Abstract
We study rank-based approaches to estimate the correlation between two right-censored variables. With end-of-study censoring, it is often impossible to nonparametrically identify the complete bivariate survival distribution, and therefore it is impossible to nonparametrically compute Spearman’s rank correlation. As a solution, we propose two measures that can be nonparametrically estimated. The first measure is Spearman’s correlation in a restricted region. The second measure is Spearman’s correlation for an altered but estimable joint distribution. We describe population parameters for these measures and illustrate how they are similar to and different from the overall Spearman’s correlation. We propose consistent estimators of these measures and study their performance through simulations. We illustrate our methods with a study assessing the correlation between the time to viral failure and the time to regimen change among persons living with HIV in Latin America who start antiretroviral therapy.
Keywords: bivariate survival, HIV, nonparametric, Spearman’s correlation, viral failure
1 |. INTRODUCTION
In many medical studies, researchers are interested in measuring the correlation between two time-to-event variables. For example, in studies of HIV/AIDS, there is interest in studying the correlation between the time from antiretroviral therapy (ART) initiation to viral failure and the time from ART initiation to regimen change. These variables should be highly correlated, and it might be important to know if the observed correlation is weaker than expected. Bivariate survival data may also come from paired subjects. For example, researchers might be interested in assessing the correlation between the time to cardiovascular disease for a patient and that for his/her parents, or between times to events in twins.
Right censoring is a defining element of bivariate survival data: One or both of the times to event may not be observed. When looking at correlation between times to events that occur in a single subject (e.g., time to viral failure and time to regimen change), the censoring time may be the same time for both outcomes. However, if the times to events are in paired subjects (e.g., times to events in twins), then the censoring times may differ. Our interest is in both scenarios, but we do not consider the setting where an event occurring in one variable causes censoring of the other, that is, competing risks.
Several different methods have been proposed to measure and test the correlation between two right-censored time-to-event variables. Clayton (1978) introduced a bivariate hazard ratio or a cross ratio as a single number summary of correlation in the context of a frailty model. Oakes (1982, 1989) suggested a test for independence based on the cross ratio, showed its relationship with Kendall’s tau, and extended its definition to a larger class of models. Fan et al. (2000) used a weighted average of the inverse of the cross ratio and a limited region Kendall’s tau. Cuzick (1982) proposed a model of correlation and several test statistics, one of which resembles Spearman’s rank correlation for censored data. Dabrowska (1986) derived generalized statistics to test the null hypothesis that the joint survival distribution is equal to the product of the marginals. Under certain assumptions, one of these statistics is related to a censored version of Spearman’s correlation, and another corresponds to a log-rank test based on martingale residuals. Shih and Louis (1996) developed two additional statistics based on martingale residuals. Shih and Louis (1995) also suggested a two-stage estimation procedure to evaluate the correlation in bivariate data using copulas (Nelsen, 2007). Other approaches for measuring correlation using copulas for bivariate survival data have been considered by Carriere (2000), Romeo et al. (2006), and Schemper et al. (2013).
Spearman’s rank correlation is ubiquitous in biomedical research because of its simple interpretation, robustness, and ability to capture nonlinear correlations. It is used much more frequently in practice than Kendall’s tau, perhaps because it closely approximates Pearson’s correlation under normality (Kruskal, 1958), and it is much easier to compute and interpret than a cross ratio. In the absence of censoring, Spearman’s correlation is simply the correlation of the ranked data. However, despite related work by Cuzick (1982), Dabrowska (1986), and Oakes (1989), there is no nonparametric estimator of Spearman’s correlation for bivariate survival data. Schemper et al. (2013) proposed a semiparametric iterative multiple imputation (IMI) method to estimate Spearman’s correlation (denoted as ρIMI throughout this paper). Their method transforms bivariate survival data into a Gaussian dependency structure using a normal copula, multiply imputes censored observations from this induced bivariate distribution, and approximates Spearman’s correlation using Pearson’s correlation of the normal deviates. This approach is semiparametric because it does not require any assumptions about the marginal distributions. However, it uses a Gaussian dependency structure, which may lead to bias due to misspecification.
The goal of this paper is to propose and study nonparametric estimators of Spearman’s correlation for right-censored data. Our methods use a nonparametric bivariate survival surface estimator. A challenge with estimating a bivariate survival surface, however, is that it may be nonparametrically estimable only within a certain region, for example, due to end-of-study censoring. This motivates us to propose two correlation estimators: one that is defined only within the restricted region and another one that implicitly assigns values outside of this region as having the highest rank.
In Section 2, we express Spearman’s correlation for time-to-event data and describe target parameters of interest. In Section 3, we address estimation and inference. In Section 4, we evaluate the performance of our estimators with several sets of simulations. In Section 5, we apply our methods to an HIV study by examining the correlation between times from treatment initiation to viral failure and regimen change. In Section 6, we discuss our methods and future directions. We have implemented our methods in the survSpearman R package (Eden et al., 2021).
2 |. POPULATION PARAMETERS
2.1 |. Notation and definitions
We are interested in estimating correlation between two time-to-event variables denoted as (TX, TY) defined on [0, ∞) × [0, ∞). Variables TX and TY can be observed on a single subject or on a pair of subjects. Each time to event can be censored. We denote time-to-censoring variables as (CX, CY) and assume independence between (TX, TY) and (CX, CY), but CX and CY can be dependent. If TX and TY are observed on a single subject, then it is likely that CX = CY. If CX = CY with probability one, then we call this univariate censoring, otherwise censoring is bivariate. In most studies, the follow-up period is bounded. We denote the maximum follow-up times, respectively, as τX and τY, and consider these to be fixed by study design. Censoring, due to the end of study, has been referred to as type I censoring. With type I censoring, no events will be observed beyond the region Ω = [0, τX) × [0, τY), or equivalently, CX ≤ τX and CY ≤ τY. For our presentation, we distinguish between strict type I censoring, where censoring occurs only at τX and τY (i.e., CX = τX, CY = τY), and generalized type I censoring (Klein and Moeschberger, 1997), where censoring may also occur prior to τX and τY (i.e., CX ≤ τX, CY ≤ τY). Strict type I censoring is rarely observed in practice, but will be helpful for explaining concepts; generalized type I censoring is quite common in practice, where follow-up time is bounded due to the length of the study, while subjects may start the study at different times or may drop out before the end of study. When the follow-up time is unbounded (τX = ∞ and τY = ∞), we refer to the censoring as unbounded.
As a result of censoring, we only observe X = min(TX, CX), Y = min(TY, CY), and event indicators , . We denote marginal and joint distribution functions of TX and TY as FX(x) = Pr(TX ≤ x), FY(y) = Pr(TY ≤ y), F(x, y) = Pr(TX ≤ x, TY ≤ y), and marginal and joint survival functions as SX(x) = Pr(TX > x), SY(y) = Pr(TY > y), S(x, y) = Pr(TX > x, TY > y). We define FX(x−) = limt↑x FX(t) and F(x−, y) = limt↑x F(t, y); FY(y−) and F(x, y−) are defined similarly.
