Abstract
Objectives
Doravirine is a recently licensed HIV-1 NNRTI with improved efficacy, pharmacokinetics and safety profile compared with efavirenz and limited cross-resistance with rilpivirine and etravirine. In this in vitro study, cross-resistance to doravirine was analysed in a representative panel of NNRTI-resistant clones.
Methods
In vitro phenotypic susceptibility to doravirine was assessed in 10 clinically derived infectious clones with intermediate- to high-level resistance to rilpivirine, etravirine, efavirenz and nevirapine, and in NL4-3 site-directed mutants harbouring K103N, Y181C, M230L or K103N/Y181C NNRTI mutations.
Results
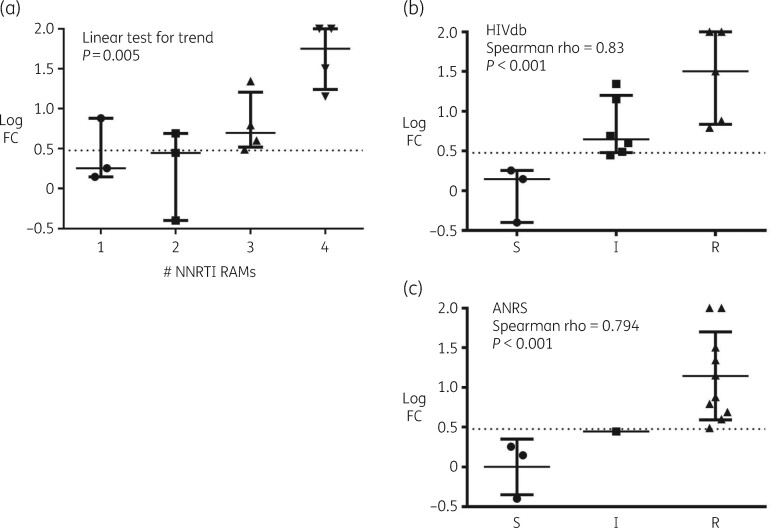
Although none of the infectious clones harboured any of the major doravirine resistance-associated mutations (RAMs) included in the IAS-USA reference list, doravirine fold change (FC) values were comparable to or higher than those calculated for other NNRTIs, particularly etravirine and rilpivirine. As expected, single NNRTI mutations K103N and Y181C did not impair doravirine susceptibility (FC 1.4 and 1.8, respectively), while reduced activity was observed with the single M230L or double K103N/Y181C mutations (FC 7.6 and 4.9, respectively). Median FC values increased significantly with increasing numbers of NNRTI RAMs (P = 0.005) and were >10 in 4/4 and 1/4 clones harbouring four and three NNRTI RAMs, respectively. FC values correlated well with predicted susceptibility as inferred by Stanford HIV Drug Resistance Database (HIVdb) and ANRS algorithms (both P < 0.001).
Conclusions
Substantial cross-resistance to doravirine was detected in NNRTI-resistant viruses harbouring complex mutational patterns, even in the absence of major IAS-USA doravirine RAMs. Therefore, based on the simple IAS-USA reference list, doravirine resistance may be underestimated in viruses harbouring multiple NNRTI mutations.
