Abstract
JC polyomavirus (JCV) is the causative agent of progressive multifocal leukoencephalopathy (PML), a central nervous system infection that mainly affects AIDS patients. The extensive application of highly active antiretroviral therapy (HAART) is leading to the appearance of “long-term” survival PML patients. A reliable and feasible qualitative-quantitative test for both the detection of JCV and follow-up of its viral burden in this emerging group of patients is clearly required. With this aim, a dual qualitative-quantitative nested PCR is presented in this study for the analysis of JCV DNA in cerebrospinal fluid (CSF). Two newly designed internal controls, one competitive and the other noncompetitive, have been constructed to adapt this PCR to either measure the JCV burden or to allow a highly confident determination of JCV presence or clearance. The analytical sensitivity of the technique allows the detection of 0.01 fg (three genomes) of JCV DNA. Its qualitative application has been evaluated by analyzing single CSF samples from a group of 17 patients with PML and a control group of 20 patients with diverse neurological conditions other than PML, yielding sensitivity and specificity values of 100 and 90%, respectively. The quantitative application has been evaluated in vitro in blind tests with samples including serial dilutions of JCV, and in all cases the samples were successfully ordered considering the JCV titer. The dual quantitative-qualitative application offered by this nested PCR may provide an answer to the new requirements for evaluating and finely monitoring PML in AIDS patients receiving HAART.
JC virus (JCV) is a ubiquitous polyomavirus probably acquired asymptomatically during childhood and latently found in most healthy adults (6, 10, 15, 16, 17). It is involved in central nervous system (CNS) infection of immunocompromised patients, causing progressive multifocal leukoencephalopathy (PML), a rapidly evolving neurological disorder that affects 2 to 10% of AIDS patients (5, 7).
Diagnosis of PML is generally presumptive and is made by imaging techniques (computerized tomography or magnetic resonance), but confirmation requires histopathological analysis of brain biopsy. Different PCR-based strategies to detect JCV DNA in the cerebrospinal fluid (CSF) have been described that use unique or successive CSF samples and single or nested PCRs, sometimes including DNA purification, and are designed to amplify different coding (VP1, small T-antigen, large T-antigen) or noncoding viral regions, with variable diagnostic yields (7, 8, 10, 14, 17, 18, 24, 25).
Before the extensive application of human immunodeficiency virus (HIV) antiretroviral combination therapies, PML had a fatal outcome within weeks or months because of the lack of effective therapies (9, 20). Since the application of highly active antiretroviral therapy (HAART), several cases of remission of PML have been reported (4, 11, 21), and at our institution we have observed prolonged survival and clinical and neurological improvement in PML patients (19).
The emergence of long-term survival PML patients among those receiving HAART stresses the need for new technical approaches capable of finely monitoring the presence and progression of this infection. In this study, we present a dual PCR suitable for both highly confident detection of the presence and clearance of JCV and quantitative follow-up of the JCV burden in PML patients.
MATERIALS AND METHODS
Patients.
CSF samples were obtained from a group of 37 patients (27 HIV positive and 10 HIV negative). Among them, 17 HIV-positive patients had PML according to clinical (diverse neurological symptoms, such as mono- or hemiparesis, speech or visual disorders, or sensorial deficits), neuroimaging (lesions suggestive of PML appeared on CT as low-attenuation zones in the white matter without contrast enhancement and no mass effect and in magnetic resonance images as areas of increased signal intensity in T2 and isotense or hypointense in relation to cortex in T1), and/or stereotactic brain biopsy criteria (based on the detection of foci of myelin destruction and abnormalities in oligodendrocytes as seen on hematoxylin and eosin staining); PML diagnosis was based on neuroimaging and clinical symptoms in 9 patients and on additional histopathological confirmation in the remaining 8 cases. The remaining 20 patients had diverse neurological conditions other than PML (10 of them were HIV positive and the other 10 HIV negative); among the 10 HIV-positive patients, diverse neurological entities were found: 5 cases of dementia-AIDS complexes, 1 case of cerebral toxoplasmosis, 1 case of enteroviral meningitis, 1 case of cytomegalovirus (CMV) encephalitis, 1 case of cerebral lymphoma, and 1 case of hepatic encephalopathy. Among the 10 HIV-negative patients, 4 had bacterial meningitis and the remaining 6 were nonimmunocompromised patients with a clinical evolution incompatible with PML.
