Summary
Background
The interleukin (IL)‐23/IL‐17 immune axis is of central importance in psoriasis. However, the impact of IL‐17 family cytokines other than IL‐17A in psoriasis has not been fully established.
Objectives
To elucidate the contribution of IL‐17 family cytokines in psoriasis.
Methods
To address the expression and localization of IL‐17 family cytokines, lesional and nonlesional skin samples from patients with psoriasis were analysed by several complementary methods, including quantitative polymerase chain reaction, immunoassays, in situ hybridization and immunohistochemistry. Mechanistic studies assessing the functional activity of IL‐17 family cytokines were performed using ex vivo cultured human skin biopsies and primary human keratinocytes.
Results
We demonstrated that IL‐17A, IL‐17F, IL‐17A/F and IL‐17C are expressed at increased levels in psoriasis lesional skin and induce overlapping gene expression responses in ex vivo cultured human skin that correlate with the transcriptomic signature of psoriasis skin. Furthermore, we showed that brodalumab, in contrast to ixekizumab, normalizes gene expression responses induced by the combination of IL‐17A, IL‐17F, IL‐17A/F and IL‐17C in human keratinocytes.
Conclusions
Several IL‐17 ligands signalling through IL‐17RA are overexpressed in psoriasis skin and induce similar psoriasis‐related inflammatory pathways demonstrating their relevance in relation to therapeutic intervention in psoriasis.
Short abstract
What is already known about this topic?
The key role of interleukin (IL)‐17A in psoriasis is well established.
Previous studies have shown that IL‐17A, IL‐17F and IL‐17C are overexpressed in psoriasis skin, whereas contradictory results have been published for IL‐17E.
IL‐17 family cytokines induce secretion of inflammatory mediators such as antimicrobial peptides, chemokines and cytokines involved in the pathophysiology of psoriasis.
What does this study add?
Levels of IL‐17A/F are increased in lesional psoriasis skin but markedly lower than IL‐17A and IL‐17F.
In ex vivo cultured human skin, a physiologically relevant model, IL‐17A, IL‐17F, IL‐17A/F and IL‐17C show functional redundancy in shaping the psoriasis transcriptome.
IL‐17RA antagonism normalizes expression of psoriasis‐related genes in keratinocytes induced by the combination of IL‐17 family cytokines.
What is the translational message?
Overexpression and functional redundancy of IL‐17 family cytokines in psoriasis may explain why some patients with psoriasis with primary or secondary failure of response to secukinumab or ixekizumab achieve a clinical response after switching to brodalumab.
Linked Comment: M. Sugaya. Br J Dermatol 2021; 185:483.
The development of targeted treatments for psoriasis has demonstrated a key role of the interleukin (IL)‐23/IL‐17 pathway.1 The IL‐17 family cytokines consist of six distinct homodimers (IL‐17A–F), as well as an IL‐17A/F heterodimer.2, 3 IL‐17 ligands signal through heterodimeric receptor complexes. Of the IL‐17 receptors (IL‐17R), IL‐17RA has emerged as a common co‐receptor for IL‐17A, IL‐17A/F, IL‐17F, IL‐17C and IL‐17E, and interacts with another receptor subunit (IL‐17RB, IL‐17RC, IL‐17RD or IL‐17RE) to confer ligand‐ and cell type‐dependent signalling specificity.4
Previous studies have shown that IL‐17A, IL‐17F and IL‐17C are overexpressed in psoriasis skin,5, 6, 7, 8, 9 whereas contradictory results have been published on whether IL‐17E is expressed or increased in psoriasis skin.6, 7, 10, 11 IL‐17A is a central pathogenic cytokine in psoriasis as shown by the substantial clinical efficacy achieved in patients treated with secukinumab and ixekizumab, antibodies specifically targeting IL‐17A.12, 13 Recently, phase III studies with bimekizumab, targeting both IL‐17A and IL‐17F, have shown high efficacy, indicating that IL‐17F also contributes to driving skin inflammation in psoriasis.14, 15 Moreover, combined inhibition of several IL‐17 family cytokines with brodalumab, targeting IL‐17RA, has been shown in clinical studies to be very efficacious.16, 17, 18
IL‐17A, IL‐17A/F and IL‐17F, primarily produced by T helper (Th) 17 cells, act on a range of tissue cells, of which keratinocytes and fibroblasts are the main target cells in the skin.19 In contrast, IL‐17C functions primarily in an autocrine manner acting on epithelial cells, including keratinocytes.20 IL‐17A, IL‐17F and IL‐17C act in synergy with other cytokines, such as tumour necrosis factor (TNF) or IL‐1, to induce expression of antimicrobial peptides, chemokines and proinflammatory cytokines, which promote innate immune responses, the recruitment of inflammatory cells, and keratinocyte activation and proliferation driving the psoriatic inflammation.1, 7, 20, 21, 22 Furthermore, interplay between IL‐17A and IL‐17F and IL‐17C has been described.23 IL‐17A and IL‐17F, as well as IL‐17C, induce expression of IL‐17C in keratinocytes.7, 21, 22 IL‐17C may thereby amplify the inflammatory response by further increasing the production of inflammatory mediators, including C‐C motif chemokine ligand 20 (CCL20), which attracts IL‐17‐producing T cells. Additionally, IL‐17C was shown directly to induce expression of IL‐17A and IL‐17F in mouse Th17 cells.24 IL‐17E has mostly been associated with type 2 immune responses, but has also been reported to affect keratinocyte proliferation and mediate the recruitment of neutrophils.10, 25, 26
In this study, we analysed the mRNA and protein expression pattern of the IL‐17 family cytokines in psoriasis skin with several complementary methods. Furthermore, we investigated the biological responses of the IL‐17 family cytokines in ex vivo cultured human skin and their modulation by brodalumab and ixekizumab in keratinocyte cultures. Our findings demonstrate that IL‐17A, IL‐17F, IL‐17A/F and IL‐17C are upregulated in psoriasis skin and show functional redundancy in driving skin inflammation in psoriasis.
