Abstract
Oils and fats of vegetable and animal origin remain an important renewable feedstock for the chemical industry. Their industrial use has increased during the last 10 years from 31 to 51 million tonnes annually. Remarkable achievements made in the field of oleochemistry in this timeframe are summarized herein, including the reduction of fatty esters to ethers, the selective oxidation and oxidative cleavage of C–C double bonds, the synthesis of alkyl‐branched fatty compounds, the isomerizing hydroformylation and alkoxycarbonylation, and olefin metathesis. The use of oleochemicals for the synthesis of a great variety of polymeric materials has increased tremendously, too. In addition to lipases and phospholipases, other enzymes have found their way into biocatalytic oleochemistry. Important achievements have also generated new oil qualities in existing crop plants or by using microorganisms optimized by metabolic engineering.
Keywords: applications, catalysis, fatty acids, renewable resources, synthesis
This Review summarizes the use of oils and fats as a renewable raw material by covering novel examples, such as selective oxidation or cleavage of C–C double bonds in fatty acids, isomerizing hydroformylation and alkoxycarbonylation, and the use of these renewables for polymer chemistry. Further topics include biocatalysis, the optimization of microorganisms by metabolic engineering, and the generation of new oil qualities in existing crop plants.
1. Introduction
Oils and fats of vegetable and animal origin are historically and currently the most important renewable feedstock of the chemical industry. In Germany, the chemical industry used about 2.6 million tonnes (Mt) of renewable feedstock in 2018. Thereof, about 1.17 Mt (46 %) consisted of oils and fats used for the production of surfactants and cosmetics (56 %), paints and coatings (7.2 %), lubricants (4 %), polymers (15 %), adhesives, and others (17.8 %).[1]
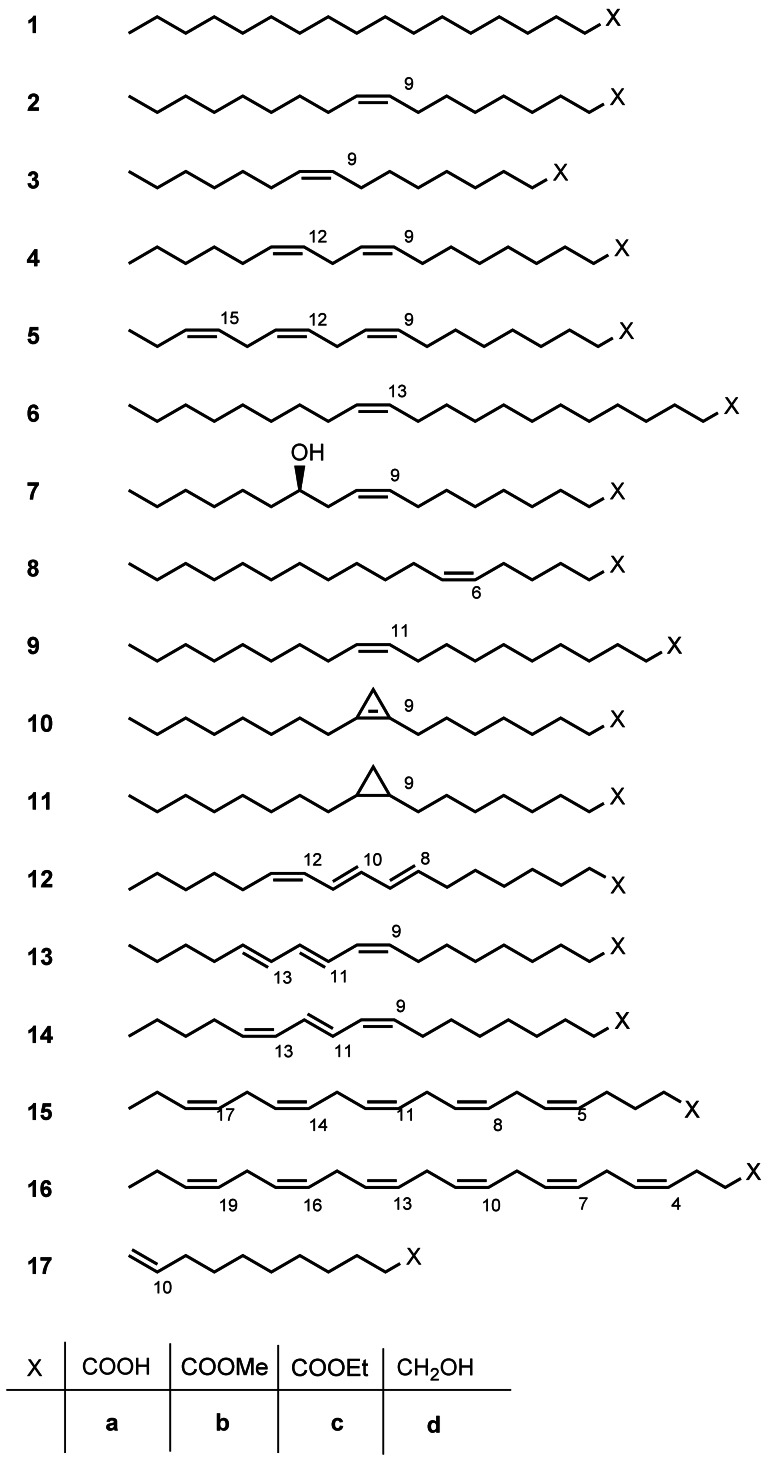
Classic oleochemistry preferentially deals with the chemistry of the carboxy group of fatty acids. Some reactions across the C–C double bond, such as hydrogenation, epoxidation, and ozonolysis are performed industrially as well.[2] During the last decades, modern organic chemistry, including enzymatic and biotechnological methods, was introduced to fatty acid chemistry.[3, 4] In the first decade of this century, important advances were made in the application of homogeneously and enzymatically catalyzed reactions applied to fatty compounds as well as microbial transformations. Fat‐based monomers were more frequently used for polymer synthesis.[4] Classic plant breeding as well as metabolic engineering was applied to improve natural oils and fats significantly, insofar as they show a more uniform and often unusual fatty acid spectrum, adjusted for various applications. Moreover, microbial oils provide interesting substrates such as 3 a, 15 a, and 16 a for chemical and biological syntheses. A broad spectrum of long‐chain fatty acids, esters, and alcohols is available for oleochemical applications (Scheme 1).
Scheme 1.
Fatty compounds as substrates for synthesis: stearic acid (1 a), oleic acid (2 a), palmitoleic acid (3 a), linoleic acid (4 a), α‐linolenic acid (5 a), erucic acid (6 a), ricinoleic acid (7 a), petroselinic acid (8 a), gondoic acid (9 a), sterculic acid (10 a), dihydrosterculic acid (11 a), calendic acid (12 a), α‐eleostearic acid (13 a), punicic acid (14 a), eicosapentaenoic acid (15 a), docosahexaenoic acid (16 a), 10‐undecenoic acid (17 a) and the respective methyl esters (1 b–17 b), ethyl esters (1 c–17 c), and alcohols (1 d–17 d).
We herein report the advances made in the chemistry and biotechnology of fatty compounds as well as improvements in the production of natural oils and fats in plants and microbials within the last ten years.
2. Production and Consumption of Oils and Fats
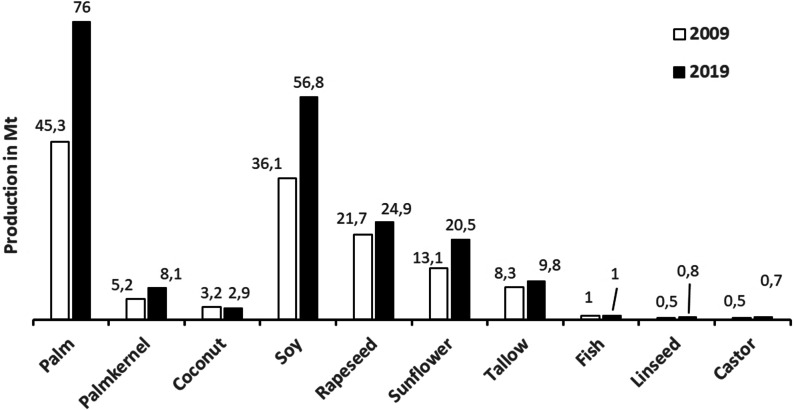
The annual global production of the major vegetable oils (palm and palm kernel, coconut, soybean, rapeseed, cotton, peanut, sunflower, and olive oil) increased by 47.8 % to 208.1 Mt in 2019,[5] compared to 140.8 Mt in 2009.[6] In addition, about 27.4 Mt of animal fats (tallow, lard, butter, and fish oil) were produced, a moderate increase of 16 % compared to 2009.
The annual global production of oils and fats suitable as oleochemical feedstock is shown in Figure 1 for 2009 and 2019. Palm oil showed the most significant growth of 68 %, followed by soybean oil of 57 %, sunflower oil of 56.5 %, and palm kernel oil of 56 %, whereas rapeseed oil and tallow showed a moderate increase of 15 % and 19 %, respectively. Castor and linseed oil, providing fatty acids 7 a and 5 a, respectively, are almost exclusively used by oleochemistry. The production of these oils increased by 23 % and 30 %, respectively.
Figure 1.
Global production of oils and fats that are important as feedstock for the oleochemical industry in 2009[6] and 2019.[5]
In 2020, this steadily increasing trend in the global oils and fats production surprisingly reversed. The production decreased by 2.2 Mt (−0.9 %) and consumption decreased by 1.7 Mt (−0.7 %).[5] Especially the production of palm oil displayed a downtrend by 2.3 Mt (−3.0 %), because of previous dryness, reduced fertilizer application provoked by the low palm oil prices in 2019, and a growing share of the oil palms being older than 20 years with declining yield potential. Thus, the yield of palm oil fell in Indonesia from 3.43 t ha−1 in 2019 to 3.35 t ha−1 in 2020. However, also effects of the COVID‐19 pandemic are curbing the production, and especially the consumption, because of the worldwide lockdowns in 2020. Palm oil consumption declined by 2 Mt in 2019/20, mainly in the food and biodiesel sector; this is the first decline in palm oil statistics ever recorded.[5]
Important to note, in order to increase the palm oil output in Indonesia and in Malaysia, it will be increasingly necessary to replant oil palms having an improved t ha−1 yield and not to depend on area expansion. This requirement is also triggered by sustainability concerns, as demand in developed countries favors deforestation‐free oils and seeks sustainability certifications for vegetable oil used as biodiesel feedstock. Several certification schemes operate and are widely used in Malaysia and Indonesia.[7]
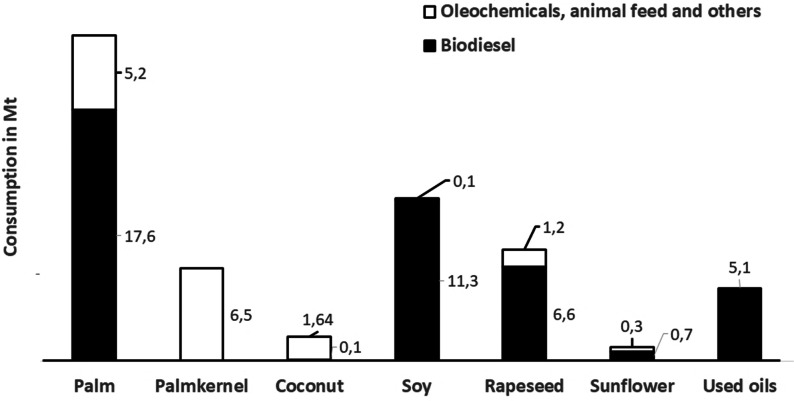
Industrial Use: In 2009/10, of the globally consumed plant oils totaling 138.5 Mt, about 108 Mt (78 %) were used as food and 31 Mt (22 %) were used industrially for the production of biodiesel, oleochemicals, animal feed, and other applications.[8a] In 2019, the food use increased to 150 Mt (74.5 %), industrial use increased to 51 Mt (25.5 %).[8b] The industrial use of the globally consumed palm oil has more than doubled from 10.1 Mt (24 %) in 2009 to 22.8 Mt (31 %) in 2019, a tremendous increase of 12.7 Mt, almost exclusively in South East Asia (Figure 2).
Figure 2.
Global industrial consumption of plant oils for biodiesel,[5] oleochemicals, animal feed, and other applications in 2019.[8b]
The industrial use of palm oil and palm kernel oil is steadily moving to Southeast Asia, mainly to Indonesia and Malaysia, close to the raw material sources.[4] Thus, in 2009, 3.1 Mt of palm oil was used industrially in this region, about 30 % of the global industrial palm oil consumption of 10.1 Mt.[8a] In 2019, the industrial consumption of palm oil had almost quadrupled to 12.1 Mt, representing 53 % of the global industrial palm oil consumption.[8b] In the same time period, the palm kernel oil consumption quadrupled from 0.6 Mt to approximately 2.5 Mt in Indonesia. In Malaysia, palm kernel oil consumption increased moderately from 1.4 Mt to 1.55 Mt in 2019. Thus, the two countries, as the main producers of palm and palm kernel oil, also accounted for more than 60 % of the global palm kernel oil use, primarily within their oleochemical industries.[5, 6] The production of biodiesel increased from ca. 16 Mt in 2009 to 46 Mt in 2019. The main feedstock was palm oil, followed by soybean and rapeseed oil (Figure 2).[5] Remarkably, in 2009 Indonesia produced 0.4 Mt (2.5 %) and in 2019 7.5 Mt (16 %) of biodiesel. Used cooking oil is a growing feedstock and contributed about 11 % to the global production in 2019, even 18.6 % in the EU (Figure 2).
3. Low‐Molecular‐Weight Products: Syntheses, New Reactions, and Platform Chemicals
The study of reactions of fatty compounds to provide interesting low molecular weight products by using modern synthetic, especially catalytic methods has developed tremendously during the last ten years. Especially fatty esters 2 b and 17 b were used as model compounds to study the scope of new catalysts for reactions across the internal and terminal C–C double bond, respectively. Synthetic transformations for the valorization of fatty derivatives were recently reviewed,[9] as well as catalytic approaches to monomers based on renewables and especially fats and oils,[10] and functional self‐assembled lipidic systems derived from renewable resources.[11]
3.1. Reduction of the Carboxy Group
The use of oils and fats as fuel and the necessity of improving the fuel properties has stimulated numerous studies on their catalytic deoxygenation to alkanes and alkenes.[12] Electroorganic synthesis has been applied for biofuel synthesis from fatty acids and triglycerides,[13] and the synthetic potential of Kolbe and non‐Kolbe electrolysis of fatty acids has been broadly discussed.[14]
3.1.1. Reduction to Alcohols
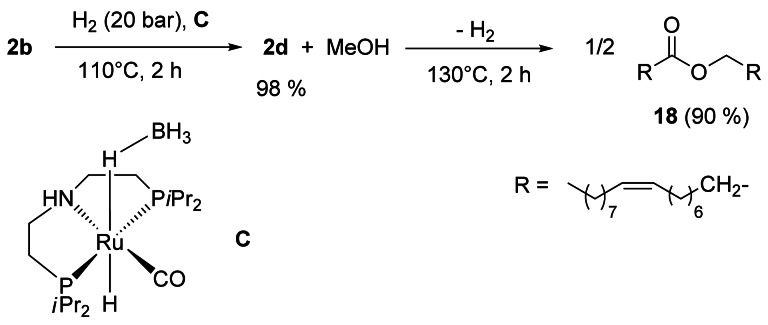
The hydrogenation of fatty esters to alcohols is efficiently promoted by pincer‐type catalysts of inter alia Ru,[15] Os[16] and, more recently, first‐row transition metals[17] such as Co[18] or Mn.[19] Most importantly, the acceptorless dehydrogenating coupling of alcohols to esters can be catalyzed as well,[20] as shown by the one‐pot, two‐step synthesis of wax ester 18 by consecutive hydrogenation–dehydrogenation reactions of 2 b using the readily available precatalyst C (Scheme 2).[21, 22]
Scheme 2.
Synthesis of oleyl oleate 18 by consecutive hydrogenation–dehydrogenation reaction of 2 b.[21]
3.1.2. Reduction to Ethers and to Amines
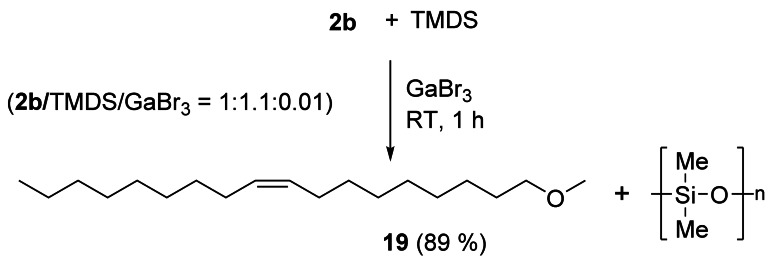
The reduction of fatty esters allows an easy access to fatty ethers avoiding the competitive reduction to alcohols. InBr3,[23] GaBr3,[24] Fe(CO)5,[25] and potassium tetrakis[(3,5‐trifluoromethyl)phenyl]borate[26] have been used as catalysts with preferentially tetramethyldisiloxane (TMDS) as the reductant to perform this reaction. The reduction of 2 b, for example, using GaBr3/TMDS without any solvent showed complete conversion of 2 b and 89 % yield of ether 19 (Scheme 3).[24]
Scheme 3.
Catalytic reduction of 2 b to give ether 19.[24]
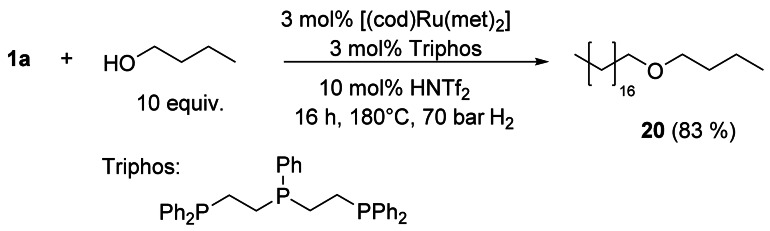
Remarkably, triglycerides were reduced to glyceryl trialkylethers[27] and long‐chain polyesters to the respective polyethers.[28] Hydrogenation of fatty acids or esters in the presence of an alcohol and catalyzed by Ru/triphos and a Lewis acid, such as Al(OTf)3,[29] or a Brønsted acid, such as trifluoromethanesulfonimide, gave the respective fatty ether in a yield of up to 83 % (Scheme 4).[30]
Scheme 4.