CSF samples were separated into aliquots and stored at −70°C immediately after extraction until analysis.
JCV DNA purification.
For JCV DNA extraction, 9 μl of lysis buffer was added to 111 μl of CSF (final concentration: 10 mM Tris-Cl, 0.5% SDS, 0.2 μg of proteinase K per μl). Lysis reactions were incubated for 2 h at 50°C and afterwards for 5 min at 100°C. Reactions were protein extracted, and DNA was ethanol precipitated. Once dried, the pellet was resuspended in 24 μl of ultrapure water and immediately used for the PCR.
JCV nested PCR.
A nested PCR which amplifies part of the region codifying the large T antigen was performed for the detection of JCV DNA. All of the PCR assays were “blind” (i.e., without clinical information on sources).
For the first amplification reaction, the primers selected were JC1 (5′-AAGTCCATTTTATCAAGCAA-3′, nucleotides [nt] 3546 to 3565) and JC2 (5′-TTGTAAAGGTGTGAATAAGGA-3′, nt 4001 to 3981). The PCR thermal profile was 5 min at 94°C, 40 amplification cycles consisting of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, and a tail end of 5 min at 72°C.
For the second nested reaction the primers were JC3 (5′-TGCTACAGTATCAACAGCCT-3′, nt 3632 to 3651) and JC4 (5′-AGGAGCATGACTTTAACCCA-3′, nt 3909 to 3890) (8). The PCR profile was 5 min at 94°C, 40 amplification cycles consisting of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C, and a tail end of 5 min at 72°C. The annealing temperature was increased by 16°C in comparison with that proposed previously (8) in order to decrease the smears that were detected when analyzing amplification products by electrophoresis; this indicates the occurrence of nonproductive unspecific amplifications.
Sequence searches in the EMBL DNA databases assured the absence of significative homologies between the primers selected and the rest of the DNA sequences compiled.
Reactions included 25 pmol of each primer, 1× PCR buffer (Boehringer Mannheim) supplemented with 0.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 1 U of Taq polymerase (Boehringer Mannheim) in a 50-μl final reaction volume. Five microliters of the extracted JCV DNA was amplified in the first reaction, and 1.5 μl of this first amplification was used as a template for the second reaction. Ten microliters of each reaction was used for electrophoretic detection of the amplicon, and NdeII digestions of the amplicon were performed to check the amplification specificity. To avoid cross-contamination, all reagents were separated into aliquots and discarded after use; water samples were processed in parallel throughout the process and were included in tubes positioned at random in the amplification plate.
The specificity of the PCR was assayed by proving the absence of cross-reactions with other neurotropic viruses. Testing was performed with serial dilutions of DNA extracted from supernatants (200 μl; following the same DNA extraction method as for JCV) from in vitro-cultured samples of CMV, herpes simplex virus (HSV), and varicella-zoster virus (VZV).
Construction of the amplification internal controls. (i) Noncompetitive internal control.
The bla (β-lactamase) gene from Escherichia coli was amplified to obtain a DNA fragment equivalent in length (416 bp) to that of the JCV region which was used as the initial template for the nested PCR. The primers selected were bla1 (5′-TGTTATCACTCATGGTTAT-3′, nt 1772 to 1809) and bla2 (5′-AAGAGTATGAGTATTCAA-3′, nt 2188 to 2171). The PCR thermal profile was 5 min at 94°C, 40 amplification cycles consisting of 1 min at at 94°C, 1 min 49°C, and 1 min at 72°C, and a tail end of 5 min at 72°C.
The 416-bp amplicon was cloned in the pGem-T vector (Promega). The recombinant plasmid pGembla (Fig. 1a) was checked by electrophoretical and restriction analysis, and the presence of the expected insert was proved by PCR with primers bla1 and bla2.