2.2 |. Spearman’s rank correlation
As shown by Liu et al. (2018), in the absence of censoring, the population parameter for Spearman’s correlation between TX and TY can be defined as:
| (1) |
When both TX and TY are continuous, the above definition becomes
the grade correlation (Kruskal, 1958). Notice that if FX and FY are estimated with their respective empirical distributions, then FX(TX) and FY(TY) are simply estimated as the ranks of TX and TY, respectively, divided by the number of points in the sample, corresponding to the well-known Spearman’s correlation estimator:
where (TX,i, TY,i) for i = 1, …, n are independent and identically distributed (iid) draws of (TX, TY). Liu et al. (2018) have shown that Equation (1) can be presented as:
| (2) |
where ; and cρ = 3 when TX and TY are continuous. The right-hand side of (2) is the covariance of probability-scale residuals (PSRs) proposed and studied by Li and Shepherd (2012) and Shepherd et al. (2016) and defined as:
where sign(tX − TX) is −1, 0, and 1 for tX < TX, tX = TX, and tX > TX, respectively. We can rewrite definition (2) in terms of survival functions:
| (3) |
Right censoring causes serious challenges for nonparametric estimation of (3). First, nonparametric estimation of S(x, y) is challenging due to nonunique solutions of the nonparametric likelihood in the presence of censoring and the fact that even consistent estimators of S(x, y) may have negative mass for some x and y (Dabrowska, 1988; Pruitt, 1991). Second, nonparametric estimation of SX(x), SY(y), and S(x, y) beyond the maximum follow-up time(s) is not possible. To overcome the latter challenge, one could focus on estimating Spearman’s correlation inside Ω. Another possible approach is to focus on Spearman’s correlation for an altered but estimable joint distribution. Population parameters for these two nonparametric approaches are presented in the next two subsections. Section 2.5 contains some examples.
2.3 |. Spearman’s rank correlation in a restricted region
Suppose a researcher is interested in a rank correlation only inside a restricted region, denoted ΩR. This correlation can be computed as Spearman’s correlation defined conditionally on ΩR, which we denote as . With failure time data, others have proposed and advocated the use of estimators in restricted regions including restricted mean survival times (Royston and Parmar, 2013) and limited region Kendall’s tau (Fan et al., 2000).
A natural choice is ΩR = Ω = [0, τX) × [0, τY), to estimate the restricted rank correlation over the region for which estimation is possible; to avoid introducing new notation, in this section we will use ΩR = Ω. However, investigators can vary ΩR depending on their research question as long as ΩR ⊆ Ω; presumably ΩR would generally be a rectangle that includes the origin (0,0). The probability of double failure happening in this rectangle is
| (4) |
We consider the conditional distribution over ΩR. An example of a conditional distribution is illustrated in the middle panel of Figure 1. Its probability mass function is
| (5) |
and its marginal survival function on the X-axis is
| (6) |
where . The marginal survival function on the Y-axis, SY(y|ΩR), is similarly defined. Spearman’s correlation in the restricted region satisfies
| (7) |
where . Note that the population parameter, , depends only on τX and τY and is invariant to the censoring distribution within ΩR.
FIGURE 1.
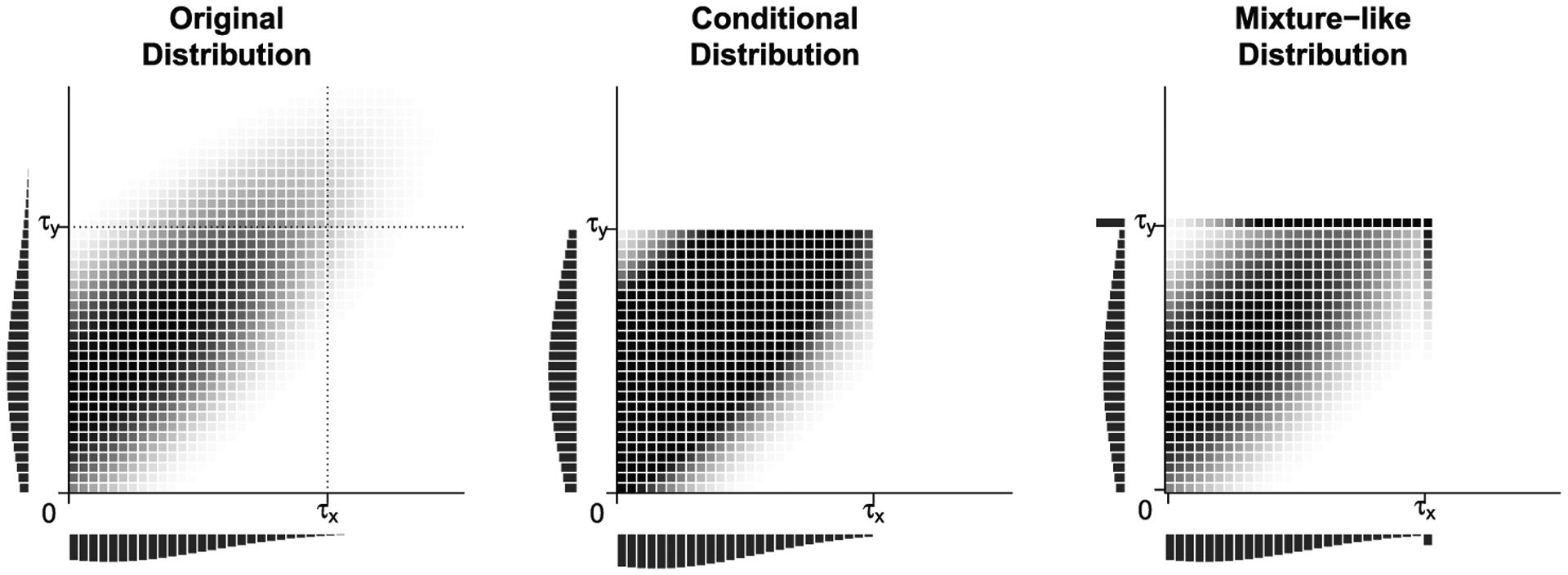
Illustration of bivariate distributions underlying the three population parameters. Left: Original distribution over [0, ∞) × [0, ∞), which has Spearman’s correlation ρS. Middle: Conditional distribution over ΩR = Ω, which has Spearman’s correlation . Right: Mixture-like distribution SH over region Ω ∪ [0, τX] × τY ∪ τX × [0, τY], which has Spearman’s correlation
Note that and ρS are different population parameters. Therefore, it is not appropriate to use to estimate ρS; rather, can be of interest on its own. For example, Fan et al. (2000) suggested a limited region Kendall’s tau to estimate over time the correlation of time to appendicitis in twins; can be used instead of the limited region Kendall’s tau for similar purposes.
2.4 |. Spearman’s rank correlation with highest ranks
Suppose now that a researcher is interested in the overall rank correlation, but the observations are only available within a restricted region. This is a typical situation in studies with a bounded follow-up time or when measurements have an upper detection limit. In this case, SX(x), SY(y), and S(x, y) are only nonparametrically estimable inside the region Ω = [0, τX) × [0, τY), where τX < ∞ and τY < ∞. Since we are interested in a rank correlation, one approach would be to define any observation censored at τX as receiving the highest rank value for TX and any observation censored at τY as receiving the highest rank value for TY, which is the same as setting TX = min(TX, τX) and TY = min(TY, τY). Such an approach is sensible because these censored observations at τX and τY do have the highest rank values and there is no information to distinguish between these highest rank values without parametric modeling assumptions. Mathematically, this approach replaces S(dx, dy) with a probability mass function SH(dx, dy), which is, S(dx, dy) inside Ω and the left-over probability mass outside of Ω. That is,
| (8) |
The new probability mass function SH(dx, dy) is depicted in the right panel of Figure 1. The part of SH(dx, dy) inside Ω is the same as S(dx, dy) but its part outside of Ω is concentrated on the borders of Ω and at point (τX, τY). The corresponding population parameter for the rank correlation of this new distribution is , which satisfies:
| (9) |
where and are the marginal survival functions of SH(x, y), and
| (10) |
In other words, is Spearman’s correlation computed by setting TX = min(TX, τX) and TY = min(TY, τY). Note that the population parameter, , depends only on τX and τY and is invariant to the censoring distribution within Ω.
For practical applications and intuitively, can be viewed as the rank correlation computed for data with an upper detection limit, where all values above the detection limit are set to a common largest value. Note that although in general, (see examples in Section 2.5), unlike , parameter is designed to take into account all observations including those outside of Ω. When the majority of the probability mass is within Ω, can be viewed as an approximation of ρS. When τX = ∞ and τY = ∞, then .
2.5 |. A few examples
Here, we illustrate how the restricted region can affect and . Having a restricted region implies type I censoring. Although the figures in this section are generated with strict type I censoring, remains the same for generalized type I censoring, as does .