Introduction
Doravirine (formerly MK-1439) is a novel once-daily NNRTI approved by the US FDA for the treatment of HIV-1 in therapy-naive patients or as a switch option in virologically suppressed patients with no history of treatment failure and no known substitutions associated with resistance to doravirine.1 The approval label of the EMA provided slightly different therapeutic indications, recommending the use of doravirine for the treatment of adults infected with HIV-1 without past or present evidence of resistance to the NNRTI class.2 In clinical studies, doravirine showed non-inferior efficacy and improved pharmacokinetics and/or safety profile, both as a switch option in virologically suppressed patients3 and as part of a first-line regimen, when compared with efavirenz and darunavir.4,5 Moreover, doravirine was effective in halting viral replication even in the presence of transmitted NNRTI mutations, such as K103N and G190A, in a small group of treatment-naive patients.6 Based on in vitro and in vivo data, the last update of the IAS-USA HIV-1 drug resistance mutations list indicates V106A/M and Y188L as major doravirine mutations and V106I/T, Y188C/H, G190E, P225H, F227C/L/R, M230L and L234I as minor mutations.7 Indeed, in vitro studies demonstrated that doravirine retained full activity against most of the single resistance-associated mutations (RAMs) selected by older NNRTIs, except for V106A, Y188L and M230L, as well as against some combinations of multiple NNRTI RAMs.8,9 A recent study conducted on a large panel of clinical isolates collected from treatment-naive patients revealed that 92.5% of samples were susceptible to doravirine, as indicated by a fold change (FC) value lower than the biological cut-off of 3-fold.10 In vitro resistance selection experiments revealed the emergence of V106A/M/I, V108I, F227C/I/L and L234I, with minimal HIV-1 subtype-related differences,11 resulting in a limited cross-resistance with rilpivirine and possibly with etravirine.12 Although three recent large surveys showed a low prevalence of doravirine RAMs in NNRTI-exposed individuals,13–15 the impact of various combinations of NNRTI RAMs and the possible cross-resistance with other NNRTIs have been poorly investigated. Importantly, one clinical trial (NCT04233216) has recently started to recruit heavily treatment-experienced patients with multidrug-resistant viruses harbouring NNRTI and NRTI RAMs to evaluate the efficacy of the doravirine/islatravir combination plus optimized background therapy. In this study, we aimed to evaluate the activity of doravirine in a reference panel of NNRTI-resistant infectious clones and in site-directed mutants including relevant NNRTI mutations.
Materials and methods
Cell lines
Lenti-X 293 T cells (Takara Bio, Kusatsu, Japan) and TZM-bl cells were cultured in high-glucose DMEM with l-glutamine, supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. The MT-2 cell line was cultured in RPMI supplemented with 2 mM l-glutamine, 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. TZM-bl and MT-2 cell lines were obtained from the Centre for AIDS Reagent of the National Institute for Biological Standards and Control. All cell culture media and relevant reagents were obtained from EuroClone (Italy).
NNRTI-resistant infectious clones
A reference panel of HIV-1 infectious clones harbouring combinations of major NNRTI RAMs was obtained from the NIH AIDS Reagent Program. These clinically derived recombinant viruses were characterized by intermediate- to high-level resistance to rilpivirine, etravirine, efavirenz and nevirapine as determined by the Phenosense Assay.16 Reverse transcriptase sequences of NNRTI-resistant clones were submitted to GenBank under accession numbers JQ814884–JQ814893. In addition, we introduced the individual NNRTI mutations K103N, Y181C, M230L and the combination of K103N/Y181C into the HIV-1 NL4-3 backbone through the QuikChange® Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The NL4-3 plasmid was obtained from the NIH AIDS Reagent Program. NL4-3 and all clones with NNRTI mutations were transfected into Lenti-X 293 T cells, propagated in MT-2 cells and titrated in TZM-bl cells as previously described.17
Phenotypic determination of susceptibility to doravirine
In vitro susceptibility to doravirine was determined in duplicate through a TZM-bl cell-based assay previously shown to correlate well with the reference phenotypic Phenosense Assay in the measurement of susceptibility to HIV-1 protease, reverse transcriptase and integrase inhibitors.17 Briefly, 10 000 TZM-bl cells/well were infected with the WT NL4-3 strain or NNRTI-resistant viruses at a multiplicity of infection of 0.03 in the presence of 5-fold dilutions of doravirine (MedChemExpress, Monmouth Junction, NJ, USA) ranging from 10 μM to 0.00512 nM. After 48 h, cells were treated with the Glo-Lysis buffer (Promega, Madison, WI, USA) and the Bright-Glo Luciferase Assay (Promega), then relative luminescence units were measured through the GloMax Discover instrument (Promega) and elaborated with GraphPad software to calculate IC50 values. FC values were calculated with respect to the IC50 value obtained with the NL4-3 WT strain.