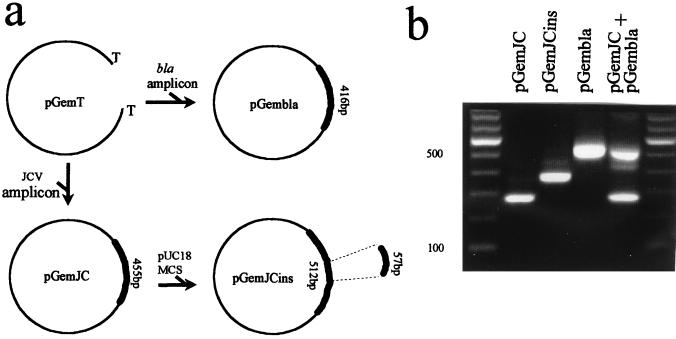
FIG. 1.
(a) Scheme of construction of the recombinant plasmids used as internal controls: pGemJC, pGemJCins, and pGembla. Cloning vector pGem-T is also included. The origin of the different DNA fragments used for cloning and their respective lengths are indicated. The region of JCV selected for cloning is defined between coordinates 3546 and 4001. pUC18 MCS, multicloning site from pUC18 vector. (b) Amplicons derived from PCR amplification of control plasmids: pGemJC (277 bp), pGemJCins (334 bp), and pGembla (539 bp). Products of pGembla and pGemJC when coamplified in the same reaction are shown in the last lane. Left lane, 100-bp ladder as a molecular weight marker.
(ii) Competitive internal control.
The same region of JCV that is used as the initial template in the nested PCR (limited by the outer JC1 and JC2 primers) was cloned into the pGem-T vector, thus leading to the recombinant plasmid pGemJC (Fig. 1a). The recombinant plasmid was checked by electrophoretic mobility and enzymatic digestion; nested amplification of pGemJC led to the same 277-bp amplicon as the one obtained when JCV was used as a template (Fig. 1b). Finally, the whole insert was sequenced (Boehringer Mannheim).
pGemJC was engineered to render a slightly enlarged amplicon compared to that obtained with the JCV target. A DNA fragment was cloned within the JC region of pGemJC: a 57-bp insert obtained from the multicloning site of pUC18. The recombinant plasmid pGemJCins (Fig. 1a) was amplified by nested PCR with JC1-JC2 and JC3-JC4, giving the expected amplicon of 334 bp, which is clearly distinguishable from that obtained with pGemJC (Fig. 1b). Finally, pGemJCins was sequenced to prove the proper entry of the insertion.
Qualitative PCR.
For qualitative PCR, 5 μl of JCV DNA extracted from CSF and 1 μl of the noncompetitive control (pGembla) were coamplified; pGembla control was included in amounts slightly above the limit of detection of the technique (0.01 fg, see Results) to ensure that minimum traces of inhibitors were able to block its amplification. Primers specific either for JCV and pGembla were included in the same reaction to guarantee the absence of competition between the amplifications of the respective templates. Primers JC1-JC2 and JC3-JC4 were included to amplify JCV, and for the amplification of the pGembla plasmid control, UNA/UNB (17-bp M13 universal primers: 5′-GTTTTCCCAGTCACGAC3′/5′-CAGGAAACAGCTATGAC-3′) for the first reaction and NCA (5′-AATTGTAATACGACTCACTATA-3′, nt 2982 to 3003) and NCB (5′-TACTCAAGCTATGCATGCAA-3′, nt 123 to 104) for the second nested reaction were included. Primers for pGembla amplification were designed to lead to an amplicon longer (539 bp) than the one obtained from pGemJC (277 bp) in order to discriminate them (Fig. 1b). The amplification profiles were the same as those previously indicated for JCV. The sensitivity limit value was the same for assays including or not including the internal control.
Positive samples should show both the JCV and pGembla amplicons. The pGembla amplicon was the only one detected in the negative samples, and no amplicon was observed in the inhibited samples. For those cases in which inhibitions were detected, DNA was additionally purified and diluted to eliminate the effect of the inhibitors.
The software package EPI INFO (CDC) was used to calculate the sensitivity, specificity, and predictive values.
Quantitative PCR.