In some settings, ρS, , and will be quite similar. For example, the values of these parameters for the distribution shown in Figure 1 are 0.635, 0.549, and 0.634, respectively. However, these parameters may be very different in some settings.
Figure 2 shows (left panel) and (right panel) as a function of ρS for different τX and τY for Frank’s copula family. In this example, ρS and are very similar. In contrast, ρS and are very different, especially when ρS is negative and ΩR is small (e.g., region defined by 0 to the 0.5 quantiles). This is because in these settings, only a small fraction of the underlying distribution is inside ΩR, and therefore shows a weak negative correlation. Figure 3 contains three additional examples; some of these distributions may not be realistic in practice, but are useful for illustrative purposes. The left panel shows an X-like distribution, for which ρS = 0, , and . Here, is similar to ρS because incorporates the probability mass in the upper left, upper right, and lower right regions. However, is very different from ρS because ΩR only contains a positively correlated subset of the distribution. The middle panel of Figure 3 shows a distribution with highly correlated values in ΩR, zero correlation in the upper right region, and no mass in the upper left and lower right regions. For this distribution, ρS = 0.66, , and . Here, and ρS are similar because only the points in the restricted region are correlated. In contrast, is quite high because the large probability mass of the upper right region is concentrated on a single highest-rank point when computing , which pulls its value upwards considerably. The right panel of Figure 3 shows a distribution with a highly negative overall correlation, for which ρS = −0.90, , and . Here, and ρS are fairly different because although the probability mass of censored observations outside of ΩR are taken into account, there is some loss of information with the highest rank assignment. The parameter is very different from ρS because only a small fraction of the negatively correlated mass is included in ΩR. Note that parametric or semiparametric approaches also struggle with many of these settings because they effectively use the information from the restricted region to impute what is occurring outside the restricted region. For example, (see Schemper et al., 2013) for the middle panel is approximately 0.95.
FIGURE 2.
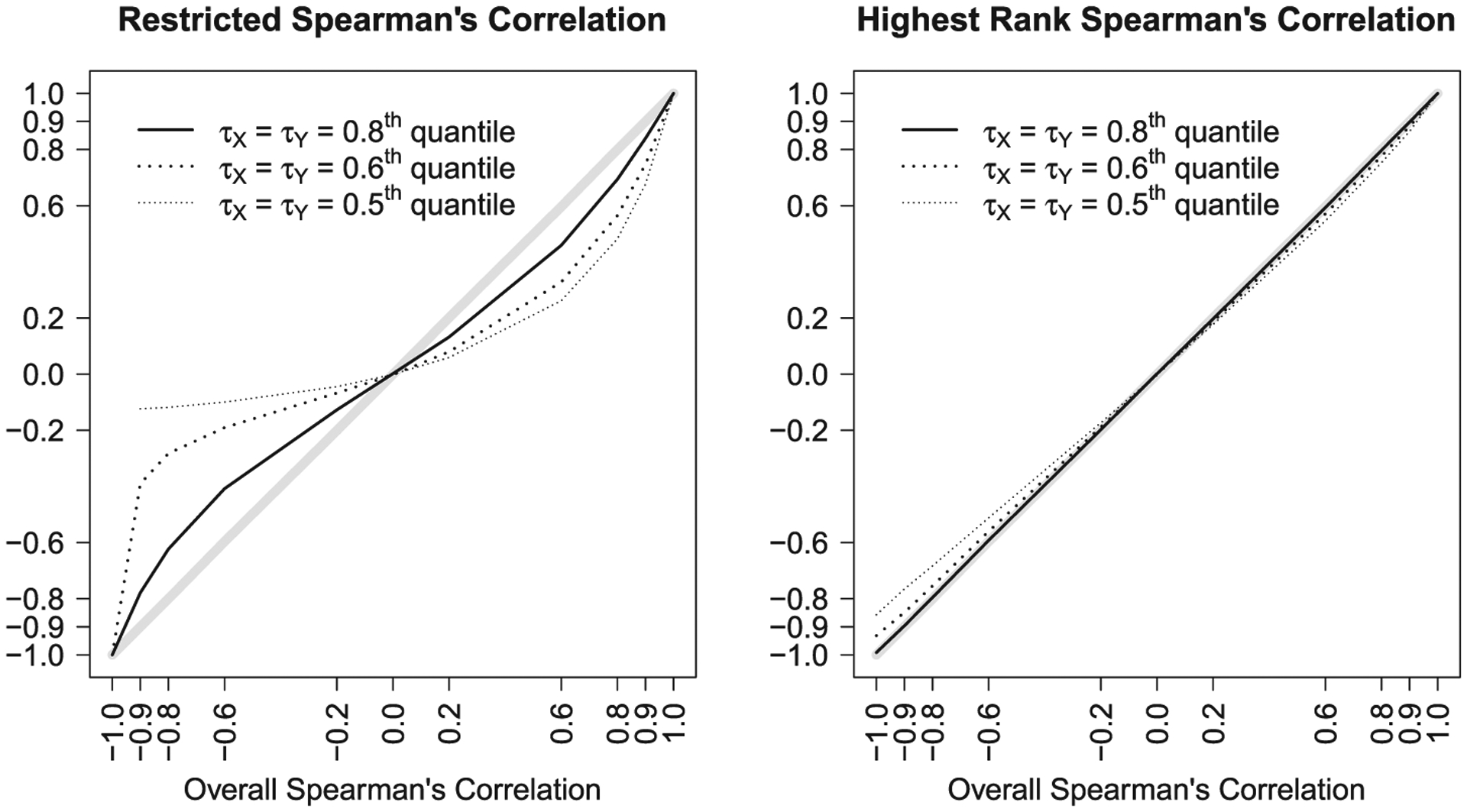
Restricted Spearman’s correlation, (left panel), and highest rank Spearman’s correlation, (right panel), for Frank’s copula family for different restricted regions defined by τX and τY (τX = τY): 0.5th (50% censored), 0.6th (40% censored), 0.8th (20% censored) quantiles. A diagonal gray line is added for reference. Although the plots are generated based on data under strict type I censoring, the population parameters are the same for generalized type I censoring and are invariant to the rate of censoring within the restricted region
FIGURE 3.
