Abstract
Pleomorphic xanthoastrocytoma (PXA) is a rare type of brain tumor that affects children and young adults. Molecular prognostic markers of PXAs remain poorly established. Similar to gangliogliomas, PXAs show prominent immune cell infiltrate, but its composition also remains unknown. In this study, we correlated DNA methylation and BRAF status with clinical outcome and explored the tumor microenvironment. We performed DNA methylation in 21 tumor samples from 18 subjects with a histological diagnosis of PXA. MethylCIBERSORT was used to deconvolute the PXA microenvironment by analyzing the associated immune cell-types. Median age at diagnosis was 16 years (range 7–32). At median follow-up of 30 months, 3-year and 5-year overall survival was 73% and 71%, respectively. Overall survival ranged from 1 to 139 months. Eleven out of 18 subjects (61%) showed disease progression. Progression-free survival ranged from 1 to 89 months. Trisomy 7 and CDKN2A/B (p16) homozygous deletion did not show any association with overall survival (p = 0.67 and p = 0.74, respectively). Decreased overall survival was observed for subjects with tumors lacking the BRAF V600E mutation (p = 0.02). PXAs had significantly increased CD8 T-cell epigenetic signatures compared with previously profiled gangliogliomas (p = 0.0019). The characterization of immune cell-types in PXAs may have implications for future development of immunotherapy.
Keywords: BRAF V600E, Brain tumor, CDKN2A/B, DNA methylation, Pleomorphic xanthoastrocytoma, Tumor immune microenvironment
INTRODUCTION
Pleomorphic xanthoastrocytoma (PXA) is an uncommon type of brain tumor mostly diagnosed in children and young adults and accounts for <1% of astrocytic tumors (1). Most commonly the tumors are classified as World Health Organization (WHO) grade II tumors. However, malignant transformation to anaplastic WHO grade III tumors can occur in 15%–20% of patients. Diagnosis of PXA is usually made by histology and immunohistochemistry. Anaplastic PXA is usually associated with increased mitosis, necrosis, infiltrative growth, and less often microvascular proliferation (2). Patients with PXAs usually have a favorable outcome with reports of 70% survival at 10 years from diagnosis, but they appear to have a higher risk of recurrence compared with patients with other types of low-grade gliomas (1). There have been novel discoveries in the pathology and molecular biology of PXAs and signs of anaplasia have been associated with a more adverse prognosis (1). PXAs are uncommon and reported cohorts are relatively small. Therefore the impact of the molecular information on clinical outcome has not been firmly established.
DNA methylation has emerged as a prominent molecular biomarker in brain tumors and involves epigenetic alterations with transfer of a methyl group to the 5′ position of the cystine ring (3,4). Altered cytosine residues are usually adjacent to the guanine nucleotides, forming CpG islands (3,4). The global distribution of CpG islands form a methylation pattern, and DNA methylation aberrations have been linked to mechanisms of carcinogenesis (3,4). Inactivation of tumor suppressor genes due to hypermethylation within promotor regions, as well as global hypomethylation, can contribute to genomic instability and formation of cancerous cells (3,4). Previous studies have also shown that DNA methylation can be used to identify specific signatures to better classify central nervous system tumors (5). Genome-wide DNA methylation profiling is emerging as a powerful tool that can define molecular signatures to aid in the classification of tumors. Few studies to date have examined genome-wide DNA methylation in PXAs (6,7).
BRAF V600E mutation is a common genetic alteration found in both PXA and anaplastic PXA and is present in ∼65% of these tumors (8). PXAs also commonly have the homozygous deletion of CDKN2A/B tumor suppressor, which encodes for p16 on chromosome 9p21 (9,10). The combination of BRAF mutation and CDKN2A/B homozygous deletion has been associated with low-grade tumors that often transform to high-grade tumors and has been associated with increased resistance to treatment (11). Recent reports have also shown that anaplastic PXA-like tumors share features with glioblastoma and epithelioid glioblastoma (12,13). In one study, histologically diagnosed glioblastoma displayed methylation profiles with similar features to PXA (13). These tumors also harbored BRAF V600E and 9p21 CDKN2A/B homozygous deletion and were shown to have a better prognosis. The advancement in molecular characterization of these tumors through DNA methylation will enable more precise tumor classification for diagnosis and to identify novel therapeutic targets.
