlation between the serotype and the genotype for the three resulting serotype groups (Fig. 4). Thus, MLEE cannot differentiate between serotypes A, D, and A/D, indicating that these serotypes are closely related genetically. Some studies (4, 6) have shown that MLEE differentiates serotypes A, D, and A/D and consider them to be genetically different. The results of our work are consistent with the preliminary results of other studies (27) indicating that strains readily switch from serotype A to serotype D via the intermediate serotype A/D.
Correlation between genotype and strain origin.
CCA for the genotypes and origins of the strains provided completely new information. The sample gave three clusters on the projection. There was a strong relationship between the genotypic information and the origin of the strain, in contrast to the results of other studies (13). This was confirmed by the significant value obtained in the permutation test (P < 0.02). The first cluster, which was genetically diverse, exclusively contained strains isolated from patients. The second cluster contained strains only from the environment (Fig. 5). The third cluster contained strains isolated either from patients or from the environment. These results are of epidemiological importance, because most of the environmental strains had particular isoenzyme profiles which were different from those of strains from patients. So, the genotypes of the strains were correlated with their origins. Some studies (11, 12) have demonstrated karyotype changes in Cryptococcus after multiple infections in the mouse. This suggests that the fungus acquires a characteristic genotype after infection. Our data suggest that environmental strains are only slightly pathogenic, if at all, for humans. The genotypes of the strains responsible for infection and those isolated from the environment were very different.
Thus, MLEE is a powerful discriminatory method. Analysis of the relationship between ETs should improve our understanding of the genetic polymorphism and the genotypic similarity of clinical and environmental C. neoformans isolates. This should help us to understand the transmission of this parasite and identify the risk factors for contamination so that more effective prevention measures can be implemented.
REFERENCES
- 1.Bellay T, Cherniak R, O’Neill E B, Kosel T. Serotyping of Cryptococcus neoformans by dot enzyme assay. J Clin Microbiol. 1996;34:466–470. doi: 10.1128/jcm.34.2.466-470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett J E, Kwon-Chung K J, Howard D H. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 3.Benzecri J-P, Benzecri F. Pratique de l’analyse des données. Analyse des correspondances. Dunot, Paris: ExposéÉlémentaire; 1980. [Google Scholar]
- 4.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W The Cryptococcal Disease Active Surveillance Group. Molecular subtype distribution of Cryptococcus neoformans in four areas in the United States. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt M E, Hutwagner L C, Kuykendall R J, Pinner R W The Cryptococcal Disease Active Surveillance Group. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J Clin Microbiol. 1993;33:1890–1895. doi: 10.1128/jcm.33.7.1890-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2923. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coustau C, Renaud F, Maillard C, Pasteur N, Delay B. Differential susceptibility to a trematode parasite among genotypes of the Mytilus edulis/galloprovincialis complex. Genet Res. 1991;57:207–212. doi: 10.1017/s0016672300029359. [DOI] [PubMed] [Google Scholar]
- 8.Dromer F, Guelho E, Ronin O, Dupont B. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J Clin Microbiol. 1993;31:359–363. doi: 10.1128/jcm.31.2.359-363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dromer F, Mathoulin S, Dupont B, Laporte A The French Cryptococcosis Study Group. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993) Clin Infect Dis. 1996;23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- 10.Drouhet E. Milestones in the history of Cryptococcus and cryptococcosis. J Mycol Med. 1997;7:10–27. [Google Scholar]
- 11.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes K A, Sullivan D J, Coleman D C, Clarke J C K, Emilianus R, Atkinson C, Cann K J. Involvement of multiple Cryptococcus neoformans strains in a single episode of cryptococcosis and reinfection with novel strains in recurrent infection demonstrated by random amplification of polymorphic DNA and DNA fingerprinting. J Clin Microbiol. 1995;33:99–102. doi: 10.1128/jcm.33.1.99-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohno S. Program and abstracts of the 3rd International Conference on Cryptococcus and Cryptococcosis. 1996. Epidemiology of cryptococcosis in Japan, Session II, abstr. 3; pp. 41–42. [Google Scholar]
- 14.Kwon-Chung K J, Polachek I, Bennett J E. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J Clin Microbiol. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin D, Lehmann P F, Hamory B H, Padhye A A, Durry E, Pinner R W, Lasker B A. Comparison of three typing methods for clinical and environmental isolates of Aspergillus fumigatus. J Clin Microbiol. 1995;33:1596–1601. doi: 10.1128/jcm.33.6.1596-1601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasteur N, Pasteur G, Bonhomme F, Catalan J, Britton-Davidian J. Manuel technique de génétique par électrophorèse des protéines. Lavoisier, Paris: Techniques et Documentation; 1987. [Google Scholar]
- 17.Perfect J R, Ketabchi N, Cox G M, Ingram C W, Beiser C L. Karyotyping of Cryptococcus neoformans as an epidemiologic tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pujol C, Reynes J, Renaud F, Mallié M, Bastide J-M. Analyse génétique de souches de Candida albicans par électrophorèse des isoenzymes. J Mycol Med. 1993;3:14–19. [Google Scholar]
- 19.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Janbon F, Mallié M, Bastide J M. The yeast Candida albicans has a clonal mode of reproduction in the population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez E, De Meeüs T, Mallié M, Renaud F, Symoens F, Mondon P, Piens M A, Lebeau B, Viviani M A, Grillot R, Nolard N, Chapuis F, Tortorano A-M, Bastide J-M. Multicentric epidemiological study of Aspergillus fumigatus isolates by multilocus enzyme electrophoresis. J Clin Microbiol. 1996;34:2559–2568. doi: 10.1128/jcm.34.10.2559-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez E, Symoens F, Mondon P, Mallié M, Piens M A, Lebeau B, Tortorano A M, Chaib F, Carlotti A, Villard J, Viviani M A, Chapuis F, Nolard N, Grillot R, Bastide J-M. Combination of three typing methods for the molecular epidemiology of Aspergillus fumigatus infections. J Med Microbiol. 1999;48:1–14. doi: 10.1099/00222615-48-2-181. [DOI] [PubMed] [Google Scholar]
- 22.Sorrell T C, Chen S C, Ruma P, Meyer W, Pfeiffer T J, Ellis D H, Brownlee A G. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplified polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ter Braak C J F. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology. 1986;67:1167–1179. [Google Scholar]
- 24.Ter Braak C J F. CANOCO—a Fortran program for canonical community ordination. Ithaca, N.Y: Microcomputer Power; 1987. [Google Scholar]
- 25.Ter Braak C J F. Canonical correspondence analysis and related methods in aquatic ecology. Aquat Sci. 1995;55:255–289. [Google Scholar]
- 26.Varma A, Swinne D, Staib F, Bennet J F, Kwon-Chung K J. Diversity of DNA fingerprints in Cryptococcus neoformans. J Clin Microbiol. 1995;33:1807–1814. doi: 10.1128/jcm.33.7.1807-1814.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viviani M A, Wen H, Roverselli A, Caldarelli-Stefano R, Cogliati M, Ferrante P, Tortorano M A. Identification by polymerase chain reaction fingerprinting of Cryptococcus serotype AD. J Med Vet Mycol. 1997;35:355–360. [PubMed] [Google Scholar]
- 28.Wilson D E, Bennett J E, Bailay J W. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127:820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]