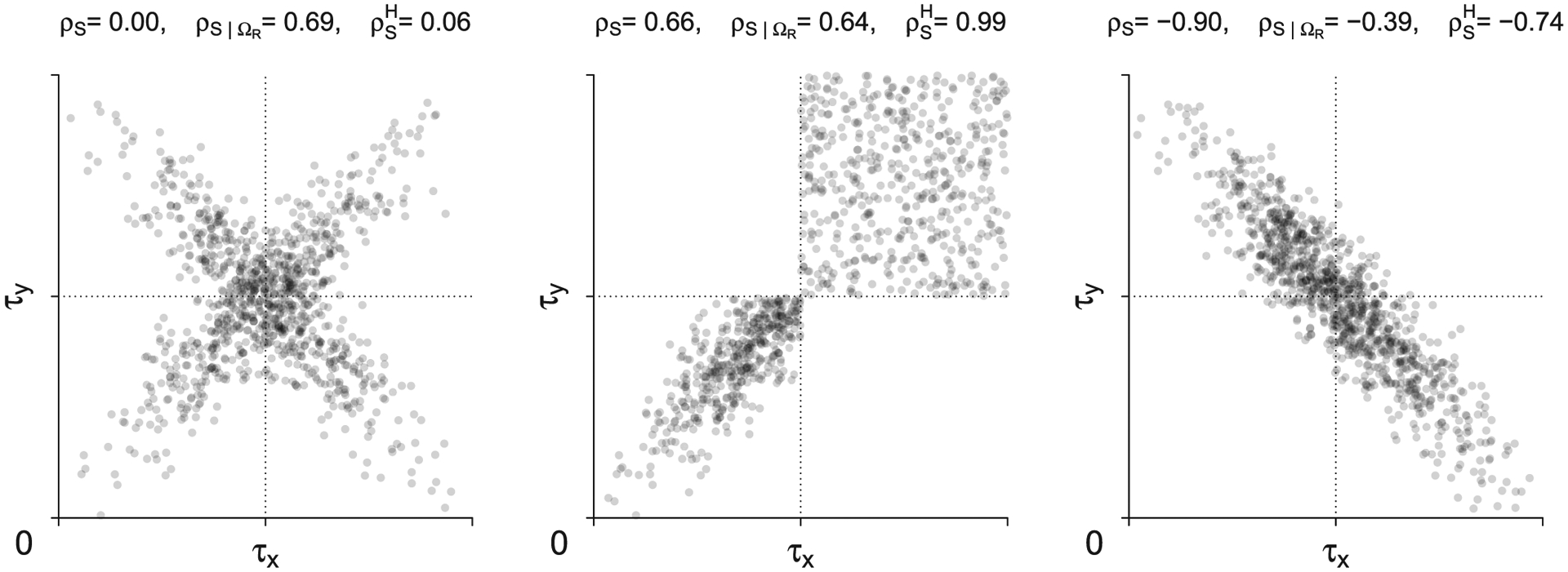
Example of bivariate distributions and their population parameters ρS with no censoring, and and with strict type I censoring with ΩR = [0, τX) × [0, τY). The proportions of observed double events in the left, middle, and right panels are 25%, 43%, and 7%, respectively. Drawn are 1000 points randomly selected from the underlying distributions. Although the plots are based on strict type I censoring, the population parameters are the same for generalized type I censoring and are invariant to the rate of censoring within ΩR
3 |. NONPARAMETRIC ESTIMATION
3.1 |. Estimation of under generalized type I censoring
We estimate using a plug-in estimator for Equation (7):
where
| (11) |
and is computed similarly. The conditional survival curves, , , and are estimated using plug-in estimators for (4), (5), and (6).
For we use the estimator of Dabrowska (1988). The marginal distributions of Dabrowska’s estimator, and , are Kaplan–Meier estimators. There are other choices for nonparametrically estimating S(dx, dy), including the estimators proposed by Prentice and Cai (1992), van der Laan (1997), Campbell (1981), and Lin and Ying (1993), to name a few. We considered the estimators of Campbell (1981) and Lin and Ying (1993) because of their computational simplicity, but ultimately chose Dabrowska’s estimator because it is consistent for S(dx, dy), straightforward to compute, and tended to result in estimates of Spearman’s correlation with better performance (see Section 4). The confidence interval (CI) of is estimated using the bootstrap. Because of the consistency and asymptotic normality of Dabrowska’s estimator, is consistent for and asymptotically normal when (see Theorem A1 in the Appendix).
A few problems can arise in practice when computing , all due to negative mass at some points in . First, in some extreme cases (e.g., small sample sizes, strong positive or negative correlation, and heavy censoring), may exceed 1; if this happens, our software sets it to and generates a warning. Second, negative mass can also lead to problems computing in (11) because or may be negative because or is negative at some points. If this happens, we correct the guilty conditional marginal probability mass estimator by assigning negative values to 0, and by normalizing the rest of the probability mass values; specifically, the corrected probability mass is . Third, negative mass may lead to a negative estimate of PR, the probability of both events occurring in ΩR (see Equation (4)); when this occurs, is not defined.
Although the number of points with negative mass does not decrease as the sample size increases (Pruitt, 1991), the amount of negative mass at each point does go to zero, which therefore reduces the possibility of these problems occurring. Also, the tendency of having negative mass is lower when a lower proportion of observations are singly or doubly censored. To give a sense of the magnitude of these problems, for a sample size of 50 with 70% bivariate censoring, or was negative for 1.3% of 1000 simulations, and was less than zero in 2.4% of simulations. With a sample size of 100 and 70% bivariate censoring, these problems occurred in 0% and 0.8% of simulations, respectively.
3.2 |. Estimation of under generalized type I censoring
Equation (9) provides a straightforward way of estimating , given a nonparametric estimate of the bivariate survival surface, :
where i* enumerates all the events for X plus τX, j* enumerates all the event for Y plus τY, and and are the plug-in estimators for (8) and (10), respectively. As before, we compute using Dabrowska’s estimator. The CI of is estimated using the bootstrap. Again, is consistent for and asymptotically normal when (see Theorem A2 in the Appendix). In practice, for some extreme cases similar to those mentioned in Section 3.1 for , may exceed 1; when this occurs, we set it to , and our software generates a warning.
3.3 |. Estimation of ρS under unbounded censoring
The estimator may also be used to estimate ρS with unbounded censoring. Under unbounded censoring, τX = τY = ∞ by definition, and one would naturally estimate by plugging , , and into (3) in a manner similar to that described above. However, if the maximum value of X, for example, is a censored event (i.e., ΔX = 0), then will not sum to 1 resulting in improper integration when using plug-in estimators in (3). A workaround is to assign the remaining mass (which is typically very little) to a point just beyond the largest observed event time of X and set τX to this point. We then estimate ρS with . Although in this setting, τX is no longer fixed but unbounded, estimation of ρS in this manner performs well (see Section 4). Under unbounded censoring, is consistent for ρS and asymptotically normal when ρS ∈ (−1, 1) (see Theorem A3 in the Appendix).
4 |. SIMULATIONS
4.1 |. Simulation set-up
We performed several simulations to investigate the finite sample performance of our estimators. The random variables TX and TY were simulated using various choices of copula families and parameters. Specifically, following Fan et al. (2000), we simulated dependent random uniform variables, U and V, from Clayton’s and Frank’s copula families; both copulas are defined by a single parameter, θ. The dependence between U and V was specified by choosing the parameter θ in such a way that the true Spearman’s correlation varied among no correlation (ρS = 0), moderate correlation (ρS = −0.2 and 0.2 for Frank’s family and 0.2 for Clayton’s family), and strong correlation (ρS = −0.6 and 0.6 for Frank’s family and 0.6 for Clayton’s family). (Clayton’s family does not permit negative correlation.) We then set TX = −log(1 − U) and TY = −log(1 − V) such that TX and TY were exponentially distributed with mean 1.
Four types of censoring scenarios were implemented: (1) unbounded univariate, (2) unbounded bivariate, (3) generalized type I univariate, and (4) generalized type I bivariate. Each censoring scenario was implemented for censoring proportions PC = {0.3, 0.7}. Bivariate unbounded censoring times CX and CY for each observation were simulated independently from an exponential distribution, Pr(CX ≤ t) = Pr(CY ≤ t) = 1 − e−λt, with λ = PC/(1 − PC). The event times TX and TY were censored if TX > CX and TY > CY, respectively. Univariate unbounded censoring was implemented in a similar manner except only one censoring event was generated per (TX, TY) pair. For generalized type I censoring, and , were first simulated as described above with probability PC and then CX and CY were defined as (, τX) and (, τY), respectively, with τX = τY set at the median survival time, . The resulting censoring proportions for generalized type I censoring were therefore higher than PC: for example, for generalized type I bivariate censoring with PC = 0.3 and 0.7, the outcomes were censored for approximately 56% and 73% of observations, respectively.
We evaluated the performance of and in the presence of unbounded and generalized type I censoring using a sample size of 200 for strong, moderate, and no correlation. For unbounded censoring, the population parameter of is the same as ρS. For generalized type I censoring, the population parameters of and were the same as ρS for Clayton’s family. For Frank’s family with the overall Spearman’s correlation of −0.6, −0.2, 0.2, and 0.6, the population parameters of were −0.098, −0.042, 0.058, and 0.261; and the population parameters of were −0.512, −0.173, 0.180, and 0.545. These population parameters were empirically estimated with a sample size of 106.