PXAs are also typically characterized by tumor lymphocytic infiltration, but the significance of the PXA immune microenvironment has not yet been well-defined. In this study, we explored the tumor microenvironment by analyzing inflammatory, endothelial, and fibroblast cell populations in PXA compared with ganglioglioma, another central nervous system tumor that commonly harbors the BRAF V600E mutation (14). We also present our findings on PXA with an update on how molecular markers identified through genome-wide methylation profiling could potentially aid in identifying patients likely to have disease recurrence or tumor progression.
MATERIALS AND METHODS
Subject Selection
Twenty-one tumor samples from 18 subjects selected from the NYU research database were diagnosed as PXA both by DNA methylation classifier and by histology. Seven subjects had a mismatch between methylation classification and histological diagnosis and were excluded from the methylation analysis. Clinical data including patient sex, age, tumor location, radiologic findings, clinical manifestations, information on surgical resection, treatment received, and survival outcomes were collected.
Histology and Immunohistochemistry
All cases were reviewed by board certified neuropathologists. Formalin-fixed paraffin-embedded tumor tissue sections were used for clinical immunohistochemistry staining including GFAP, synaptophysin, p53, S100, and BRAF V600E. Distinction between PXA (WHO grade II) and anaplastic PXA (WHO grade III) were based on 2016 WHO classification guidelines for grading criteria (≥5 mitoses per 10 high-power field). Histological information regarding infiltrative growth, mitotic activity, Ki-67 labeling index, necrosis, and microvascular proliferation was noted.
Radiological Analysis
Preoperative brain MRI from 9 subjects that were diagnosed as PXA both by histology and by methylation were available. All MR images were reviewed by a board certified neuroradiologist with subspecialized training and experience in pediatric neuroradiology. Each tumor was evaluated for enhancement, edema, reduced diffusion, and tumor texture (cystic, mixed, or solid). Imaging characteristics were compared between those subjects that had progression versus those that did not have progression.
DNA Methylation
Genome-wide methylation was profiled in 21 tumor samples from 18 subjects and had a diagnosis of PXA both by DNA methylation and by histology. DNA was extracted from banked tumor tissue using the automated Maxwell system (Promega, Madison, WI). DNA is bisulfite-converted using the DNA methylation kit from Zymo Research (Irvine, CA). Genome-wide methylation patterns were analyzed using the Illumina Human Methylation 450k or EPIC BeadChip Array (Illumina, San Diego, CA) according to manufacture instructions as previously described (15). Tumors were classified using the methylation classifier previously developed for central nervous system tumors (5), which outputs the tumor class and the copy number profile. The gains and losses were noted relevant to baseline, as previously described (16–21). “Gain” or “amplification” in the copy number variation profiles was determined by log2 ≥ 0.3. The data were analyzed using R package in Bioconductor.
MethylCIBERSORT Analysis
MethylCIBERSORT was used to deconvolute inflammatory, endothelial, and fibroblast cell populations with genome-wide DNA methylation data. We used a reference-based approach for tumor deconvolution with DNA methylation (22). Using methylCIBERSORT, we compared the PXA cohort with previously profiled cohorts of ganglioglioma samples.
Statistical Analysis
Methylation analysis using beta distribution, CpG island analysis, and hierarchical clustering was performed. Associated methylation profiles were correlated with clinical characteristics and outcome. Cumulative overall survival probabilities were calculated using the Kaplan-Meier method and log rank test to compare different groups. Data were analyzed using GraphPad Prism software. p-values <0.05 were considered statistically significant.
RESULTS
Clinicopathological Characteristics
The clinical characteristics of the subjects that had a “complete match” (i.e. were diagnosed as PXA by histopathology and classified as PXA by DNA methylation) are illustrated in Table 1. Of the 18 matched subjects, the median age at diagnosis was 16 years (range 7–32). Tumors were mainly located in the temporal (33%), parietal (28%), and frontal regions (28%). At median follow-up of 30 months, 3-year and 5-year overall survival was 73% and 71%, respectively. Overall survival ranged from 1 to 139 months. The 4 subjects that died in the study all harbored anaplastic PXAs. Eleven out of 18 subjects (61%) showed disease progression. Progression-free survival ranged from 1 to 89 months. Nine of those subjects had gross total resection and 9 had subtotal resection. All the subjects with nonanaplastic PXA were treated with surgery alone, while the anaplastic PXA subjects received either ancillary chemotherapy, radiation, or both.
TABLE 1.