Results and discussion
According to the IAS-USA drug resistance mutations list,7 none of the clones harboured any major doravirine RAMs (namely V106A/M and Y188L), while two clones included the minor doravirine RAM M230L. As described in Table 1, 8 out of the 10 NNRTI-resistant clones and the mutant K103N/Y181C NL4-3 clone have an FC value higher than the biological cut-off. The highest FC values (>100) were found in samples 12225 and 12237, harbouring mutations E138G/H221Y/F227L/M230L and V106I/Y181C/G190A/H221Y, respectively, while the other sample harbouring M230L together with L100I and V179D (ID 12243) showed an FC value of 6.2. Other clones with Y181C and additional NNRTI RAMs (ID 12231, 12235 and 12239) had FC values of 14.2–31.8, suggesting a considerably reduced susceptibility to doravirine, while the absence of Y181C and doravirine RAMs (clones 12227, 12229, 12233 and 12241) resulted in FC values from 0.4 to 3.1, suggesting no or minimal impact on doravirine susceptibility. The single K103N or Y181C mutations within the NL4-3 backbone did not reduce doravirine susceptibility (FC values 1.4 and 1.8, respectively), while the double mutant K103N/Y181C showed a reduced susceptibility (FC 4.9). These data were comparable to those already described for viruses harbouring single K103N or Y181C and the K103N/Y181C mutations, showing mean FC values of 1.5, 2.5 and 4.3, respectively.8 Interestingly, clone 12231 harbouring mutations K103N/V179F/Y181C showed an FC value of 22.1, suggesting that the non-polymorphic V179F substitution by itself or in combination with other uncharacterized amino acid variations can further decrease doravirine susceptibility. Similarly, the site-directed NL4-3/M230L mutant showed an FC value of 7.6, comparable to those observed in clone 12243 but significantly lower than in clone 12225, harbouring the additional doravirine RAM F227L together with NNRTI RAMs E138G and H221Y. Overall, doravirine FC values were comparable to or lower than those calculated with other licensed NNRTIs in clones 12227, 12229, 12233, 12241 and 12243, while in the remaining five clones doravirine FC values were higher than those of other NNRTIs, particularly etravirine and rilpivirine. Irrespective of the type of NNRTI RAMs, a higher number of NNRTI RAMs correlated significantly with increasing median FC values (linear test for trend, P = 0.005) (Figure 1a). Indeed, all of the eight viruses with ≥3 NNRTI mutations showed FC values >3, including clones 12231, 12235, 12237 and 12239 with no canonical doravirine mutations. By comparing FC values and predicted susceptibility to doravirine, both the HIV Drug Resistance Database (HIVdb; version 8.9-1) and the ANRS algorithm (version 30) could estimate doravirine activity with good accuracy, indicating that these two algorithms can be reliably used to evaluate doravirine susceptibility for possible use in patients harbouring NNRTI RAMs (Figure 1b and c).
Table 1.
