Abstract
Motivation
It is a fundamental task to identify microRNAs (miRNAs) targets and accurately locate their target sites. Genome-scale experiments for miRNA target site detection are still costly. The prediction accuracies of existing computational algorithms and tools are often not up to the expectation due to a large number of false positives. One major obstacle to achieve a higher accuracy is the lack of knowledge of the target binding features of miRNAs. The published high-throughput experimental data provide an opportunity to analyze position-wise preference of miRNAs in terms of target binding, which can be an important feature in miRNA target prediction algorithms.
Results
We developed a Markov model to characterize position-wise pairing patterns of miRNA–target interactions. We further integrated this model as a scoring method and developed a dynamic programming (DP) algorithm, MDPS (Markov model-scored Dynamic Programming algorithm for miRNA target site Selection) that can screen putative target sites of miRNA-target binding. The MDPS algorithm thus can take into account both the dependency of neighboring pairing positions and the global pairing information. Based on the trained Markov models from both miRNA-specific and general datasets, we discovered that the position-wise binding information specific to a given miRNA would benefit its target prediction. We also found that miRNAs maintain region-wise similarity in their target binding patterns. Combining MDPS with existing methods significantly improves their precision while only slightly reduces their recall. Therefore, position-wise pairing patterns have the promise to improve target prediction if incorporated into existing software tools.
Availability and implementation
The source code and tool to calculate MDPS score is available at http://hulab.ucf.edu/research/projects/MDPS/index.html.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
MicroRNAs (miRNAs) are a type of small (about 22 nucleotides long) non-coding RNAs. They play important regulatory roles in various cellular processes and their dysfunction associates with complex diseases (Barham et al., 2019; Li and Kowdley, 2012; Wang et al., 2017, 2020). miRNAs may target messenger RNAs (mRNAs), long non-coding RNAs, transfer RNAs, circular RNAs etc. (Burroughs et al., 2011). They bind their targets through imperfect complementary matching, which regulates the amount of protein translated and/or causes the destabilization of the targets (Axtell et al., 2011; Bartel, 2004; Wang et al., 2011). Thus, the identification of miRNA targets is critical for the functional characterization of miRNAs and their involvement in various biological processes.
Early experiments have identified the canonical rule of seed matching during miRNA binding. This canonical rule requires that a miRNA–target interaction involves extensive binding between the miRNA seed region (Positions 2–8) and the mRNA 3′ untranslated regions (UTRs; Brennecke et al., 2005). Later this canonical rule was given a bit of leeway, allowing non-canonical seeds (one mismatch or wobble in the seed region) and the binding in miRNA 3′ regions centered on Positions 13–16, along with other features such as target accessibility (Kertesz et al., 2007), local AU content (Grimson et al., 2007), folding energy (Enright et al., 2003; Grimson et al., 2007), conservation (Helwak et al., 2013) etc. Dozens of tools developed focus primarily on these features (Ding et al., 2015, 2016, 2018).
In the past several years, next-generation sequencing (NGS)-based technologies have significantly advanced the study of miRNA targets. Chi et al. (2009) applied NGS techniques with the cross-linking and immunoprecipitation (CLIP) to directly identify miRNA targets (Chou et al., 2016). Hafner et al. (2010) used photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP) to increase the resolution of the original CLIP-seq method. Helwak et al. (2013) performed cross-linking, ligation and sequencing of hybrids (CLASH) experiments to detect miRNA-target pairs as chimeric reads in NGS data. Moore et al. (2015) improved the CLASH experiments with the covalent ligation of endogenous Argonaute-bound RNAs-CLIP (CLEAR-CLIP) experiments. The CLASH and CLEAR-CLIP experiments ultimately presented a transcriptome-wide dataset containing more than 18 000 and 30 000, respectively, high-confidence miRNA–target interactions, revealing the prevalence of seed and non-seed interactions and the diversity of in vivo miRNA targets in mRNA 3′ UTR, 5′ UTR and coding DNA sequence regions. The interactions are of different stability and have different free folding energy (ranging from 1.5 to 32 kcal/mol). With NGS datasets from these studies, a number of new tools have been developed for miRNA target prediction based on the aforementioned features together with new features learned from NGS data (Ding et al., 2016; Li and Hu, 2019; Lu and Leslie, 2016; Wang, 2016).