Catalytic reductive etherification of 1 a.[30]
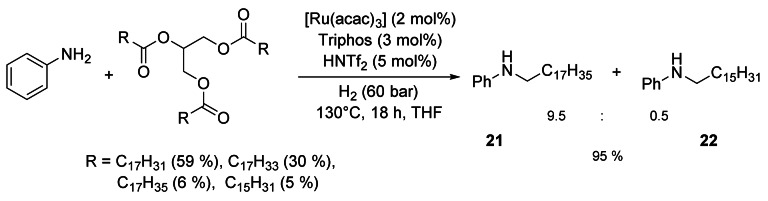
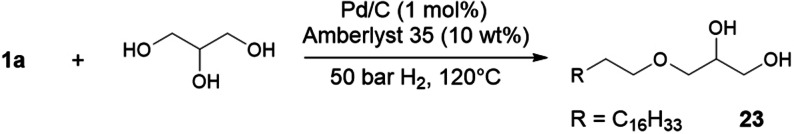
Because all C–C double bonds are hydrogenated, rapeseed oil, which consists of various unsaturated C18 fatty acids, can be directly converted with, for example, butanol to ether 20. The catalyst Ru/triphos/HNTf2 also enabled the selective N‐monoalkylation of a variety of primary and secondary amines with triglycerides such as sunflower oil (Scheme 5).[31]1‐O‐monoalkyl ethers of glycerol were obtained with high selectivity by catalytic reductive alkylation of fatty acids with glycerol using Pd/C as the catalyst and an acid ion‐exchange resin as the cocatalyst. (Scheme 6).[32, 33]
Scheme 5.
Ru‐catalyzed N‐alkylation of aniline with sunflower oil.[31]
Scheme 6.
Reductive alkylation of 1 a with glycerol (molar ratio 1:40).[32]
3.2. Reactions Across the C–C Double Bond of Unsaturated Fatty Compounds
3.2.1. Oxidations
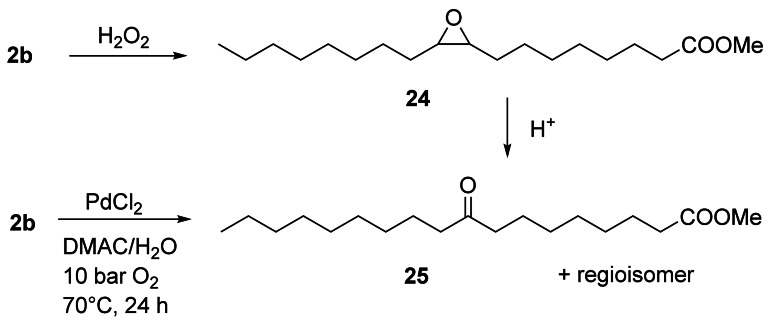
Ester 2 b was oxidized to methyl 9(10)‐ketostearate 25 in 85 % yield by a co‐catalyst‐free Wacker oxidation employing oxygen as the re‐oxidant and utilizing PdCl2 in dimethylacetamide (DMAC) (Scheme 7).[34] Furthermore, when benzoquinone/Fe (phthalocyanine) was used as a cocatalyst for the reoxidation of Pd, the reaction could be performed at room temperature and 1 bar O2 to give 25 in 79 % yield.[35] Ketone 25 was also obtained by rearrangement of epoxide 24 catalyzed by acidic resins (Scheme 7).[36] Methyl 12‐ketostearate was obtained by Pd‐catalyzed isomerization of the homoallylic alcohol 7 b.[37] Remarkably, an aldehyde‐selective Wacker‐type oxidation allowed the oxidation of 17 b to methyl 11‐oxoundecanoate with 79 % selectivity.[38]
Scheme 7.
Two‐step synthesis of 25 via epoxide 24 by acid‐catalyzed Meinwald rearrangement[36] and a direct approach to 25 by Wacker oxidation of 2 b.[34]
3.2.1.1. Epoxidation and Follow‐Up Products
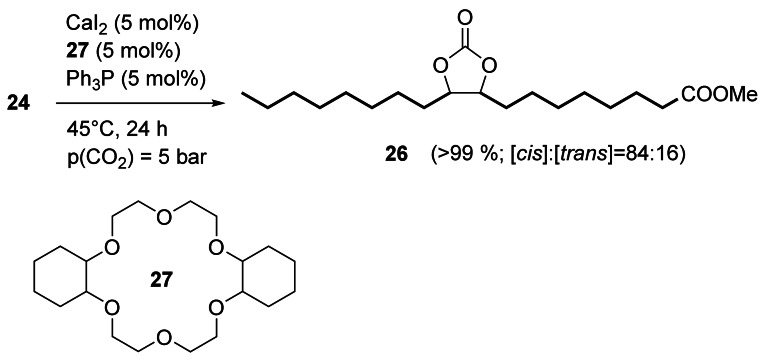
The epoxidation of unsaturated fatty compounds has recently been reviewed with an emphasis on catalytic[39] and chemoenzymatic epoxidation.[40] Follow‐up products of fatty epoxides, especially cyclic carbonates, are of steadily increasing importance, for example, as plasticizers[41] and as monomers for non‐isocyanate polyurethanes (NIPUs). The catalytic transformation of epoxides to carbonates was recently reviewed.[42] Werner et al. developed a high‐yielding transformation of fatty epoxides to the respective cyclic carbonates under mild reaction conditions using metal catalysts such as CaI2/dicyclohexyl 18‐crown‐6 (27)[43] as well as organocatalysts such as phenolic phosphonium salts.[44] Mono‐, di‐, and triepoxides as well as epoxidized triglycerides could be transformed with high conversion and yield, however, with low diastereoselectivity. For instance, the cis‐configured epoxide 24 yielded a mixture of cis‐ and trans‐carbonate 26 (Scheme 8).[43] Kleij et al. reported a protocol using a binary Al complex/bis(triphenylphosphine)iminium chloride catalyst and obtained cis‐26 with high diastereoselectivity (97:3).[45]
Scheme 8.
Reaction of epoxide 24 with CO2 to give the cyclic carbonate 26.[43]
3.2.1.2. Oxidative Cleavage of C–C Double Bonds
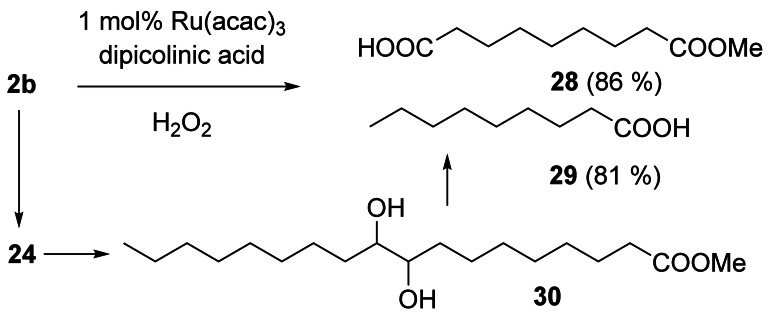
The ozonolysis of 2 a to give azelaic and pelargonic acid is an established industrial process.[2] A multitude of methods have been reported to replace ozone with a safer oxidant. However, a practical alternative was only found about 10 years ago,[46] correlated with the development of catalytic methods for the oxidative cleavage with H2O2, which has the potential for industrial application.[47, 48, 49] Behr et al. reported on the oxidative cleavage of 2 b with H2O2 (35 %, 8 equiv.) catalyzed by ruthenium/dipicolinic acid (Scheme 9).[50] A two‐step process was industrialized by Matrica.[51] 2 b is oxidized with H2O2, catalyzed by tungstic acid, to give vic‐diol 30, which is cleaved with oxygen, catalyzed by cobalt acetate.[52]
Scheme 9.
Oxidative cleavage of 2 b (tBuOH:H2O=3:1, 80 °C, 20 h) via tandem epoxidation, hydrolysis of 24, and oxidative cleavage of the vic‐diol 30.[50]
3.2.2. Selective Hydrogenation
The selective hydrogenation of the complex mixture of natural polyunsaturated acids, such as 4 a, 5 a or the more complex 14 a and 15 a, to monounsaturated fatty acids is a continuing challenge.[53] Good results were obtained with Pd nanoparticles in polar organic solvents such as 1,2‐propylenecarbonate[54] or PEG‐4000 as complexing solvent.[55] A series of non‐food oil derived methyl esters were selectively hydrogenated over Cu/SiO2.[56] All these selective hydrogenations occur with stereoisomerization of the cis‐configured double bonds. Sels et al. discussed the design of novel hydrogenation catalysts to enable the production of trans‐free hydrogenated products for the food sector.[57]
3.2.3. Alkyl‐Branched Fatty Compounds and Cycloadditions
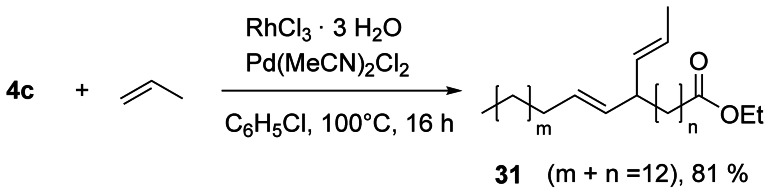
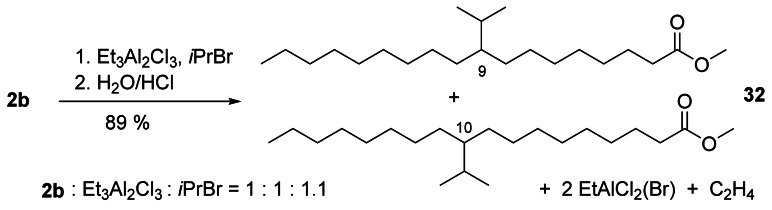
The synthesis as well as properties and industrial applications of alkyl‐branched fatty compounds have been reviewed.[58] Gooßen et al. used a Rh/Pd catalyst to propenylate 4 c giving product 31 as a mixture of isomers (Scheme 10). Pd catalyzes the conjugation of the two double bonds, which are propenylated with Rh catalysis.[59] The protocol for the synthesis of well‐defined alkyl‐branched oleochemicals was considerably improved by using haloalkanes instead of chlorocarbonates[4] to give the hydroalkylation products in excellent to good yields (Scheme 11).[60] Importantly, the reaction protocol was scalable (>500 g 2 b) and was also applied to substrates with two or more double bonds as well as to triglycerides.
Scheme 10.
Catalytic codimerization of 4 c with propene.[59]
Scheme 11.
Et3Al2Cl3‐mediated hydroisopropylation of 2 b.[60]
The skeletal isomerization of unsaturated fatty acids such as 2 a was performed in the presence of H+‐ferrierite zeolite catalysts to give, after hydrogenation, methyl‐branched iso‐fatty acids with a remarkable selectivity of up to about 80 %[61, 62] in contrast to the present industrial process.[58]
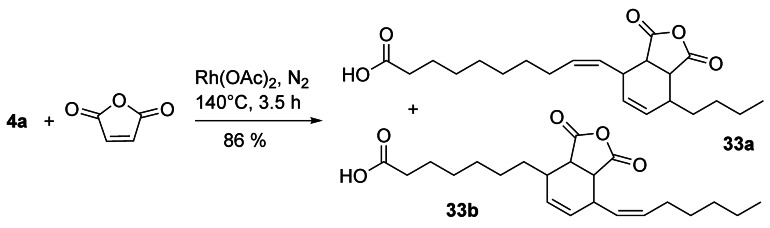
Addition products of unsaturated fatty derivatives and maleic anhydride as well as various follow‐up products are of interest because of their versatile properties in numerous applications.[63] Interestingly, the Rh‐catalyzed reaction of 4 a and maleic anhydride gave the cycloaddition products 33 a and 33 b in a 1:1 ratio (Scheme 12),[64] which are identical to the Diels–Alder addition products of 12 a and 13 a.[4]
Scheme 12.
Rh‐catalyzed cycloaddition of 4 a and maleic anhydride.[64]
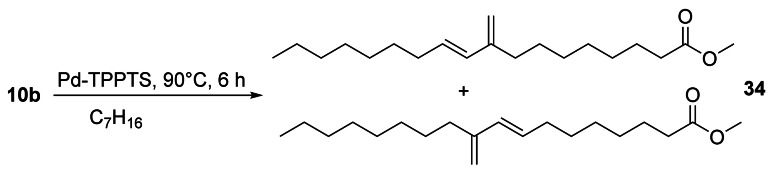
Compound 10 b, derived from the seed oil of Sterculia foetida L, was rearranged to the conjugated diene 34 with a methylene branch as a mixture of two regioisomers with a selectivity of 99 % under Pd catalysis (Scheme 13).[65]
Scheme 13.
Pd‐catalyzed rearrangement of 10 b.[65]
3.2.4. Hydroformylation[66] and Hydroaminomethylation[67]
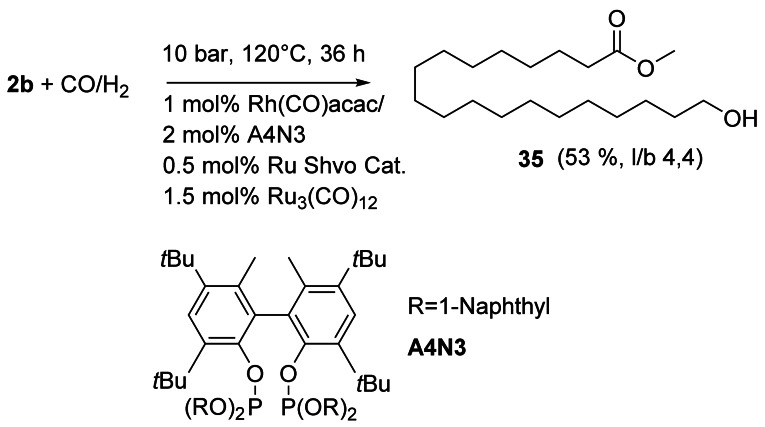
The isomerizing hydroformylation[4, 68] of 2 b was considerably improved using a palladium isomerization and a rhodium hydroformylation catalyst simultaneously, affording the α,ω‐aldehyde ester.[69] The corresponding alcohol 35 was directly obtained using a Rh/Ru/Ru ternary tandem catalytic system with Shvo's hydrogenation catalyst (Scheme 14).[70]
Scheme 14.
Isomerizing tandem hydroformylation of 2 b and in situ hydrogenation.[70]
The important task of recycling the homogeneous hydroformylation rhodium catalyst has been addressed by organic solvent nanofiltration,[71] selective product crystallization,[72] and especially by thermomorphic solvent systems,[73] for example, a combination of water/1‐butanol for the hydroformylation of 17 b [74] and 2 b.[75] Most interesting are new developments in the reactor setup for gas–liquid–liquid multiphase catalysis, for example, using a jet‐loop reactor to improve the aqueous biphasic hydroformylation of 17 b and 2 b.[76]
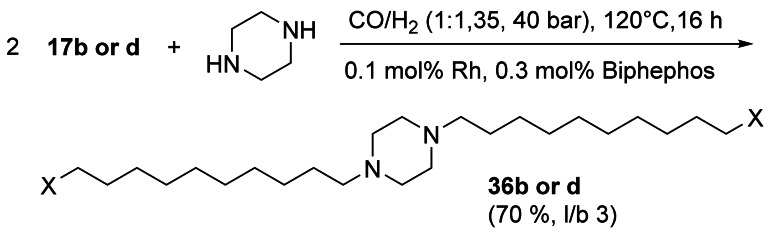
Bis‐hydroaminoalkylation of 17 b and 17 d with piperazine yielded long‐chain linear diester 36 b and diol 36 d, respectively, interesting substrates for polyesters (Scheme 15). Key to success was the selective crystallization of the product from the reaction mixture in >98 % purity.[77]
Scheme 15.
Bis‐hydroaminomethylation of 17 b and 17 d.[77]
3.2.5. Alkoxycarbonylation and Hydroxycarbonylation
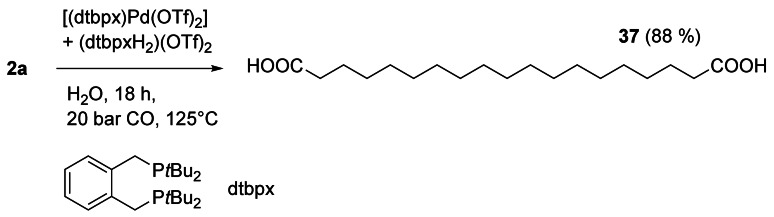
The mechanism of the isomerizing alkoxycarbonylation of unsaturated fatty compounds was studied in detail, including computational studies.[78, 79, 80] Mecking et al. fully analyzed the products of the methoxycarbonylation of 2 b with the well‐defined precatalyst [(dtbpx)Pd(OTf)2],[81] and found the formation of the linear α,ω‐diester with a selectivity of 90.6 %, besides all possible branched diesters in minor amounts.[82] The diester was obtained in high purity by simple recrystallization from methanol. Most importantly, the robust catalytic system is capable of transforming not only pure 2 b, but also technical grade plant oils[83, 84] and microbial oils.[85] The catalyst can also be used for selective terminal carbonylation of the internal double bond of 2 a with water as a nucleophile to open a direct access to α,ω‐dicarboxylic acids such as 37 (Scheme 16).[86] The recycling and reuse of the catalyst has been studied as well.[87, 88]
Scheme 16.
Isomerizing hydroxycarbonylation of 2 a.[86]
3.2.6. Olefin Metathesis
The metathesis reaction of unsaturated fatty compounds has been studied intensively over the past ten years and has been thoroughly reviewed.[89, 90, 91, 92] Studies have focused on the important topics self‐metathesis and ethenolysis,[93, 94, 95] cross‐metathesis with functionalized alkenes, and isomerizing metathesis.[96]
3.2.6.1. Self‐Metathesis and Ethenolysis
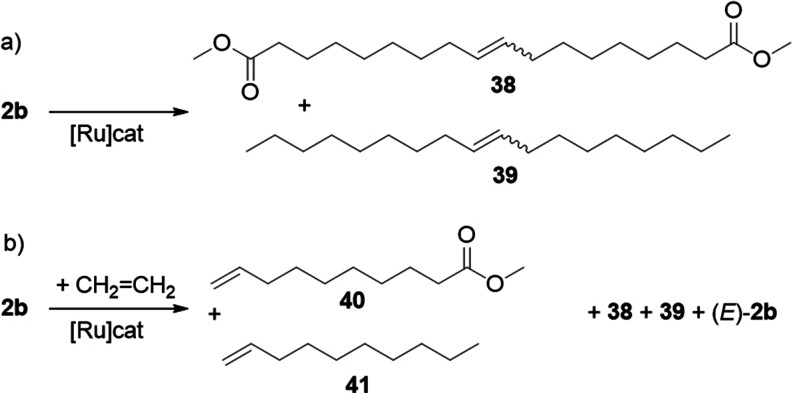
Self‐metathesis of 2 b gives the long‐chain diester 38 and alkene 39 in only moderate yield in equilibrium, since olefin metathesis is a reversible reaction (Scheme 17 a). However, self‐metathesis of polyunsaturated fatty compounds, such as 5 b, allows full conversion to diester 38 and others, since volatile metathesis products, such as cyclohexa‐1,4‐diene or hex‐3‐ene, can be easily removed in order to shift the equilibrium.[97, 98, 99]
Scheme 17.
a) Self‐metathesis and b) ethenolysis of 2 b.