Summary of Clinical Characteristics of Subjects That Had a Diagnosis of PXA Both by Histology and by DNA Methylation
| Variable | Number of Matched Subjects (n = 18) | Percentage |
|---|---|---|
| Age (median in years) | 16 (7–32) | |
| Sex | ||
| Male | 9 | 50% |
| Female | 9 | 50% |
| Histopathology | ||
| Nonanaplastic PXA | 6 | 33% |
| Anaplastic PXA | 12 | 67% |
| Extent of surgery | ||
| Gross total resection | 9 | 50% |
| Subtotal resection | 9 | 50% |
| Radiation therapy | ||
| Adjuvant radiation | 6 | 33% |
| No radiation | 12 | 67% |
| Chemotherapy | ||
| Not received | 13 | 72% |
| Received | 5 | 28% |
| Location of tumor | ||
| Temporal | 6 | 33% |
| Parietal | 5 | 28% |
| Frontal | 5 | 28% |
| Frontal-parietal | 1 | 6% |
| Temporal-parietal | 1 | 6% |
| Progression | ||
| Progression | 11 | 61% |
| No progression | 7 | 39% |
| Survival | ||
| Alive | 14 | 78% |
| Deceased | 4 | 22% |
Radiological Analysis
Out of 9 subjects with available pre-operative imaging, 5 (56%) had tumors with avid enhancement, 4 subjects (44%) had reduced diffusion, and 6 subjects (66%) had moderate or mild edema. Out of 5 reviewed subjects that had tumor progression, 4 (80%) had tumors with avid enhancement and 4 (80%) had tumors with reduced diffusion. All tumors that had subsequent progression were noted to be dural-based and had solid components. Out of 4 tumors without subsequent progression, one (25%) had avid enhancement, two (50%) were associated with mild or moderate edema (50%), and no subjects were noted to have reduced diffusion. There was a high rate of reduced diffusion in 4 out of 5 subjects (80%) with anaplastic tumors.
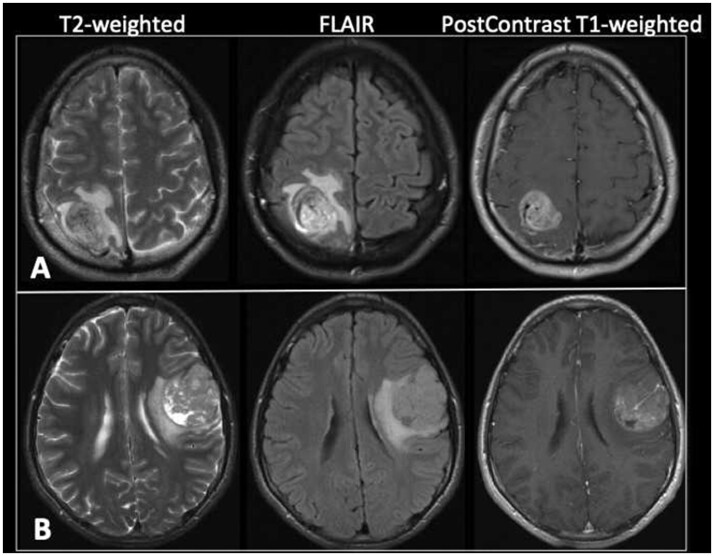
Diffusion restriction was significantly greater in patients whose tumors progressed versus those that did not progress (p = 0.048). Conversely, avid tumor enhancement and peritumoral edema was not significantly different between the 2 groups (p = 0.20 and 0.17, respectively). The reduced diffusion in PXA subjects that had tumor progression suggests increased cellularity, which is commonly observed in anaplastic PXAs (23). Representative preoperative MR images of 2 selected subjects with anaplastic PXA (subjects 3 and 12) are illustrated in Figure 6.
FIGURE 6.
Preoperative MR images of subjects with anaplastic PXA that had tumor progression. (A) Axial T2-weighted, FLAIR, and postcontrast T1-weighted images of subject 3 at the time of initial presentation demonstrate a heterogeneously enhancing right parietal lobe mass with surrounding vasogenic edema and mild local mass effect. A prominent vessel is noted coursing through the mass. (B) Axial T2-weighted, FLAIR, and postcontrast T1-weighted images of subject 12 at the time of initial presentation demonstrate a heterogeneous predominantly enhancing mass in the posterior left frontal lobe. There are a few small cystic areas within the mass. A 5-mm-thick area of increased T2 signal deep to the tumor is consistent with vasogenic edema. There is mild regional mass effect including on the left lateral ventricle. A prominent vessel is seen within the mass.