Despite the existence of various studies on miRNAs, it is still challenging to predict miRNA targets. High-throughput experimental approaches are still costly and may not be able to carry out under certain conditions. Computational methods often have low accuracy especially low precision although they are indispensable. The low accuracy of available computational methods may be partially due to our limited knowledge of the characteristics of miRNA target sites. Several studies thus investigated new features of miRNA binding sites. Among them, a Markov chain-based method started to model the base pairings between the entire mature miRNAs and their targets (Fu et al., 2009). Although only two states, forming a matching base pair or not, were considered in this Markov model, this study demonstrated the value of considering flexible matching patterns instead of the canonical seed matching when identifying miRNA target sites.
In this study, we created Markov models for miRNA–target interactions based on the position-wise pairing information (match, mismatch, bulge and wobble). We also evaluated the importance of the pairing patterns of a miRNA beyond its seed region for target prediction. From the model learning, we identified position-wise pairing patterns of a mature miRNA as a valuable feature for miRNA target site prediction. We also found region-specific correlations between miRNAs in terms of target binding. We further developed a Markov model-scored Dynamic Programming algorithm for miRNA target site Selection (MDPS) using the position-wise pairing information. Combining MDPS with three existing tools, we demonstrated that incorporating the discovered position-wise pairing information into existing target prediction pipelines has the potential to improve the accuracy of current miRNA target predictions.
2 Materials and methods
2.1 Training and testing
Using the miRNA–mRNA interactions reported in the CLASH study (Helwak et al., 2013), we designed two training datasets. One set contained 77 miRNAs with at least 50 targets each in the CLASH experiments. We named this set of interaction as ‘target-enriched dataset’. The other set included 122 miRNAs with at least 20 targets each, where the miRNA–mRNA folding energy was at least −15 kcal/mol. We termed this set as the ‘energy-filtered dataset’ (Supplementary Material S1).
For each of these CLASH interaction sets, we randomly chosen 80% of the interactions as the training data and kept the remaining 20% for testing. We did 10-fold cross-validation on the training data to obtain the best prediction model which was later used to make the prediction on the testing data of the corresponding interaction set. In addition, we also used an independent experimentally validated miRNA target dataset generated by a CLEAR-CLIP study (Moore et al., 2015) to test MDPS, as this dataset also provides miRNA-target binding information based on each position of an miRNA. To be consistent, we filtered out interactions in CLASH and CLEAR-CLIP that did not map to any mRNA transcript from ENSEMBL version 75 (Supplementary Material S1). To obtain the position-wise alignment information, we aligned the reported target sequences and the miRNA sequences using the RNAhybrid tool (Kruger and Rehmsmeier, 2006), as in the CLASH study (Helwak et al., 2013). The number of miRNAs and their corresponding targets for these datasets are given in Table 1.
Table 1.
Training and test datasets
| Total |
Target-enriched dataset |
Energy-filtered dataset |
||||
|---|---|---|---|---|---|---|
| MiRNAs | Targets | MiRNAs | Targets | MiRNAs | Targets | |
| CLASH | 399 | 18 041 | 77 | 15 390 | 122 | 16 209 |
| CLEAR-CLIP | 451 | 20 094 | — | — | — | — |
Note: We randomly selected 80% of the CLASH interactions to train a model using 10-fold cross-validation. We then tested the model on the 20% of the remaining CLASH interactions. We also tested the model on the independent CLEAR-CLIP interactions.
2.2 Different states of miRNA–target interactions in MDPS
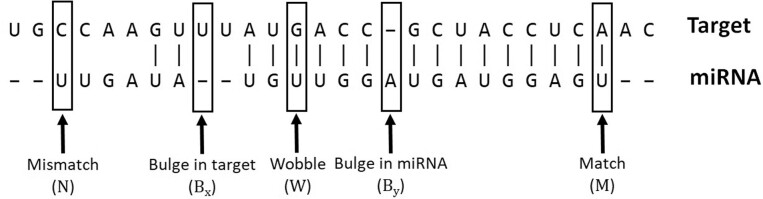
Considering the position dependency of neighboring pairings, we used a Markov model to learn the position-wise binding patterns for a given miRNA and its targets. We first defined the five states for the pairings in the alignment of a given miRNA sequence and one of its target sequences: match (M), mismatch (N), G-U wobble match (W), bulge in target (Bx) and bulge in miRNA (By; Fig. 1).