Self‐metathesis has to be suppressed as far as possible in an ethenolysis reaction (Scheme 17 b). N‐heterocyclic carbene (NHC) ruthenium catalysts, such as C1 (Scheme 18), provide selectivities as high as 95 % for the kinetic ethenolysis products 40 and 41, unfortunately with a relatively low TON of up to 5000 (Scheme 17 b).[100]
Scheme 18.
Ruthenium‐based metathesis precatalysts used for the transformation of fatty acid derivatives.
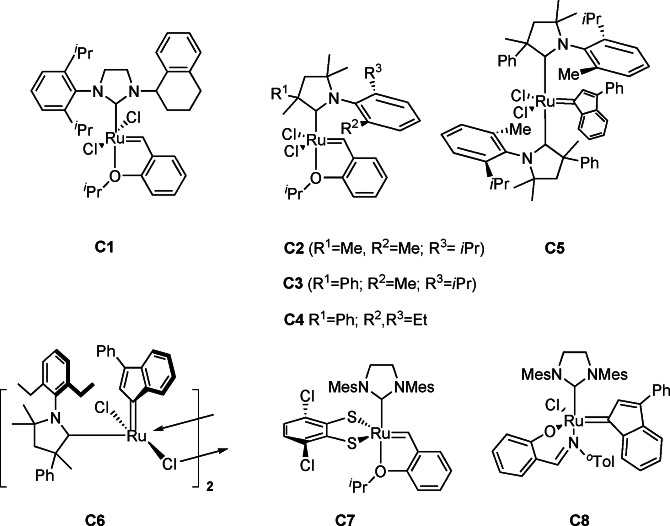
It is well known that methylene‐Ru intermediates, formed in the catalytic cycle of ethenolysis, are less stable than alkylidene intermediates, thus reducing the TON in comparison to that for alkenolysis with 1‐alkenes and internal alkenes.[93, 94, 95] Hence, butenolysis of 2 b is performed industrially.[93c] To obtain an economically viable process, it was stated that a TON >50 000 is required for the ethenolysis of 2 b, based on the cost of the homogeneous catalyst.[93a] The most promising results have been obtained with Ru catalysts such as C2 and C3 bearing cyclic (amino)alkylcarbene (CAAC) ligands (Scheme 18).[101] TONs of 180 000 using 99.95 % pure ethene have been achieved at a catalyst loading of 3 ppm. These impressive results were obtained only when a glovebox was used.[102] The easily synthesized bis(CAAC)Ru catalyst C5 shows similar efficiencies,[103] and a dimeric indenylidene CAAC Ru complex C6 as well, though being more robust to standard‐grade (99.9 %) ethene.[104] Grela et al. focused on practicability issues, such as reaction in air, ethene with 99.9 % purity, and undistilled 2 b with 90–95 % purity.[105] The CAAC‐bearing catalyst C4 was most efficient. The selectivity was 95 % and the TON goes up to 28 000.[106]
3.2.6.2. Cross‐Metathesis with Functionalized Alkenes
Cross‐metathesis of unsaturated fatty compounds with functionalized alkenes, such as methyl acrylate,[107] dimethyl maleate,[108] maleic acid,[109] allyl acetate and cis‐1,4‐diacetoxy‐2‐butene,[110, 111, 112] acrylonitrile,[113, 114] or acrolein[115] gives straightforward access to interesting bifunctional unsaturated compounds with typically high (E)‐stereoselectivity.
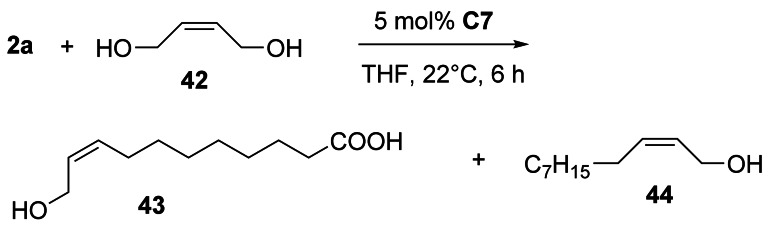
In contrast, the cross‐metathesis of 2 a or 2 d with (Z)‐2‐butene‐1,4‐diol 42, using catalyst C7, yielded (Z)‐allyl alcohols 43 and 44 with high stereoselectivity ([Z]:[E]=94:6) in 60–65 % yield (Scheme 19).[116]
Scheme 19.
(Z)‐Stereoselective cross‐metathesis of 2 a and diol 42.[116]
3.2.6.3. Isomerizing Metathesis
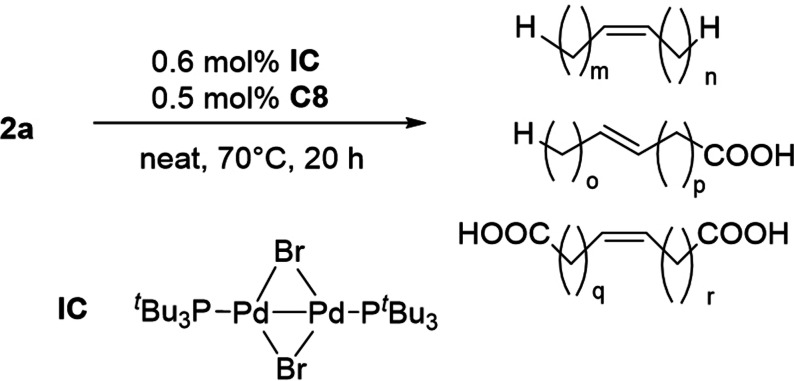
The cooperative action of the Pd isomerization catalyst IC and the metathesis catalyst C8 (Scheme 18) enabled the transformation of unsaturated fatty compounds such as 2 a or 2 b into complex product blends having defined, tuneable properties, as shown by Gooßen et al. (Scheme 20).[117] Alkenes and unsaturated mono‐ and diacids were obtained in adjustable distributions. The method was also applied to improve rape seed oil biodiesel with ethene (≤1 bar), in order to obtain boiling properties similar to conventional diesel fuel.[118] Biodiesel was also modified via self‐ and cross‐metathesis with 1‐hexene, resulting in similarly improved boiling curves.[119]
Scheme 20.
Isomerizing self‐metathesis of 2 a.[117]
4. Polymers
100 years after Staudinger's first publication on polymerization, sustainability has become one of the most important topics for polymer scientists,[120] and of course for our society. Fatty acid derivatives have long been used for applications in polymer science;[4, 121, 122] detailed summaries with different foci are available.[10, 123, 124, 125, 126, 127] It is important to note that we had to be very selective in covering this flourishing and very large field of research in this brief overview, and thus, composites and blends, cross‐linked systems (i.e. thermosets), copolymerization with other (renewable) monomers, obviously unsustainable procedures, as well as other directions could not be included or discussed in detail.
4.1. Step‐Growth Polymerization
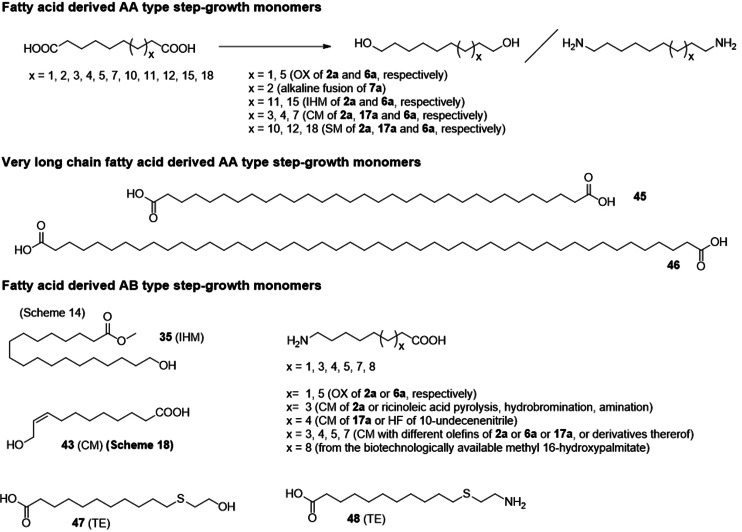
The use of fatty acid based monomers for step‐growth polymerization has significantly increased within the last 10 years and relates in large part to the efficient transformation of 2 a, 5 a, and 17 a as well as their derivatives to prepare AA‐ as well as AB‐type monomers (see also Section 3 for important synthesis routes).
4.1.1. Linear Polymers from Step‐Growth Reactions
Typical AB‐ and AA‐type monomers obtained from fatty acid derivatives that lead to linear, unbranched polyesters and polyamides are depicted in Scheme 21 and are predominantly derived via oxidative cleavage (Section 3.2.1), olefin metathesis (Section 3.2.6), isomerizing hydroformylation (Section 3.2.4), and thiol‐ene addition.[128]
Scheme 21.
Unbranched AB‐ and AA‐type monomers derived from fatty acid derivatives. Saturated methyl esters are depicted for clarity, but carboxylic acids and/or unsaturated derivatives are described and used as well. The respective synthesis route are given: SM: self‐metathesis; CM: cross‐metathesis; (I)HF: (isomerizing) hydroformylation; TE: thiol–ene; OX: oxidative cleavage.
An important advance is the development of long‐chain polyesters from such starting materials that can mimic important properties of polyethylene, while offering degradability.[129, 130] Remarkably, polyesters 19,19 and 23,23 were prepared on 100 g scale; the necessary dimethyl‐1,19‐nonadecanedioate monomer was directly prepared from technical‐grade high oleic sunflower oil. Such polyesters offer suitable mechanical properties (elongation at break >600 %; Young's modulus of 400 MPa) for non‐wovens or extruded transparent films.[131] Similar long‐chain polyesters can be obtained via different routes, for instance from biotechnologically derived monomers[132, 133] or ring‐opening polymerization of macrolactones.[134]
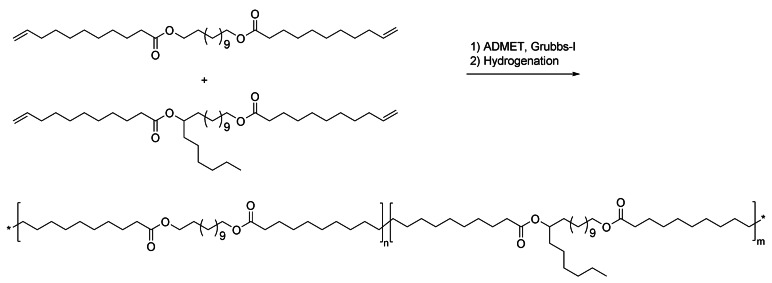
Two highly remarkable monomers, the very long‐chain diesters 45 and 46, were prepared by Mecking et al. from 38 (Scheme 17) by isomerizing the central double bond selectively to the statistically disfavored α,β‐position in a catalytic dynamic isomerizing crystallization approach. Subsequently, “chain doubling” was achieved by an additional cross‐metathesis step to obtain ultra‐long‐chain α,ω‐difunctional building blocks of up to 48 carbons in length.[135] Polyester 48,48, derived from 46, showed an unprecedentedly high melting point of 120 °C.
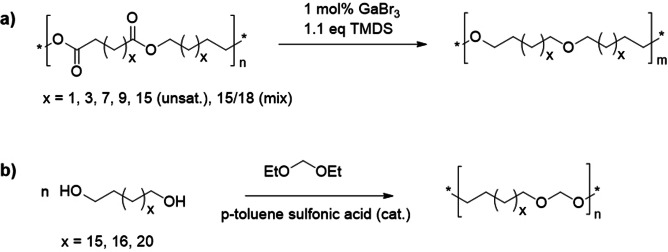
Apart from polyesters, polyurethanes, polyamides, and polyacetals have been prepared from the monomers described in Scheme 21, of course after appropriate chemical modification. A novel development in this context is the synthesis of polyethers via either catalytic reduction of the corresponding polyesters (Scheme 22 a),[28] or the catalytic reduction of the respective ester monomers and subsequent acyclic diene metathesis (ADMET) or thiol–ene polymerization.[136] This route offers access to so far unknown long‐chain polyethers. Compared to the respective polyesters, the polyethers showed lower meting points, whereby the difference in melting point decreased with increased spacing of the functional groups.[28] A detailed review on the manifold possibilities and advances offered by such different types of fatty acid derived polymer classes, from synthesis to properties and possible applications, is available.[137] Fatty acid based polyamides and polyurethanes were thoroughly reviewed recently, also addressing sustainability aspects.[138] Polyacetals should be specifically mentioned and can be obtained from medium‐ and long‐chain fatty acid derived diols in an acid‐catalyzed process with 1) an excess of diethoxymethane to yield the diacetals, which are polymerized after isolation and purification,[139] or 2) via direct polymerization of diols with an excess of diethoxymethane (Scheme 22 b).[140] This route has been termed acetal metathesis polymerization (AMP).]139a] Studying the properties of the novel polyacetals revealed that the solid polymers display higher stability against hydrolytic degradation compared to their shorter‐chain counterparts.[140]
Scheme 22.
a) Polyethers obtained via catalytic reduction of polyesters;[28] b) polyacetals obtained via so‐called acetal metathesis polymerization (the example shown is from ref. [140]).[139, 140]
Very recently, an important step towards practical application was demonstrated for long‐chain polyethylenes such as polyester 18,18 as well as the polycarbonate 18 via closed loop chemical recycling.[141]
Last but not least, it is worth mentioning a few more selected examples of similar long‐chain and linear polyesters,[142] polyamides,[143] polyethers,[136a] polycarbonates,[144] or (non‐isocyanate) polyurethanes[145] that can also be obtained via ADMET, thiol–ene, or other polymerization techniques.
4.1.2. Branched Polymers from Step‐Growth Reactions
4.1.2.1. Branched Monomers
Fatty acids offer the possibility to design branched monomers or hyperbranched polymers with interesting and unique properties. One such branched monomer precursor, which can be obtained via several catalytic and non‐catalytic routes (see Section 3.2.1 for selected examples) is keto fatty acid ester 25 (Scheme 7), which is suitable for the synthesis of branched polyamides (after reductive amination of 25) with interesting properties, such as an increased hydrophobicity and improved elongation at break when copolymerized with Nylon 6.6.[34] Similar monomers can also be obtained via thiol–ene addition.[128f] Following the oxidation and amination route, methyl 10‐aminoundecanoate and methyl 10‐hydroxyundecanoate can be obtained from 17 b, resulting in a methyl‐branched polyamide or polyester, respectively.[146] In both cases, steric hindrance resulted in more difficult polymerization and poorer thermal properties. A very different route to polyesters with various degrees of branching was described using ADMET polymerization of linear and branched α,ω‐diene monomers (Scheme 23).[147] After hydrogenation, thermo‐mechanical properties in accordance to some types of polyethylene were observed, offering a sustainable alternative to olefin‐based elastomers, especially for specific applications requiring degradability. Atypical monomers were obtained by forming ester enolates from saturated fatty acid methyl esters, which reacted with dimethyl carbonate to form malonates with different chain lengths (C6–C16) and bearing saturated alkyl branches at C2 of the malonate.[148] Polyesters as well as polyamides were derived from these malonate monomers, and the thermal properties could be tuned owing to the direct correlation to the alkyl chain length. The route via malonates was also adopted for the synthesis of fatty acid monomers bearing six‐membered cyclic carbonates, which were used for the synthesis of non‐isocyanate polyurethanes.[149] Moreover, branched polyacetals were prepared via the abovementioned acetal metathesis route (see Scheme 22), in this case derived from heptanal and 17 a.[150]
Scheme 23.
Branched polyesters, derived from castor and vernonia oil, mimicking the structure and properties of olefin‐based elastomers.[147]
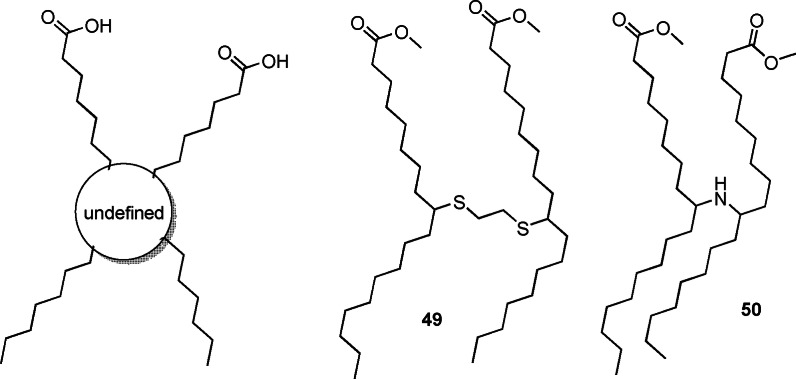
A special case of highly branched fatty acid derived monomers are the so‐called dimer fatty acids (Scheme 24), classically obtained and industrially available since the 1950s via the intermolecular coupling of unsaturated fatty acids and their esters.[151] This process mainly proceeds via isomerization followed by a Diels–Alder reaction. The industrially obtained dimer acids are a mixture of several compounds (monomer, dimer, trimer, cyclic, aromatic structures), requiring extensive purification before polymerization. Such dimer acids generally provide high flexibility, pronounced hydrophobicity, and hydrolysis stability, for instance to polyurethanes.[152] Their use in polyamides, for example, as hot‐melt adhesives or as reactive components, typically leads to lower crystallinity as well as lower melting temperatures.[153] Also applications in polyurethane foams,[154] coatings,[155] and controlled release materials[156] have been described. Over the years, as summarized in ref. [157], attempts have been made to improve their synthesis. More recently, hydrobromination of oleic acid, followed by nucleophilic substitution with dithiols, resulted in new dimer structures.[158] Interestingly, 2 b can be dimerized directly with ethane‐1,2‐dithiol in a simple UV‐induced one‐pot procedure to obtain the defined dimer acid 49 (Scheme 24) in one reaction step, which significantly reduces water uptake when used for polyamide synthesis.[157]
Scheme 24.
Industrially available (left) and new, more defined dimer fatty acid derivatives (right); please note that 49 and 50 are each mixtures of three regioisomers.
A large variety of structurally defined dimer acids, some fully renewable (i.e. 50, Scheme 24), were prepared via catalytic Wacker‐type oxidation of 2 b, 6 b, or 17 b, leading to the respective keto fatty acid derivatives, which were subsequently dimerized via reductive amination.[159] When these compounds were copolymerized with 1,10‐diaminodecane, high‐molecular‐weight polyamides with low glass transition and melting temperatures were obtained.