Materials and methods
Skin specimens from healthy participants and patients with psoriasis
Anonymized human skin samples were obtained from healthy participants following reconstructive surgery and from four different cohorts of patients with moderate‐to‐severe psoriasis (in addition, the second cohort also included a patient with mild psoriasis). Skin samples were donated after informed written consent was provided. All skin material was sampled in accordance with national legislation in the country of origin. Paired biopsies of lesional and nonlesional psoriasis skin were used for gene expression analysis by quantitative polymerase chain reaction [qPCR; cohort 1 (n = 10) and cohort 3 (n = 6)], cytokine protein analysis (cohorts 2 and 4, both n = 8) and histological analysis (cohort 2, n = 8). Each punch biopsy in cohort 2 was divided in half and used for cytokine and histological analysis.
Human skin cultures and cytokine stimulation
For ex vivo culture, skin samples from healthy participants were stored at 4–8 °C and processed within 24 h of surgery. Full‐thickness (3‐mm) punch biopsies (five replicates per treatment) were excised and placed in supplemented EpiLife medium (ThermoFisher Scientific, Waltham, MA, USA) in 96‐well tissue‐culture plates in a humidified incubator at 37 °C with 5% CO2. Stimulation was performed for 24 h with IL‐17 family cytokines as indicated.
Primary human keratinocytes were isolated from the skin of healthy adult donors (Appendix S1; see Supporting Information) and cultured in supplemented EpiLife medium (ThermoFisher Scientific) in a humidified incubator at 37 °C with 5% CO2.
Cells were stimulated for 48 h with a combination of IL‐17 family cytokines in the presence of TNF, as indicated. For all samples treated with brodalumab, cells were preincubated with brodalumab for 30 min at 37 °C prior to cytokine addition. For all samples treated with ixekizumab, ixekizumab was preincubated with cytokine mixtures for 30 min at 37 °C before addition to the cells. Recombinant human cytokines were purchased from R&D Systems [Minneapolis, MN, USA (IL‐17A: #317‐ILB; IL‐17C: #1234‐IL; IL‐17E: # 1258‐IL; IL‐17F: #1335‐IL; IL‐17A/F: #5194‐IL; TNF: 210‐TA)].
Histology on psoriasis biopsies
Single‐ and triple‐fluorescence in situ hybridization (ISH) were performed with RNAscope® (Bio‐Techne, Minneapolis, MN, USA). For multiplex fluorescence, tyramide signal amplification plus conjugated fluorophores was applied: Cy5 (CD3), Cy3 (IL17F) and fluorescein isothiocyanate (IL17A). Immunohistochemistry (IHC) was performed with goat antihuman IL‐17A (R&D Systems), goat antihuman IL‐17C (R&D Systems) and mouse antihuman neutrophil elastase (Agilent, Santa Clara, CA, USA). The primary antibodies for IL‐17A and IL‐17C were detected with BOND Polymer Refine Detection with rabbit antigoat (Agilent) as the secondary antibody and DAB as the chromogen. Neutrophil elastase was detected with BOND Polymer Refine RED Detection, with fast red as the chromogen. All stains were performed on a Leica BOND RX (Leica Biosystems, Wetzlar, Germany). Whole‐slide image analysis was performed with Visiopharm Integrator Software (Visiopharm, Hoersholm, Denmark). Further details can be found in Appendix S1.
Cytokine protein analysis on psoriasis biopsies
IL‐17F protein levels were measured by enzyme‐linked immunosorbent assay (ELISA), using the Human IL‐17F DuoSet ELISA Kit (R&D Systems). The MSD platform (Meso Scale Discovery, Rockville, MD, USA) was used for measurement of IL‐17A/F and IL‐17E (Human U‐PLEX, individual assay for IL‐17A/F and IL‐17E) and IL‐17A and IL‐17C (Human U‐PLEX, multiplex assay). Further details can be found in Appendix S1.
Quantitative real‐time polymerase chain reaction analysis
RNA extraction from lesional and nonlesional psoriasis skin and from keratinocyte cultures was followed by cDNA synthesis, and amplification of cDNA by quantitative real‐time PCR using Taqman® Gene Expression Assays (see Table S1).
Gene array and bioinformatics analyses
Gene expression profiling of ex vivo cultured human skin biopsies was carried out on whole‐transcript arrays (GeneChip™ Human Gene 2·1 ST Array; Thermo Fisher Scientific). Differential expression analysis was performed with the respective contrasts stimulated vs. unstimulated, using the moderated t‐test of the R/limma package. Correlation analysis was carried out using the Pearson correlation coefficients (r) of the R/psych package. Further details are available in Appendix S1.
Results
Increased expression of interleukin (IL)‐17A, IL‐17F, IL‐17A/F and IL‐17C in lesional psoriasis skin
To elucidate the relevance of individual IL‐17 family cytokines in psoriasis, their expression levels were investigated in lesional and nonlesional psoriasis skin at both the mRNA and protein level. Lesional skin had increased mRNA levels of IL17A, IL17C and IL17F and decreased mRNA levels of IL17B, IL17D and IL17E (IL25) vs. nonlesional skin as shown in two independent patient cohorts (Figure 1a and Figure S1a; see Supporting Information). IL17E and its specific receptor subunit IL17RB had the lowest expression in lesional skin of the IL‐17 ligands and the IL17R subunits, respectively (Figure 1a and Figure S1b).
Figure 1.