Histology and Immunohistochemistry
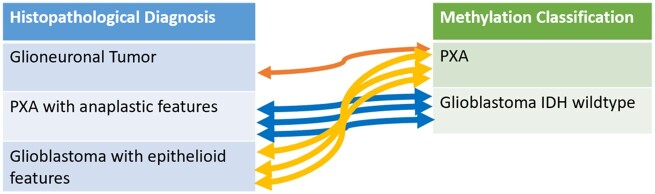
Using the WHO criteria, nonanaplastic PXAs were classified as Grade II and anaplastic PXAs were classified as Grade III. Thirteen of the 18 subjects had anaplastic PXA at initial diagnosis. Eight tumors were also shown to have p53 positivity and were noted to be anaplastic. Eleven out of the 13 subjects (83%) that had immunohistochemistry testing for BRAF V600E showed the mutation. BRAF V600E mutations were evaluated by immunohistochemistry staining in all cases and 4 subjects with the BRAF V600E mutation also confirmed positivity for the mutation with next generation sequencing. A summary of the histological and immunohistochemistry features of the subjects that matched as PXA by methylation classification and histological diagnosis is shown in Table 2. Of the 7 excluded subjects, 3 subjects were classified by methylation as PXA but diagnosed by histology as epithelioid glioblastoma and 3 subjects were classified by methylation as IDH-wildtype glioblastoma but were diagnosed by histology as anaplastic PXA (Fig. 1). The histological and immunohistochemistry features of the 7 excluded subjects are shown in Table 3.
TABLE 2.
Histological and Immunohistochemical Features of Subjects That Had a Diagnosis of PXA Both by Histology and by DNA Methylation
| Subject# | Sex | Age (Years) | Classification Score | Tumor Location | Anaplastic Features | Eosinophilic Granular Bodies | Pericellular Reticulin | Necrosis | Mitoses | Microvascular Proliferation | Ki-67 Index | Immunohistochemistry | BRAF V600E Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 17 | 0.99 | Right temporal lobe | No | Yes | N/A | No | 4/10 HPF | No | 7% | GFAP+, p53+ | Positive |
| 2 | Male | 18 | 0.99 | Left parasagittal frontal lobe | No | No | None | No | Rare | No | 5% | GFAP+ Synaptophysin+ | Positive |
| 3 | Male | 23 | 0.93 | Right parietal lobe | Yes | Yes | None | Yes | >5/10 HPF | Yes | 1–2%, focally 7.1% | GFAP+, S100+ | Positive |
| 4 | Male | 10 | 0.99 | Right temporal lobe | Yes | Yes | N/A | No | 7/10 HPF | No | “Elevated” | GFAP+, Synaptophysin+, p53+ | Negative |
| 5 | Female | 16 | 0.99 | Left frontal lobe | Yes | Yes | N/A | Yes | N/A | Yes | “Elevated” | GFAP+, Synaptophysin+ | Positive |
| 6 | Female | 10 | 0.99 | Left temporal lobe | Yes | Yes | None | No | 5/10 HPF | No | N/A | GFAP+, p53+ | Negative |
| 7 | Female | 32 | 0.99 | Right temporal parietal mass | Yes | Yes | N/A | No | 6/10 HPF | No | 10% | GFAP+, Synaptophysin– | Positive |
| 8 | Male | 25 | 0.99 | Left temporal region | No | No | N/A | No | None | No | 4% | GFAP+, Synaptophysin+, p53– | Positive |
| 9 | Male | 8 | 0.99 | Right temporal lobe | No | Yes | N/A | No | Rare | No | 10–15% | GFAP+, Synaptophysin– | Positive |
| 10 | Female | 23 | 0.99 | Right parietal lobe | Yes | Yes | N/A | Yes | <5/10 HPF | No | 15% | GFAP+, Synaptophysin+, p53+ | Positive |
| 11 | Male | 11 | 0.99 | Left frontoparietal | Yes | No | Focally+ | No | 10/10 HPF | No | 40–50% | GFAP+, Synaptophysin+, S100+, p53+ | Positive |
| 12 | Male | 15 | 0.88 | Left frontal lobe | Yes | Yes | None | Yes | 5/10 HPF | Yes | 1–3%, focally 5–10% | GFAP+, Synaptophysin+ | Positive |
| 13 | Male | 16 | 0.62 | Right temporal lobe | Yes | Yes | Diffusely+ | No | >5/10 HPF | No | 5–10% | GFAP+, S100+, p53+ | N/A |
| 14 | Female | 16 | 0.99 | Right Parietal/superficial | Yes | Yes | Diffusely+ | No | >5/10 HPF | No | 5–20% | GFAP+, p53+ | N/A |
| 15 | Female | 9 | 0.99 | Right frontal lobe | Yes | Yes | Diffusely+ | No | >5/10 HPF | No | 10–15% | GFAP+, S100+, p53+ | N/A |
| 16 | Male | 7 | 0.91 | Left frontal lobe | Yes | Yes | Focally+ | Yes | >5/10 HPF | No | up to 20% | GFAP+, Synaptophysin+, p53+ | Positive |
| 17 | Female | 10 | 0.99 | Left parietal lobe | No | Yes | Diffusely+ | No | 3/10 HPF | No | <5% | GFAP+, S100+, p53 ≤ 5% | N/A |
| 18 | Female | 7 | 0.99 | Right parietal lobe | No | Yes | Patchy+ | No | <1/10 HPF | No | <5% | GFAP+, Synaptophysin+, p53– | N/A |
FIGURE 1.