Fig. 1.
Five states in a miRNA–target interaction
With the five states, we designed a 5 × 5 transition matrix t that describes the transition probabilities of the five states and a weight matrix w to describe the probability of a state that a miRNA position prefers. For a miRNA sequence of length n, its weight matrix w is a 4 by n matrix, in which each column corresponds to one position in this miRNA, each row corresponds to one of the following four states: M, N, W, By and each number in the matrix gives the probability that the corresponding miRNA position prefers the corresponding state. The state Bx does not correspond to any miRNA position and thus was not considered in the weight matrix w. We calculated the transition and the weight matrices using the two training datasets. In brief, to create the weight matrix, we counted the number of the occurrences of each of the four states at each miRNA position in all miRNA–target interactions in a dataset. To create the transition matrix, we calculated the number of times each transition occurred in the interactions. We added a small pseudo count of 0.0001 to every entry in the matrices and then normalized the numbers in each row so that the sum of the numbers in a row to be 1. Both w and t were calculated from 5′ to 3′ direction of miRNAs, with the aligned miRNA-target sequences in the training data.
We defined two types of models: miRNA-specific and miRNA-general model. The miRNA-specific model was learned by calculating the transition and weight matrices given the pairing information of a specific miRNA and its targets. The miRNA-general model was trained by the pairing information of all available miRNAs and their targets. Note that, a miRNA-general model was parametrized by only one transition matrix and one weight matrix. The transition and weight matrices were the unweighted average of the transition and weight matrices of all the involved miRNA-specific models, respectively.
2.3 MDPS scoring strategy
MDPS selects miRNA target sites by scoring miRNA–target interactions using a dynamic programming (DP) algorithm. For a given miRNA and a calculated weight matrix and transition matrix, we have the following DP algorithm to score a target RNA sequence to determine whether it may contain a potential target site of this miRNA.
Here, we first define two notations, and state(i, j). We define as the best score of the alignment between and , with the last alignment position is at the kth posture. Here, represents the miRNA sequence from the Position 1 to the Position i. Similarly, represents the target sequence from the Position 1 to the Position j. There are three different possibilities for the last alignment position. When k = 0, it means the last alignment position is at the states M, N or W, which we call Posture 0. When k = 1, it means the last alignment position is at the Posture 1 and the state is By. When k = 2, it means the last alignment position is at the Posture 2 and the state is Bx. We also define state(i, j) as the state of the pairing of the ith miRNA position and the jth mRNA position. Since two actual base pairs are involved, state(i, j) can only be one of the states: M, N and W.
With the two notations, it is evident that , if i = 0 or j = 0. We also have for any j > 0, where means the -entry of the weight matrix of this miRNA. In addition, we have for any i > 1. With these initializations, we have the following iteration formula to calculate for any i > 1 and j > 1:
Similarly, we calculate by the following iteration with initialization and for i > 1 and any j.
Similarly, we initialize by , and calculate for any i and j > 1 by
With the above three types of iterations, we obtain the maximum of , for any j and k, and for n being the length of the miRNA under consideration. This maximum value is regarded as the score of the alignment of this miRNA and the target RNA sequence under consideration. The actual alignment resulted in this score describes the pairing between this miRNA and this target RNA. For a better understanding of the DP method, we show an example of the DP score calculation method in the Supplementary Material S2.
Using the above CLASH training datasets, we generated the MDPS models that consisted of the w matrices, the t matrices and the corresponding score cutoffs that gave the best predictions on the CLASH training dataset for different miRNAs. We generated these miRNA-specific models from the target-enriched dataset and the energy-filtered dataset separately. In addition to these miRNA-specific models, we also generated the general models for all miRNAs by average these miRNA-specific models from the target-enriched dataset and the energy-filtered dataset separately. Since the column size of the w matrix was the length of the corresponding miRNAs. The column size of the average w matrix in the models was the length of the longest miRNAs in the training datasets. If the score was larger than a given cutoff, this sequence was called the target of this miRNA. We tested five different cutoffs and chosen the Average score + 2 * SD as the final cutoff for the final MDPS models, where the Average score and the SD are the mean and the SD of the alignment scores of the miRNA-target duplexes in the training datasets, respectively.