For quantitative PCR, 5 μl of the extracted JCV DNA was coamplified with 1 μl of the competitive control (pGemJCins; slightly above the sensitivity limit of detection [see Results]). The primers included were JC1-JC2 and JC3-JC4 for the first and second reactions, respectively, and were shared for the competitive amplification of both JCV and pGemJCins. The 277-bp JCV amplicon was observed when the JCV burden was above the competitive control titer, whereas the 334-bp control amplicon was observed when it was below this titer.
A set of seven dilutions (from 1 to 10−6 pg) of the competitive control were coamplified with the sample to be measured. Competition was detected when equivalent amounts of JCV and control were coamplified. Therefore, the titer of target in the sample (JCV DNA) was determined by the input amount of competitive control included in the first dilution whose amplification is competed with.
RESULTS
Analytical evaluation of the nested PCR.
As an initial step in the evaluation of our test, the analytical specificity of this PCR was assessed by proving the absence of cross-reactions with other agents commonly involved in CNS infections, such as enterovirus, CMV, VZV, and HSV. No amplicons were detected when DNA extracted from supernatants corresponding to in vitro-cultured samples of these viruses (data not shown) was used as a template.
A preliminary characterization of the product amplified by this PCR was performed by using CSF from one patient with biopsy-confirmed PML. A band with an electrophoretic mobility corresponding to the expected size (277 bp) was observed (Fig. 2a). When the PCR product was digested with NdeII, two restriction products (107 and 170 bp) were obtained, confirming the expected presence of a site for that enzyme in the amplified region (Fig. 2a). The amplicon was sequenced, and the full identity with the known nucleotidic composition was proved.
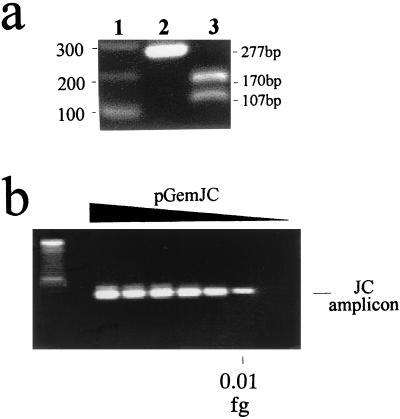
FIG. 2.
(a) Characterization of the nested-PCR amplicon. Lanes: 1, 100-bp ladder as a molecular weight marker; 2, 277-bp band corresponding to the product of the nested-PCR; 3, restriction products after digestion of the 277-bp amplicon with NdeII. Band mobilities are indicated on the sides of the gel. (b) Amplification products of the nested PCR with serial dilutions of pGemJC as a template. The sensitivity limit is indicated. Left lane: 100-bp ladder as a molecular weight marker.
The analytical sensitivity limit of this nested PCR corresponded to 0.01 fg (equivalent to three JCV genomes) (Fig. 2b) in successive determinations. It was calculated by amplification of serial limiting dilutions of cloned JCV DNA (pGemJC; Fig. 1, Materials and Methods) in CSF that previously had been tested as JCV-free.
Qualitative PCR.
The qualitative application of this PCR for PML diagnosis was evaluated by blind analysis of a group of 37 patients. All 17 patients fulfilling the diagnostic criteria for PML were PCR positive for JCV. Among the 20 patients (10 HIV positive and 10 HIV negative) with other clinical entities, PCR for JCV was negative in 18 cases and positive in the remaining 2. In all cases the amplification control pGembla (Fig. 1) was included to ensure the absence of laboratory contamination. These two cases were HIV-positive patients with an alternative CNS diagnosis in which a secondary infection by JCV could not be fully ruled out. Therefore, the qualitative JCV PCR detection rendered the following diagnostic values: sensitivity, 100%; specificity, 90%; positive predictive value of 89.5% and negative predictive value of 100%.
Among those cases which had been PCR positive for JCV, a second sample of CSF was obtained in seven patients receiving HAART with a favorable clinical outcome, according to neuroimaging and neurological criteria (19). The second PCR determination was negative for six of seven patients. To rule out the role of potential inhibitors causing false-negative results, an internal amplification control was constructed (pGembla; Materials and Methods, Fig. 1a) and included in all assays. The amplification of the internal control in all cases indicated that PCR-negative results were a result of the clearance of JC virus from the CSF of those patients. In all assays corresponding to follow-up patients, at least one baseline sample was included to rule out the presence of slight methodological deviations that could be responsible for false-negative results.