Subjects excluded due to a mismatch between histological diagnosis and DNA methylation classification. Three subjects were classified by methylation as PXA but diagnosed by histology as epithelioid glioblastoma. Three subjects were classified by methylation as IDH-wildtype glioblastoma but diagnosed by histology as anaplastic PXA.
TABLE 3.
Histological and Immunohistochemical Features of Subjects That Had a Mismatch Between Histological Diagnosis and DNA Methylation Classification
| Subject Number | Sex | Age (years) | Classifier Score | Methylation Classification | Histological Diagnosis | Tumor Location | Eosinophilic Granular Bodies | Pericellular Reticulin | Necrosis | Mitoses | Microvascular Proliferation | Ki-67 Index | Immunohistochemistry | BRAF Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | Female | 23 | 0.99 | PXA | Epithelioid glioblastoma | Right mesial temporal lesion | Yes | No | No | “Frequent” | No | 20% | GFAP+, S100+ | Positive |
| 20 | Male | 22 | 0.97 | PXA | Glioneuronal tumor, with features of ganglioglioma and PXA | Right temporal lobe | Yes | No | No | None | No | 4% | GFAP+, Synaptophysin+, p53+ | Positive |
| 21 | Male | 52 | 0.99 | Glioblastoma IDH wildtype | PXA with anaplastic features | Right parietal lobe | Yes | N/A | No | 5/10 HPF | No | 8% | GFAP+, Synaptophysin+, rare cells p53+ | Positive |
| 22 | Female | 25 | 0.99 | Glioblastoma IDH wildtype | PXA with anaplastic features | Right temporal lobe | Yes | N/A | Yes | “Frequent” | N/A | 40% | GFAP+, Synaptophysin+, S100+ | Negative |
| 23 | Male | 65 | 0.99 | Glioblastoma IDH wildtype | PXA with anaplastic features | Left temporal lobe | Yes | “focal” | Yes | 3/10 HPF | Yes | 10% | GFAP+, rare cells Synaptophysin + | N/A |
| 24 | Male | 24 | 0.54 | PXA | Epithelioid glioblastoma | Left temporo-occipital parietal lobe | N/A | N/A | Yes | “Frequent” | Yes | 70% | GFAP+, p53+ | Negative |
| 25 | Male | 16 | 0.99 | PXA | Epithelioid glioblastoma | Right parietal lobe | No | N/A | Yes | “Frequent” | Yes | 30% | GFAP+, Synaptophysin+, p53+ | Positive |
DNA Methylation and Correlation With Clinical Outcomes