2.4 Combining MDPS scores with existing tools
Existing target prediction algorithms emphasize the miRNA-target pairing in the seed regions (Agarwal et al., 2015; Friedman et al., 2008; Grimson et al., 2007; Lewis et al., 2003), and/or do not consider the dependence of the neighboring pairings (Enright et al., 2003). The alignment scores measured by MDPS may thus provide additional features for assessing miRNA–target interactions. If so, by combining the alignment scores from MDPS with the existing tools, the precision of the miRNA-target prediction may be improved. To test this hypothesis, we combined the MDPS scores with three popular methods, miRanda, RNA22 and TargetScan (Agarwal et al., 2015; Betel et al., 2010; Friedman et al., 2008; Lewis et al., 2003; Miranda et al., 2006). First, we generated the predictions on a given miRNA and target sequences using these tools, by running miRanda 3.3a and TargetScanHuman 7.0 and using the existing predictions of RNA22 (ENSEMBL 65, miRbase 18). Then we applied MDPS on the prediction of these tools. We compared the performance of the combined methods with that of the original methods without the MDPS alignment scores on the two test datasets (Table 1).
3 Results
3.1 Non-seed regions may be important for miRNA–target interactions
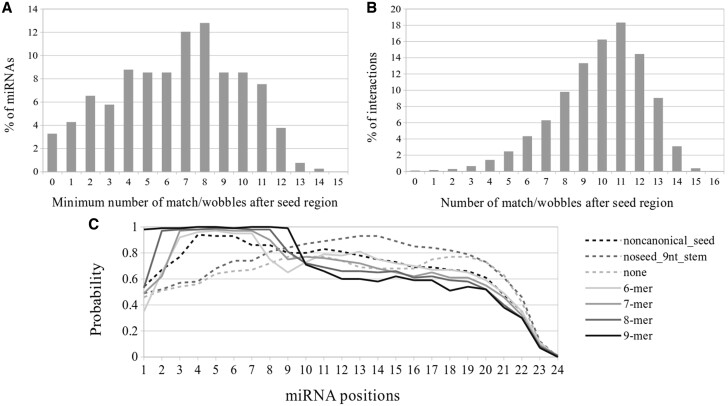
To evaluate the importance of the miRNA positions outside seed region in target binding, we analyzed the 18 041 CLASH interactions to see how many miRNAs and how many interactions had pairings (match/wobble states) outside the seed regions. We considered miRNA Positions 1–8 as seed region in this section. Figure 2A shows the percentage of miRNAs in CLASH data having different minimum number of match/wobble pairings outside the seed regions. More than 12% of miRNAs had at least eight matches/wobbles after the eighth position in the interactions they were involved. For the 399 miRNAs listed in the CLASH study (Helwak et al., 2013), 386 (97%) miRNAs had interactions with at least one match/wobble pairing outside the seed regions. Figure 2B shows the distribution of the number of match/wobble pairing outside the seed regions among the 18 041 CLASH interactions. Only 14 interactions had no match/wobble pairing outside the seed region.
Fig. 2.
Non-seed regions may be important for miRNA–target interactions. (A) Percentage of miRNAs with the different lowest number of match/wobble pairings after the Position 8 in the 18 041 CLASH interactions. (B) Percentage of the 18 041 CLASH interactions having different number of match/wobble pairing after the Position 8. (C) The frequency of match/wobble pairing at different miRNA positions for different types of CLASH interactions
We further studied whether miRNA–target interactions with seed matching had match/wobble pairings outside the seed regions. Similar to the CLASH study (Helwak et al., 2013), we considered the 6mer, 7mer, 8mer and 9mer interactions as interactions with seed matching, which had 6, 7, 8 and 9 continuous matches from the miRNA Position 1, respectively. We found that there were indeed many match/wobble pairings after the seed regions, even for the 9-mer interactions (Fig. 2C). From Figure 2, it is thus evident that it may be valuable to consider miRNA-target pairings after seed regions.