Mean IC50 ± SD, fold change values for doravirine and other licensed NNRTIs against infectious clones harbouring NNRTI resistance-associated mutations (RAMs) and levels of doravirine activity predicted by the Stanford HIVdb and ANRS prediction algorithms
| Virus ID | NNRTI RAMs | NRTI RAMs | Other mutations | Mean doravirine IC50 ± SD (nM) | Fold change (FC) |
Predicted doravirine activityd |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Doravirineb | Nevirapinec | Efavirenzc | Etravirinec | Rilpivirinec | Stanford HIVdb | ANRS | |||||
| 12225a | E138G, H221Y, F227L, M230L | M41L, L210W, T215Y | K122E, D177E, I178L, R211K, V245M, I293V | >100 | >100 | >200 | 15 | 21 | 18 | R | R |
| 12227a | K101P, K103N | M41L, T215Y | A98S, K102Q, D123E, K166R, D177E, D192N, R211K, V245K, K277R, R284K, T286A, E297K | 0.4 ± 0.1 | 0.4 | >200 | >200 | 5.8 | 92 | S | S |
| 12229a | L100I, K103N, H221Y | M41L, L74V, M184V, T215Y | R83K, D177E, T200A, R211Q, K281R, R284K, I293V, E297A | 4.8 ± 0.3 | 4.0 | >200 | >200 | 6.8 | 6.3 | I | R |
| 12231a | K103N, V179F, Y181C | M41L, T215F | K49R, K82R, A98S, D177E, G196E, Q207K, R211A, L228R, A272P, I293V, K311R | 26.5 ± 14.4 | 22.1 | >200 | 90 | 8.8 | 2.3 | I | R |
| 12233a | K101E, Y181V | K70R, M184V, T215F | T27S, E28K, K32E, V60I, T69N, R83K, V179I, T200A, Q207D, L228R, V245E, T286A, E297K | 3.3 ± 0.5 | 2.8 | >200 | 2.1 | 27 | 24 | I | I |
| 12235a | A98G, K101E, Y181C, G190A | M41L, E44D, D67N, T69D, L74I, L210W, T215Y | T39E, V118I, K122E, I135T, G196E, E203K, R211K, A272S, V276T, K277R, Q278E, L283I, I293V, E297K | 38.2 ± 5.8 | 31.8 | >200 | >200 | 15 | 22 | R | R |
| 12237a | V106I, Y181C, G190A, H221Y | none | V35I, S68G, A98S, D121E, K122E, I135V, R211K, F214L, V245E, D250E, A272P, E297K | >100 | >100 | >200 | 26 | 6 | 3.5 | R | R |
| 12239a | A98G, K101E, E138K, Y181C | M41L, T215D | K122E, I135T, S162Y, T200A, L210F, P243T, V245E, D250E, A272P, I274V, Q278H, K281R, T286A, A288S, K311R | 17.0 ± 4.0 | 14.2 | >200 | 3.6 | 10 | 9.2 | I | R |
| 12241a | K101E, E138G, G190S | none | V35M, R211K, V245E, S251I | 3.8 ± 1.4 | 3.1 | >200 | >200 | 3.2 | 2.6 | I | R |
| 12243a | L100I, V179D, M230L | M41L, D67G, L74I, M184V, T215Y | V35I, K103R, K122E, I202V, R211K, T240K | 7.5 ± 0.6 | 6.2 | >200 | >200 | 95 | 13 | R | R |
| NL4-3/103N | K103N | 2.0 ± 1.2 | 1.4 | NA | NA | NA | NA | S | S | ||
| NL4-3/181C | Y181C | 2.6 ± 1.1 | 1.8 | NA | NA | NA | NA | S | S | ||
| NL4-3/ 103N/181C | K103N, Y181C | 6.9 ± 2.8 | 4.9 | NA | NA | NA | NA | I | R | ||
| NL4-3/230L | M230L | 10.6 ± 4.7 | 7.6 | NA | NA | NA | NA | R | R | ||
NA, not available.
NIH AIDS Reagent Program catalogue number.
Doravirine FC values calculated according to the NL4-3 strain IC50 value of 1.4 ± 0.7 nM.
FC values determined through the Phenosense Assay.16
The HIVdb five-level grading was collapsed to three levels as indicated by the HIValg release notes at https://hivdb.stanford.edu/page/release-notes/. S, susceptible; I, intermediate resistance; R, high-level resistance.
Figure 1.