The noncompetitive internal control was also included to rule out inhibitors in all of the tests that were performed with the patients selected for the evaluation of the technique (see above). The efficiency of this control as a sensor of amplification inhibitors was proved by the disappearance of its amplicon when hemoglobin dilutions were added to the PCR sample (data not shown).
Quantitative PCR.
In order to devise a test suitable for a finely graded follow-up of those PML patients who remain PCR positive for JCV, a quantitative test based on the same nested PCR was developed. The evaluation of this quantitative application was performed in vitro by blind tests with a series of laboratory-prepared samples that included serial dilutions of JCV cloned DNA. Our purpose was to measure the JCV titers in those samples to order them according to their dilution factor.
To develop the quantitative application of the nested PCR, a competitive internal control was constructed (pGemJCins; Fig. 1, Materials and Methods), and titration assays were performed for its evaluation. Serial dilutions of pGemJCins were coamplified with samples including increasing amounts of cloned JCV DNA (pGemJC) (Fig. 3a). Cloned JCV was used as a target, instead of the true JCV genomes from biological samples, in order to be able to finely adjust the amount of DNA for each assay. The expected competition between the respective amplifications of pGemJC and pGemJCins was observed, because they consume the same pairs of primers. The JCins amplicon was observed when the pGemJCins burden was above that of pGemJC. Therefore, pGemJC samples were able to compete with JCins amplification up to the corresponding equivalent dilution of pGemJCins (Fig. 3a). In summary, the titer of the target (JCV DNA) in a sample corresponds to the amount of internal control (pGemJCins) included in the first dilution whose amplification is competed with.
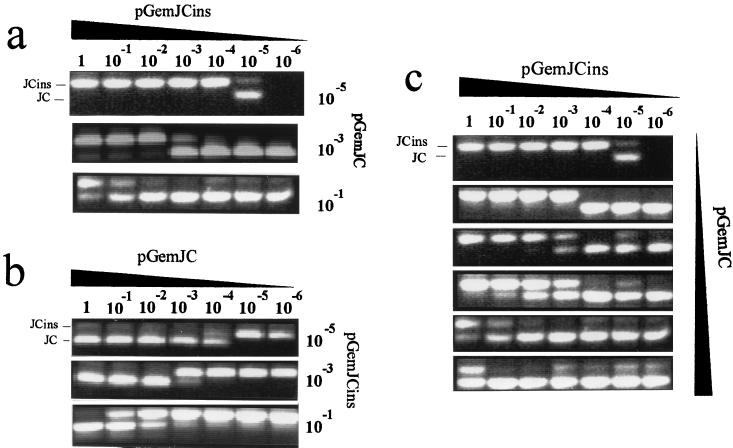
FIG. 3.
Titration assays to evaluate the quantitative-competitive PCR. Mobilities corresponding to pGemJC and pGemJCins amplicons are indicated. The dilution factors for each sample are also indicated; 1 corresponds to 1 pg of cloned JCV DNA. Dilutions increase from 1 to 10−6. Panels: a, serial dilutions of pGemJCins competed with increasing titers of pGemJC; b, serial dilutions of pGemJC competed with increasing titers of pGemJCins; and c, blind test of quantification with serial dilutions of pGemJCins. Tests corresponding to the different samples are ordered according to their concentration, which was measured by their ability to compete with dilutions of the internal control.
The titration assay was also performed in the opposite sense, that is, selecting pGemJCins to compete with a set of serial dilutions of pGemJC. All of the observations made above were also valid for this case (Fig. 3b). This indicates that the only factor relevant in the competition dynamics is the ratio between the two competing molecules; the relative length of the respective targets does not have a significant role.