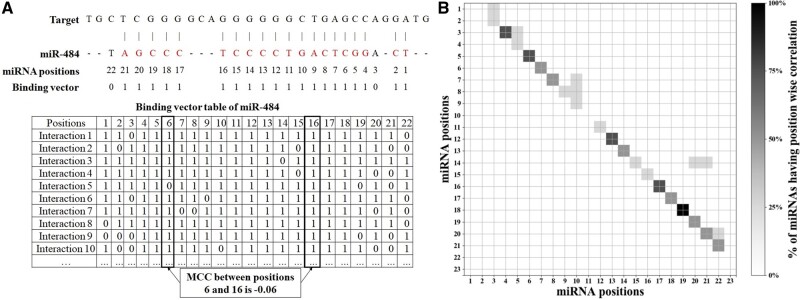
We also studied the dependency of pairings in miRNA–target interactions. When two miRNA-target pairings (match/wobble) occur side-by-side, the strength of one pairing might help to stabilize the pairing by its side. To study the dependency between neighboring pairing states, for each position of a miRNA, we labeled each ‘Match’ or ‘Wobble’ state with a ‘1’ and each ‘Mismatch’ or ‘bulge’ with a ‘0’. In this way, for each miRNA position, we had a binary binding vector representing the binding states of that miRNA position in the interactions. The size of this binding vector reflected the number of miRNA interactions (Fig. 3A). We then applied Matthews correlation coefficient (MCC) formula on the two binary vectors for each pair of positions of the miRNA to find the correlations between different positions of the same miRNA (Fig. 3A). We found that only the neighboring positions tend to have positive correlation (MCC ). We also found that the Regions 2–9, 11–14 and 16–21 of a large number of miRNAs tend to show the correlations (Fig. 3B). This suggested potential dependency or cooperation between adjacent binding positions of a miRNA and its targets. Together with the above studies on positions after the seed regions, it was clear that we may need to consider the pairing of all miRNA positions and their dependency for miRNA–target interactions, which was exactly done in the MDPS scores.
Fig. 3.
Correlated pairs of miRNA positions. (A) An illustration of how MCC is calculated for miR-484. (B) The percentage of miRNAs having correlated position pairs (MCC ). The heatmap has miRNA positions in the axes and the percentage of correlated miRNAs are shown for every pair of positions
3.2 Different miRNAs share correlated target binding patterns
Since many miRNA–target interactions involve seed regions and the pairing at different miRNA positions are dependent, we hypothesized that many miRNAs may have similar or correlated target binding patterns. We tested this hypothesis with the obtained weight matrices and transition matrices and found that many miRNAs indeed share correlated binding patterns.
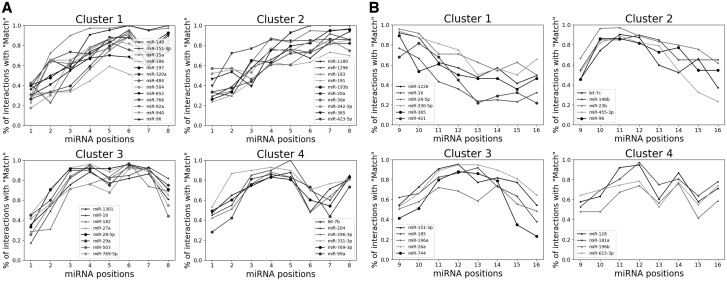
To investigate whether different miRNAs have similar or correlated binding patterns, we divided a miRNA sequence into two equal size regions, Positions 1–8 and 9–16. We illustrated our study with the energy-filtered dataset here and the conclusion was similar for the target-enriched dataset. For each of the 122 miRNAs in the energy-filtered dataset, we obtained its position-wise ‘Match’ and ‘Mismatch’ probabilities from the learned weight matrix. We then computed the Spearman’s correlation between each pair of miRNAs based on their position-wise ‘Match’ and ‘Mismatch’ probabilities in the two regions separately. We did not consider the G-U wobble and bulge states in this calculation, as the probabilities of these two states were very low at most positions of a miRNA. We ignored the miRNAs that belonged to the same family here, as these miRNAs had high sequence similarity and thus strong correlations. We performed clique-finding-based clustering (29; correlation cutoff = 0.75) and identified 17 distinct clusters of miRNAs that were correlated in terms of ‘Match’ state probabilities at Positions 1–8. The largest 8 clusters had 50.88% of the total 122 miRNAs (Fig. 4A shows four different exclusive clusters). When considering the Positions 9–16 of a miRNA, we got 29 distinct clusters correlated on ‘Match’ state probabilities (Fig. 4B). The largest 10 clusters had only 29.82% of the total 122 miRNAs considering ‘Match’ probabilities. These statistics suggested that the seed regions (Positions 1–8) of miRNAs were more correlated than the non-seed region (Positions 9–16), which supported the current practice of considering seed matching for miRNA targeting.