4.1.2.2. Hyperbranched Polymers
Hyperbranched polymers derived from fatty acids are far less frequently described, yet offer interesting structures and properties. For instance, 7 b was transformed to an AB2 monomer by thiol–ene addition of 2‐mercaptoethanol in a solvent‐free reaction.[160] The resulting hyperbranched polyesters showed lower complex viscosities than their linear analogues and low glass transition temperatures. Similarly, 17 b was used as a starting material to obtain an AB2 monomer by reaction with thioglycerol,[128e, 161] resulting in hyperbranched polyesters with a high degree of branching and low inherent viscosity.[161] AB2 and AB3 monomers for hyperbranched polyesters were also prepared via epoxidation and ring opening of 2 b, 6 b, 7 b, and 17 b.[162] Interestingly, the degree of branching depended on the type of monomer as well as on the conversion and the applied catalyst. A different approach uses trifunctional (A3 or B3) fatty acid derived monomers in combination with the respective B2 or A2 monomers in a Diels–Alder reaction between furans (A) and maleimides (B).[163] A2B and AB2 systems were also described. Most interestingly, the Diels–Alder approach allowed for the thermoreversibility of the polymerization reaction. Highly branched polyesters could also be obtained by the so‐called acyclic triene metathesis of high oleic sunflower oil[164] or the highly unsaturated Plukenetia conophora oil.[165] The oils could be polymerized directly under bulk conditions without any pretreatment.
4.2. Chain‐Growth Polymerization
To obtain fully renewable monomers for chain‐growth polymerization from fatty acid derivatives is more difficult than the approaches discussed in Section 4.1, and have thus been reported less frequently.
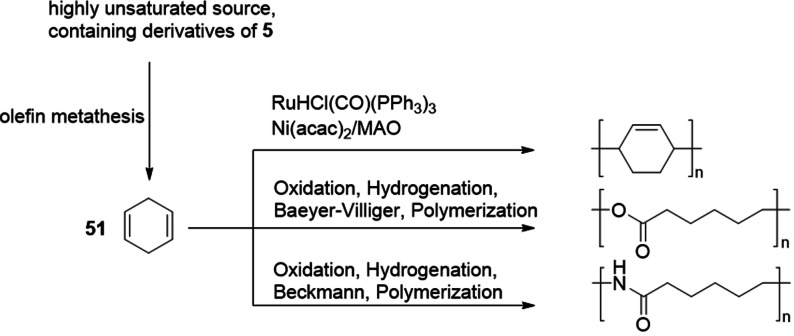
As described in Section 3.2.6, 1,4‐cyclohexadiene 51 can be obtained efficiently by self‐metathesis of highly unsaturated plant oils or fatty acid methyl esters.[97, 98] Compound 51 was shown to be a suitable renewable platform molecule (Scheme 25) for the synthesis of poly(cyclohexadiene) via in situ isomerization and polymerization[166] as well as for the synthesis of polyamide 6[167] and poly(caprolactone).[168] The latter two commercially important polymers could also be functionalized to tune their properties. 1,4‐Cyclohexadiene oxide was furthermore used as a comonomer for the synthesis of unsaturated polyesters as well as polycarbonates.[169] Ring‐opening polymerization (ROP) was also used for the solvent‐free copolymerization of methyl 9,10‐epoxystearate with different cyclic anhydrides using a (salen)CrIIICl catalyst and n‐Bu4NCl as co‐catalyst to yield polyesters with low glass transition temperatures.[170] Another interesting class of fatty acid based monomers for cationic ROP are 2‐oxazolines, leading to materials suitable for the preparation of amphiphilic structures as well as for low‐surface‐energy and low‐adhesive coatings.[171]
Scheme 25.
1,4‐Cyclohexadiene 51 as a renewable platform chemical for the synthesis of various polymers.
Furthermore, a variety of monomers for radical polymerizations are described, leading to either cross‐linked or copolymer systems that are outside the scope of this focused summary. A few interesting new monomers and the corresponding fatty acid based homopolymers are worth mentioning. Most typically, polymerizable moieties such as acrylates or methacrylates are attached to fatty alcohols or 7 b,[172] leading to partially renewable monomers via often unbenign synthesis routes.[173] These monomers will not be discussed herein, as they were very recently reviewed.[172]
Compounds 2 a and 4 a were transvinylated with vinyl acetate using 1 mol % [Ir(cod)Cl]2 to yield fatty acid vinyl esters.[174] Subsequent free radical polymerization led to materials possibly useful as paints or varnishes via the oxidative drying of the unsaturation remaining in the side chains of the polymers. Interestingly, the even less activated olefinic double bond of 17 b could be directly copolymerized with vinyl acetate (up to 50 %) using a preformed alkylcobalt(III) acetylacetonate (R‐Co(acac)2) complex, where the alkyl group acts as a radical initiator and the Co(acac)2 as the controlling agent.[175] Molecular weights above 10 kDa could be achieved while maintaining relatively low dispersities. Furthermore, a sustainable purification via supercritical CO2 extraction was introduced, allowing the recovery of unreacted 17 b. Finally worth mentioning is a one‐pot catalytic copolymerization of 2 b or plant oils with ethylene using an isomerization catalyst and a Brookhart polymerization catalyst at the same time.[176]
5. Enzymatic and Microbial Transformations
The use of isolated (immobilized) enzymes for the modification of fats, oils, and related lipids for oleochemistry is well documented in the literature.[4, 177] Biocatalysis using enzymes offers important advantages such as chemo‐, regio‐, and stereoselectivity under mild reaction conditions. Enzymes address some aspects of green chemistry, since they are biodegradable, use mostly water as a solvent, and most importantly, they can catalyze a broad variety of reactions. Very recently, two reviews have been published on the industrial applications of biocatalysts.[178]
Since the publication of our last review,[4] in addition to lipases, phospholipases (PLA) as well as a few other enzymes, such as P450‐monooxygenases, have found their way into oleochemistry. Lipases have been widely applied for many decades to prepare structured triglycerides, margarine, and biodiesel. Phospholipases, on the other hand, are the biocatalysts of choice for degumming. Here, especially the use of PLC was established[179] as an alternative to PLA1 or PLA2. Examples of other enzymes are given in the subsequent sections. For microbial biotransformations, substantial progress in molecular biology and metabolic engineering allows the design of entirely new pathways in microorganisms. This has opened new opportunities for lipid modification and enables the transformation of simple—often renewable—precursor molecules into valuable lipid‐related products.[177b] Modern methods for enzyme discovery as well as advanced protein engineering tools have helped expand the biocatalytic toolbox and adjust enzymes for large‐scale processing.[180] Furthermore, cascade reactions[181] open up new opportunities for the use of enzymes in oleochemistry.
5.1. Novel Enzymes/New Chemistry
Typically, lipases are used for the synthesis of esters (e.g. biodiesel production), as the presence of water will result in hydrolysis. Although it has been known for more than a decade that some lipases (or lipase‐like enzymes) also have acyltransferase activity, only recently has substantial progress been reported. This includes lipase CAL‐A and an enzyme from Candida parapsilosis (CpLIP2). Both enable the synthesis of esters, such as fatty acid ethyl esters (FAEE), in the presence of a bulk aqueous phase. Through the use of rational protein design, the acyltransferase activity of lipase CAL‐A could be substantially improved by a single point mutation (D122L) and up to 95 % FAEE could be produced from palm kernel oil in the presence of 5–10 % water in EtOH.[182] The group led by Subileau and Dubreucq improved CpLIP2 and identified a broad range of further enzymes.[183] Very recently, the structural and molecular reasons for this phenomenon were elucidated.[184] High acyltransferase activity correlates well with the hydrophobicity of the substrate‐binding pocket and a scoring system was proven to accurately predict the promiscuous acyltransferase activity from protein sequences in databases, where more than a thousand novel candidates were identified; several of them have been experimentally verified.[184, 185]
Lipases have been successfully subjected to protein engineering in numerous reports.[180a] More recently, the fatty acid selectivity of lipase CAL‐A was improved to enable the enrichment of erucic acid 6 a.[186] 6 a is also present in Crambe abyssinica seed oil (≈59 %) and its concentration needs to be elevated for oleochemical applications. A detailed protein engineering study led to the identification of a double mutant (V238L/V290L) with which the level of 6 a could be increased to 74 %.[186b] Similarly, the selectivity for gondoic acid 9 a present in Camelina sativa oil was enhanced using CAL‐A variants to enable its enrichment.[186a, 186c]
A very interesting enzyme class are the flavin‐dependent hydratases, which add water to double bonds and thus form hydroxyl groups.[187] Mostly oleate hydratases have been studied, which convert oleic acid in a regio‐ and stereoselective manner to the corresponding (R)‐10‐hydroxystearic acid (up to 100 g L−1).[188] OAs have also been included in cascade reactions to functionalize fatty acid derivatives, for instance to obtain long‐chain aliphatic amines (see below).[189] In 2013, the first crystal structure of a linoleic acid hydratase was reported by the Feussner group,[190] followed by that of an oleate hydratase from Elizabethkingia meningoseptica.[191] Besides insights into the mechanism, this provided the basis for protein engineering to expand the substrate scope of OA, which can now catalyze the asymmetric hydration of terminal and internal alkenes (notably lacking a carboxylic group) also on preparative scale (up to 93 % conversion, >99 % ee, >95 % regioselectivity).[192]
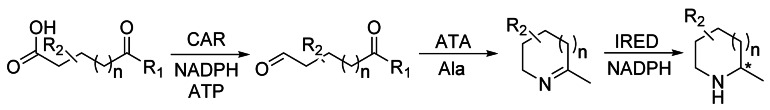
Carboxylic acid reductases (CARs) are useful biocatalysts for the conversion of carboxylic acids into aldehydes under mild reaction conditions without overreduction to the corresponding alcohol. Aldehydes are especially important for the flavor and fragrance industry, as well as for various other applications in chemistry as summarized in recent reviews, which also discuss the current status on the discovery, structures, and applications of CARs.[193] For their catalytic activity, CARs require coexpression of a suitable phosphopantetheinyl transferase (PPTase) and the cofactors NADPH and ATP in stoichiometric amounts. Thanks to the recent the elucidation of the first structures of the CARs from Nocardia iowensis and Segniliparus rugosus,[194] and to the recent development of a high‐throughput assay, improved variants have already been generated by protein engineering.[195] Practically, often whole‐cell systems are used and the cofactor ATP can be readily recycled. On the other hand, the formed aldehydes are usually toxic to the host, e.g. E. coli. Thus, Turner and co‐workers used isolated enzymes in a cascade reaction (Scheme 26).[196]
Scheme 26.
A combination of a carboxylic acid reductase (CAR), a transaminase (ATA), and an imine reductase (IRED) makes it possible to convert (short‐chain) fatty acids and their analogues into useful heterocycles (n=0, 1).[196]
CARs have also been used for the reduction of dicarboxylic acids, yielding diols such as 1,4‐butanediol or 1,6‐hexanediol.[197] Carboxylic acids can also directly be converted into the corresponding carboxylic amides in the presence of ATP and an amine nucleophile, but NADPH must be excluded and thus only the adenylation domain of the CAR is used in this case.[198] The recycling of the expensive ATP from the formed AMP can be achieved in vitro with a polyphosphate kinase using cheap pyrophosphate (PPi).[199] This concept was first shown for SAM‐dependent methyltransferases, now allowing larger scale applications of CARs.[200]
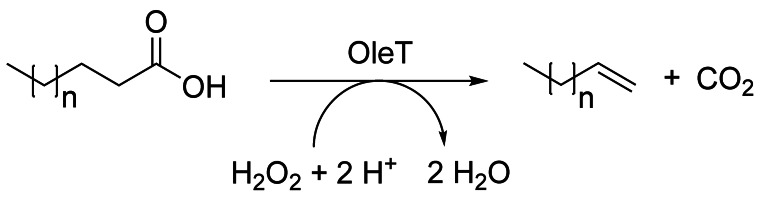
Very interesting precursors for oleochemistry are, at least in principle, accessible by decarboxylases, which can form terminal alkenes (with release of CO2, Scheme 27) directly from fatty acids.[201] Three enzymatic systems have been studied in more detail: the fatty acid decarboxylase OleT, the medium‐chain fatty acid decarboxylase UndA, and the decarboxylase UndB. As the heme‐dependent OleT requires electrons for in vitro applications, different strategies have been developed using the addition or in situ formation of H2O2, coupling with the CamA/CamB system to use NAD(P)H or protein fusions, for instance with a catalase.[202] Obstacles to overcome remain the typically low yields due to the instability of these enzymes. For OleT, total turnover numbers of >2000 and product titers of up to 0.93 g L−1 have been reported.[203] The Kourist group has furthermore demonstrated a light‐driven conversion of ω‐functionalized fatty acids using OleT in combination with a Ru‐based metathesis catalyst in a sequential chemoenzymatic cascade in a buffer/isooctane system.[204]
Scheme 27.
OleT‐catalyzed decarboxylation of carboxylic acids yields terminal alkenes (n=0–4).[201]
5.2. Cascade Reactions (Enzymatic and Microbial Systems)
Combining several enzymes that catalyze different reactions in cascades enables a more efficient access to complex products. In 2013, a cascade comprising an oleate hydratase, an alcohol dehydrogenase, a Baeyer–Villiger monoxygenase and finally an esterase was reported to yield ω‐hydroxy carboxylic acid or α,ω‐dicarboxylic acids directly from 2 a.[205] This biocatalytic route is an alternative to the oleochemical processes, where either pyrolysis or ozonolysis are traditionally applied. Such a cascade was extended to make 11‐aminoundecanoic acid from 7 a, leading to 77 % product from 300 mm 7 a.[206] The synthesis of ω‐aminododecanoic acid from lauric acid on a pilot scale was also reported using a whole‐cell system for enzymatic oxidation and transamination.[207]
Nowadays, complex products are produced in suitable microbial hosts that have been designed by metabolic engineering, for example, by introducing artificial or improving existing pathways. Thus, biofuels, fatty acid derivatives, and surfactants such as sophorolipids can be made directly in microorganisms. As an example, an engineered pathway to produce the polyunsaturated fatty acid EPA (15 a)—commonly obtained from fish oil—in the oleagineous yeast Yarrowia lipolytica was reported.[208] 15 a was formed in up to 15 % yeast cell dry weight, whereby >56 % of the total fatty acids was 15 a. To achieve this titer, nine chimeric genes were newly introduced into the yeast; a Δ17‐desaturase was shown to be the most important, besides a mutation in the peroxisome.
More recently, a Rhodococcus opacus strain was engineered to produce free fatty acids (FFA) or FAEE and long‐chain hydrocarbons.[209] First, the formation of triglycerides from glucose was improved (82.9 g L−1). Next, an engineered strain with acyl‐coenzyme A synthetase genes deleted and overexpressing three lipases produced 50.2 g L−1 of FFAs. Another engineered strain, in which also heterologous aldehyde/alcohol dehydrogenase and wax ester synthase were overexpressed, made 50.2 g L−1 FAEEs. Finally, a third construct enabled the formation of 5.2 g L−1 of hydrocarbons. All these values are substantially higher than those of previously reported E. coli‐based systems.[210] Thus, metabolic engineering strategies can contribute to oleaginous biorefinery platforms for the sustainable production of chemicals and fuels.
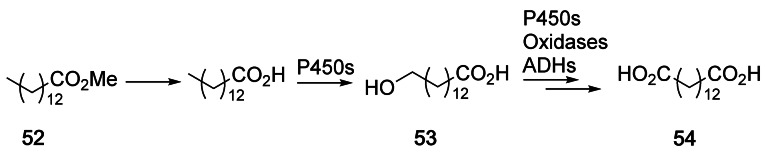
Selective oxidation of non‐activated C−H bonds is a very challenging reaction in organic synthesis, but it can be achieved efficiently using biocatalysis. The oxidation of long‐chain alkanes or fatty acids to α,ω‐diacids by Candida tropicalis, a natural degrader of these compounds, was initially already reported in the 1990ies.[133, 211] More recently, the competing β‐oxidation pathway in the strain C. tropicalis H5343 was targeted and after deletion of four genes, high yields of tetradecanedioic acid (54, 210 g L−1) from methyl myristate 52 were achieved (Scheme 28).[211]
Scheme 28.
Conversion of methyl myristate 52 to the corresponding ω‐hydroxyacid 53 or α,ω‐diacid 54 by Candida tropicalis using P450s, oxidases, and alcohol dehydrogenases (ADHs).[211]
In another study, 16 genes involved in the oxidation of the ω‐hydroxy acids were deleted in the C. tropicalis DP428 strain. This led to the formation of 174 g L−1 of 14‐hydroxytetradecanoic acid 53 by biotransformation of 52 (200 g L−1) within 148 h.[211] C. tropicalis is currently implemented on a commercial scale (40,000 tonnes per year).[212] These examples illustrate the high efficiency of P450s for the conversion of the native substrates in the native host.
6. Improvement in the Production of Natural Oils and Fats in Plants
Microbial and plant oils are valuable and diverse feedstocks for industry.[213] To allow for an efficient and increasingly sustainable supply of fatty acids and thus to promote the above discussed possible applications, improvements in the production of fats and oils are of high importance. While oil production with crop plants on a large scale is very well established, the use of microbial and algal fermentation is still restricted to high‐value products in specialized organisms like Yarrowia, Mortierella, Chlorella, and Schizochytrium.[214] Until now, more than 450 different fatty acids of plant origin have been described (Scheme 1). During the last decade, two databases have been published summarizing data on the different fatty acids that can be found in plants and algae.[215, 216]
The biosynthesis of oils and storage lipids in plants has been established over the last decades and this knowledge now serves as a basis for developing tailor‐made oils with the help of modern biotechnology.[213, 217, 218, 219] The production may be optimized and/or manipulated in three steps (Figure 3): 1) Oil biosynthesis, starting with fatty acid biosynthesis, which yields the backbone and chain lengths of up to 18 carbons. 2) Next, functional groups may be introduced, such as double bonds, hydroxyl groups as in 7 a, or cyclopropane rings as in 10 a, or the backbone can be further elongated up to C36.[213, 220] 3) Finally, these fatty acids need to be transferred into triacylglycerols (TAG) or wax esters (WE) by specific acyltransferases.[221]
Figure 3.