Distribution of doravirine fold change (FC) values measured with clones harbouring NNRTI resistance-associated mutations (RAMs), as grouped by (a) number of NNRTI RAMs (linear test for trend, P = 0.005); (b) predicted doravirine susceptibility by the Stanford HIVdb algorithm; (c) predicted doravirine susceptibility by the ANRS algorithm. S, susceptible; I, intermediate resistance; R, high-level resistance. The dotted line indicates the biological FC cut-off (3-fold) recently established for doravirine.10
The 10 clinically derived NNRTI-resistant clones were originally conceived to include combinations of NNRTI mutations commonly observed among sequences stored in the Stanford HIVdb and causing intermediate- to high-level resistance to nevirapine, efavirenz, rilpivirine and etravirine.16 This study completes the NNRTI susceptibility profile for this reference panel of clones, which is publicly available and ideal to inform further NNRTI development. Partly contrary to expectations, we found that doravirine activity was overall similar to that of the second-generation NNRTIs etravirine and rilpivirine, indicating that doravirine may only partially overcome NNRTI resistance. While previous in vitro testing had been mostly carried out on viruses harbouring one or two NNRTI mutations,8,9 the key result of this study is that multiple mutations selected by older NNRTIs can confer substantial cross-resistance to doravirine even in the absence of major IAS-USA doravirine RAMs. It must also be noted that clones derived from clinical isolates accommodate in vivo selected minor or compensatory changes which cannot be recapitulated in site-directed mutants, yet may play a relevant role in modulating resistance. For example, patient-derived viruses harbouring G190S or L100I + K103N without any canonical doravirine mutation showed FC values in the range of 1.5–11 and 2.7–19, respectively.8
A relevant consequence of these findings is that prediction of resistance simply based on the presence of IAS-USA doravirine RAMs may overestimate doravirine activity. Genotypic interpretation systems such as the Stanford HIVdb or ANRS should be preferred in the context of multiple NNRTI mutations. Indeed, a recent study showed that the Stanford HIVdb detected more transmitted resistance to doravirine with respect to the IAS-USA list.15 Importantly, clinical use of doravirine as part of salvage therapy in heavily treatment-experienced patients still needs to be informed by expanded in vitro genotype–phenotype correlation analysis, coupled with in vivo data allowing the establishment of clinical FC cut-offs.
Acknowledgements
This study was presented at the 17th European AIDS Conference, 6–9 November 2019, Basel, Switzerland, abstract PS5/6.
Funding
The authors acknowledge the contribution of the CARE Consortium funded by the European Union’s Horizon 2020 programme and the Ministry of Science and Higher Education of the Russian Federation. In addition, the study was partially supported by ViiV Healthcare for the project ‘HIV multidrug resistance pathways in EuResist Integrated DataBase’.
Transparency declarations
M.Z. reports consultancy fees for ViiV Healthcare, Gilead Sciences and Janssen-Cilag, and grants for his institution from ViiV Healthcare and Gilead outside the submitted work. R.W.S. received research funding from Janssen Pharmaceuticals and Vela Diagnostics outside the submitted work. All other authors have none to declare.
References
- 1.US Food and Drug Administration. PIFELTRO (Doravirine) Prescribing Information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/210806s003lbl.pdf.
- 2.European Medicines Agency. Pifeltro Product Information, Annex I—Summary of Product Characteristics. https://www.ema.europa.eu/en/documents/product-information/pifeltro-epar-product-information_en.pdf.