To prove the quantitative application of this competitive nested PCR, a series of samples with different pGemJC burdens were blind tested (Fig. 3c). A set of serial dilutions of pGemJCins was prepared to be coamplified with the different JC problem samples. Samples with a low burden of target should be those whose amplification can only compete with the more diluted preparations of pGemJCins, whereas those with higher titers should be able to compete even with the more concentrated control samples. As indicated in Fig. 3c, it was possible to correctly order the problem samples according to their degree of dilution.
DISCUSSION
PML is a very severe CNS infection caused by JCV and characterized until recently by a fatal outcome (9, 20). The clinical symptoms and neuroimaging presentation are suggestive of the disease, but diagnostic confirmation requires histopathological analysis of brain biopsy.
Prior attempts to detect JCV in CSF have indicated the need to perform successive determinations for each patient to take advantage of the increased amount of virus released into CSF as PML progresses (7, 8), since the amounts of free JC virions in the CSF of early-PML patients are extremely low (7, 8, 14). Up to 20 to 30% of false-negative results have been reported when such early determinations are obtained (7, 14). In our case, a high diagnostic sensitivity (100%) was obtained despite performing only a single determination for each patient and immediately after the appearance of the first neurological symptoms. This corresponds to our analytical sensitivity limit (0.01 fg), which is higher than usual, as a result of working with a genome region not subjected to sequence variability (1, 2, 13) and the inclusion of strict viral lysis methods and DNA purification procedures.
Most of the patients in the control group (each of the 10 HIV-negative and 8 of the 10 HIV-positive patients) offered negative PCR results, and no cross-reactions were observed with samples of neurotropic viruses other than JCV. This indicates the proper specificity of our approach. To interpret the meaning of a PCR-positive result for two patients in this group, it is worth noting that autopsy analysis has indicated that JCV DNA can be found in 31% of HIV-positive patients (12, 22), a much higher proportion than the 5% of AIDS patients considered to have PML. In our study these two patients corresponded to HIV-positive cases with low CD4 counts. In both patients the PCR was repetitively positive and the internal amplification control was included in all of the determinations to ensure that laboratory contamination did not occur. Therefore, we interpret these data as a detection of subclinical presences of JCV as a result of the high sensitivity of this technique, rather than as false-positive PCR results due to laboratory contamination. Ongoing experiments are being performed with the quantitative JCV to try to define a “cut-off” for the JCV burden that could discriminate the subclinical presence of JCV from the viral titers associated with PML.
The recent reports of “long-term” survival and clinical improvement in patients with PML receiving HAART (4, 11, 19, 21) stresses the need for new technical approaches that allow for the monitoring of JCV in CSF. We have proved the usefulness of our qualitative PCR for highly confident detection of JCV clearances from the CSF. The technique has been applied to a group of seven PML patients receiving HAART, with a good clinical and neurological outcome, and in six of them viral clearance was documented (19).
The variable levels of JCV burden expected either for PML progression (7, 8) or for regression in those emerging “long-term” survival PML patients require the development of more-precise quantitative determinations. Several attempts have been made to quantify JCV in CSF samples. Serial limiting dilution assays or measurements of the signal resulting from the hybridization of amplicons with labeled probes have been the most common approaches (3, 8). Here, we have successfully evaluated in vitro a much more precise quantification method based on the coamplification of the JCV target with serial dilutions of a newly designed competitive internal control. This quantifying approach has clear advantages: (i) the internal control allows us to detect false-negative samples, and (ii) the cutoff in the assay can be selected by varying the amount of internal control used as competitor (23). The latter could be suitable for a correct interpretation of those cases in which traces of JCV are expected to be due either to subclinical infections, such as those previously commented upon, or to an efficient response to therapy.
Clinical samples have recently been tested with the quantitative application, and it has been possible to measure logarithmic differences among CSF samples belonging to different PML patients. This proves that the resolution of this technique is able not only to efficiently measure the JCV titer in laboratory-calibrated samples but also to detect biological variations in the JCV burden in CSF samples.
The dual qualitative-quantitative PCR for analysis of JCV in CSF presented here could help to fill the diagnostic gaps which are appearing in the diagnosis and follow-up of the emergent group of PML patients with a favorable clinical outcome.
ACKNOWLEDGMENT
We are indebted to Thomas O’Boyle for his help in the preparation of the manuscript.