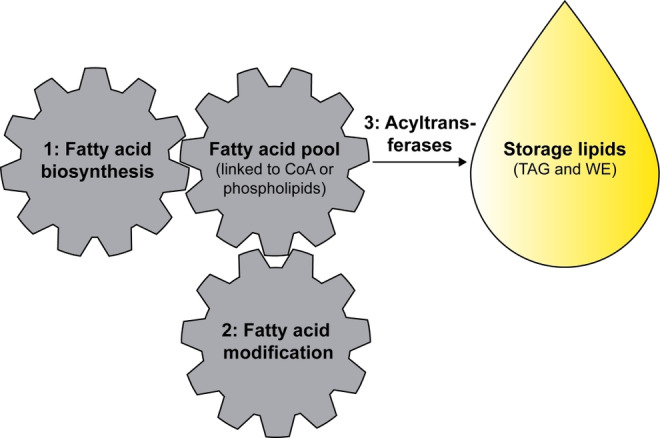
Oil synthesis in a living cell may depend on three steps.
6.1. Improving Existing Oil Qualities in Crop Plants
While classical breeding aims at improving the yield and composition of existing plant oils, modern biotechnological methods can transfer foreign genes to crop plants to improve the yield and/or to generate new oil qualities even beyond natural limits.[217] Over the last decade, the “push/pull concept” has been developed to increase oil yield in crop plants.[222] Here, fatty acid biosynthesis is increased by overexpressing the transcription factor WRINKLED1 (WRI1), which “pushes” additional substrate into the pathway. This is combined by expressing the final enzyme ACYL‐COA:DIACYLGLYCEROL ACYLTRANSFERASE 1 (DGAT1) of the oil‐forming pathway, which “pulls” fatty acids out of the membranes into the storage lipid pool. So far, the overexpression of WRI1 alone in soybean led to an increase of up to 3 % in the oil content under field conditions.[223] In corn, a natural mutation leading to an amino acid exchange in DGAT1 resulted in an increase in oil and 2 a contents by up to 41 % and 107 %, respectively.[224] Besides increasing the general flux into the oil, the assembly of the triacylglycerol molecules can also often limit the yield. In the case of the high production of erucic acid (6 a) in rapeseed, a natural limit is about 50 % 6 a, because in many commercial varieties a specific acyl transferase is missing, which transfers 6 a into the sn2‐position of the triglyceride. This limitation was overcome by overexpressing the endogenous rapeseed FATTY ACID ELONGASE 1 (FAE1, “push” approach) in combination with an ACYL‐COA:LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE (LPAAT, “pull” approach) gene from Limnanthes douglasii that had a high substrate preference for 6 a. Indeed, the obtained rapeseed oil from these plants contained up to 72 % of 6 a.[225] Another approach to produce 6 a is to establish alternative industrial oil crops like Crambe abyssinica that already have higher endogenous amounts of this fatty acid in their seed oil. This led to the production of 77 % of 6 a.[226]
6.2. Establishing New Oil Qualities in Crop Plants and Algae
Over the last decade, important achievements have been made to generate new oil qualities in existing crop plants. The prerequisites for the production of 7 a in rapeseed are currently analyzed by transferring all necessary genes, step by step, into the model brassicaceae Arabidopsis thaliana (thale cress). The expression of the FATTY ACID HYDROXYLASE12 (FAH12) gene from castor bean alone led to a drastic reduction of the oil content and accumulation of 7 a was limited to about 13 % in A. thaliana.[227] However, the additional expression of three acyltransferase genes, which specifically incorporate 7 a at each stereochemical position of the TAG molecule, resulted in normal oil accumulation, which now contained up to 44 % of 7 a.[228] The production of the cyclopropane fatty acid dihydrosterculic acid 11 a was recently established in the biofuel crop Camelina sativa (false flax).[229, 230, 231] Here, expression of an enzyme from E. coli resulted in an accumulation of about 10 % of 11 a in the seed oil of an high oleic acid line.[232] An additional increase to more than 15 % was achieved by introducing a LPAAT gene from Sterculia foetida (skunk tree). The formation of the conjugated fatty acids α‐eleostearic acid 13 a and punicic acid 14 a in soybean, tobacco, and rapeseed by introducing the corresponding conjugases alone is currently limited to about 15 %.[233, 234, 235] In summary, the picture emerges that the introduction of only one single enzyme catalyzing the formation of a new fatty acid results in the accumulation at levels between 10 and 15 % in crop plants. In many cases, this is due to a limited transfer of the fatty acid from the membrane lipid pool to the storage lipid pool.[217, 221, 236] Future work is therefore needed to identify the necessary acyltransferases.[237]
While the oil qualities discussed above are primarily used as feedstock for the chemical industry, the production of very long chain ω3‐fatty acids in crop plants is for feed and food purposes. Here, between three and seven foreign genes have to be transferred into crop plants.[218, 238] Similar to the production of 7 a and 11 a discussed above, their yield depends not only on their effective biosynthesis and transfer into the storage lipid fraction (steps 2 and 3 in Figure 3), but also on the oil composition of the crop plant that is used as the production platform. While the minimal set of three genes needed for the production of eicosapentaenoic acid 15 a led to the formation of about 1 % in linseed, because this plant oil is already rich in the ω3‐fatty acid 5 a (about 50 %) as starting material, the same construct led only to the formation of half of the amount in rapeseed.[239, 240] Consequently another crop plant rich in 5 a, C. sativa, is currently used as the platform for the production of 15 a and docosahexaenoic acid 16 a. In order to further optimize this oil as a starting material, its DGAT1 gene, which confers the transfer of saturated and monounsaturated fatty acids into the seed oil, was downregulated leading to an increase in α‐linolenic acid to about 56 %.[241] In an independent approach, all three FAE1 genes that are responsible for chain elongation longer than C18 were mutagenized, resulting in a α‐linolenic acid content of about 50 %.[242] Recently, C. sativa plants transformed with a seven‐gene construct were shown to produce about 10 % of each 15 a and 16 a in field trials.[243] This oil composition mirrors closely the quality of fish oil. Through the use of another combination of seven genes, also about 10 % of 16 a was produced in field trails in rapeseed.[244] In the case of this trial, it should be noted that similar approaches were used to optimize the production of 15 a and 16 a in the marine diatom Phaeodactylum tricornutum.[245]
To produce biofuels, lubricants, or other industrial feedstocks from plant oils, often only the fatty acids are used and the remaining glycerol might be a leftover. In order to address this problem, two different oil qualities are under development. A specific DGAT enzyme introduces an acetate group to the sn3‐position of TAG molecules to yield so‐called acetyl‐TAG species. This oil quality has the advantage that it can be used directly as a fuel or lubricant. Introducing an enzyme from Euonymus alatus (burning bush) into Camelina yielded about 70 % the corresponding acetyl‐TAGs in the seed oil in field trials.[246] Introducing the same enzyme into Camelina lines producing high amounts of medium‐chain fatty acids (C8–C14) in their seed oil even improved the properties of the oil.[247] The second strategy aims to replace TAG molecules by wax ester (WE) molecules in the seed oil of crop plants. Here, a fatty acid is directly esterified to a fatty alcohol by a wax synthase. This acyltransferase and a fatty acid reductase are introduced, the latter providing the enzyme with fatty alcohols.[248, 249] So far, between 30 and 50 % of the TAG in the seed oil was replaced by WE. The resulting oil quality ranged from wax esters derived from 6 a and 6 c in Crambe abyssinica and 2 a and 2 d to even shorter chain lengths in Camelina, depending on the corresponding fatty acid pattern of the genetic background used.[250, 251, 252]
6.3. Oil Production in Vegetative Plant Tissues and Algae
Although the yield of oilseed crops has increased over the last decades, it is still a challenge to meet the steadily growing demand for plant oils as renewable resources or for high‐energy feed and food applications (compare also Section 2).[217, 231, 236, 249, 253] One strategy to solve this problem may be the production of oil in all vegetative tissues instead of seeds only. This can be achieved by driving the expression of the whole oil biosynthesis machinery under the control of senescence‐specific promotors at the end of a plant's annual life cycle, when its biomass has reached its maximum (Figure 3). The oilseed crop plant tobacco serves as a model to develop strategies to reach this goal and the “push and pull” concept was further extended to “push, pull, and protect”.[236, 254] The latter point addresses appropriate packaging and unwanted degradation of the product in the cells of the sink tissue. More than 15 % of TAG accumulated in tobacco leaves by the co‐expression of three genes: WRI1, DGAT1, and oleosin (“protect”).[255] By additional silencing of the TAG degrading lipase SDP1, accumulation could be increased up to 33 % in tobacco and up to 8.4 % in sorghum leaves.[256, 257] Again similar approaches were used to optimize oil production in the marine diatom Phaeodactylum tricornutum, in the heterokont oleaginous microalga Nannochloropsis oceanica, and in the oleaginous fungus Mortierella alpina.[258, 259, 260, 261]
7. Conclusion and Outlook
Oils and fats of vegetable and animal origin have been and probably will remain the most important renewable feedstock of the chemical industry. It can be expected that the observed geographical shift of oleochemical production to South East Asia will continue during the next decade. Selective catalytic reactions across the double bond has made important advances. The catalytic oxidative cleavage of the C–C double bond as well as the butenolysis of 2 b has been industrialized. It may be assumed that more reactions will be commercialized, for example ethenolysis and isomerizing alkoxycarbonylation. C–H activation of the saturated alkyl chain has not been tackled in this review and should be more intensively addressed in the coming decade. Microbial oils have been used increasingly as substrates for catalytic transformations. More examples will be applied specifically to these substrates that are becoming more readily available. The new fatty acid derivatives discussed are important as substrates for the synthesis, and, hopefully, production of a great variety of polyesters, polyamides, and polyurethanes. Equally importantly, enzyme or whole‐cell catalysis for the modification of fats and oils as well as the de novo synthesis of various fatty acids from abundantly available renewable crop plants will contribute to further advancing the field.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Ursula Biermann studied food chemistry in Hannover and Munich (Germany). She received her Ph.D. in 1979 at the Technical University of Munich. Since 1987, she has been a research fellow at the Institute of Chemistry of the University of Oldenburg under the direction of J. O. Metzger, where she worked on the synthesis of novel fatty compounds using natural oils and fats as chemical raw materials until she retired in 2018.

Biographical Information
Uwe T. Bornscheuer studied chemistry and received his Ph.D. in 1993 at Hannover University (Germany) followed by a postdoc at Nagoya University (Japan). In 1998, he completed his Habilitation at Stuttgart University on the use of enzymes in organic synthesis. He has been Professor at the Institute of Biochemistry at Greifswald University since 1999. In addition to other awards, he received the BioCat2008 Award in 2008. He was recently named a Chemistry Europe Fellow. His current research interests focus on the discovery, engineering, and applications of enzymes.

Biographical Information
Ivo Feussner studied chemistry in Marburg (Germany) and received his doctorate in 1993 under H. Kindl. During 1993–1999 he conducted research at the Martin‐Luther‐University and at the Institute for Plant Biochemistry in Halle/Saale. In 2000 he gained his Habilitation and moved to the Institute for Plant Genetics and Crop Plant Research in Gatersleben. Since 2002, he has been a Full Professor for Plant Biochemistry at the Georg‐August‐University of Göttingen. Since 2009 he has been a fellow of the Saxonian Academy of Sciences, Leipzig, and since 2012 a fellow of the Academy of Sciences, Göttingen.

Biographical Information
Michael A. R. Meier studied chemistry in Regensburg (Germany) and received his Ph.D. from the Eindhoven University of Technology (The Netherlands) in 2006. After further stays in Emden and Potsdam, he was appointed as full professor at the Karlsruhe institute of Technology (KIT) in 2010. He has received several awards and is associate editor of ACS Sustainable Chemistry & Engineering. His research interests include the sustainable use and derivatization of renewable resources for polymer chemistry as well as the design of novel highly defined macromolecular architectures.

Biographical Information
Jürgen O. Metzger studied chemistry and received his Ph.D. under the supervision of H. Sinn in 1970 at the University of Hamburg (Germany). In 1991, he was appointed Professor of Organic Chemistry at the University of Oldenburg, and he retired in 2006. He is chairman of abiosus e.V., a non‐profit association for the advancement of research on renewable raw materials. His research areas include sustainability in chemistry, and fats and oils as renewable raw materials.

Acknowledgements
The authors are grateful for financial support from the Deutsche Forschungsgemeinschaft (DFG), Bundesministerium für Bildung und Forschung (BMBF), Bundesministerium für Ernährung und Landwirtschaft (BMEL), the European Union, and the Fonds der Chemischen Industrie (FCI). Open access funding enabled and organized by Projekt DEAL.
U. Biermann, U. T. Bornscheuer, I. Feussner, M. A. R. Meier, J. O. Metzger, Angew. Chem. Int. Ed. 2021, 60, 20144.
Dedicated to equal opportunities and a diverse scientific workforce
References
- 1.Fachagentur Nachwachsende Rohstoffe e.V., Anbau und Verwendung nachwachsender Rohstoffe in Deutschland, 2020, https://mediathek.fnr.de/grafiken/daten-und-fakten/biobasierte-produkte/chemische-industrie/stoffliche-einsatzmengen-nachwachsender-rohstoffe-in-der-chemischen-industrie-in-deutschland.html, visited September 14, 2020.
- 2.Baumann H., Bühler M., Fochem H., Hirsinger F., Zoebelein H., Falbe J., Angew. Chem. Int. Ed. Engl. 1988, 27, 41–62; [Google Scholar]; Angew. Chem. 1988, 100, 41–62. [Google Scholar]
- 3.Biermann U., Friedt W., Lang S., Lühs W., Machmüller G., Metzger J. O., Klaas R. M., Schäfer H. J., Schneider M. P., Angew. Chem. Int. Ed. 2000, 39, 2206–2224; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2000, 112, 2292–2310. [Google Scholar]
- 4.Biermann U., Bornscheuer U., Meier M. A. R., Metzger J. O., Schäfer H. J., Angew. Chem. Int. Ed. 2011, 50, 3854–3871; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 3938–3956. [Google Scholar]
- 5.Oil World Annual 2020, ISTA Mielke GmbH, Hamburg, 2020.
- 6.Oil World Annual 2012, ISTA Mielke GmbH, Hamburg, 2012.
- 7.OECD/FAO (2020), OECD-FAO Agricultural Outlook 2020–2029, FAO, Rome/OECD Publishing, Paris, p. 148, 10.1787/1112c23b-en. [DOI]
- 8.
- 8a.Oilseeds: World Markets and Trade, USDA, December 2009;
- 8b.Oilseeds: World Markets and Trade, USDA, September 2020, https://www.fas.usda.gov/data/oilseeds-world-markets-and-trade.
- 9.Fraile J. M., Garcia J. I., Herrerias C. I., Pires E., Synthesis 2017, 49, 1444–1460. [Google Scholar]
- 10.Stadler B. M., Wulf C., Werner T., Tin S., de Vries J. G., ACS Catal. 2019, 9, 8012–8067. [Google Scholar]
- 11.Silverman J. R., Samateh M., John G., Eur. J. Lipid Sci. Technol. 2016, 118, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a.Mittelbach M., Eur. J. Lipid Sci. Technol. 2015, 117, 1832–1846; [Google Scholar]
- 12b.Dawes G. J. S., Scott E. L., Le Notre J., Sanders J. P. M., Bitter J. H., Green Chem. 2015, 17, 3231–3250; [Google Scholar]
- 12c.Chatterjee A., Eliasson S. H. H., Jensen V. R., Catal. Sci. Technol. 2018, 8, 1487–1499; [Google Scholar]
- 12d.Zhang X., Jordan F., Szostak M., Org. Chem. Front. 2018, 5, 2515–2521. [Google Scholar]
- 13.
- 13a.dos Santos T. R., Harnisch F., Nilges P., Schroder U., ChemSusChem 2015, 8, 886–893; [DOI] [PubMed] [Google Scholar]
- 13b.Yuan G., Wang L., Zhang X. W., Luque R., Wang Q. F., Green Chem. 2020, 22, 525–531. [Google Scholar]
- 14.
- 14a.Schäfer H. J., Eur. J. Lipid Sci. Technol. 2012, 114, 2–9; [Google Scholar]
- 14b.Holzhäuser F. J., Mensah J. B., Palkovits R., Green Chem. 2020, 22, 286–301. [Google Scholar]
- 15.Gunanathan C., Milstein D., Chem. Rev. 2014, 114, 12024–12087. [DOI] [PubMed] [Google Scholar]
- 16.Morris S. A., Gusev D. G., Angew. Chem. Int. Ed. 2017, 56, 6228–6231; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 6324–6327. [Google Scholar]
- 17.Alig L., Fritz M., Schneider S., Chem. Rev. 2019, 119, 2681–2751. [DOI] [PubMed] [Google Scholar]
- 18.Junge K., Wendt B., Cingolani A., Spannenberg A., Wei Z. H., Jiao H. J., Beller M., Chem. Eur. J. 2018, 24, 1046–1052. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee A., Milstein D., ACS Catal. 2018, 8, 11435–11469. [Google Scholar]
- 20.Werkmeister S., Neumann J., Junge K., Beller M., Chem. Eur. J. 2015, 21, 12226–12250. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen D. H., Raffa G., Morin Y., Desset S., Capet F., Nardello-Rataj V., Dumeignil F., Gauvin R. M., Green Chem. 2017, 19, 5665–5673. [Google Scholar]
- 22.Nguyen D. H., Trivelli X., Capet F., Swesi Y., Favre-Reguillon A., Vanoye L., Dumeignil F., Gauvin R. M., ACS Catal. 2018, 8, 4719–4734. [Google Scholar]
- 23.Sakai N., Moriya T., Fujii K., Konakahara T., Synthesis 2008, 3533–3536. [Google Scholar]
- 24.Biermann U., Metzger J. O., ChemSusChem 2014, 7, 644–649. [DOI] [PubMed] [Google Scholar]
- 25.Das S., Li Y., Junge K., Beller M., Chem. Commun. 2012, 48, 10742–10744. [DOI] [PubMed] [Google Scholar]
- 26.Rysak V., Dixit R., Trivelli X., Merle N., Agbossou-Niedercorn F., Vanka K., Michon C., Catal. Sci. Technol. 2020, 10, 4586–4592. [Google Scholar]
- 27.Biermann U., Metzger J. O., Eur. J. Lipid Sci. Technol. 2014, 116, 74–79. [Google Scholar]
- 28.Dannecker P.-K., Biermann U., von Czapiewski M., Metzger J. O., Meier M. A. R., Angew. Chem. Int. Ed. 2018, 57, 8775–8779; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 8911–8915. [Google Scholar]
- 29.Li Y., Topf C., Cui X., Junge K., Beller M., Angew. Chem. Int. Ed. 2015, 54, 5196–5200; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 5285–5289. [Google Scholar]
- 30.Erb B., Risto E., Wendling T., Gooßen L. J., ChemSusChem 2016, 9, 1442–1448. [DOI] [PubMed] [Google Scholar]
- 31.