The bias, root mean squared error (RMSE), type I error rate, and power, computed as the proportion of times that bootstrap CIs (based on 1000 bootstrap samples) did not include zero, were also evaluated for sample sizes of 100 and 200 under unbounded censoring for moderate and no correlation. The performance of was compared to estimator proposed by Schemper et al. (2013) for these simulation scenarios. We also evaluated the performance of as an estimator of ρS using survival surfaces proposed by Lin and Ying (1993) and Campbell (1981).
Lastly, we did some additional comparisons of to semiparametric estimators and (maximum likelihood estimator assuming Frank’s copula dependency structure). These comparisons were made in the context of a well-behaved dependency structure induced by Frank’s copula and in the context of a complex dependency structure, a mixture of 60% highly negatively correlated data (ρS = −0.8, Frank’s copula with θ = −8) and 40% perfectly correlated data (ρS = 1) with the overall Spearman’s correlation being about −0.0813. This simulation scenario was loosely motivated by data from site B in our real data analysis presented in Section 5; Supplementary Figure S5 shows the scatter plot of the uncensored data. Sample sizes of 200, 500, and 1000 were used and unbounded univariate censoring with PC = 0.5 was applied as described above. The goal of these comparisons was to show better efficiency of compared to under a correctly specified model and to demonstrate greater accuracy of compared to and under model misspecification.
All simulations used 1000 replications.
4.2 |. Simulation results
Figure 4 shows the mean point estimates and the 0.025th and 0.975th quantiles of estimators under unbounded censoring (row 1), the semiparametric estimator proposed by Schemper et al. (2013) under unbounded censoring (row 2), under generalized type I censoring (row 3), and under generalized type I censoring (row 4). The sample size was 200, the censoring was bivariate with varying censoring proportions, and and were computed using Dabrowska’s survival surface estimator.
FIGURE 4.
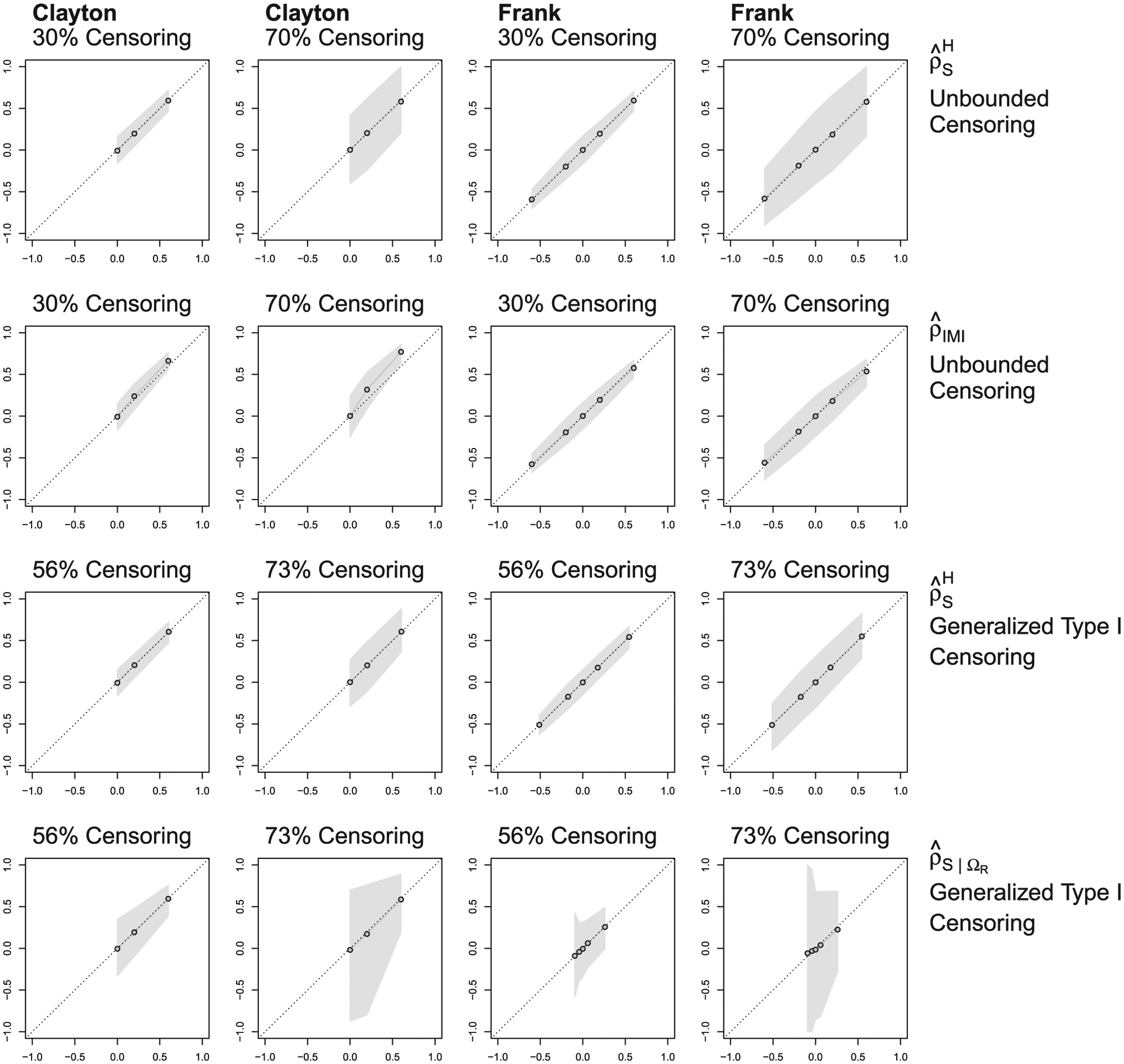
Point estimates (X-axis) vs population parameters (Y-axis) under different bivariate censoring scenarios. The top and second rows are and as estimators of the overall Spearman’s correlation, ρS. The third row is as an estimator of . The bottom row is as an estimator of . The columns represent Clayton’s and Frank’s copulas. The population parameters for Clayton’s family are 0, 0.2, and 0.6 for all estimates. For Frank’s family, the population parameters of ρS are −0.6, −0.2, 0.2, and 0.6; the population parameters of are −0.512, −0.173, 0.180, and 0.545; the population parameters of are −0.098, −0.042, 0.058, and 0.261. The dots are the mean point estimates based on 1000 simulations. The shaded areas represent the 0.025th and 0.975th quantiles. For generalized type I censoring, the restricted region, ΩR, was defined by the median survival times
For Clayton’s and Frank’s families under unbounded censoring (row 1), the mean of was very close to the true population parameter verifying the consistency of for ρS. When data were generated under Frank’s copula, the semiparametric estimator, (row 2), was similarly unbiased for unbounded censoring, and it tended to be less variable than . However, when data were generated using Clayton’s copula, was biased for ρS, also noted by Schemper et al. (2013).
Supplementary Tables S1 and S2 provide more details and additional comparisons between and under unbounded censoring for different sample sizes and censoring proportions in terms of bias, RMSE, type I error rate and power. In short, the bias of for ρS was low, even with fairly small numbers of events; both the bias and RMSE decreased as the number of events increased. In general, compared favorably to .
Under generalized type I censoring (the third and fourth rows of Figure 4), the means of and are very close to their true population parameters, suggesting with n = 200, these estimators are essentially unbiased for and respectively. The variance of our estimators naturally increased as the probability of censoring increased. The variance of was greater than that of under generalized type I censoring (rows 3 and 4), presumably because only uses the events inside Ω = ΩR, whereas assigns probability mass to values outside of Ω. Note that the variance of under unbounded censoring was slightly larger than that under generalized type I censoring in spite of the lighter censoring. This is probably because under generalized type I censoring, the probability mass, , calculated outside of Ω is concentrated on the same points, making its variance smaller compared to the case of unbounded censoring. Results for univariate censoring were very similar to those for bivariate censoring, except the estimators were slightly less variable (see Supplementary Figure S1).