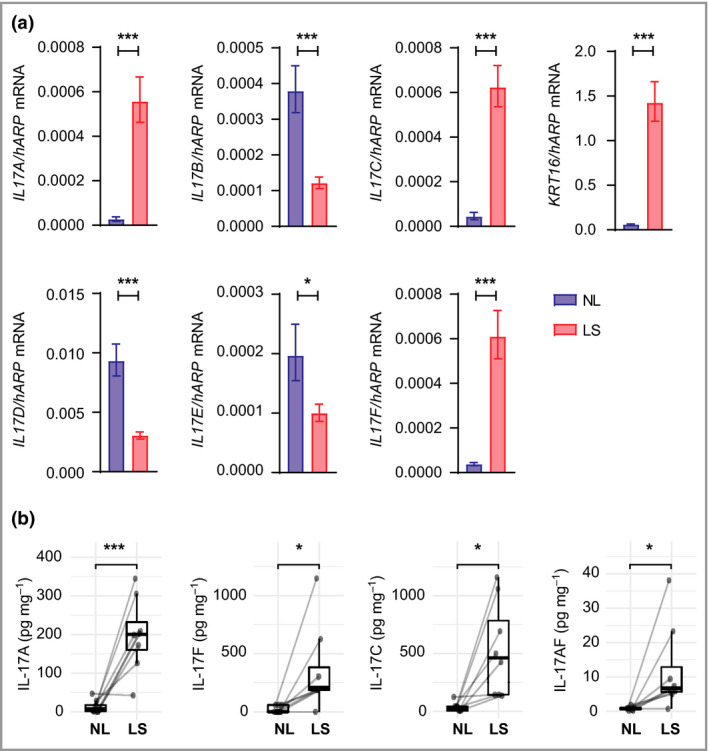
Interleukin (IL)‐17A, IL‐17F, IL‐17A/F and IL‐17C are overexpressed in lesional psoriasis skin. (a) Gene expression of IL17 family cytokines and KRT16 (positive control) in paired samples of lesional and nonlesional skin (n = 10) analysed by quantitative polymerase chain reaction. hARP (RPLP0) was used as the reference gene for normalization. Bars show mean mRNA expression (2ΔCt) (SEM). P‐values were calculated via marginal means (i.e. least square means) and were Benjamini–Hochberg adjusted for multiple testing. (b) Protein levels of IL‐17 family cytokines measured in paired samples of lesional and nonlesional skin (n = 8) using enzyme‐linked immunosorbent assay or MSD kits (Meso Scale Discovery). Protein levels (pg mg–1 total protein) are plotted as means with lower and upper hinges corresponding to the 25th and 75th percentiles. P‐values were calculated using a linear mixed‐effects model. *P < 0·05, **P < 0·01 and ***P < 0·001. LS, lesional skin; NL, nonlesional skin.
In line with the mRNA expression pattern, protein levels of IL‐17A, IL‐17C and IL‐17F, as well as IL‐17A/F, were significantly increased in lesional skin compared with nonlesional skin (Figure 1b and Figure S1c). While the levels of IL‐17A, IL‐17F and IL‐17C varied between the patients, IL‐17C levels were typically higher than those of IL‐17A and IL‐17F. In addition, protein levels of IL‐17A/F were markedly lower than those of IL‐17A or IL‐17F in all patients. We were unable to detect IL‐17E protein in any of these samples, likely owing to low IL17E mRNA levels.
Distinct localization patterns of interleukin‐17 family cytokines in psoriatic skin
Next, we investigated the localization and identity of the cells producing IL‐17RA and all associated IL‐17 ligands in psoriasis skin by ISH. IL17RA mRNA was expressed by keratinocytes in all viable layers of the epidermis, and also present in the papillary and reticular dermis (Figure 2a). IL17A and IL17F mRNA were predominantly present in cells in the epidermis and papillary dermis, and only occasionally in inflammatory infiltrates in the reticular dermis of lesional skin. The IL17A‐ and IL17F‐expressing cells were few in number, showing high levels of mRNA for the cytokine and morphologically resembling T cells (Figure 2b, c). T‐cell identity was confirmed by triple‐fluorescence ISH for IL17A, IL17F and CD3 (four patients), with the vast majority of the IL17A‐ and IL17F‐expressing cells showing co‐expression of CD3 (Figure 2d). Furthermore, IL17F was almost always co‐expressed with IL17A, whereas IL17A, to a lesser extent, was co‐expressed with IL17F in the psoriasis skin samples (data not shown). IL17C mRNA was highly expressed in lesional skin by clusters of differentiated keratinocytes in the upper layers of epidermis (Figure 2e). Interestingly, we found that IL17E mRNA was only expressed by few keratinocytes often associated with hair follicles in lesional (Figure 2f) and nonlesional skin. Quantitative analysis of the ISH staining indicated very low expression of IL17A, IL17C, IL17E and IL17F in nonlesional skin and a significant increase in IL17A, IL17F and IL17C, but not IL17E, mRNA in lesional skin (Figure 2g), as was also observed by qPCR analysis (Figure 1a and Figure S1a). Furthermore, we found a strong correlation between mRNA levels by ISH and protein levels in lesional skin samples from the same patients, indicating that the elevated mRNA expression of IL17A, IL17F and IL17C directly led to increased cytokine production in the skin (Figure 3a). Interestingly, when we compared IL17A mRNA and protein expression on consecutive sections of lesional skin (n = 8), we observed that while IL17A mRNA and protein are found in few cells in the epidermis and papillary dermis, the majority of cells that stained positive for IL‐17A protein localized to the reticular dermis (Figure 3b, c). In addition, neutrophils (i.e. in Munro abscesses) staining positive for neutrophil elastase also stained positive for IL‐17A protein in the absence of IL17A mRNA (Figure 3d–f). However, IL17C mRNA and protein co‐localized in keratinocytes in the upper epidermis (Figure 3g, h).
Figure 2.