- 31a.Adam R., Cabrero-Antonino J. R., Junge K., Jackstell R., Beller M., Angew. Chem. Int. Ed. 2016, 55, 11049–11053; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 11215–11219; [Google Scholar]
- 31b.Cabrero-Antonino J. R., Adam R., Beller M., Angew. Chem. Int. Ed. 2019, 58, 12820–12838; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 12950–12968; [Google Scholar]
- 31c.Shi Y., Kamer P. C. J., Cole-Hamilton D. J., Green Chem. 2017, 19, 5460–5466. [Google Scholar]
- 32.Sutter M., Dayoub W., Métay E., Raoul Y., Lemaire M., ChemSusChem 2012, 5, 2397–2409. [DOI] [PubMed] [Google Scholar]
- 33.Sutter M., Da Silva E., Duguet N., Raoul Y., Métay E., Lemaire M., Chem. Rev. 2015, 115, 8609–8651. [DOI] [PubMed] [Google Scholar]
- 34.Winkler M., Meier M. A. R., Green Chem. 2014, 16, 1784–1788. [Google Scholar]
- 35.Morandi B., Wickens Z. K., Grubbs R. H., Angew. Chem. Int. Ed. 2013, 52, 2944–2948; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 3016–3020. [Google Scholar]
- 36.
- 36a.Rios L. A., Llano B. A., Hoelderich W. F., Appl. Catal. A 2012, 445, 346–350; [Google Scholar]
- 36b.Dorado V., Gil L., Mayoral J. A., Herrerias C. I., Fraile J. M., Catal. Sci. Technol. 2020, 10, 1789–1795. [Google Scholar]
- 37.Sereinig N., Janssen M. C. C., de Vries J. G., WO 2013/013990.
- 38.Wickens Z. K., Morandi B., Grubbs R. H., Angew. Chem. Int. Ed. 2013, 52, 11257–11260; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 11467–11470. [Google Scholar]
- 39.Danov S. M., Kazantsev O. A., Esipovich A. L., Belousov A. S., Rogozhin A. E., Kanakov E. A., Catal. Sci. Technol. 2017, 7, 3659–3675. [Google Scholar]
- 40.Milchert E., Malarczyk K., Klos M., Molecules 2015, 20, 21481–21493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schäffner B., Blug M., Kruse D., Polyakov M., Kockritz A., Martin A., Rajagopalan P., Bentrup U., Bruckner A., Jung S., Agar D., Rungeler B., Pfennig A., Muller K., Arlt W., Woldt B., Grass M., Buchholz S., ChemSusChem 2014, 7, 1133–1139. [DOI] [PubMed] [Google Scholar]
- 42.Büttner H., Longwitz L., Steinbauer J., Wulf C., Werner T., Top. Curr. Chem. 2017, 375, 50. [DOI] [PubMed] [Google Scholar]
- 43.Longwitz L., Steinhauer J., Spannenberg A., Werner T., ACS Catal. 2018, 8, 665–672. [Google Scholar]
- 44.Büttner H., Steinbauer J., Wulf C., Dindaroglu M., Schmalz H. G., Werner T., ChemSusChem 2017, 10, 1076–1079. [DOI] [PubMed] [Google Scholar]
- 45.Peña Carrodeguas L., Cristofol A., Fraile J. M., Mayoral J. A., Dorado V., Herrerias C. I., Kleij A. W., Green Chem. 2017, 19, 3535–3541. [Google Scholar]
- 46.Köckritz A., Martin A., Eur. J. Lipid Sci. Technol. 2011, 113, 83–91. [Google Scholar]
- 47.Enferadi Kerenkan A., Beland F., Do T. O., Catal. Sci. Technol. 2016, 6, 971–987. [Google Scholar]
- 48.Spannring P., Bruijnincx P. C. A., Weckhuysen B. M., Gebbink R., Catal. Sci. Technol. 2014, 4, 2182–2209. [Google Scholar]
- 49.Teong S. P., Li X. K., Zhang Y. G., Green Chem. 2019, 21, 5753–5780. [Google Scholar]
- 50.
- 50a.Behr A., Tenhumberg N., Wintzer A., RSC Adv. 2013, 3, 172–180; [Google Scholar]
- 50b.Behr A., Tenhumberg N., Wintzer A., Eur. J. Lipid Sci. Technol. 2012, 114, 905–910. [Google Scholar]
- 51.Soutelo-Maria A., Dubois J. L., Couturier J. L., Cravotto G., Catalysts 2018, 8, 464. [Google Scholar]
- 52.Bieser A., Borsotti G., Digioia F., Ferrari A., Pirocco A. (Novamont S. P. A.), U.S. Patent 8,846,962 (B2), 2011.
- 53.Hu C. Q., Creaser D., Siahrostami S., Gronbeck H., Ojagh H., Skoglundh M., Catal. Sci. Technol. 2014, 4, 2427–2444. [Google Scholar]
- 54.Behr A., Doring N., Durowiczheil S., Ellenberg B., Kozik C., Lohr C., Schmidke H., Fett Wiss. Technol. 1993, 95, 2–11. [Google Scholar]
- 55.Liu W., Xu L. G., Lu G. H., Zhang H., ACS Sustainable Chem. Eng. 2017, 5, 1368–1375. [Google Scholar]
- 56.Zaccheria F., Psaro R., Ravasio N., Bondioli P., Eur. J. Lipid Sci. Technol. 2012, 114, 24–30. [Google Scholar]
- 57.Philippaerts A., Jacobs P. A., Sels B. F., Angew. Chem. Int. Ed. 2013, 52, 5220–5226; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 5328–5334. [Google Scholar]
- 58.Haßelberg J., Behr A., Eur. J. Lipid Sci. Technol. 2016, 118, 36–46. [Google Scholar]
- 59.Erb B., Dierker M., Ohlmann D. M., Gooßen L. J., Eur. J. Lipid Sci. Technol. 2016, 118, 111–116. [Google Scholar]
- 60.Biermann U., Metzger J. O., Eur. J. Lipid Sci. Technol. 2018, 120, 1700318. [Google Scholar]
- 61.
- 61a.Ngo H. L., Dunn R. O., Sharma B., Foglia T. A., Eur. J. Lipid Sci. Technol. 2011, 113, 180–188; [Google Scholar]
- 61b.Sarker M. I., Latona R. J., Moreau R. A., Micheroni D., Jones K. C., Ngo H., Eur. J. Lipid Sci. Technol. 2017, 119, 1700262. [Google Scholar]
- 62.
- 62a.Wiedemann S. C. C., Stewart J. A., Soulimani F., van Bergen-Brenkman T., Langelaar S., Wels B., de Peinder P., Bruijnincx P. C. A., Weckhuysen B. M., J. Catal. 2014, 316, 24–35; [Google Scholar]
- 62b.Wiedemann S. C. C., Ristanović Z., Whiting G. T., Reddy Marthala V. R., Kärger J., Weitkamp J., Wels B., Bruijnincx P. C. A., Weckhuysen B. M., Chem. Eur. J. 2016, 22, 199–210. [DOI] [PubMed] [Google Scholar]
- 63.Biermann U., Jungbauer A., Metzger J. O., Eur. J. Lipid Sci. Technol. 2012, 114, 49–54. [Google Scholar]
- 64.Eschig S., Philipp C., Salthammer T., Eur. J. Lipid Sci. Technol. 2013, 115, 101–110. [Google Scholar]
- 65.Manurung R., Daniel L., van de Bovenkamp H. H., Buntara T., Maemunah S., Kraai G., Makertihartha I. G. B. N., Broekhuis A. A., Heeres H. J., Eur. J. Lipid Sci. Technol. 2012, 114, 31–48. [Google Scholar]
- 66.
- 66a.Franke R., Selent D., Borner A., Chem. Rev. 2012, 112, 5675–5732; [DOI] [PubMed] [Google Scholar]
- 66b.Vanbésien T., Monflier E., Hapiot F., Eur. J. Lipid Sci. Technol. 2016, 118, 26–35. [Google Scholar]
- 67.Vanbésien T., Le Notre J., Monflier E., Hapiot F., Eur. J. Lipid Sci. Technol. 2018, 120, 1700190. [Google Scholar]
- 68.Goldbach V., Roesle P., Mecking S., ACS Catal. 2015, 5, 5951–5972. [Google Scholar]
- 69.Gaide T., Bianga J., Schlipkoter K., Behr A., Vorholt A. J., ACS Catal. 2017, 7, 4163–4171. [Google Scholar]
- 70.Yuki Y., Takahashi K., Tanaka Y., Nozaki K., J. Am. Chem. Soc. 2013, 135, 17393–17400. [DOI] [PubMed] [Google Scholar]
- 71.Dreimann J. M., Skiborowski M., Behr A., Vorholt A. J., ChemCatChem 2016, 8, 3330–3333. [Google Scholar]
- 72.Furst M. R. L., Korkmaz V., Gaide T., Seidensticker T., Behr A., Vorholt A. J., ChemCatChem 2017, 9, 4319–4323. [Google Scholar]
- 73.Bianga J., Knnemann K. U., Gaide T., Vorholt A. J., Seidensticker T., Dreimann J. M., Vogt D., Chem. Eur. J. 2019, 25, 11586–11608. [DOI] [PubMed] [Google Scholar]
- 74.
- 74a.Gaide T., Dreimann J. M., Behr A., Vorholt A. J., Angew. Chem. Int. Ed. 2016, 55, 2924–2928; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 2977–2981; [Google Scholar]
- 74b.Bianga J., Herrmann N., Schurm L., Gaide T., Dreimann J. M., Vogt D., Seidensticker T., Eur. J. Lipid Sci. Technol. 2020, 122, 1900317. [Google Scholar]
- 75.Herrmann N., Bianga J., Gaide T., Drewing M., Vogt D., Seidensticker T., Green Chem. 2019, 21, 6738–6745. [Google Scholar]
- 76.Herrmann N., Bianga J., Palten M., Riemer T., Vogt D., Dreimann J. M., Seidensticker T., Eur. J. Lipid Sci. Technol. 2020, 122, 1900166. [Google Scholar]
- 77.Seidensticker T., Busch H., Diederichs C., von Dincklage J. J., Vorholt A. J., ChemCatChem 2016, 8, 2890–2893. [Google Scholar]
- 78.
- 78a.Roesle P., Durr C. J., Moller H. M., Cavallo L., Caporaso L., Mecking S., J. Am. Chem. Soc. 2012, 134, 17696–17703; [DOI] [PubMed] [Google Scholar]
- 78b.Roesle P., Caporaso L., Schnitte M., Goldbach V., Cavallo L., Mecking S., J. Am. Chem. Soc. 2014, 136, 16871–16881. [DOI] [PubMed] [Google Scholar]
- 79.Walther G., Knopke L. R., Rabeah J., Checinski M. P., Jiao H. J., Bentrup U., Bruckner A., Martin A., Kockritz A., J. Catal. 2013, 297, 44–55. [Google Scholar]
- 80.Jameel F., Kohls E., Stein M., ChemCatChem 2019, 11, 4894–4906. [Google Scholar]
- 81.Stempfle F., Quinzler D., Heckler I., Mecking S., Macromolecules 2011, 44, 4159–4166. [Google Scholar]
- 82.Christl J. T., Roesle P., Stempfle F., Wucher P., Gottker-Schnetmann I., Muller G., Mecking S., Chem. Eur. J. 2013, 19, 17131–17140. [DOI] [PubMed] [Google Scholar]
- 83.Walther G., Deutsch J., Martin A., Baumann F. E., Fridag D., Franke R., Köckritz A., ChemSusChem 2011, 4, 1052–1054. [DOI] [PubMed] [Google Scholar]
- 84.Furst M. R. L., Le Goff R., Quinzler D., Mecking S., Botting C. H., Cole-Hamilton D. J., Green Chem. 2012, 14, 472–477. [Google Scholar]
- 85.
- 85a.Roesle P., Stempfle F., Hess S. K., Zimmerer J., Bartulos C. R., Lepetit B., Eckert A., Kroth P. G., Mecking S., Angew. Chem. Int. Ed. 2014, 53, 6800–6804; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 6918–6922; [Google Scholar]
- 85b.Hess S. K., Schunck N. S., Goldbach V., Ewe D., Kroth P. G., Mecking S., J. Am. Chem. Soc. 2017, 139, 13487–13491; [DOI] [PubMed] [Google Scholar]
- 85c.Hess S. K., Lepetit B., Kroth P. G., Mecking S., Eur. J. Lipid Sci. Technol. 2018, 120, 1700152. [Google Scholar]
- 86.Goldbach V., Falivene L., Caporaso L., Cavallo L., Mecking S., ACS Catal. 2016, 6, 8229–8238. [Google Scholar]
- 87.Gaide T., Behr A., Arns A., Benski F., Vorholt A. J., Chem. Eng. Process. 2016, 99, 197–204. [Google Scholar]
- 88.Terhorst M., Heider C., Vorholt A., Vogt D., Seidensticker T., ACS Sustainable Chem. Eng. 2020, 8, 10633–10638. [Google Scholar]
- 89.de Espinosa L. M., Meier M. A. R., in Organometallics and Renewables. Topics in Organometallic Chemistry, Vol. 39 (Eds.: Meier M. A. R., Weckhuysen B., Bruijnincx P.), Springer, Berlin, Heidelberg, 2012, pp. 1–44. [Google Scholar]
- 90.Chikkali S., Mecking S., Angew. Chem. Int. Ed. 2012, 51, 5802–5808; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 5902–5909. [Google Scholar]
- 91.Bruneau C., Fischmeister C., in Organometallics for Green Catalysis, Vol. 63 (Eds.: Dixneuf P. H., Soule J. F.), 2019, pp. 77–102. [Google Scholar]
- 92.Yelchuri V., Srikanth K., Prasad R. B. N., Karuna M. S. L., J. Chem. Sci. 2019, 131, 39. [Google Scholar]
- 93.
- 93a.Nickel A., Pederson R. L., in Olefin Metathesis: Theory and Practice (Ed.: Grela K.), Wiley & Sons, Hoboken, 2014, p. 335–348; [Google Scholar]
- 93b.Nickel A., Ung T., Mkrtumyan G., Uy J., Lee C. W., Stoianova D., Papazian J., Wei W. H., Mallari A., Schrodi Y., Pederson R. L., Top. Catal. 2012, 55, 518–523; [Google Scholar]
- 93c.https://elevance.com/technology/, visited 28.12.2020.
- 94.Bidange J., Fischmeister C., Bruneau C., Chem. Eur. J. 2016, 22, 12226–12244. [DOI] [PubMed] [Google Scholar]
- 95.Spekreijse J., Sanders J. P. M., Bitter J. H., Scott E. L., ChemSusChem 2017, 10, 470–482. [DOI] [PubMed] [Google Scholar]
- 96.Pollini J., Pankau W. M., Goossen L. J., Chem. Eur. J. 2019, 25, 7416–7425. [DOI] [PubMed] [Google Scholar]
- 97.Mathers T., Shreve M. J., Meyler E., Damodaran K., Iwig D. F., Kelley D. J., Macromol. Rapid Commun. 2011, 32, 1338–1342. [DOI] [PubMed] [Google Scholar]
- 98.Mutlu H., Hofsass R., Montenegro R. E., Meier M. A. R., RSC Adv. 2013, 3, 4927–4934. [Google Scholar]
- 99.Pingen D., Zimmerer J., Klinkenberg N., Mecking S., Green Chem. 2018, 20, 1874–1878. [Google Scholar]
- 100.Thomas M., Keitz B. K., Champagne T. M., Grubbs R. H., J. Am. Chem. Soc. 2011, 133, 7490–7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J., Song S. F., Wang X., Jiao J. J., Shi M., Chem. Commun. 2013, 49, 9491–9493. [DOI] [PubMed] [Google Scholar]
- 102.Marx V. M., Sullivan A. H., Melaimi M., Virgil S. C., Keitz B. K., Weinberger D. S., Bertrand G., Grubbs R. H., Angew. Chem. Int. Ed. 2015, 54, 1919–1923; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 1939–1943. [Google Scholar]
- 103.Gawin R., Kozakiewicz A., Gunka P. A., Dabrowski P., Skowerski K., Angew. Chem. Int. Ed. 2017, 56, 981–986; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 1001–1006. [Google Scholar]
- 104.Nascimento D. L., Gawin A., Gawin R., Gunka P. A., Zachara J., Skowerski K., Fogg D. E., J. Am. Chem. Soc. 2019, 141, 10626–10631; see also [DOI] [PubMed] [Google Scholar]; Nascimento D. L., Fogg D. E., J. Am. Chem. Soc. 2019, 141, 19236–19240. [DOI] [PubMed] [Google Scholar]
- 105.Wyrebek P., Malecki P., Sytniczuk A., Kosnik W., Gawin A., Kostrzewa J., Kajetanowicz A., Grela K., ACS Omega 2018, 3, 18481–18488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kajetanowicz A., Chwalba M., Gawin A., Tracz A., Grela K., Eur. J. Lipid Sci. Technol. 2020, 122, 1900263. [Google Scholar]
- 107.Biermann U., Meier M. A. R., Butte W., Metzger J. O., Eur. J. Lipid Sci. Technol. 2011, 113, 39–45. [Google Scholar]
- 108.
- 108a.Behr A., Toepell S., Harmuth S., RSC Adv. 2014, 4, 16320–16326; [Google Scholar]
- 108b.Behr A., Gomes J. P., Bayrak Z., Eur. J. Lipid Sci. Technol. 2011, 113, 189–196. [Google Scholar]
- 109.Ferreira L. A., Schrekker H. S., Catal. Sci. Technol. 2016, 6, 8138–8147. [Google Scholar]
- 110.Behr A., Toepell S., J. Am. Oil Chem. Soc. 2015, 92, 603–611. [Google Scholar]
- 111.von Czapiewski M., Kreye O., Mutlu H., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2013, 115, 76–85. [Google Scholar]
- 112.Kajetanowicz A., Sytniczuk A., Grela K., Green Chem. 2014, 16, 1579–1585. [Google Scholar]
- 113.Ref. [91], pp. 95–99.