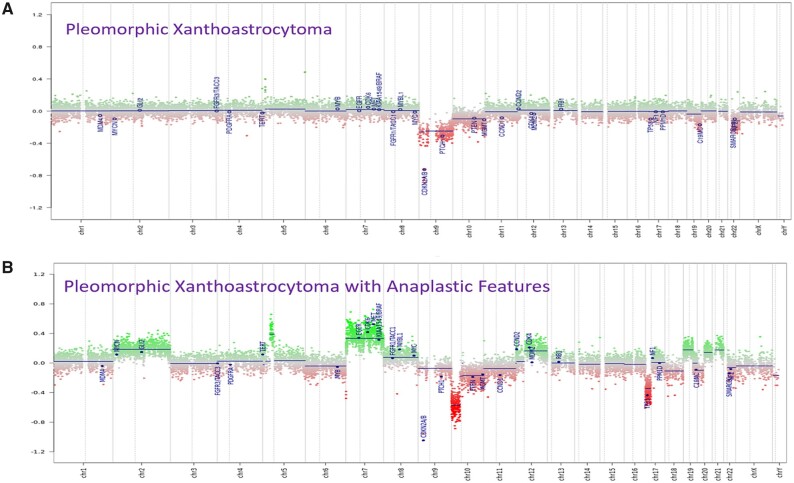
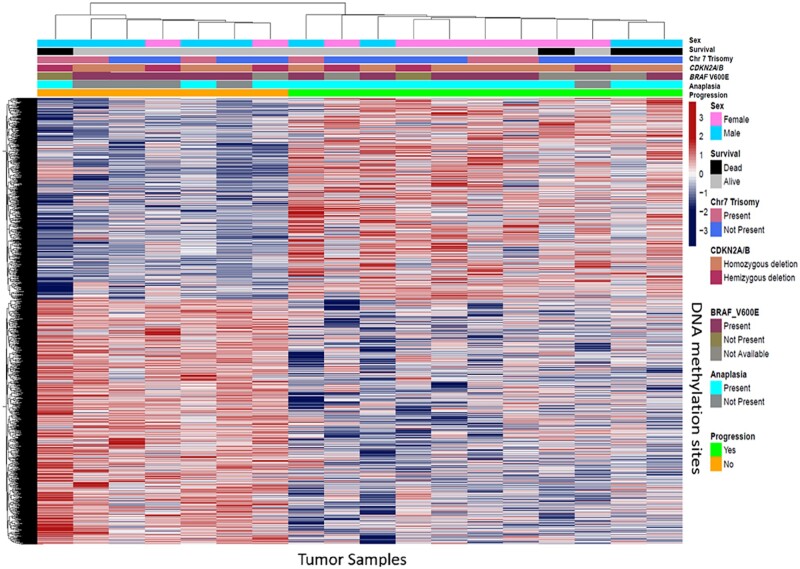
Copy number profiles generated by DNA methylation array showed that anaplastic PXAs have increased copy number variation compared with tumors without anaplasia (Fig. 2). For anaplastic PXAs, the most common chromosomal aberrations were noted to be gain of chromosome 7 and loss of chromosomes 9 and 10, which was similarly found in a recent study (24). DNA loss on chromosome 9 was previously reported to be most common in PXAs (10). CDKN2A/B homozygous deletion was noted in 12 subjects. Nine anaplastic PXA subjects also had the CDKN2A/B homozygous deletion (75%). A supervised hierarchical clustering of the DNA methylation regions shows methylation patterns noted in subjects that progressed compared with subjects that did not progress (Fig. 3).
FIGURE 2.
Genome-wide copy number profile generated by DNA methylation data showing chromosomal gains and losses of a representative (A) nonanaplastic PXA tumor sample and (B) anaplastic PXA tumor sample.
FIGURE 3.
Heat map of DNA methylated regions of PXA samples using the top 10 000 most differentially methylated probes. Hierarchical supervised clustering was based on disease presence or absence of progression, anaplasia, BRAF V600E mutation, CDKN2A/B homozygous deletion, and Trisomy 7. Methylation beta-values are illustrated according to the color scale.
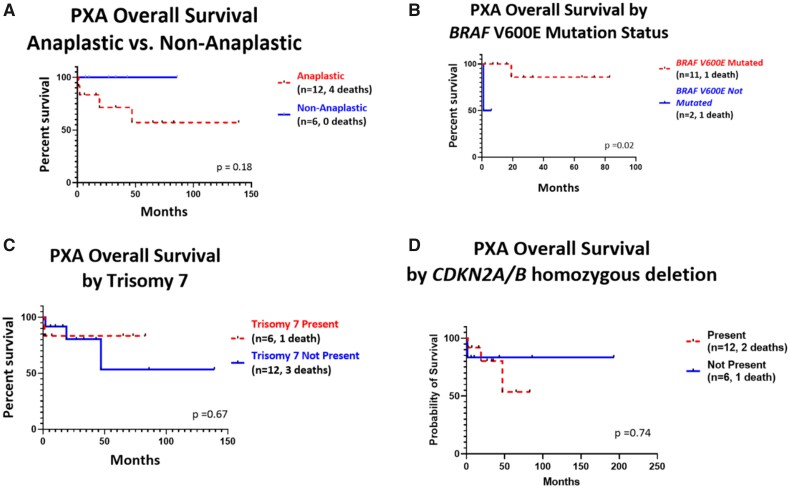
Overall survival of subjects that were diagnosed PXAs both by methylation and histology were analyzed (Fig. 4). All 4 of the subjects that died were diagnosed with anaplastic PXAs. However, in our cohort the absence of anaplasia was not significantly associated with overall survival (p = 0.18, Fig. 4A). Decreased overall survival was observed for subjects with tumors lacking the BRAF V600E mutation (p = 0.02, Fig. 4B). Trisomy 7 was also not shown to have any significant association with overall survival (p = 0.67, respectively, Fig. 4C). CDKN2A/B homozygous deletion did not show any significant association with overall survival (p = 0.74, Fig. 4D).