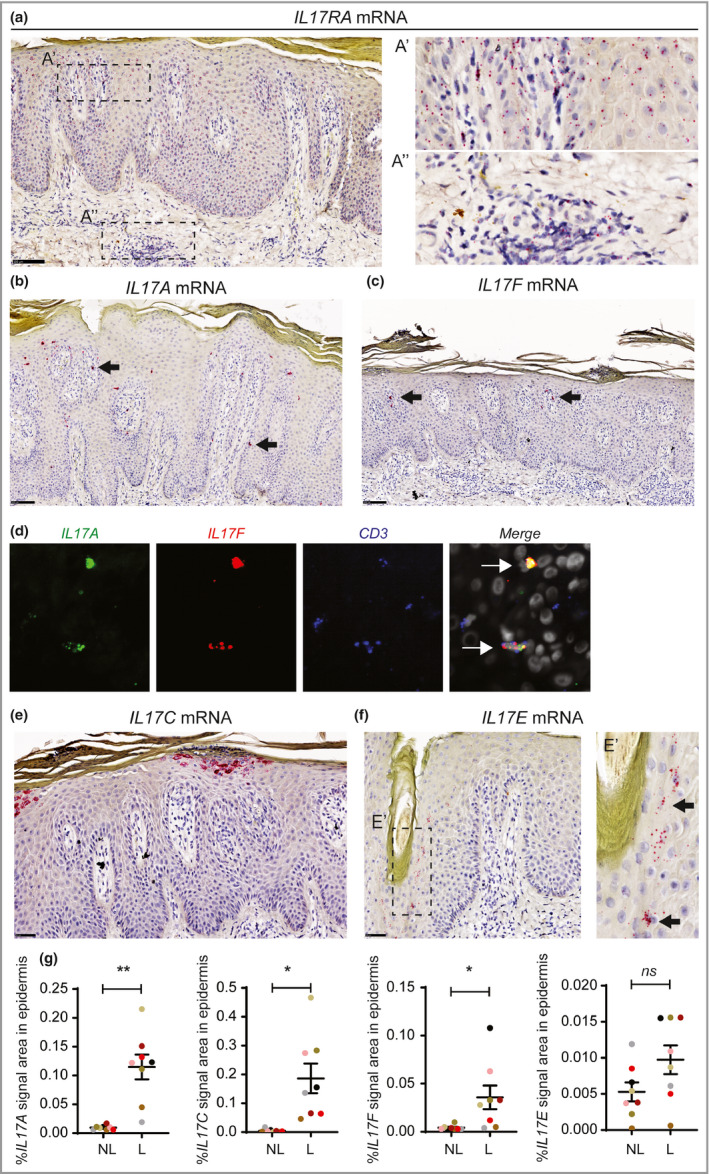
Cells expressing IL17A, IL17F and IL17C mRNA are predominantly localized in the epidermis of psoriatic skin. In situ hybridization (ISH) on lesional skin sections detecting (a) IL17RA mRNA, (b) IL17A mRNA and (c) IL17F mRNA. (d) Co‐expression of IL17A and IL17F by CD3+ T cells, as illustrated by triple‐fluorescence ISH. (e, f) As in (a–c), but for IL17C and IL17E mRNA, respectively. (g) Quantification of IL17A, IL17C, IL17F and IL17E mRNA expression by ISH in lesional and nonlesional psoriasis skin, as percentage signal area in the epidermis. Mean values (SEM) were calculated, and statistical analysis was performed by two‐sided paired t‐tests. Scale bars for (a–c) and (e, f) are 100 µm and 50 µm, respectively. Arrows in (b–d and f) point to locations with positive cells. *P < 0·05 and **P < 0·01. L, lesional; NL, nonlesional.
Figure 3.
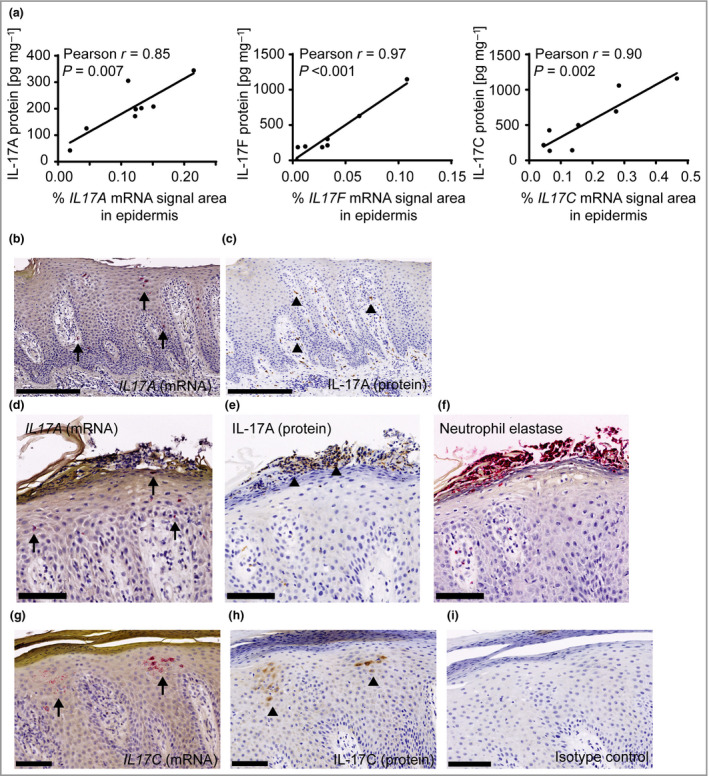
Expression of interleukin (IL)‐17 family cytokines in psoriasis skin by in situ hybridization (ISH) and immunohistochemistry (IHC). (a) Correlation analysis between cytokine levels measured by quantification of ISH and protein concentrations in skin lysates from matched samples (n = 8). (b) IL17A mRNA expression by ISH. (c) Consecutive section of (b) showing IL‐17A protein expression by IHC. (d) IL17A mRNA expression by ISH. (e) Consecutive section of (d) showing IL‐17A protein expression by IHC. (f) Consecutive section of (d and e) showing neutrophil elastase expression by IHC. (g) IL17C mRNA expression by ISH. (h) Consecutive section of (g) showing IL‐17C protein expression by IHC. (i) Negative control staining using an isotype control antibody. Scale bars for (b) and (c) are 250 µm, and scale bars for (d–i) are 100 µm. Arrows (IL17 mRNA) and arrowheads (IL‐17protein) in (b–i) point to locations with positive cells.
Global gene expression induced by interleukin (IL)‐17A, IL‐17F, IL‐17A/F and IL‐17C in human skin overlaps with the psoriasis gene signature
To investigate the contribution of the different IL‐17 family cytokines in psoriasis, we stimulated ex vivo cultured human skin biopsies for 24 h with the cytokines as indicated (Figure 4a). Global gene expression profiling was performed using Affymetrix arrays to allow direct comparison of the cytokine‐induced differentially expressed genes (DEGs; vs. untreated controls) with psoriasis DEGs (comparing lesional to nonlesional skin) using the meta‐analysis derived psoriasis (MAD3‐PSO) transcriptome.27 IL‐17A, IL‐17A/F and IL‐17F induced dose‐dependent and qualitatively comparable gene expression responses, and increased expression of most of the top‐50 upregulated MAD3‐PSO genes (Figure 4a and Table S2; see Supporting Information). IL‐17C induced a more modest response, but, overall, affected the same upregulated MAD3‐PSO genes as for IL‐17A, IL‐17A/F, and IL‐17F. When we correlated all upregulated MAD3‐PSO genes (log2 fold change > 1) to the cytokine‐induced gene expression profiles (using the highest concentration at 100 ng mL–1), a significant correlation was found for IL‐17A (r = 0·35), IL‐17F (r = 0·39), IL‐17A/F (r = 0·40) and IL‐17C (r = 0·26; all P < 0·001) (Figure 4b). As exemplified by a panel of selected genes, the dose‐dependent responses illustrate the difference in potency among the IL‐17 family cytokines, with IL‐17A showing the highest potency (Figure 4c–h).