- 114.Miao X. W., Fischmeister C., Bruneau C., Dixneuf P. H., Dubois J. L., Couturier J. L., ChemSusChem 2012, 5, 1410–1414. [DOI] [PubMed] [Google Scholar]
- 115.Bonin H., Keraani A., Dubois J. L., Brandhorst M., Fischmeister C., Bruneau C., Eur. J. Lipid Sci. Technol. 2015, 117, 209–216. [Google Scholar]
- 116.Koh M. J., Khan R. K. M., Torker S., Yu M., Mikus M. S., Hoveyda A. H., Nature 2015, 517, 181–186. [DOI] [PubMed] [Google Scholar]
- 117.Ohlmann D. M., Tschauder N., Stockis J. P., Goossen K., Dierker M., Gooßen L. J., J. Am. Chem. Soc. 2012, 134, 13716–13729. [DOI] [PubMed] [Google Scholar]
- 118.Pfister K. F., Baader S., Baader M., Berndt S., Gooßen L. J., Sci. Adv. 2017, 3, e1602624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Montenegro R. E., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2012, 114, 55–62. [Google Scholar]
- 120.Abd-El-Aziz A. S., Antonietti M., Barner-Kowollik C., Binder W. H., Böker A., Boyer C., Buchmeiser M. R., Cheng S. Z. D., D′Agosto F., Floudas G., Frey H., Galli G., Genzer J., Hartmann L., Hoogenboom R., Ishizone T., Kaplan D. L., Leclerc M., Lendlein A., Liu B., Long T. E., Ludwigs S., Lutz J.-F., Matyjaszewski K., Meier M. A. R., Müllen K., Müllner M., Rieger B., Russell T. P., Savin D. A., Schlüter A. D., Schubert U. S., Seiffert S., Severing K., Soares J. B. P., Staffilani M., Sumerlin B. S., Sun Y., Tang B. Z., Tang C., Theato P., Tirelli N., Tsui O. K. C., Unterlass M. M., Vana P., Voit B., Vyazovkin S., Weder C., Wiesner U., Wong W.-Y., Wu C., Yagci Y., Yuan J., Zhang G., Macromol. Chem. Phys. 2020, 221, 2000216. [Google Scholar]
- 121.Meier M. A. R., Metzger J. O., Schubert U. S., Chem. Soc. Rev. 2007, 36, 1788–1802. [DOI] [PubMed] [Google Scholar]
- 122.Montero de Espinosa L., Meier M. A. R., Eur. Polym. J. 2011, 47, 837–852. [Google Scholar]
- 123.Zhang C., Garrison T. F., Madbouly S. A., Kessler M. R., Prog. Polym. Sci. 2017, 71, 91–143. [Google Scholar]
- 124.Llevot A., J. Am. Oil Chem. Soc. 2017, 94, 169–186. [Google Scholar]
- 125.Kristufek S. L., Wacker K. T., Tsao Y.-Y. T., Suand L., Wooley K. L., Nat. Prod. Rep. 2017, 34, 433. [DOI] [PubMed] [Google Scholar]
- 126.Llevot A., Dannecker P.-K., von Czapiewski M., Over L. C., Söyler Z., Meier M. A. R., Chem. Eur. J. 2016, 22, 11510–11521. [DOI] [PubMed] [Google Scholar]
- 127.Maisonneuve L., Lebarbé T., Grau E., Cramail H., Polym. Chem. 2013, 4, 5472–5517. [Google Scholar]
- 128.
- 128a.Türünç O., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2013, 115, 41–54; [Google Scholar]
- 128b.Gandini A., Lacerda T. M., in Polymers from Plant Oils, Scrivener Publishing LLC, 2018, pp. 109–134; [Google Scholar]
- 128c.Lligadas G., Ronda J. C., Galià M., Cádiz V., J. Polym. Sci. Part A J. Polym. Sci. A 2013, 51, 2111–2124; [Google Scholar]
- 128d.Nguyen P. H., Spoljaric S., Seppälä J., Polymer 2018, 153, 83–192; [Google Scholar]
- 128e.Türünç O., Meier M. A. R., Macromol. Rapid Commun. 2010, 31, 1822–1826; [DOI] [PubMed] [Google Scholar]
- 128f.Türünç O., Firdaus M., Klein G., Meier M. A. R., Green Chem. 2012, 14, 2577–2583; [Google Scholar]
- 128g.van den Berg O., Dispinar T., Hommez B., Du Prez F. E., Eur. Polym. J. 2013, 49, 804–812; [Google Scholar]
- 128h.Smith A. D., Tennyson A. G., Smith R. C., Sustainable Chem. 2020, 1, 209–237. [Google Scholar]
- 129.Mecking S., Philos. Trans. R. Soc. A 2020, 378, 20190266. [DOI] [PubMed] [Google Scholar]
- 130.Stempfle F., Ortmann P., Mecking S., Macromol. Rapid Commun. 2013, 34, 47–50. [DOI] [PubMed] [Google Scholar]
- 131.Stempfle F., Ritter B. S., Mülhaupt R., Mecking S., Green Chem. 2014, 16, 2008–2014. [Google Scholar]
- 132.Kolb N., Winkler M., Syldatk C., Meier M. A. R., Eur. Polym. J. 2014, 51, 159–166. [Google Scholar]
- 133.Lu W., Ness J. E., Xie W., Zhang X., Minshull J., Gross R. A., J. Am. Chem. Soc. 2010, 132, 15451–15455. [DOI] [PubMed] [Google Scholar]
- 134.
- 134a.Bouyahyi M., Pepels M. P. F., Heise A., Duchateau R., Macromolecules 2012, 45, 3356–3366; [Google Scholar]
- 134b.Witt T., Häußler M., Mecking S., Macromol. Rapid Commun. 2017, 38, 1600638; [DOI] [PubMed] [Google Scholar]
- 134c.Pepels M. P. F., Koeken R. A. C., van der Linden S. J. J., Heise A., Duchateau R., Macromolecules 2015, 48, 4779–4792. [Google Scholar]
- 135.Witt T., Häußler M., Kulpa S., Mecking S., Angew. Chem. Int. Ed. 2017, 56, 7589–7594; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 7697–7702. [Google Scholar]
- 136.
- 136a.Dannecker P.-K., Biermann U., Sink A., Bloesser F. R., Metzger J. O., Meier M. A. R., Macromol. Chem. Phys. 2019, 220, 1800440; [Google Scholar]
- 136b.Firdaus M., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2014, 116, 31–36. [Google Scholar]
- 137.Stempfle F., Ortmann P., Mecking S., Chem. Rev. 2016, 116, 4597–4641. [DOI] [PubMed] [Google Scholar]
- 138.
- 138a.Meier M. A. R., Macromol. Rapid Commun. 2019, 40, 1800524; [DOI] [PubMed] [Google Scholar]
- 138b.Llevot A., Meier M. A. R., Polym. Int. 2019, 68, 826–831. [Google Scholar]
- 139.
- 139a.Pemba A. G., Flores J. A., Miller S. A., Green Chem. 2013, 15, 325–329; [Google Scholar]
- 139b.Chikkali S., Stempfle F., Mecking S., Macromol. Rapid Commun. 2012, 33, 1126–1129. [DOI] [PubMed] [Google Scholar]
- 140.Ortmann P., Heckler I., Mecking S., Green Chem. 2014, 16, 1816–1827. [Google Scholar]
- 141.Häußler M., Eck M., Rothauer D., Mecking S., Nature 2021, 590, 423–427. [DOI] [PubMed] [Google Scholar]
- 142.
- 142a.Nomura K., Chaijaroen P., Abdellatif M. M., ACS Omega 2020, 5, 18301–18312; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142b.Le D., Samart C., Kongparakul S., Nomura K., RSC Adv. 2019, 9, 10245–10252; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142c.Türünç O., Meier M. A. R., Green Chem. 2011, 13, 314–320; [Google Scholar]
- 142d.Moser B. R., Vermillion K. E., Banks B. N., Doll K. M., J. Am. Oil Chem. Soc. 2020, 97, 517–530; [Google Scholar]
- 142e.Sehlinger A., Schneider R., Meier M. A. R., Eur. Polym. J. 2014, 50, 150–157; [Google Scholar]
- 142f.Moser B. R., Doll K. M., Peterson S. C., J. Am. Oil Chem. Soc. 2019, 96, 825–837. [Google Scholar]
- 143.
- 143a.Nguyen P. H., Spoljaric S., Seppälä J., Eur. Polym. J. 2018, 109, 16–25; [Google Scholar]
- 143b.Kreye O., Türünç O., Sehlinger A., Rackwitz J., Meier M. A. R., Chem. Eur. J. 2012, 18, 5767–5776. [DOI] [PubMed] [Google Scholar]
- 144.More A. S., Palaskar D. V., Cloutet E., Gadenne B., Alfosc C., Cramail H., Polym. Chem. 2011, 2, 2796–2803. [Google Scholar]
- 145.
- 145a.Lebarbé T., More A. S., Sane P. S., Grau E., Alfos C., Cramail H., Macromol. Rapid Commun. 2014, 35, 479–483; [DOI] [PubMed] [Google Scholar]
- 145b.Lamarzelle O., Hibert G., Lecommandoux S., Grau E., Cramail H., Polym. Chem. 2017, 8, 3438–3447. [Google Scholar]
- 146.von Czapiewski M., Meier M. A. R., Macromol. Chem. Phys. 2017, 218, 1700153. [Google Scholar]
- 147.Lebarbé T., Neqal M., Grau E., Alfos C., Cramail H., Green Chem. 2014, 16, 1755–1758. [Google Scholar]
- 148.Kolb N., Meier M. A. R., Green Chem. 2012, 14, 2429–2435. [Google Scholar]
- 149.Maisonneuve L., Wirotius A.-L., Alfos C., Grau E., Cramail H., Polym. Chem. 2014, 5, 6142–6147. [Google Scholar]
- 150.Moreno A., Lligadas G., Ronda J. C., Galià M., Cádiz V., Eur. Polym. J. 2018, 108, 348–356. [Google Scholar]
- 151.Leonard E. C., J. Am. Oil Chem. Soc. 1979, 56, 782A–785A. [Google Scholar]
- 152.Daute P., Grützmacher R., Höfer R., Westfechtel A., Fett Wiss. Technol. 1993, 95, 91–94. [Google Scholar]
- 153.
- 153a.Freitas R. F. R., Klein C., Pereira M. P., Duczinski R. B., Einloft S., Seferin M., Ligabue R., J. Adhes. Sci. Technol. 2015, 29, 1860; [Google Scholar]
- 153b.Reulier M., Avérous L., Eur. Polym. J. 2015, 67, 418–427; [Google Scholar]
- 153c.Hablot E., Donnio B., Bouquey M., Avérous L., Polymer 2010, 51, 5895–5902. [Google Scholar]
- 154.Yang R., Wang B., Xu L., Zhao C., Zhang X., Li J., Polym. Bull. 2019, 76, 3753–3768. [Google Scholar]
- 155.Paraskar P. M., Kulkarni R. D., J. Polym. Environ. 2021, 29, 54–70. [Google Scholar]
- 156.Zhang X., Jiang Y., Jia C., Lu P., Chen H., Polym. Plast. Technol. Eng. 2019, 58, 1571–1584. [Google Scholar]
- 157.Unverferth M., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2016, 118, 1470–1474. [Google Scholar]
- 158.Goh S. C., Chin S. Y., Lee L. M. S., Yarmo M. A., Nik Yusof N. I., Adv. Mater. Res. 2012, 364, 95–99. [Google Scholar]
- 159.von Czapiewski M., Meier M. A. R., Eur. J. Lipid Sci. Technol. 2018, 120, 1700350. [Google Scholar]
- 160.Shan P., Chen N., Hao C., Liu H., Zhang X., Ind. Crops Prod. 2020, 148, 112302. [Google Scholar]
- 161.Bao Y., He J., Li Y., Polym. Int. 2013, 62, 1457–1464. [Google Scholar]
- 162.Testud B., Pintori D., Grau E., Taton D., Cramail H., Green Chem. 2017, 19, 259–269. [Google Scholar]
- 163.Vilela C., Silvestre A. J. D., Gandini A., J. Polym. Sci. Part A 2013, 51, 2260–2270. [Google Scholar]
- 164.Biermann U., Metzger J. O., Meier M. A. R., Macromol. Chem. Phys. 2010, 211, 854–862. [Google Scholar]
- 165.Akintayo C. O., Mutlu H., Kempf M., Wilhelm M., Meier M. A. R., Macromol. Chem. Phys. 2012, 213, 87–96. [Google Scholar]
- 166.Mathers R. T., Shreve M. J., Meyler E., Damodaran K., Iwig D. F., Macromol. Rapid Commun. 2011, 32, 1338–1342. [DOI] [PubMed] [Google Scholar]
- 167.Oelmann S., Meier M. A. R., Macromol. Chem. Phys. 2015, 216, 1972–1981. [Google Scholar]
- 168.Winkler M., Raupp Y., Köhl L., Wagner H., Meier M. A. R., Macromolecules 2014, 47, 2842–2846. [Google Scholar]
- 169.Winkler M., Romain C., Meier M. A. R., Williams C. K., Green Chem. 2015, 17, 300–306. [Google Scholar]
- 170.Biermann U., Sehlinger A., Meier M. A. R., Metzger J. O., Eur. J. Lipid Sci. Technol. 2016, 118, 104–110. [Google Scholar]
- 171.Hoogenboom R., Eur. J. Lipid Sci. Technol. 2011, 113, 59–71. [Google Scholar]
- 172.Lomège J., Lapinte V., Negrell C., Robin J.-J., Caillol S., Biomacromolecules 2019, 20, 4–26. [DOI] [PubMed] [Google Scholar]
- 173.Scholten P. B. V., Moatsou D., Detrembleur C., Meier M. A. R., Macromol. Rapid Commun. 2020, 41, 2000266. [DOI] [PubMed] [Google Scholar]
- 174.Vilela C., Armando R. R., Silvestre J. D., Gandini A., Ind. Crops Prod. 2010, 32, 97–104. [Google Scholar]
- 175.Scholten P. B. V., Detrembleur C., Meier M. A. R., ACS Sustainable Chem. Eng. 2019, 7, 2751–2762. [Google Scholar]
- 176.Fleckhaus A., Fokou P. A., Klaassen G., Klaas M. R., Eur. J. Lipid Sci. Technol. 2019, 121, 1700429. [Google Scholar]
- 177.
- 177a.Bornscheuer U. T., Annu. Rev. Food Sci. Technol. 2018, 9, 85–103; [DOI] [PubMed] [Google Scholar]
- 177b.Lipid modification by enzymes and engineered microbes (Ed.: Bornscheuer U. T.), AOCS Press/ Academic Press, Champaign/ London, 2018. [Google Scholar]
- 178.
- 178a.Wu S., Snajdrova R., Moore J. C., Baldenius K., Bornscheuer U. T., Angew. Chem. Int. Ed. 2021, 60, 88–119; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 89–123; [Google Scholar]
- 178b.Hauer B., ACS Catal. 2020, 10, 8418–8427. [Google Scholar]
- 179.Hitchman T., Oil Mill Gazet. 2009, 115, 2–5. [Google Scholar]
- 180.
- 180a.Zorn K., Oroz-Guinea I., Brundiek B., Bornscheuer U. T., Prog. Lipid Res. 2016, 63, 153–164; [DOI] [PubMed] [Google Scholar]
- 180b.Bornscheuer U. T., Huisman G. W., Kazlauskas R. J., Lutz S., Moore J. C., Robins K., Nature 2012, 485, 185–194; [DOI] [PubMed] [Google Scholar]
- 180c.Arnold F. H., Angew. Chem. Int. Ed. 2019, 58, 14420–14426; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 14558–14565. [Google Scholar]
- 181.
- 181a.Rudroff F., Mihovilovic M. D., Gröger H., Snajdrova R., Iding H., Bornscheuer U. T., Nat. Catal. 2018, 1, 12–22; [Google Scholar]
- 181b.Muschiol J., Peters C., Oberleitner N., Mihovilovic M. D., Bornscheuer U. T., Rudroff F., Chem. Commun. 2015, 51, 5798–5811; [DOI] [PubMed] [Google Scholar]
- 181c.Sperl J. M., Sieber V., ACS Catal. 2018, 8, 2385–2396; [Google Scholar]
- 181d.Cascade Biocatalysis (Eds.: Riva S., Fessner W. D.), Wiley-VCH, Weinheim, 2014. [Google Scholar]
- 182.
- 182a.Müller J., Sowa M. A., Fredrich B., Brundiek H., Bornscheuer U. T., ChemBioChem 2015, 16, 1791–1796; [DOI] [PubMed] [Google Scholar]
- 182b.Müller J., Sowa M. A., Dörr M., Bornscheuer U. T., Eur. J. Lipid Sci. Technol. 2015, 117, 1903–1907. [Google Scholar]
- 183.
- 183a.Jan A. H., Dubreucq E., Subileau M., ChemBioChem 2017, 18, 941–950; [DOI] [PubMed] [Google Scholar]
- 183b.Jan A. H., Subileau M., Deyrieux C., Perrier V., Dubreucq E., Biochim. Biophys. Acta Proteins Proteomics 2016, 1864, 187–194. [DOI] [PubMed] [Google Scholar]
- 184.Müller H., Becker A.-K., Palm G. J., Berndt L., Badenhorst C. P. S., Godehard S. P., Reisky L., Lammers M., Bornscheuer U. T., Angew. Chem. Int. Ed. 2020, 59, 11607–11612; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 11704–11709. [Google Scholar]
- 185.Godehard S. P., Badenhorst C. P. S., Müller H., Bornscheuer U. T., ACS Catal. 2020, 10, 7552–7562. [Google Scholar]
- 186.
- 186a.Zorn K., Oroz-Guinea I., Brundiek H., Dörr M., Bornscheuer U. T., Adv. Synth. Catal. 2018, 360, 4115–4131; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186b.Zorn K., Oroz-Guinea I., Bornscheuer U. T., Process Biochem. 2019, 79, 65–73; [Google Scholar]
- 186c.Oroz-Guinea I., Zorn K., Bornscheuer U. T., J. Am. Oil Chem. Soc. 2019, 96, 1327–1335. [Google Scholar]
- 187.
- 187a.Volkov A., Liavonchanka A., Kamneva O., Fiedler T., Göbel C., Kreikemeyer B., Feussner I., J. Biol. Chem. 2010, 285, 10353–10361; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187b.Engleder M., Pichler H., Appl. Microbiol. Biotechnol. 2018, 102, 5841–5858; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187c.Ogawa J., Takeuchi M., Kishino S., in Lipid Modification by Enzymes and Engineered Microbes, Vol. 1 (Ed.: Bornscheuer U. T.), AOCS Press/Academic Press, London, 2018, pp. 119–137. [Google Scholar]
- 188.Schütz R., Rawlings A. V., Wandeler E., Jackson E., Trevisan S., Monneuse J.-M., Bendik I., Massironi M., Imfeld D., Int. J. Cosmet. Sci. 2019, 41, 240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.