Fig. 4.
Clusters of miRNAs with similar ‘Match’ patterns in specific regions. The X-axis of a cluster plot shows the positions of the miRNAs in that cluster and the Y-axis of the plot shows the percentage of interactions having ‘Match’ in corresponding miRNA positions (A) Clusters of miRNAs correlated with the ‘Match’ state probability from Positions 1 to 8. (B) Clusters of miRNAs correlated with the ‘Match’ state probability from Positions 9 to 16
3.3 miRNA-general models showed better performance on target site prediction than miRNA-specific models
Many miRNAs have similar or correlated target binding patterns, as demonstrated in Section 3.2. We thus hypothesized that the model learned from all miRNAs and their corresponding targets would work better than the models learned for individual miRNAs, where we trained the models with the corresponding individual miRNA and its targets. In the miRNA-general model, we learned a common weight matrix and a common transition matrix for all miRNAs together. In the miRNA-specific model, we had a unique weight matrix and a unique transition matrix for each individual miRNA with a decent number of targets (). The models were learned with the 10-fold cross-validation based on two training datasets.
We found that the miRNA-general model worked better than the miRNA-specific models for individual miRNAs with a specific model. In the target-enriched datasets, the miRNA-general model identified 93.49% of the CLASH interactions correctly while the miRNA-specific models identified 87.56% of the CLASH interactions. Similarly, in the energy-filtered datasets, the miRNA-general model identified 91.59% of the CLASH interactions while the miRNA-specific models identified only 85.91% of the interactions (Supplementary Material S3). We did not strive to study the false positive predictions here, as we did not have the negative datasets here. In addition, our goal was to reduce the false positive predictions in existing tools.
The miRNA-general models worked better, probably because of the following reasons. First, as demonstrated above, miRNAs do share similar or correlated patterns in terms of target binding, which enables the miRNA-general model caught the ‘key’ or ‘conserved’ characteristics of miRNA–target interactions; Second, there were much more training data to train a miRNA-general model than that to train a miRNA-specific model; Third, because of the number of targets a miRNA had was still small in our training datasets, the miRNA-specific model might encountered ‘overfitting’. Note that since the 10-fold cross-validation accuracy on the 10 groups of untrained datasets was similar (Supplementary Material S1), it was unlikely that the miRNA-specific models were overfitted or not well-trained. Therefore, the general models worked better highly likely because of the similarity of the binding patterns of different miRNAs.
Despite of the overall better performance of the miRNA-general models, for certain miRNAs, their miRNA-specific models did work better. For instance, for miR-10a, the miRNA-specific model predicted 100% of its target sites correctly, whereas the miRNA-general model predicted 86% of its target sites correctly. This miRNA had 51 targets in the energy-filtered training dataset. Note that, it was the miRNA-specific binding patterns, not the number of target sites in the training dataset that resulted in the different performance of the miRNA-specific models and the miRNA-general models. For example, in case of miR-186, the miRNA-general model did not perform better, even though it had 81 training target sites. On the other hand, the miRNA-specific model performed better for miR-1301, although it only had 26 training target sites.