REFERENCES
- 1.Agostini H T, Ryschkewitsch C F, Singer E J, Stoner G L. JC virus regulatory region rearrangements and genotypes in progressive multifocal leucoencephalopathy: two independent aspects of virus variation. J Gen Virol. 1997;78:659–664. doi: 10.1099/0022-1317-78-3-659. [DOI] [PubMed] [Google Scholar]
- 2.Agostini H T, Ryschkewitsch C F, Mory R, Singer E J, Stoner G. JC virus (JCV) genotypes in brain tissue from patients with progressive multifocal leucoencephalopathy (PML) and in urine from controls without PML: increased frequency of JCV type 2 in PML. J Infect Dis. 1997;176:1–8. doi: 10.1086/514010. [DOI] [PubMed] [Google Scholar]
- 3.Antinori A, De Luca A, Ammasari A, Cingolani A, Murri R, Colosimo G, Roselli R, Scerrati M, Tamburrini E. Failure of cytarabine and increased JC virus-DNA burden in the cerebrospinal fluid of patients with AIDS-related progressive multifocal leucoencephalopathy. AIDS. 1994;8:1022–1024. [PubMed] [Google Scholar]
- 4.Baqi M, Kucharczyk W, Walmsley S L. Regression of progressive multifocal encephalopathy with highly active antiretroviral therapy. AIDS. 1997;11:1526–1527. [PubMed] [Google Scholar]
- 5.Berger J R, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 6.Chesters P M, Heritage J, McCance D J. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 7.Cinque P, Scaperllini P, Vago L, Linde A, Lazzarin A. Diagnosis of central nervous system complications in HIV-infected patients: cerebrospinal fluid analysis by the polymerase chain reaction. AIDS. 1997;11:1–17. doi: 10.1097/00002030-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 8.De Luca, A., A. Cingolani, A. Linzalone, A. Ammassari, R. Murri, M. L. Giancola, G. Maiuro, and A. Antinori. 1996. Improved detection of JC virus DNA in cerebrospinal fluid for diagnosis of AIDS-related progressive multifocal leukoencephalopathy. J. Clin. Microbiol. 1334–1346. [DOI] [PMC free article] [PubMed]
- 9.De Truchis P, Flament-Saillour M, Urtizberea J A, Hassine D, Clair B. Inefficacy of cytarabine in progressive multifocal leucoencephalopathy in AIDS. Lancet. 1993;342:622–623. doi: 10.1016/0140-6736(93)91453-s. [DOI] [PubMed] [Google Scholar]
- 10.Dubois V, Lafon M E, Ragnaud J M, Pellegrin J L, Damasio F, Baudouin C, Michaud V, Fleury J A. Detection of JC virus DNA in the peripheral blood leukocytes of HIV-infected patients. AIDS. 1996;10:353–358. doi: 10.1097/00002030-199604000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Elliot B, Aromin I, Gold R, Flanigan T, Mileno M. 2.5 year remission of AIDS-associated progressive multifocal leucoencephalopathy with combined antiretroviral therapy. Lancet. 1997;349:850–851. doi: 10.1016/S0140-6736(05)61753-9. [DOI] [PubMed] [Google Scholar]
- 12.Ferrante P, Caldarelli-Stefano R, Omodeo-Zorini B, Vago L, Boldorini R, Constanzi G. PCR detection of JC virus DNA in brain tissue of patients with progressive multifocal leucoencephalopathy. J Med Virol. 1995;47:219–225. doi: 10.1002/jmv.1890470306. [DOI] [PubMed] [Google Scholar]
- 13.Guo J, Kitamura T, Ebihara H, Sugimoto C, Kunitake T, Takehisa J, Na Y Q, Al-Ahdal M N, Hallin A, Kawabe K, Taguchi F, Togo Y. Geographic distribution of the human polyomavirus JC virus types A and B and isolation of a new type from Ghana. J Gen Virol. 1996;77:919–927. doi: 10.1099/0022-1317-77-5-919. [DOI] [PubMed] [Google Scholar]