- 189a.Song J.-W., Jeon E.-Y., Song D.-H., Jang H.-Y., Bornscheuer U. T., Oh D.-K., Park J.-B., Angew. Chem. Int. Ed. 2013, 52, 2534–2537; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 2594–2597; [Google Scholar]
- 189b.Lee D.-S., Song J.-W., Voss M., Schuiten E., Akula R. K., Kwon Y.-U., Bornscheuer U., Park J.-B., Adv. Synth. Catal. 2019, 361, 1359–1367. [Google Scholar]
- 190.Volkov A., Khoshnevis S., Neumann P., Herrfurth C., Wohlwend D., Ficner R., Feussner I., Acta Crystallogr. Sect. D 2013, 69, 648–657. [DOI] [PubMed] [Google Scholar]
- 191.Engleder M., Pavkov-Keller T., Emmerstorfer A., Hromic A., Schrempf S., Steinkellner G., Wriessnegger T., Leitner E., Strohmeier G. A., Kaluzna I., Mink D., Schürmann M., Wallner S., Macheroux P., Gruber K., Pichler H., ChemBioChem 2015, 16, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.
- 192a.Demming R. M., Hammer S. C., Nestl B. M., Gergel S., Fademrecht S., Pleiss J., Hauer B., Angew. Chem. Int. Ed. 2019, 58, 173–177; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 179–183; [Google Scholar]
- 192b.Demming R. M., Otte K. B., Nestl B. M., Hauer B., ChemCatChem 2017, 9, 758–766. [Google Scholar]
- 193.
- 193a.Finnigan W., Thomas A., Cromar H., Gough B., Snajdrova R., Adams J. P., Littlechild J. A., Harmer N. J., ChemCatChem 2017, 9, 1005–1017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193b.Derrington S. R., Turner N. J., France S. P., J. Biotechnol. 2019, 304, 78–88; [DOI] [PubMed] [Google Scholar]
- 193c.Qu G., Guo J., Yang D., Sun Z., Green. Chem. 2018, 20, 777–792; [Google Scholar]
- 193d.Winkler M., Curr. Opin. Chem. Biol. 2018, 43, 23–29. [DOI] [PubMed] [Google Scholar]
- 194.Gahloth D., Dunstan M. S., Quaglia D., Klumbys E., Lockhart-Cairns M. P., Hill A. M., Derrington S. R., Scrutton N. S., Turner N. J., Leys D., Nat. Chem. Biol. 2017, 13, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.
- 195a.Ressmann A. K., Schwendenwein D., Leonhartsberger S., Mihovilovic M. D., Bornscheuer U. T., Winkler M., Rudroff F., Adv. Synth. Catal. 2019, 361, 2538–2543; [Google Scholar]
- 195b.Schwendenwein D., Ressmann A. K., Doerr M., Hoehne M., Bornscheuer U. T., Mihovilovic M. D., Rudroff F., Winkler M., Adv. Synth. Catal. 2019, 361, 2544–2549. [Google Scholar]
- 196.France S. P., Hussain S., Hill A. M., Hepworth L. J., Howard R. M., Mulholland K. R., Flitsch S. L., Turner N. J., ACS Catal. 2016, 6, 3753–3759. [Google Scholar]
- 197.Khusnutdinova A. N., Flick R., Popovic A., Brown G., Tchigvintsev A., Nocek B., Correia K., Joo J. C., Mahadevan R., Yakunin A. F., Biotechnol. J. 2017, 12, 1600751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wood A. J. L., Weise N. J., Frampton J. D., Dunstan M. S., Hollas M. A., Derrington S. R., Lloyd R. C., Quaglia D., Parmeggiani F., Leys D., Turner N. J., Flitsch S. L., Angew. Chem. Int. Ed. 2017, 56, 14498–14501; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 14690–14693. [Google Scholar]
- 199.Strohmeier G. A., Eiteljörg U. C., Schwarz A., Winkler M., Chem. Eur. J. 2019, 25, 6119–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mordhorst S., Siegrist J., Muller M., Richter M., Andexer J. N., Angew. Chem. Int. Ed. 2017, 56, 4037–4041; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 4095–4099. [Google Scholar]
- 201.Kourist R., Schweiger A., Büchenschütz H., in Lipid modification by enzymes and engineered microbes (Ed.: Bornscheuer U. T.), AOCS Press/ Academic Press, Champaign/ London, 2018, pp. 89–118. [Google Scholar]
- 202.Andre C., Kim S. W., Yu X.-H., Shanklin J., Proc. Natl. Acad. Sci. USA 2013, 110, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dennig A., Kuhn M., Tassoti S., Thiessenhusen A., Gilch S., Bülter T., Haas T., Hall M., Faber K., Angew. Chem. Int. Ed. 2015, 54, 8819–8822; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 8943–8946. [Google Scholar]
- 204.Bojarra S., Reichert D., Grote M., Baraibar Á. G., Dennig A., Nidetzky B., Mügge C., Kourist R., ChemCatChem 2018, 10, 1192–1201. [Google Scholar]
- 205.Song J.-W., Jeon E.-Y., Song D.-H., Jang H.-Y., Bornscheuer U. T., Oh D.-K., Park J.-B., Angew. Chem. Int. Ed. 2013, 52, 2534–2537; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 2594–2597. [Google Scholar]
- 206.Kim T.-H., Kang S.-H., Han J.-E., Seo E.-J., Jeon E.-Y., Choi G.-E., Park J.-B., Oh D.-K., ACS Catal. 2020, 10, 4871–4878. [Google Scholar]
- 207.
- 207a.Schrewe M., Ladkau N., Bühler B., Schmid A., Adv. Synth. Catal. 2013, 355, 1693–1697; [Google Scholar]
- 207b.Gröger H., Appl. Microbiol. Biotechnol. 2019, 103, 83–95. [DOI] [PubMed] [Google Scholar]
- 208.Xue Z., Sharpe P. L., Hong S. P., Yadav N. S., Xie D., Short D. R., Damude H. G., Rupert R. A., Seip J. E., Wang J., Pollak D. W., Bostick M. W., Bosak M. D., Macool D. J., Hollerbach D. H., Zhang H., Arcilla D. M., Bledsoe S. A., Croker K., McCord E. F., Tyreus B. D., Jackson E. N., Zhu Q., Nat. Biotechnol. 2013, 31, 734–740. [DOI] [PubMed] [Google Scholar]
- 209.Kim H. M., Chae T. U., Choi S. Y., Kim W. J., Lee S. Y., Nat. Chem. Biol. 2019, 15, 721–729. [DOI] [PubMed] [Google Scholar]
- 210.
- 210a.Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D., Nature 2010, 463, 559–562; [DOI] [PubMed] [Google Scholar]
- 210b.Kassab E., Mehlmer N., Brück T., Front. Bioeng. Biotechnol. 2019, 7, 408; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210c.Kassab E., Fuchs M., Haack M., Mehlmer N., Brück T. B., Microb. Cell Fact. 2019, 18, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Picataggio S., Rohrer T., Deanda K., Lanning D., Reynolds R., Mielenz J., Eirich L. D., Nat. Biotechnol. 1992, 10, 894–898. [DOI] [PubMed] [Google Scholar]
- 212.
- 212a.Xu M., Yang C., Qin B., Li N., Liu X., (Cathay), EP3533879A1, 2019;
- 212b.Liu W., Xu M., Chou H., Liu X., (Cathay), EP3550014A1, 2019;
- 212c.Qin B., Yang Y., Liu X., (Cathay), WO2018010057A1, 2018.
- 213.Dyer J. M., Stymne S., Green A. G., Carlsson A. S., Plant J. 2008, 54, 640–655. [DOI] [PubMed] [Google Scholar]
- 214.Jones A. D., Boundy-Mills K. L., Barla G. F., Kumar S., Ubanwa B., Balan V., in Microbial Lipid Production: Methods and Protocols (Ed.: Balan V.), Springer, New York, 2019, pp. 1–32. [DOI] [PubMed] [Google Scholar]
- 215.Lang I., Hodac L., Friedl T., Feussner I., BMC Plant Biol. 2011, 11, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ohlrogge J., Thrower N., Mhaske V., Stymne S., Baxter M., Yang W., Liu J., Shaw K., Shorrosh B., Zhang M., Wilkerson C., Matthäus B., Plant J. 2018, 96, 1299–1308. [DOI] [PubMed] [Google Scholar]
- 217.Ortiz R., Geleta M., Gustafsson C., Lager I., Hofvander P., Löfstedt C., Cahoon E. B., Minina E., Bozhkov P., Stymne S., Curr. Opin. Plant Biol. 2020, 56, 181–189. [DOI] [PubMed] [Google Scholar]
- 218.Singh S. P., Zhou X.-R., Liu Q., Stymne S., Green A. G., Curr. Opin. Plant Biol. 2005, 8, 197–203. [DOI] [PubMed] [Google Scholar]
- 219.Li-Beisson Y., Shorrosh B., Beisson F., Andersson M. X., Arondel V., Bates P. D., Baud S., Bird D., DeBono A., Durrett T. P., Franke R. B., Graham I. A., Katayama K., Kelly A. A., Larson T., Markham J. E., Miquel M., Molina I., Nishida I., Rowland O., Samuels L., Schmid K. M., Wada H., Welti R., Xu C., Zallot R., Ohlrogge J., in The Arabidopsis Book (Eds.: Somerville C., Meyerowitz E.), The American Society of Plant Biologists, Rockville, MD, 2013, p. e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Shanklin J., Cahoon E. B., Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 611–641. [DOI] [PubMed] [Google Scholar]
- 221.Bates P. D., Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1214–1225. [Google Scholar]
- 222.Vanhercke T., El Tahchy A., Shrestha P., Zhou X.-R., Singh S. P., Petrie J. R., FEBS Lett. 2013, 587, 364–369. [DOI] [PubMed] [Google Scholar]
- 223.Guo W., Chen L., Chen H., Yang H., You Q., Bao A., Chen S., Hao Q., Huang Y., Qiu D., Shan Z., Yang Z., Yuan S., Zhang C., Zhang X., Jiao Y., Tran L.-S. P., Zhou X., Cao D., Plant Biotechnol. J. 2020, 18, 1639–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zheng P., Allen W. B., Roesler K., Williams M. E., Zhang S., Li J., Glassman K., Ranch J., Nubel D., Solawetz W., Bhattramakki D., Llaca V., Deschamps S., Zhong G.-Y., Tarczynski M. C., Shen B., Nat. Genet. 2008, 40, 367–372. [DOI] [PubMed] [Google Scholar]
- 225.Nath U. K., Wilmer J. A., Wallington E. J., Becker H. C., Möllers C., Theor. Appl. Genet. 2009, 118, 765–773. [DOI] [PubMed] [Google Scholar]
- 226.Li X., van Loo E. N., Gruber J., Fan J., Guan R., Frentzen M., Stymne S., Zhu L.-H., Plant Biotechnol. J. 2012, 10, 862–870. [DOI] [PubMed] [Google Scholar]
- 227.Kumar R., Wallis J. G., Skidmore C., Browse J., Plant J. 2006, 48, 920–932. [DOI] [PubMed] [Google Scholar]
- 228.Lunn D., Wallis J. G., Browse J., Plant Physiol. 2019, 179, 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yu X. H., Cahoon R. E., Horn P. J., Shi H., Prakash R. R., Cai Y., Hearney M., Chapman K. D., Cahoon E. B., Schwender J., Shanklin J., Plant Biotechnol. J. 2018, 16, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Feussner I., Eur. J. Lipid Sci. Technol. 2015, 117, 271–273. [Google Scholar]
- 231.Beaudoin F., Sayanova O., Haslam R. P., Bancroft I., Napier J. A., Ol. Corps Gras Lipides 2014, 21, D606. [Google Scholar]
- 232.Nguyen H. T., Silva J. E., Podicheti R., Macrander J., Yang W., Nazarenus T. J., Nam J.-W., Jaworski J. G., Lu C., Scheffler B. E., Mockaitis K., Cahoon E. B., Plant Biotechnol. J. 2013, 11, 759–769. [DOI] [PubMed] [Google Scholar]
- 233.Xu Y., Mietkiewska E., Shah S., Weselake R. J., Chen G., Metab. Eng. 2020, 62, 20–29. [DOI] [PubMed] [Google Scholar]
- 234.Hornung E., Krueger C., Pernstich C., Gipmans M., Porzel A., Feussner I., Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2005, 1738, 105–114. [DOI] [PubMed] [Google Scholar]
- 235.Cahoon E. B., Dietrich C. R., Meyer K., Damude H. G., Dyer J. M., Kinney A. J., Phytochemistry 2006, 67, 1166–1176. [DOI] [PubMed] [Google Scholar]
- 236.Vanhercke T., Dyer J. M., Mullen R. T., Kilaru A., Rahman M. M., Petrie J. R., Green A. G., Yurchenko O., Singh S. P., Prog. Lipid Res. 2019, 74, 103–129. [DOI] [PubMed] [Google Scholar]
- 237.Cahoon E. B., Li-Beisson Y., Curr. Opin. Plant Biol. 2020, 55, 66–73. [DOI] [PubMed] [Google Scholar]
- 238.Napier J. A., Sayanova O., Proc. Nutr. Soc. 2005, 64, 387–393. [DOI] [PubMed] [Google Scholar]
- 239.Abbadi A., Domergue F., Bauer J., Napier J. A., Welti R., Zähringer U., Cirpus P., Heinz E., Plant Cell 2004, 16, 2734–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Abbadi A., Fulda M., Feussner I., Orsini J., Weyen J., Frauen M., Leckband G., in Proceedings—The 12th International Rapeseed Congress Sustainable Development in Cruciferous Oilseed Crops Production—Vol. II, Biotechnology, Vol. II, (Eds.: Meng J., Lu C., Zhao J., Liu C., Hua W.), Science Press USA Inc., Monmouth Junction, NJ, 2007, pp. 152–155. [Google Scholar]
- 241.Marmon S., Sturtevant D., Herrfurth C., Chapman K., Stymne S., Feussner I., Plant Physiol. 2017, 173, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ozseyhan M. E., Kang J., Mu X., Lu C., Plant Physiol. Biochem. 2018, 123, 1–7. [DOI] [PubMed] [Google Scholar]
- 243.Han L., Usher S., Sandgrind S., Hassall K., Sayanova O., Michaelson L. V., Haslam R. P., Napier J. A., Plant Biotechnol. J. 2020, 18, 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Petrie J. R., Zhou X.-R., Leonforte A., McAllister J., Shrestha P., Kennedy Y., Belide S., Buzza G., Gororo N., Gao W., Lester G., Mansour M. P., Mulder R. J., Liu Q., Tian L., Silva C., Cogan N. O. I., Nichols P. D., Green A. G., de Feyter R., Devine M. D., Singh S. P., Front. Plant Sci. 2020, 11, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Haslam R. P., Hamilton M. L., Economou C. K., Smith R., Hassall K. L., Napier J. A., Sayanova O., Biotechnol. Biofuels 2020, 13, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu J., Tjellström H., McGlew K., Shaw V., Rice A., Simpson J., Kosma D., Ma W., Yang W., Strawsine M., Cahoon E., Durrett T. P., Ohlrogge J., Ind. Crops Prod. 2015, 65, 259–268. [Google Scholar]
- 247.Bansal S., Kim H. J., Na G., Hamilton M. E., Cahoon E. B., Lu C., Durrett T. P., J. Exp. Bot. 2018, 69, 4395–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lassner M. W., Lardizabal K., Metz J. G., in Perspectives on new crops and new uses (Ed.: Janick J.), ASHS Press, Alexandria, VA, 1999, pp. 220–224. [Google Scholar]
- 249.Carlsson A. S., Yilmaz J. L., Green A. G., Stymne S., Hofvander P., Eur. J. Lipid Sci. Technol. 2011, 113, 812–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhu L.-H., Krens F., Smith M. A., Li X., Qi W., van Loo E. N., Iven T., Feussner I., Nazarenus T. J., Huai D., Taylor D. C., Zhou X.-R., Green A. G., Shockey J., Klasson K. T., Mullen R. T., Huang B., Dyer J. M., Cahoon E. B., Sci. Rep. 2016, 6, 22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ruiz-Lopez N., Broughton R., Usher S., Salas J. J., Haslam R. P., Napier J. A., Beaudoin F., Plant Biotechnol. J. 2017, 15, 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Yu D., Hornung E., Iven T., Feussner I., Biotechnol. Biofuels 2018, 11, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ohlrogge J., Allen D., Berguson B., DellaPenna D., Shachar-Hill Y., Stymne S., Science 2009, 324, 1019–1020. [DOI] [PubMed] [Google Scholar]
- 254.Vanhercke T., Petrie J. R., Singh S. P., Biocatal. Agric. Biotechnol. 2014, 3, 75–80. [Google Scholar]
- 255.Vanhercke T., El Tahchy A., Liu Q., Zhou X.-R., Shrestha P., Divi U. K., Ral J.-P., Mansour M. P., Nichols P. D., James C. N., Horn P. J., Chapman K. D., Beaudoin F., Ruiz-López N., Larkin P. J., de Feyter R. C., Singh S. P., Petrie J. R., Plant Biotechnol. J. 2014, 12, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Vanhercke T., Divi U. K., El Tahchy A., Liu Q., Mitchell M., Taylor M. C., Eastmond P. J., Bryant F., Mechanicos A., Blundell C., Zhi Y., Belide S., Shrestha P., Zhou X.-R., Ral J.-P., White R. G., Green A., Singh S. P., Petrie J. R., Metab. Eng. 2017, 39, 237–246. [DOI] [PubMed] [Google Scholar]
- 257.Vanhercke T., Belide S., Taylor M. C., El Tahchy A., Okada S., Rolland V., Liu Q., Mitchell M., Shrestha P., Venables I., Ma L., Blundell C., Mathew A., Ziolkowski L., Niesner N., Hussain D., Dong B., Liu G., Godwin I. D., Lee J., Rug M., Zhou X.-R., Singh S. P., Petrie J. R., Plant Biotechnol. J. 2019, 17, 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zulu N. N., Popko J., Zienkiewicz K., Tarazona P., Herrfurth C., Feussner I., Biotechnol. Biofuels 2017, 10, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yoneda K., Yoshida M., Suzuki I., Watanabe M. M., J. Appl. Phycol. 2018, 30, 2793–2802. [Google Scholar]
- 260.Li D.-W., Cen S.-Y., Liu Y.-H., Balamurugan S., Zheng X.-Y., Alimujiang A., Yang W.-D., Liu J.-S., Li H.-Y., J. Biotechnol. 2016, 229, 65–71. [DOI] [PubMed] [Google Scholar]
- 261.Yang H., Chen H., Hao G., Mei T., Zhang H., Chen W., Chen Y. Q., Eur. J. Lipid Sci. Technol. 2017, 119, 1600113. [Google Scholar]