Figure 4.
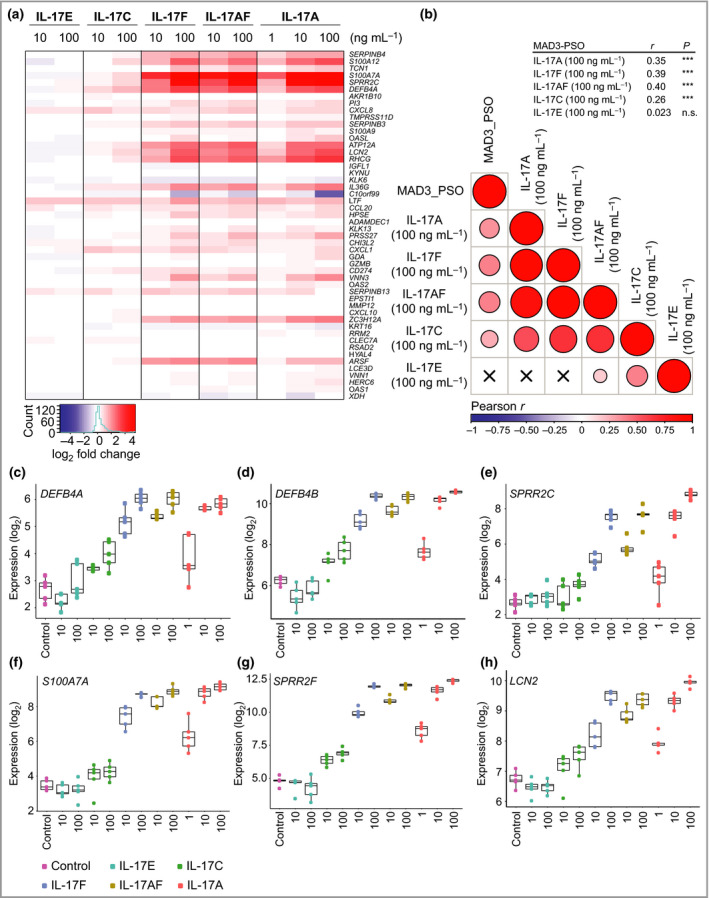
Interleukin (IL)‐17A, IL‐17F, IL‐17A/F and IL‐17C are redundant in shaping the psoriasis transcriptome. Ex vivo cultured human skin biopsies (five replicates per treatment condition) were incubated for 24 h with individual IL‐17 family cytokines at the indicated concentrations and subjected to global gene expression analysis using Affymetrix Human Gene ST 2·1 arrays. (a) Ranked heatmap of the top‐50 MAD3‐PSO genes and their expression (log2 fold change vs. untreated ex vivo skin cultures). (b) A correlation matrix of gene expression signatures induced by individual IL‐17 family cytokines (at 100 ng mL–1) in ex vivo cultured human skin vs. upregulated transcripts in the meta‐analysis derived psoriasis transcriptome (MAD3‐PSO) dataset (log2 fold change > 1). The exact correlation score (r) and significance can be seen at the top right of the table. Crosses indicate that no significant correlation was found. (c–h) Examples of genes induced by individual IL‐17 family cytokines in ex vivo cultured human skin: DEFB4A, DEFB4B, SPRR2C, S100A7A, SPRR2F and LCN2, respectively. ***P < 0·001. n.s., not significant.
With regard to IL‐17E, no clear effect on gene expression was seen in ex vivo cultured human skin (Figure 4a and Table S3; see Supporting Information), but, as expected, IL‐17E was able to induce effects in peripheral blood mononuclear cells (Figure S2a; see Supporting Information). In alignment with this finding, expression of IL17RB was markedly lower than the other IL17R subunits in the ex vivo skin cultures (Figure S2b, c), as was also found in psoriasis skin biopsies (Figure S1b).
Taken together, our data suggest that IL‐17A, IL‐17A/F, IL‐17F and IL‐17C qualitatively regulate the same subset of psoriasis‐relevant genes and can thus be considered functionally redundant in shaping the psoriasis transcriptome.
Interleukin (IL)‐17 receptor A antagonism normalizes psoriasis‐relevant IL‐17 family cytokine signalling
Based on the overexpression and functional redundancy of IL‐17A, IL‐17F, IL‐17A/F and IL‐17C in psoriasis, we next investigated the impact of IL‐17RA antagonism vs. neutralization of IL‐17A on gene expression induced by the combination of IL‐17 family cytokines (using approximately equipotent concentrations) and TNF in primary human keratinocytes (Figure 5). The combined stimulation with IL‐17 family cytokines in the presence of TNF led to a strong induction of DEFB4, S100A7A, CXCL8, LCN2 and SPRR2F (Figure 5). Treatment with ixekizumab (anti‐IL‐17A antibody) was able to inhibit gene expression to a comparable level induced by the combination of IL‐17F, IL‐17C and TNF. However, treatment with brodalumab (anti‐IL‐17RA antibody) blocked the signalling of all the IL‐17 family cytokines and was able to inhibit their induced gene expression approximately to the levels induced by TNF alone (Figure 5). These findings indicate that IL‐17A, IL‐17F, IL‐17A/F and IL‐17C make use of IL‐17RA as functional receptor and illustrate the differences in mode of action of IL‐17A neutralizing antibodies (e.g. ixekizumab) and the IL‐17RA blocking antibody brodalumab.
Figure 5.