With unbounded censoring of 50% and n = 200, the correctly specified semiparametric was more efficient than (relative efficiency in terms of variance ranging from 1.19 (for ρS = 0) to 1.60 (for ρS = 0.6), Supplementary Figure S4); both approaches yielded unbiased estimates of ρS. In contrast, when data were generated using a mixture of positively and negatively correlated bivariate distributions, the misspecified semiparametric estimators and were substantially biased and this bias did not decrease with increasing sample size. On the other hand, the nonparametric was unbiased for ρS (see Supplementary Figure S6). In this more complicated setting, estimates of with ΩR being defined using the median survival times were also unbiased (Supplementary Figure 7).
The simulations reported above incorporated 1000 bootstrap replications; in general, CI coverage and width were stable and adequate with as few as 200 bootstrap replications (see Supplementary Figure S8).
We also evaluated the performance of with survival surfaces of Campbell (1981) for univariate and bivariate censoring and of Lin and Ying (1993) for univariate censoring only (see Supplementary Figures S2 and S3). With univariate censoring, using the estimator of Lin and Ying (1993) resulted in unbiased estimation although with larger variance than that using Dabrowska’s estimator. Estimator computed using the survival surface estimator of Campbell (1981) was visibly biased for the sample size of 200 under heavy censoring.
5 |. APPLICATION
We apply our methods to a study of 6691 HIV-positive adults starting ART in Latin America. We are interested in estimating the correlation between two right-censored variables: (1) the time from ART initiation to viral failure and (2) the time from ART initiation to major regimen change. Patients experience viral failure when the amount of virus circulating in their blood (their viral load) is above a certain threshold, which may make them infectious and vulnerable to HIV-related diseases. Viral failure may be caused by many factors including poor adherence or drug resistance; it often triggers changing a patient’s ART regimen. However, patients may also change their ART regimens for reasons other than viral failure (e.g., poor tolerability, discovery of a simpler regimen, or patient/provider choice).
Our study uses data from the Caribbean, Central, and South America network for HIV Epidemiology (CCASAnet). The definitions of viral failure and regimen change were the same as those used in a prior CCASAnet study (Cesar et al., 2015; CCASAnet, 2020). In short, viral failure was defined as a single viral load > 1000 copies/mL or two viral loads > 400 copies/mL after a person’s virus had been suppressed or they had been on ART long enough that it should have been suppressed (i.e., 6 months). Regimen change was limited to major changes such that the patient switched drug classes or changed multiple drugs. Each study subject may have had one, both, or neither of these events. Follow-up ended at the last clinic visit; censoring was univariate. Our analysis data set includes patients from Brazil, Chile, Honduras, Mexico, and Peru; sites have been anonymized for presentation. After a median follow-up of 4.1 years (ranging from 1 day to 18.2 years), 1916 persons (28.6%) had a viral failure and 1895 persons (28.3%) changed regimens. Approximately 16.1% of patients had both events over the follow-up period, 12.2% changed regimens but did not have viral failure, 12.5% had viral failure but did not change regimens, and 59.1% were not observed to have either event. The upper left panel of Figure 5 shows Kaplan–Meier estimates for the marginal probabilities of viral failure and regimen change as a function of time since ART initiation. Ten years after ART initiation, the estimated probability of not having viral failure was 0.58 and the estimated probability of remaining on the initial regimen was 0.51. The upper right panel shows the estimated joint bivariate probability mass function, , based on Dabrowska’s estimator; estimated marginal probability mass functions, and , are also included. Note that because a large proportion of patients experienced only one or neither event, a large amount of mass has been assigned to τX = 18 and τY = 17 years.
FIGURE 5.
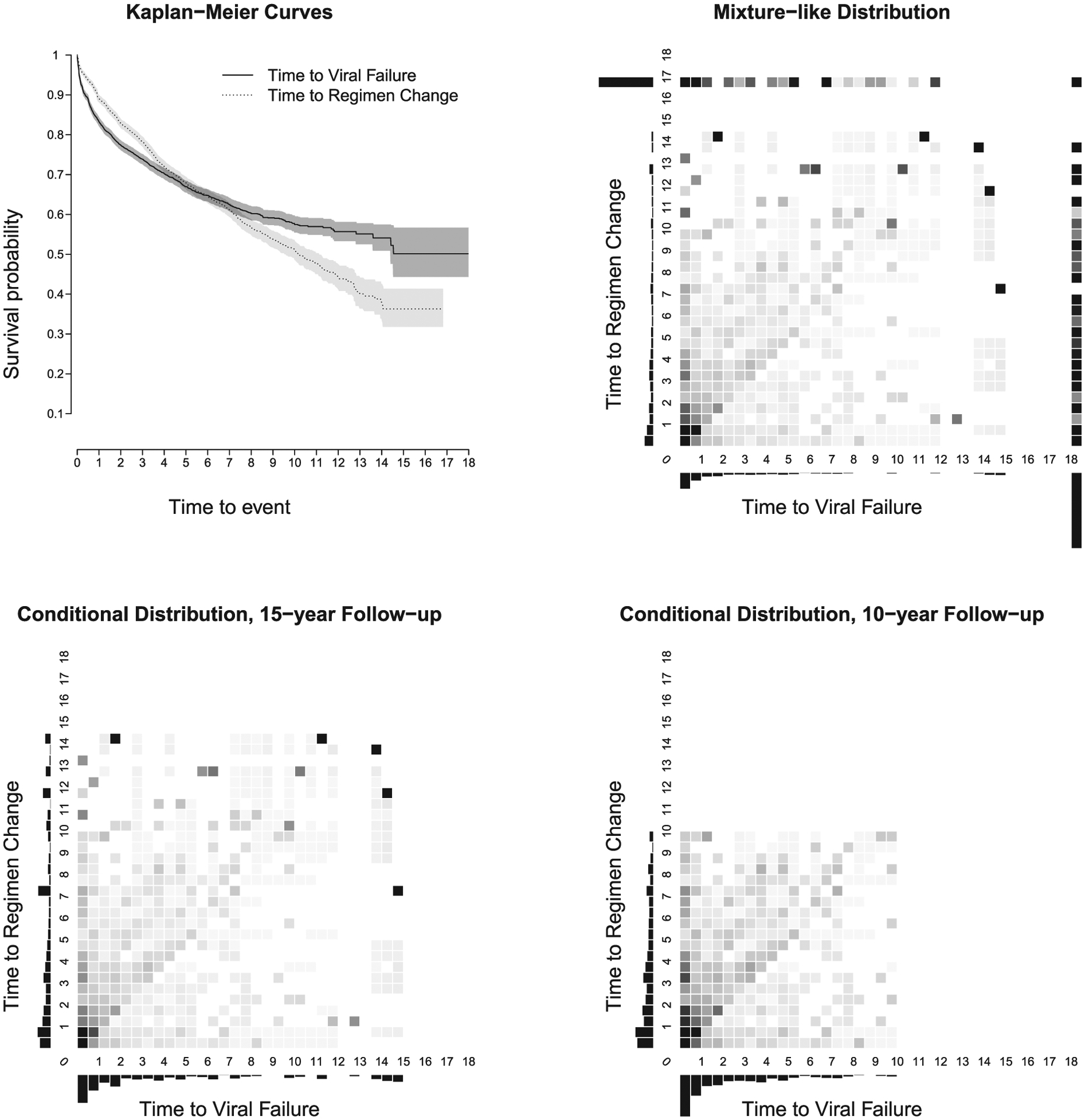
Upper left: Kaplan–Meier curves for time to viral failure and time to regimen change, where time is measured in years. Upper right: bivariate probability mass function for the mixture-like distribution, . Lower left: conditional bivariate probability mass function for 15-year follow-up. Lower right: conditional bivariate probability mass function for 10-year follow-up. For probability mass functions, the bars on the left and on the bottom represent histograms of the univariate survival mass for each event. The probability mass function was computed from the Dabrowska’s survival surface and then aggregated over half-year bivariate time periods. After aggregation, any negative values were set to 0. Lighter shade represents smaller values
The estimated highest rank correlation, , between time to viral failure and time to regimen change was 0.35 with a 95% CI based on 1000 bootstrap replications of 0.27 to 0.44. This result is fairly similar, albeit with a wider CI, to the estimator of Schemper et al., , that imputes censored values: 0.37, 95% CI = [0.33, 0.41].