FIGURE 4.
Kaplan-Meier curves showing overall survival of PXAs in our cohort. (A) Overall survival curve based on presence or absence of anaplasia. In our cohort, the absence of anaplasia was not statistically significantly associated with overall survival (p = 0.18). (B) Overall survival curve based on BRAF V600E mutation. Decreased overall survival was observed for subjects with tumors lacking the BRAF V600E mutation (p = 0.02). (C) Overall survival curve based on presence or absence of Trisomy 7. Trisomy 7 was not shown to be associated with overall survival (p = 0.67). (D) Overall survival based on CDKN2A/B deletion. CDKN2A/B homozygous deletion was not shown to be associated with overall survival (p = 0.74) compared with hemizygous deletion.
MethylCIBERSORT Analysis
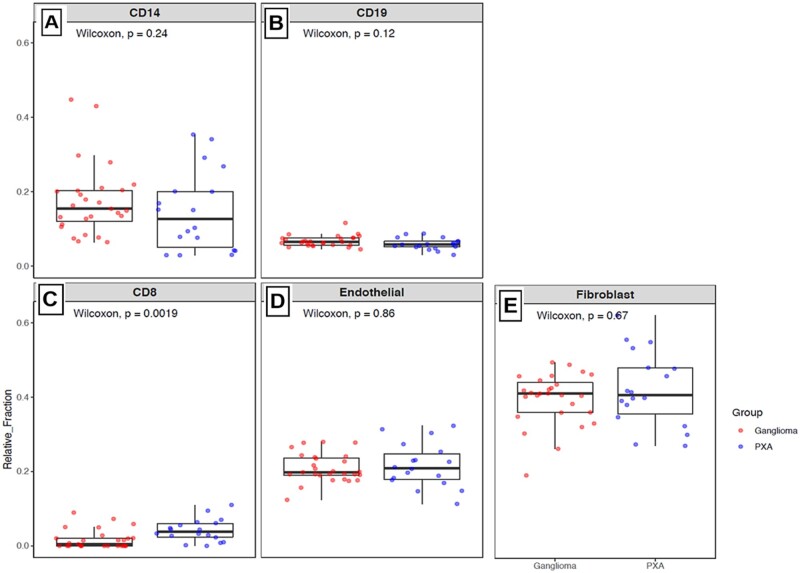
To analyze the tumor microenvironment using DNA methylation data, we used methylCIBERSORT to infer specific tumor cellular proportions in PXA tumors. We used a reference-based approach for tumor deconvolution with DNA methylation (22). We compared the PXA cohort with previously profiled ganglioglioma samples. CIBERSORT-based deconvolution of genome-wide DNA methylation data was performed to compare the tumor microenvironment in PXA versus ganglioglioma, another biphenotypic BRAF driven tumor (Fig. 5). PXAs were observed to have significantly increased CD8 T-cell epigenetic signatures compared with gangliogliomas (p = 0.0019, Fig. 5C). Compared with ganglioglioma, there was no significant difference in PXA epigenetic signatures noted in CD14 macrophage cells (p = 0.24, Fig. 5A), CD19 B-cells (p = 0.12, Fig. 5B), endothelial cells (p = 0.86, Fig. 5D), and fibroblastic cells (p = 0.67, Fig. 6E).
FIGURE 5.
Box plots showing CIBERSORT-based deconvolution of genome-wide DNA methylation data comparing PXA with ganglioglioma. Relative abundance of epigenetic signatures in (A) CD14 cells (monocyte lineage), (B) CD19 cells (B-lymphocytes), (C) CD8 cells (cytotoxic T-lymphocytes), (D) endothelial cells, and (E) fibroblastic cells is illustrated on each plot. Wilcoxon’s test was used to test differences between the immune clusters in PXA versus ganglioglioma.
DISCUSSION
Prognostic markers of PXA have not been firmly established. The frequency of CDKN2A/B deletion and BRAF V600E has been shown to be common in the genetic landscape of PXAs. In a study involving comprehensive genomic profiling of PXAs on 19 cases, CDKN2A/B homozygous deletion and BRAF V600E mutation were commonly found (24). Zou et al. also looked at next-generation sequencing of 295 genes to molecularly profile 13 cases of PXA (25). BRAF V600E was present in 5 out of 13 (38%) and CDKN2A/B homozygous deletion was found to be more frequent in PXAs than glioblastoma. The authors also concluded that BRAF and CDKN2A/B were considered good biomarkers and can aid in differentiating PXAs from glioblastomas.