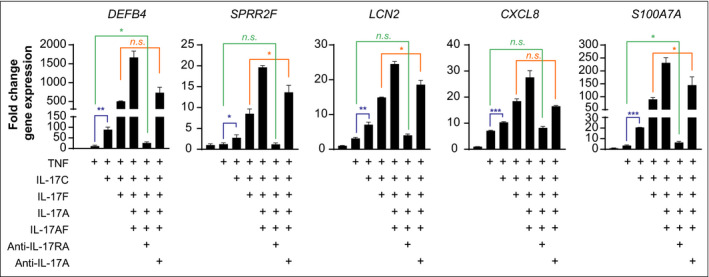
Interleukin (IL)‐17RA antagonism normalizes psoriasis‐relevant IL‐17 family cytokine signalling in vitro. Gene expression of DEFB4, SPRR2F, LCN2, CXCL8 and S100A7A by quantitative polymerase chain reaction of primary human keratinocytes treated for 48 h with a combination of IL‐17 family cytokines (IL‐17A, 1 ng mL–1; IL‐17F, 10 ng mL–1; IL‐17A/F, 10 ng mL–1; IL‐17C, 100 ng mL–1) and tumour necrosis factor (10 ng mL–1) in the absence or presence of brodalumab (anti‐IL‐17RA, 500 µg mL–1) or ixekizumab (anti‐IL‐17A, 500 µg mL–1) as indicated. RPLP0, PPIA and ACTB were used as reference genes for normalization. Bars show mean (SD) fold change in gene expression vs. unstimulated samples (2ΔΔCt). Statistical analysis was performed by unpaired t‐tests. *P < 0·05, **P < 0·01 and ***P < 0·001. n.s., not significant.
Discussion
Our study provides a comprehensive analysis by using several complementary methods (qPCR, immunoassays, ISH and IHC) to investigate thoroughly the expression and localization of IL‐17 family cytokines in psoriasis skin. Notably, for the first time, the gene and protein levels of all IL‐17RA‐associated IL‐17 ligands were assessed on the same psoriasis skin biopsies. By the different methods employed, we found that levels of IL‐17A, IL‐17F and IL‐17C, but not IL‐17E, were significantly increased in lesional psoriasis skin, and correlated at the mRNA and protein levels. In our analysis, protein levels of IL‐17C were typically higher than those of IL‐17A and IL‐17F, whereas levels of IL‐17A/F were markedly lower in lesional skin biopsies. While protein levels of IL‐17A/F in psoriasis skin have not been published previously, other studies have described increased levels of IL‐17A, IL‐17F and IL‐17C at the mRNA and/or protein level, although protein levels of these cytokines in psoriasis skin vary widely between studies.5, 6, 7, 8, 9 This may be explained by differences related to local disease severity of the biopsy site, processing of the biopsies (e.g. protein recovery), detection systems and data normalization.
Although we were able to detect modest IL17E mRNA expression by ISH in keratinocytes, mostly in locations next to hair follicles, we did not find increased levels of IL‐17E in lesional psoriasis skin by any of our methods, which is in contrast to findings from two independent studies.10, 11 The reason for this discrepancy is not entirely clear but is possibly related to different psoriasis subtypes, sample processing and/or specificity of detection methods used in the studies. Levels of IL‐17B and IL‐17D were only assessed by qPCR and, in line with previous reports, were found to be downregulated in lesional psoriasis skin biopsies.6, 7
Using multiplex ISH staining, we confirmed CD3+ T cells as the main IL17A and IL17F expressing cells. Interestingly, by comparing ISH and IHC analysis of consecutive skin sections, we found that neutrophils and cells in the dermis stained positive for IL‐17A protein in the absence of IL17A mRNA expression. While still being a matter of controversy, our findings, although based on a limited number of psoriasis skin sections, suggest that neutrophils accumulate and possibly internalize IL‐17A but seem unable to produce IL‐17A, as also supported by other studies.28, 29, 30
Transcriptional profiling of IL‐17 family cytokines have been addressed to some extent in previous studies using keratinocyte monolayer cultures or reconstructed epidermis models, but, to our knowledge, no global gene expression analysis has been generated that directly compares the different IL‐17 ligands in a skin‐relevant model system. We performed a global gene expression analysis comparing the IL‐17 ligands in ex vivo cultured human skin biopsies. The use of whole‐skin tissue allowed us to capture gene responses from different cells present in the skin and permits potential integrative or interactive effects between the different cells and cytokines that are induced. We found that the global gene expression profiles induced by IL‐17A, IL‐17A/F, IL‐17F and, to a lower extent, IL‐17C, showed a qualitatively comparable induction of genes, encoding inflammatory mediators such as antimicrobial peptides, chemokines and cytokines involved in the pathophysiology of psoriasis. Moreover, these gene expression signatures were very similar to most common upregulated genes in psoriasis and correlated significantly with the MAD3‐PSO dataset.27 The relatively weak responses induced by IL‐17C in vitro were also reported previously, not the least by comparison with the effects of IL‐17C shown in mouse models.7 While whole‐skin tissue is a more physiologically relevant model system for determining the effects of IL‐17 family cytokines in cutaneous biology, a limitation is the lack of activated immune cells. Thus, in this model system, keratinocyte‐induced mediators [e.g. CCL20 and C‐X‐C motif chemokine ligand 8 (CXCL8)] do not permit amplification of the inflammatory response by leucocytes, thus the induced phenotype may under‐represent the actual psoriasis‐like response that is generated in response to IL‐17 family cytokines.
Although specifically targeting IL‐17A can be a beneficial way to interrupt effectively the vicious circle of inflammation in a large proportion of patients with psoriasis, this approach may not sufficiently suppress the inflammatory pathways in all patients owing to overexpression and functional redundancy of IL‐17 family cytokines. This is supported by our mechanistic study in keratinocytes, which showed that brodalumab, but not ixekizumab, was able to normalize expression of psoriasis‐related genes induced by the combined IL‐17 family cytokines overexpressed in psoriasis. Translating this to a clinical setting, it may explain why some patients with psoriasis with primary or secondary failure of response to secukinumab or ixekizumab achieved a clinical response after switching to brodalumab, as reported in several studies.31, 32, 33, 34 A broader inhibition of IL‐17 family cytokines, provided by brodalumab, as well as bimekizumab, could provide a favourable mechanism of action in psoriasis treatment options.