The estimated rank correlation over the restricted region, , with ΩR = [0, 15) × [0, 15) was substantially higher, 0.65, with a wide 95% CI = [0.30, 0.97]. This can be explained by a careful look at the conditional probability mass over ΩR (lower left plot of Figure 5). Notice that there are several points in the upper right corner of this surface with large probability mass. Large amounts of mass are assigned to these points because a few events occurred at later follow-up times when there were fairly small numbers of patients remaining in the risk set. This is also seen in the Kaplan–Meier estimates, where relatively large drops in the marginal survival curves are noted between 10 and 15 years. These points pushed the probability mass function closer to the diagonal, leading to a larger estimated rank correlation. In addition, since there were only a few points with substantial probability mass, their inclusion/exclusion in various bootstrap samples led to wide variation in CIs. This example serves as a nice illustration of the potential perils of estimating rank correlations over restricted regions that include tail areas with small numbers of events. Perhaps a more reliable rank correlation would be over the restricted region, ΩR = [0, 10) × [0, 10), in which case was 0.26 (95% CI = [0.17, 0.36]); the lower right plot of Figure 5 shows the conditional probability mass over this smaller region.
In addition to showing estimates of overall correlation, Table 1 shows estimates based on sex and study site. For the most part, is fairly close to , except for those sites with small sample sizes (i.e., A, B, and D). Rank correlations over the restricted region, with ΩR = [0, 15) × [0, 15) were generally more variable and typically higher than those over ΩR = [0, 10) × [0, 10).
TABLE 1.
Correlation of time to viral failure and time to regimen change in CCASAnet cohort measured using three estimators: , , and . n is the number of subjects for each subgroup. Columns PVR, , , and show the percent of subjects having both events (PVR), having regimen change event but censored viral failure (), having viral failure event but censored regimen change (), and having both events censored (). For and , the numbers inside brackets are 95% confidence intervals (CIs) estimated from 1000 bootstrap samples. Column shows the estimates of Schemper et al. (2013) and their CIs
| Subgroup | n | P VR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| All | 6691 | 16.1 | 12.2 | 12.5 | 59.1 | 0.35 [0.27, 0.44] | 0.65 [0.30, 0.97] | 0.26 [0.17, 0.36] | 0.37 [0.33, 0.41] |
| Male | 5185 | 14.8 | 12.0 | 12.3 | 60.9 | 0.32 [0.22, 0.42] | 0.57 [0.14, 1.00] | 0.32 [0.18, 0.44] | 0.36 [0.30, 0.41] |
| Female | 1506 | 20.7 | 13.1 | 13.3 | 52.9 | 0.45 [0.30, 0.57] | 0.70 [0.20, 1.00] | 0.13 [−0.04, 0.32] | 0.40 [0.32, 0.48] |
| A | 1040 | 19.2 | 23.0 | 11.5 | 46.2 | −0.06 [−0.21, 0.30] | 0.73 [−0.24, 1.00] | 0.19 [0.03, 0.34] | 0.26 [0.00, 0.37] |
| B | 975 | 13.7 | 16.3 | 11.9 | 58.1 | 0.52 [0.14, 0.74] | −0.39 [−0.54, 0.41] | −0.43 [−1.00, 0.08] | 0.25 [0.10, 0.38] |
| C | 2225 | 8.9 | 5.5 | 12.1 | 73.4 | 0.45 [0.37, 0.53] | 0.69 [0.42, 0.93] | 0.58 [0.34, 0.80] | 0.55 [0.43, 0.66] |
| D | 138 | 18.1 | 17.4 | 10.1 | 54.3 | −0.18 [−0.53, 0.44] | 0.21 [−1.00, 1.00] | 0.50 [−0.09, 0.96] | 0.29 [0.00, 0.55] |
| E | 2313 | 22.4 | 11.8 | 13.8 | 51.9 | 0.45 [0.36, 0.53] | 0.42 [0.10, 0.73] | 0.17 [−0.01, 0.34] | 0.36 [0.30, 0.42] |
6 |. DISCUSSION
We have proposed two nonparametric methods of quantifying correlation with bivariate right-censored data. One estimator, computes Spearman’s correlation within a restricted region. The other estimator, , computes Spearman’s correlation for an estimable bivariate distribution, which is analogous to assigning data censored beyond the estimable region to the highest rank values. Under unbounded censoring, is consistent for the overall Spearman’s correlation. Under generalized type I censoring, with the majority of events happening in the restricted region, can be viewed as an approximation of the overall Spearman’s correlation. Because our methods assume neither marginal nor joint parametric distributions, they have potential advantages over parametric and semiparametric methods.
The main limitations of our estimators stem from challenges with nonparametrically estimating the bivariate survival surface. The first challenge is that with generalized type I censoring, the bivariate survival surface, and hence Spearman’s rank correlation ρS, cannot be identified beyond the region of data support without parametric assumptions. Hence, our inference targets were the estimable parameters, and . Although under generalized type I censoring they do not equal the overall Spearman’s correlation, both parameters have sensible interpretations. The alternative, using parametric and semiparametric models to estimate Spearman’s correlation—an inherently nonparametric statistic— has other limitations. Parametric and semiparametric approaches assume a dependency structure; and as seen in our simulations, semiparametric estimators such as proposed by Schemper et al. (2013), may be biased when the dependency structure is misspecified. In addition, they implicitly assume that the dependency structure outside the region of observation is the same as that seen inside the observation region. Although in the real data example, appeared more stable than our nonparametric estimators, particularly for small sample sizes, with complex real data there is a real possibility of model misspecification. As with most statistics, there may be settings where one might prefer the nonparametric estimators over the semiparametric estimators, or vice versa.
The second challenge is that even in regions where the bivariate survival curve is identifiable, nonparametric estimators of the survival surface may have negative mass, which leads to potential downstream problems with estimation of and . Researchers have grappled with nonparametric approaches to avoid negative mass, for example, van der Laan (1996). Thankfully, in our simulations, only a minor proportion of our estimates encountered problems due to negative mass, the problems go away as the numbers of events increase, and there are typically workaround solutions that appear to behave reasonably.
In our approach we considered a rectangular restricted region, [0, τX] × [0, τY]. An anonymous associate editor correctly pointed out that the top right corner of this rectangle is not always identifiable nonparametrically. Alternatively, one could consider defining estimators using the identifiable region ΩR = {(tX, tY) : TX,i ≥ tX, TY,i ≥ tY for some subject i} (see Prentice and Zhao, 2019). Although this idea is intriguing, we decided to keep the definition of ΩR as [0, τX] × [0, τY] due to easier interpretation. Note that Dabrowska’s estimator can be computed and is consistent for all points inside [0, τX] × [0, τY].
Another limitation of our method is its computational time complexity, up to , where n is the sample size and m is the number of bootstrap samples. For example, on a MacBook Air with 1.7 GHz Intel Core i7, the computational time for n = 100, 200, 500, 1000 and m = 200 is approximately 0.2, 0.7, 5.1, and 30.8 min, respectively.
Although not studied here, our approaches can be directly applied to settings where only one of the variables is right censored. This special case allows nonparametric estimation of the bivariate survival surface without negative mass (Stute, 1993). In future work, we plan to study semiparametric methods for estimating covariate-adjusted partial and conditional Spearman’s correlation for bivariate survival data.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by the U.S. National Institutes of Health, grants R01 AI093234 and U01 AI069923. We thank members of CCASAnet for allowing us to present their data, and Cathy Jenkins for help constructing the analysis data set.
APPENDIX
Theorem A1.