- 14.Hammarin A L, Bogdanovic G, Svedhem V, Pirskanen R, Morfeldt L, Grandien M. Analysis of PCR as a tool for detection of JC virus DNA in cerebrospinal fluid for diagnosis of progressive multifocal leucoencephalopathy. J Clin Microbiol. 1996;34:2929–2932. doi: 10.1128/jcm.34.12.2929-2932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura T, Aso Y, Kuniyoshi K, Hara K, Yogo Y. High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura T, Sugimoto C, Kato A, Ebihara H, Suzuki M, Taguchi F, Kawabe K, Yogo Y. Persistent JC virus (JCV) infection is demonstrated by continuous shedding of the same JCV strains. J Clin Microbiol. 1997;35:1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsiota-Bernard P, De Truchis P, Gray F, Flament-Saillour M, Voyatzakis E, Nauciel C. JC virus detection in the cerebrospinal fluid of AIDS patients with progressive multifocal leucoencephalopathy and monitoring of the antiviral treatment by a PCR method. J Med Microbiol. 1997;46:256–259. doi: 10.1099/00222615-46-3-256. [DOI] [PubMed] [Google Scholar]
- 18.McGuire D, Barhite S, Hollander H, Milles M. JC virus DNA in cerebrospinal fluid of human immunodeficiency virus-infected patients: predictive value for progressive multifocal leucoencephalopathy. Ann Neurol. 1995;37:395–399. doi: 10.1002/ana.410370316. [DOI] [PubMed] [Google Scholar]
- 19.Miralles P, Berenguer J, García de Viedma D, Padilla B, Cosín J, López-Bernaldo de Quirós J C, Muñoz L, Moreno S, Bouza E. Treatment of AIDS-associated progressive multifocal leucoencephalopathy with highly active antiretroviral therapy. AIDS. 1998;12:2467–2472. doi: 10.1097/00002030-199818000-00016. [DOI] [PubMed] [Google Scholar]
- 20.Moreno S, Miralles P, Díaz M D, Berenguer J, López Bernaldo de Quirós J C, Blázquez R, Cosín J, Bouza E. Cytarabine therapy for progressive multifocal leucoencephalopathy in patients with AIDS. Clin Infect Dis. 1996;23:1066–1068. doi: 10.1093/clinids/23.5.1066. [DOI] [PubMed] [Google Scholar]
- 21.Power C, Nath A, Aoki F Y, Del Bigio M. Remission of progressive multifocal leucoencephalopathy following splenectomy and antiretroviral therapy in a patient with HIV infection. New Engl J Med. 1997;336:661–662. doi: 10.1056/NEJM199702273360914. [DOI] [PubMed] [Google Scholar]
- 22.Quinlivan E B, Norris M, Bouldin T W, Suzuki K, Meeker R, Smith M S, Hall C, Kenney S. Subclinical central nervous system infection with JC virus in patients with AIDS. J Infect Dis. 1992;166:80–85. doi: 10.1093/infdis/166.1.80. [DOI] [PubMed] [Google Scholar]
- 23.Siebert P D, Kellogg D E. PCR MIMICs: competitive DNA fragments for use in quantitative PCR. In: McPherson M J, Hames B D, Taylor G R, editors. PCR2: a practical approach. Oxford, United Kingdom: IRL Press/Oxford University Press; 1995. [Google Scholar]
- 24.Sugimoto C, Ito D, Kanada K, Matsuda H, Saito H, Sakai H, Fujihara K, Itoyama Y, Yamada T, Kira J, Matsumoto R, Mori M, Nagashima K, Yogo Y. Amplification of JC virus regulatory DNA sequences from cerebrospinal fluid: diagnostic value for progressive multifocal leucoencephalopathy. Arch Virol. 1998;143:249–262. doi: 10.1007/s007050050284. [DOI] [PubMed] [Google Scholar]
- 25.Weber T, Turner R W, Frye S, Luke W, Kretzschmar H A, Luer W, Hunsmann G. Progressive multifocal leukoencephalopathy diagnosed by amplification of JC virus-specific DNA from cerebrospinal fluid. AIDS. 1994;8:49–57. doi: 10.1097/00002030-199401000-00008. [DOI] [PubMed] [Google Scholar]