Recently, a potential mechanism of IL‐17RA‐independent signalling for IL‐17F has been suggested.35 As brodalumab is able to inhibit fully IL‐17A‐, IL‐17F‐, IL‐17A/F‐ and IL‐17C‐induced gene expression in vitro and normalizes gene expression signatures in patients with psoriasis,16 the biological ramifications of noncanonical IL‐17 signalling pathways independent of IL‐17RA are currently unclear. Nevertheless, future studies to delineate the function of individual IL‐17 family cytokines, and through which receptors and under what circumstances they are able to signal, will yield valuable additional information on the molecular mechanisms of signalling by IL‐17 family cytokines in health and disease.
Author Contribution
Maxim A.X. Tollenaere: Data curation (lead); Formal analysis (equal); Investigation (equal); Methodology (equal); Visualization (lead); Writing‐original draft (equal); Writing‐review & editing (equal). Josephine Hebsgaard: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Resources (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). David Adrian Ewald: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Resources (equal); Software (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Paola Lovato: Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Sandra Garcet: Data curation (equal); Formal analysis (equal); Investigation (equal); Writing‐review & editing (supporting). Xuan Li: Investigation (equal); Methodology (equal); Writing‐review & editing (supporting). Steven Dennis Pilger: Formal analysis (equal); Methodology (supporting); Software (equal); Visualization (equal); Writing‐review & editing (supporting). Minna L Tiirikainen: Investigation (supporting); Writing‐review & editing (supporting). Malene Bertelsen: Formal analysis (supporting); Investigation (supporting); Methodology (supporting); Resources (supporting); Writing‐review & editing (supporting). James G Krueger: Conceptualization (equal); Investigation (supporting); Methodology (equal); Resources (supporting); Supervision (supporting); Writing‐original draft (supporting); Writing‐review & editing (supporting). Hanne Norsgaard: Conceptualization (lead); Data curation (equal); Investigation (supporting); Methodology (supporting); Project administration (lead); Supervision (equal); Writing‐original draft (lead); Writing‐review & editing (lead).
Supporting information
Appendix S1 Supplementary materials and methods.
Figure S1 Expression of interleukin‐17 family cytokines and receptors in psoriasis skin biopsies.
Figure S2 Bioactivity of interleukin‐17E in peripheral blood mononuclear cells and IL17R expression levels in ex vivo cultured human skin.
Table S1 Probes and primers used in quantitative polymerase chain reaction‐based gene expression analysis.
Table S2 Regulation of the top‐50 upregulated MAD3‐PSO genes by interleukin‐17 family cytokines.
Table S3 Differential gene expression in ex vivo cultured skin stimulated with interleukin‐17 family cytokines.
Acknowledgments
The authors would like to thank the technicians of the in vitro biology department and histology team at LEO Pharma Skin Research for technical assistance.
Funding sources LEO Pharma A/S funded this study.
Conflicts of interest M.A.X.T., J.H., D.A.E., P.L., S.D.P., M.L.T., M.B. and H.N. are employees of LEO Pharma. J.G.K. received grants paid to his institution from Novartis, Pfizer, Amgen, Lilly, Boehringer, Innovaderm, BMS, Janssen, AbbVie, Paraxel, LEO Pharma, Vitae, Akros, Regeneron, Allergan, Novan, Biogen MA, Sienna, UCB, Celgene, Botanix, Incyte, Avillion and Exicure; and personal fees from Novartis, Pfizer, Amgen, Lilly, Boehringer, Biogen Idec, AbbVie, LEO Pharma, Escalier, Valeant, Aurigene, Allergan, Asana, UCB, Sienna, Celgene, Nimbus, Menlo, Aristea, Sanofi, Sun Pharma, Almirall, Arena and BMS.
Data Availability Statement The gene array dataset described in this publication has been deposited in NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE158448 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158448).
References
- 1.Hawkes JE, Yan BY, Chan TCet al. Discovery of the IL‐23/IL‐17 signaling pathway and the treatment of psoriasis. J Immunol 2018; 201:1605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright JF, Guo Y, Quazi Aet al. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J Biol Chem 2007; 282:13447–55. [DOI] [PubMed] [Google Scholar]
- 3.Chang SH, Dong C. A novel heterodimeric cytokine consisting of IL‐17 and IL‐17F regulates inflammatory responses. Cell Res 2007; 17:435–40. [DOI] [PubMed] [Google Scholar]
- 4.Monin L, Gaffen SL. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harb Perspect Biol 2018; 10:a028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson NJ, Boniface K, Chan JRet al. Development, cytokine profile and function of human interleukin 17‐producing helper T cells. Nat Immunol 2007; 8:950–7. [DOI] [PubMed] [Google Scholar]
- 6.Johansen C, Usher PA, Kjellerup RBet al. Characterization of the interleukin‐17 isoforms and receptors in lesional psoriatic skin. Br J Dermatol 2009; 160:319–24. [DOI] [PubMed] [Google Scholar]
- 7.Johnston A, Fritz Y, Dawes SMet al. Keratinocyte overexpression of IL‐17C promotes psoriasiform skin inflammation. J Immunol 2013; 190:2252–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soderstrom C, Berstein G, Zhang Wet al. Ultra‐sensitive measurement of IL‐17A and IL‐17F in psoriasis patient serum and skin. AAPS J 2017; 19:1218–22. [DOI] [PubMed] [Google Scholar]
- 9.Vandeghinste N, Klattig J, Jagerschmidt Cet al. Neutralization of IL‐17C reduces skin inflammation in mouse models of psoriasis and atopic dermatitis. J Invest Dermatol 2018; 138:1555–63. [DOI] [PubMed] [Google Scholar]
- 10.Senra L, Stalder R, Alvarez Martinez Det al. Keratinocyte‐derived IL‐17E contributes to inflammation in psoriasis. J Invest Dermatol 2016; 136:1970–80. [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Lu H, Lee YHet al. An interleukin‐25‐mediated autoregulatory circuit in keratinocytes plays a pivotal role in psoriatic skin inflammation. Immunity 2018; 48:787–98. [DOI] [PubMed] [Google Scholar]