3.4 Combining the MDPS scores with existing tools improved their accuracy
Since the existing tools did not consider the entire miRNA regions for miRNA–target interaction prediction, and/or did not consider the dependency among different pairing positions in miRNA–target interactions, we hypothesized that by combining the MDPS scores with the existing tools, we may be able to improve the accuracy of the existing tools. We found that it was indeed the case and the MDPS scores facilitated more accurate prediction of miRNA target sites.
We combined the MDPS scores with three existing tools, miRanda, RNA22 and TargetScan. To combine MDPS scores with these tools, we applied these tools to predict miRNA–target interactions first. We then calculated the MDPS scores for the predicted targets and determined whether the predicted target sites were true or false based on the default MDPS score cutoff from the trained general models. We tested the combined tools on the untrained 20% CLASH dataset and the independent CLEAR-CLIP dataset, for both the MDPS scores trained with the target-enriched dataset and the MDPS scores trained with the energy-filtered dataset. We found that the precision of the combined tools was significantly increased while the recall of the combined tools was slightly decreased, compared with the original tools (Table 2 and Supplementary Material S3). Overall, the F1 score of the combined tool was improved. For instance, the recall, precision and F1 score of RNA22 on the CLEAR-CLIP data were increased by −9.35%, 22.71% and 22.46%, respectively, when combined with the MDPS model trained on the energy-filtered dataset. This analysis demonstrated that the MDPS score as an additional feature for miRNA target site prediction decreased the false positive predictions by the existing tools.
Table 2.
Performance comparison of the combined tools with the original tools
| Miranda |
RNA22 |
TargetScan |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| F1 | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | |
| Target-enriched model on CLASH | 18.88 | 23.64 | −5.76 | 25.24 | 26.62 | −5.22 | 20.78 | 23.37 | −4.19 |
| Target-enriched model on CLEAR-CLIP | 15.36 | 15.67 | −7.12 | 22.46 | 22.71 | −9.35 | 18.11 | 18.28 | −7.16 |
| Energy-filtered model on CLASH | 17.82 | 20.85 | −7.62 | 24.52 | 25.66 | −7.10 | 23.21 | 24.97 | −4.81 |
| Energy-filtered model on CLEAR-CLIP | 15.52 | 15.89 | −10.68 | 21.15 | 21.40 | −10.57 | 15.81 | 15.96 | −7.64 |
Note: Each number is the increased percentage when comparing the performance of the combined tool with the performance of the original tool. The detailed performance numbers are in the Supplementary Material S3.
4 Discussion
Recent experimental data on miRNA–target interactions provide insights into miRNA binding rules. Studies on these newly generated datasets have shown potential involvement of non-seed regions of miRNAs in the binding activities. However, the importance of non-seed regions for miRNA target binding has not been thoroughly studied; neither did the dependency among positions and regions of miRNA–target interactions. The MDPS algorithm was developed to learn miRNA-target pairing patterns, especially in the non-seed regions of miRNA binding, by utilizing the genome-wide CLASH datasets. MDPS takes into account the dependency of neighboring pairing positions using a Markov model. Utilizing the weight and transition matrices of the trained Markov model, MDPS is then able to score each potential miRNA binding site to pre-select/predict putative candidate miRNA–target interactions. By combining the MDPS scores with existing tools, we showed that the precision of the combined tools has been greatly improved.
The DP used in MDPS is different from the one used in miRanda (Enright et al., 2003), which applies a standard DP algorithm to perform pair-wise alignment between a miRNA and a potential target. The alignment score is then used as a criterion together with site conservation and binding energy scores to predict miRNA target sites. There are at least two important differences between the miRanda DP algorithm and the MDPS one. One is the scoring schema for miRNA-target alignments, for which miRanda uses a fixed scoring schema, such as a score of +5 for G:C and A:T pairs, +2 for G:U wobble pairs etc. (Betel et al., 2010), whereas MDPS uses a probabilistic scoring schema based on the CLASH training data. The other is, MDPS considers neighboring pairing positions in the alignments, whereas miRanda assumes the independence of neighboring pairing positions.