- 3.Johnson M, Kumar P, Molina JM et al. Switching to doravirine/lamivudine/tenofovir disoproxil fumarate (DOR/3TC/TDF) maintains HIV-1 virologic suppression through 48 weeks: results of the DRIVE-SHIFT trial. J Acquir Immune Defic Syndr 2019; 81: 463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin C, Squires KE, Molina JM et al. Doravirine/lamivudine/tenofovir disoproxil fumarate is non-inferior to efavirenz/emtricitabine/tenofovir disoproxil fumarate in treatment-naive adults with human immunodeficiency virus-1 infection: week 48 results of the DRIVE-AHEAD trial. Clin Infect Dis 2019; 68: 535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molina JM, Squires K, Sax PE et al. Doravirine versus ritonavir-boosted darunavir in antiretroviral-naive adults with HIV-1 (DRIVE-FORWARD): 96-week results of a randomised, double-blind, non-inferiority, phase 3 trial. Lancet HIV 2020; 7: e16–26. [DOI] [PubMed] [Google Scholar]
- 6.Wong A, Goldstein D, Mallolas J et al. Efficacy and safety of doravirine/lamivudine/tenofovir disoproxil fumarate (DOR/3TC/TDF) in treatment-naive adults with HIV-1 and transmitted nonnucleoside reverse transcriptase inhibitor resistance mutations. J Acquir Immune Defic Syndr 2019; 82: e47–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wensing AM, Calvez V, Ceccherini-Silberstein F et al. 2019 Update of the drug resistance mutations in HIV-1. Top Antivir Med 2019; 27: 111–21. [PMC free article] [PubMed] [Google Scholar]
- 8.Lai MT, Feng M, Falgueyret JP et al. In vitro characterization of MK-1439, a novel HIV-1 nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother 2014; 58: 1652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng M, Sachs NA, Xu M et al. Doravirine suppresses common nonnucleoside reverse transcriptase inhibitor-associated mutants at clinically relevant concentrations. Antimicrob Agents Chemother 2016; 60: 2241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asante-Appiah E, Lai J, Li Q et al. Doravirine resistance profile in clinical isolates and impact of baseline NNRTI resistance-associated mutations observed in treatment-naïve participants from phase 3 clinical trials. Oral Abstracts from the 23rd International AIDS Conference, 6–10 July 2020. Abstract PDB0406. https://onlinelibrary.wiley.com/doi/10.1002/jia2.25547.
- 11.Feng M, Wang D, Grobler JA et al. In vitro resistance selection with doravirine (MK-1439), a novel nonnucleoside reverse transcriptase inhibitor with distinct mutation development pathways. Antimicrob Agents Chemother 2015; 59: 590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith SJ, Pauly GT, Akram A et al. Rilpivirine and doravirine have complementary efficacies against NNRTI-resistant HIV-1 mutants. J Acquir Immune Defic Syndr 2016; 72: 485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterrantino G, Borghi V, Callegaro AP et al. Prevalence of predicted resistance to doravirine in HIV-1-positive patients after exposure to non-nucleoside reverse transcriptase inhibitors. Int J Antimicrob Agents 2019; 53: 515–19. [DOI] [PubMed] [Google Scholar]
- 14.Soulie C, Santoro MM, Storto A et al. Prevalence of doravirine-associated resistance mutations in HIV-1-infected antiretroviral-experienced patients from two large databases in France and Italy. J Antimicrob Chemother 2020; 75: 1026–30. [DOI] [PubMed] [Google Scholar]
- 15.Guerrero-Beltrán C, Martínez-Sanz J, Álvarez M et al. The algorithm used for the interpretation of doravirine transmitted drug resistance strongly influences clinical practice and guideline recommendations. J Antimicrob Chemother 2020; 75: 1294–300. [DOI] [PubMed] [Google Scholar]
- 16.Balamane M, Varghese V, Melikian GL et al. Panel of prototypical recombinant infectious molecular clones resistant to nevirapine, efavirenz, etravirine, and rilpivirine. Antimicrob Agents Chemother 2012; 56: 4522–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saladini F, Giannini A, Boccuto A et al. Agreement between an in-house replication competent and a reference replication defective recombinant virus assay for measuring phenotypic resistance to HIV-1 protease, reverse transcriptase, and integrase inhibitors. J Clin Lab Anal 2018; 32: e22206.. [DOI] [PMC free article] [PubMed] [Google Scholar]