- 12.Langley RG, Elewski BE, Lebwohl Met al. Secukinumab in plaque psoriasis – results of two phase 3 trials. N Engl J Med 2014; 371:326–38. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths CE, Reich K, Lebwohl Met al. Comparison of ixekizumab with etanercept or placebo in moderate‐to‐severe psoriasis (UNCOVER‐2 and UNCOVER‐3): results from two phase 3 randomised trials. Lancet 2015; 386:541–51. [DOI] [PubMed] [Google Scholar]
- 14.Gordon KB, Foley P, Krueger JGet al. Bimekizumab efficacy and safety in moderate to severe plaque psoriasis (BE READY): a multicentre, double‐blind, placebo‐controlled, randomised withdrawal phase 3 trial. Lancet 2021; 397:475–86. [DOI] [PubMed] [Google Scholar]
- 15.Reich K, Papp KA, Blauvelt Aet al. Bimekizumab versus ustekinumab for the treatment of moderate to severe plaque psoriasis (BE VIVID): efficacy and safety from a 52‐week, multicentre, double‐blind, active comparator and placebo controlled phase 3 trial. Lancet 2021; 397:487–98. [DOI] [PubMed] [Google Scholar]
- 16.Tomalin LE, Russell CB, Garcet Set al. Short‐term transcriptional response to IL‐17 receptor‐A antagonism in the treatment of psoriasis. J Allergy Clin Immunol 2020; 145:922–32. [DOI] [PubMed] [Google Scholar]
- 17.Lebwohl M, Strober B, Menter Aet al. Phase 3 studies comparing brodalumab with ustekinumab in psoriasis. N Engl J Med 2015; 373:1318–28. [DOI] [PubMed] [Google Scholar]
- 18.Papp KA, Reich K, Paul Cet al. A prospective phase III, randomized, double‐blind, placebo‐controlled study of brodalumab in patients with moderate‐to‐severe plaque psoriasis. Br J Dermatol 2016; 175:273–86. [DOI] [PubMed] [Google Scholar]
- 19.Krueger JG, Brunner PM. Interleukin‐17 alters the biology of many cell types involved in the genesis of psoriasis, systemic inflammation and associated comorbidities. Exp Dermatol 2018; 27:115–23. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez‐Carrozzi V, Sambandam A, Luis Eet al. IL‐17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat Immunol 2011; 12:1159–66. [DOI] [PubMed] [Google Scholar]
- 21.Chiricozzi A, Guttman‐Yassky E, Suarez‐Farinas Met al. Integrative responses to IL‐17 and TNF‐alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol 2011; 131:677–87. [DOI] [PubMed] [Google Scholar]
- 22.Lauffer F, Jargosch M, Baghin Vet al. IL‐17C amplifies epithelial inflammation in human psoriasis and atopic eczema. J Eur Acad Dermatol Venereol 2020; 34:800–9. [DOI] [PubMed] [Google Scholar]
- 23.Guttman‐Yassky E, Krueger JG. IL‐17C: a unique epithelial cytokine with potential for targeting across the spectrum of atopic dermatitis and psoriasis. J Invest Dermatol 2018; 138:1467–9. [DOI] [PubMed] [Google Scholar]
- 24.Chang SH, Reynolds JM, Pappu BPet al. Interleukin‐17C promotes Th17 cell responses and autoimmune disease via interleukin‐17 receptor E. Immunity 2011; 35:611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borowczyk J, Buerger C, Tadjrischi Net al. IL‐17E (IL‐25) and IL‐17A differentially affect the functions of human keratinocytes. J Invest Dermatol 2020; 140:1379–89. [DOI] [PubMed] [Google Scholar]
- 26.Senra L, Mylonas A, Kavanagh RDet al. IL‐17E (IL‐25) enhances innate immune responses during skin inflammation. J Invest Dermatol 2019; 139:1732–42. [DOI] [PubMed] [Google Scholar]
- 27.Tian S, Krueger JG, Li Ket al. Meta‐analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PLOS ONE 2012; 7:e44274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich K, Papp KA, Matheson RTet al. Evidence that a neutrophil‐keratinocyte crosstalk is an early target of IL‐17A inhibition in psoriasis. Exp Dermatol 2015; 24:529–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamassia N, Arruda‐Silva F, Calzetti Fet al. A reappraisal on the potential ability of human neutrophils to express and produce IL‐17 family members in vitro: failure to reproducibly detect it. Front Immunol 2018; 9:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamarozzi F, Wright HL, Thomas HBet al. A lack of confirmation with alternative assays questions the validity of IL‐17A expression in human neutrophils using immunohistochemistry. Immunol Lett 2014; 162:194–8. [DOI] [PubMed] [Google Scholar]
- 31.Gasslitter I, Kirsten N, Augustin Met al. Successful intra‐class switching among IL‐17 antagonists: a multicentre, multinational, retrospective study. Arch Dermatol Res 2019; 311:421–4. [DOI] [PubMed] [Google Scholar]
- 32.Kimmel G, Chima M, Kim HJet al. Brodalumab in the treatment of moderate to severe psoriasis in patients when previous anti‐interleukin 17A therapies have failed. J Am Acad Dermatol 2019; 81:857–9. [DOI] [PubMed] [Google Scholar]
- 33.Kromer C, Wilsmann‐Theis D, Gerdes Set al. Changing within the same class: efficacy of brodalumab in plaque psoriasis after treatment with an IL‐17A blocker – a retrospective multicenter study. J Dermatolog Treat 2020; 10.1080/09546634.2020.1716932. [DOI] [PubMed] [Google Scholar]
- 34.Yeung J, Vender R, Turchin Iet al. Brodalumab success in patients with moderate‐to‐severe psoriasis who failed previous IL17A inhibitors. J Am Acad Dermatol 2021; 84:1169–71. [DOI] [PubMed] [Google Scholar]
- 35.Goepfert A, Lehmann S, Blank Jet al. Structural analysis reveals that the cytokine IL‐17F forms a homodimeric complex with receptor IL‐17RC to drive IL‐17RA‐independent signaling. Immunity 2020; 52:499–512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary materials and methods.
Figure S1 Expression of interleukin‐17 family cytokines and receptors in psoriasis skin biopsies.
Figure S2 Bioactivity of interleukin‐17E in peripheral blood mononuclear cells and IL17R expression levels in ex vivo cultured human skin.
Table S1 Probes and primers used in quantitative polymerase chain reaction‐based gene expression analysis.
Table S2 Regulation of the top‐50 upregulated MAD3‐PSO genes by interleukin‐17 family cytokines.
Table S3 Differential gene expression in ex vivo cultured skin stimulated with interleukin‐17 family cytokines.