Under uninformative and generalized type I censoring and for a nonempty rectangular region ΩR such that ΩR ⊆ Ω, is a consistent estimator of . When , estimator is also asymptotically normal, and the bootstrap estimation of its variance is asymptotically valid.
Proof.
The consistency of follows from the consistency of Dabrowska’s estimator in Ω under uninformative and generalized type I censoring and the fact that is a continuous function of . To prove asymptotic normality, we note that under uninformative and generalized type I censoring, , , and are Hadamard differentiable estimators that converge to Gaussian processes; see Dabrowska (1989) and van der Vaart and Wellner (1996). Because estimator is a continuous function of , , and , it follows from the chain rule that it is also Hadamard differentiable and, therefore, it is asymptotically normal by the functional delta method (van der Vaart and Wellner, 1996). In addition, its bootstrap estimator converges to the same Gaussian process as , justifying the use of the bootstrap to construct CIs (van der Vaart and Wellner, 1996). □
Theorem A2.
Under uninformative and generalized type I censoring, estimator is consistent. When, it is also asymptotically normal, and the bootstrap estimation of its variance is asymptotically valid.
Proof.
Similar to Theorem A1. □
Theorem A3.
Under uninformative and unbounded censoring, is a consistent estimator of the overall Spearman’s correlation, ρS. Estimator is also asymptotically normal when ρS ∈ (−1, 1), and the bootstrap estimation of its variance is asymptotically valid.
Proof.
When the maximum of X and Y are observed events, then , so the consistency of for ρS follows from the consistency of Dabrowska’s estimator and the continuous mapping theorem. When the maximum of either X or Y is censored, the plug-in estimator of (9), , can be defined as a sum of four terms A, B, C, and D that are functions of the joint and conditional survival surface estimators (Theorem A.1, Supplementary Appendix). Because of Lemma A.1 (Supplementary Appendix), B, C, and . The proofs of the asymptotic normality and bootstrap validity are similar to Theorem A1. □
Footnotes
SUPPORTING INFORMATION
All analyses were performed in statistical language R (R Core Team, 2017). Complete simulation and analysis code is posted at https://biostat.app.vumc.org/ArchivedAnalyses. Example code, Web Appendices, Tables, and Figures referenced in Sections 3.3, 4.1, 4.2 are available with this paper at the Biometrics website on Wiley Online Library.
DATA AVAILABILITY STATEMENT
The deidentified data used in this article are openly available at https://biostat.app.vumc.org/ArchivedAnalyses.
REFERENCES
- Campbell G (1981) Nonparametric bivariate estimation with randomly censored data. Biometrika, 68, 417–422. [Google Scholar]
- Carriere JF (2000) Bivariate survival models for coupled lives. Scandinavian Actuarial Journal, 2000, 17–32. [Google Scholar]
- CCASAnet (2020). Times to viral failure and regimen change data. Anonymized for presentation, https://biostat.app.vumc.org/ArchivedAnalyses accessed March 17, 2021.
- Cesar C, Jenkins CA, Shepherd BE, Padgett D, Mejía F, Ribeiro SR, et al. (2015) Incidence of virological failure and major regimen change of initial combination antiretroviral therapy in the Latin America and the Caribbean: an observational cohort study. The Lancet HIV, 2, e492–e500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DG (1978) A model for association in bivariate life tables and its application in epidemiological studies of familial tendency in chronic disease incidence. Biometrika, 65, 141–151. [Google Scholar]
- Cuzick J (1982) Rank tests for association with right censored data. Biometrika, 69, 351–364. [Google Scholar]
- Dabrowska DM (1986) Rank tests for independence for bivariate censored data. The Annals of Statistics, 14, 250–264. [Google Scholar]
- Dabrowska DM (1988) Kaplan–Meier estimate on the plane. The Annals of Statistics, 16, 1475–1489. [Google Scholar]
- Dabrowska DM (1989) Kaplan–Meier estimate on the plane: weak convergence, LIL, and the bootstrap. Journal of Multivariate Analysis, 29, 308–325. [Google Scholar]
- Eden SK, Li C and Shepherd BE (2021). survSpearman: Nonparametric Spearman’s Correlation for Survival Data. R package version 1.0.0, https://CRAN.R-project.org/package=survSpearman accessed March 17, 2021.
- Fan J, Hsu L and Prentice RL (2000) Dependence estimation over a finite bivariate failure time region. Lifetime Data Analysis, 6, 343–355. [DOI] [PubMed] [Google Scholar]
- Klein JP and Moeschberger ML (1997). Survival Analysis: Techniques for Censored and Truncated Data. New York: Springer Science & Business Media. [Google Scholar]
- Kruskal WH (1958) Ordinal measures of association. Journal of the American Statistical Association, 53, 814–861. [Google Scholar]
- Li C and Shepherd BE (2012) A new residual for ordinal outcomes. Biometrika, 99, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D and Ying Z (1993) A simple nonparametric estimator of the bivariate survival function under univariate censoring. Biometrika, 80, 573–581. [Google Scholar]
- Liu Q, Li C, Wanga V and Shepherd BE (2018) Covariate-adjusted Spearman’s rank correlation with probability-scale residuals. Biometrics, 74, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen RB (2007). An Introduction to Copulas. New York: Springer Science & Business Media. [Google Scholar]
- Oakes D (1982) A model for association in bivariate survival data. Journal of the Royal Statistical Society: Series B (Methodological), 44, 414–422. [Google Scholar]
- Oakes D (1989) Bivariate survival models induced by frailties. Journal of the American Statistical Association, 84, 487–493. [Google Scholar]
- Prentice RL and Cai J (1992) Covariance and survivor function estimation using censored multivariate failure time data. Biometrika, 79, 495–512. [Google Scholar]
- Prentice RL and Zhao S (2019). The Statistical Analysis of Multivariate Failure Time Data: A Marginal Modeling Approach. Boca Raton, FL: Chapman and Hall/CRC. [Google Scholar]
- Pruitt RC (1991) On negative mass assigned by the bivariate Kaplan–Meier estimator. The Annals of Statistics, 19, 443–453. [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Version 3.4.3., https://www.R-project.org/ (accessed July 9, 2019)] [Google Scholar]
- Romeo JS, Tanaka NI and Pedroso-de Lima AC (2006) Bivariate survival modeling: a Bayesian approach based on copulas. Lifetime Data Analysis, 12, 205–222. [DOI] [PubMed] [Google Scholar]
- Royston P and Parmar MK (2013) Restricted mean survival time: an alternative to the hazard ratio for the design and analysis of randomized trials with a time-to-event outcome. BMC Medical Research Methodology, 13, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemper M, Kaider A, Wakounig S and Heinze G (2013) Estimating the correlation of bivariate failure times under censoring. Statistics in Medicine, 32, 4781–4790. [DOI] [PubMed] [Google Scholar]
- Shepherd BE, Li C and Liu Q (2016) Probability-scale residuals for continuous, discrete, and censored data. Canadian Journal of Statistics, 44, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JH and Louis TA (1995) Inferences on the association parameter in copula models for bivariate survival data. Biometrics, 51, 1384–1399. [PubMed] [Google Scholar]
- Shih JH and Louis TA (1996) Tests of independence for bivariate survival data. Biometrics, 52, 1440–1449. [PubMed] [Google Scholar]
- Stute W (1993) Consistent estimation under random censorship when covariables are present. Journal of Multivariate Analysis, 45, 89–103. [Google Scholar]
- van der Laan M (1997) Nonparametric estimators of the bivariate survival function under random censoring. Statistica Neerlandica, 51, 178–200. [Google Scholar]
- van der Laan MJ (1996) Efficient estimation in the bivariate censoring model and repairing NPMLE. The Annals of Statistics, 24, 596–627. [Google Scholar]
- van der Vaart AW and Wellner JA (1996). Weak Convergence and Empirical Processes. Springer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The deidentified data used in this article are openly available at https://biostat.app.vumc.org/ArchivedAnalyses.