Through the investigation of the Markov models learned from both target-enriched datasets and energy-filtered datasets, we were able to make interesting findings on position-wise binding patterns of miRNA–target interactions. We found subsets of miRNAs had correlated binding patterns in specific sub-regions. We also found both seed and non-seed regions contribute to the specific miRNAs’ binding patterns. Besides seed region binding, the length of the continuous pairings outside the seed region, the gap between two continuous pairings, the number and position of G-C pairing in an interaction are also some of the important features that can play a part in miRNA target prediction. The position-wise knowledge of a miRNA target binding, the continuous paring patterns, the number and position of the G-C bonds along with the canonical seed preference rule can help us to find a target prediction algorithm with less bias, better sensitivity and specificity.
Although the MDPS scores can help to improve the miRNA target site prediction, we are unsure whether these selected target sites are functional. In other words, although the miRNAs may indeed bind to the corresponding selected target sites, the miRNAs may not suppress the expression level of the target RNAs. These selected sites can only be considered as potential target sites and their functional effects need to be further investigated by experiments.
The current version of MDPS was not developed to be a tool for miRNA target prediction. Many features such as sequence conservation, binding energy and target site abundance need to be considered to confidently predict miRNA target sites. However, the study here based on MDPS shows that the dependency of neighboring pairing for miRNA binding to targets and global pairing information of miRNA–target interactions is important for target site selection. The incorporation of MDPS either as a feature or an additional step in existing miRNA target prediction pipelines has the promise to enhance their overall performance of miRNA target prediction tools.
Funding
This work was supported by the United States National Science Foundation [1356524, 1149955 and 1661414] and the United States National Institutes of Health [R15GM123407].
Conflict of Interest: none declared.
Supplementary Material
References
- Agarwal V. et al. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. et al. (2011) Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol., 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barham C. et al. (2019). Application of deep learning models to MicroRNA transcription start site identification. In: 2019 IEEE 7th International Conference on Bioinformatics and Computational Biology (ICBCB). IEEE, Hangzhou, China, pp. 22–28.
- Bartel D.P. (2004) MicroRNAs. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Betel D. et al. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol., 11, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J. et al. (2005) Principles of microRNA–target recognition. PLoS Biol., 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs A.M. et al. (2011) Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol., 8, 158–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W. et al. (2009) Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature, 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-H. et al. (2016) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res., 44, D239–D247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. et al. (2015) MicroRNA modules prefer to bind weak and unconventional target sites. Bioinformatics, 31, 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. et al. (2016) TarPmiR: a new approach for microRNA target site prediction. Bioinformatics, 32, 2768–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. et al. (2018) CCmiR: a computational approach for competitive and cooperative microRNA binding prediction. Bioinformatics, 34, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J. et al. (2003) MicroRNA targets in drosophila. Genome Biol., 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C. et al. (2008) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H.-Y. et al. (2009) Assessing potential miRNA targets based on a Markov model. Genet. Mol. Res., 8, 848–860. [DOI] [PubMed] [Google Scholar]
- Grimson A. et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell, 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M. et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A. et al. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell, 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M. et al. (2007) The role of site accessibility in microRNA target recognition. Nat. Genet., 39, 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kruger J., Rehmsmeier M. (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res., 34, W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P. et al. (2003) Prediction of mammalian microRNA targets. Cell, 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Li X., Hu H. (2019). Improving miRNA target prediction using CLASH data. In: Methods in Molecular Biology, pp. 75–83. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Li Y., Kowdley K.V. (2012) MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics, 10, 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Leslie C.S. (2016) Learning to predict miRNA-mRNA interactions from AGO CLIP sequencing and CLASH data. PLoS Comput. Biol., 12, e1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K.C. et al. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell, 126, 1203–1217. [DOI] [PubMed] [Google Scholar]
- Moore M.J. et al. (2015) miRNA–target chimeras reveal miRNA 3′-end pairing as a major determinant of argonaute target specificity. Nat. Commun., 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. (2020) Computational annotation of miRNA transcription start sites. Brief. Bioinform., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2016) Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics, 32, 1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2011) Transcriptional regulation of co-expressed microRNA target genes. Genomics, 98, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2017) Prognostic cancer gene signatures share common regulatory motifs. Sci. Rep., 7, 4750. doi: 10.1038/s41598-017-05035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.