PXAs and glioblastoma are highly related, with similar histological features, and it can be at times diagnostically challenging to distinguish them by histology alone. In our cohort, epithelioid glioblastoma usually featured large epithelioid cells with abundant eosinophilic cytoplasm and multiple mitotic figures with noted necrosis. Distinguishing histological features of PXA were the presence of eosinophilic granular bodies and increased reticulin. We have limited our study to tumors that were diagnosed as PXA both by pathology and DNA methylation. We excluded 7 subjects that had a mismatch between methylation classification and histological diagnosis. Three tumor samples that were classified by methylation as PXA but were diagnosed by histology as glioblastoma with epithelioid morphology. Those tumor samples were noted to have similar histological features as commonly found in anaplastic PXAs including frequent mitotic figures, presence of eosinophilic granule bodies, and an elevated Ki-67 index. Three other tumor samples were classified by methylation as IDH wildtype glioblastoma but were diagnosed by histology as anaplastic PXA. It has previously been shown that PXAs and epithelioid glioblastomas demonstrate similar molecular features, including wild-type IDH1 and MGMT promoter hypermethylation. A previous study by Alexandrescu et al. showed that there is a possibility that a subset of anaplastic PXAs are closely related to epithelioid glioblastoma, which is a rare aggressive IDH wildtype variant (26). PXAs with anaplastic transformation were shown to have indistinguishable histologic, immunohistochemical, molecular, clinical, and imaging features compared with epithelioid glioblastoma. Alexandrescu et al. used methylation profiling and showed that epithelioid glioblastomas clustered with anaplastic PXA and concluded that the 2 entities were highly related. This may explain why there was a discrepancy between those patients that were classified by methylation as glioblastomas but were diagnosed by histology as anaplastic PXAs, and may explain why those patients that were classified by methylation as PXAs but were diagnosed by histology as glioblastoma. One subject in our cohort had morphological features of both PXA and epithelioid glioblastoma and harbored the BRAF V600E mutation, which is commonly found in both types of tumors. The subject was diagnosed as anaplastic PXA by histology due to the focally the Ki-67 index of 7%. This subject was also classified as PXA by methylation. This confirms that DNA methylation can aid in more precise WHO classification of anaplastic PXAs, particularly for cases that have histological features difficult to distinguish from epithelioid glioblastoma.
In our study, we also correlated PXA copy number variation profiles derived from DNA methylation data with clinical outcome. Our supervised hierarchical clustering analysis of DNA methylated regions of PXA samples suggests that methylation patterns can potentially be related to progression independent of the presence of anaplastic features. The presence of CDKN2A/B homozygous deletion was not observed to be significantly associated with overall survival. However, our cohort was perhaps not sufficiently powered to determine this association. In a previous study looking at copy number variation profiles and overall survival of patients with PXAs, CDKN2A/B homozygous deletion was also not associated with overall survival, but their analysis was also limited due to the small number of cases, with 33 found to have the CDKN2A/B homozygous deletion and 5 cases without the deletion (9). In our study, BRAF mutant tumors also were associated with longer survival, which was also similarly reported in another retrospective study that evaluated BRAF V600E mutation in 74 patients with PXA (8). In another study examining PXAs in patients older than 16 years of age, PXAs with BRAF V600E was associated with lower Ki67, OLIG2 expression, and lack of p16 expression. It was also reported that BRAF mutated PXAs were associated with greater progression-free survival (27). Larger cohorts are necessary to establish whether DNA methylation signature can identify tumors with high risk of progression.
PXAs and gangliogliomas are typically characterized by lymphocytic infiltration. In malignant brain tumors T-lymphocytes fail to infiltrate the tumor, and finding ways to boost the immune response has been a challenge (28,29). T-cell function is compromised in the brain tumor microenvironment due to presence of common cytokines (28,29). The DNA methylation-based approach to understanding tumor microenvironment composition was previously described by Chakravarthy et al (22). We used DNA methylation data for CIBERSORT-based deconvolution of tumor tissues and analyze immune cell-types. To the best of our knowledge, the immune microenvironment in PXAs has not been previously analyzed with use of methylation-based data. To explore the tumor immune microenvironment in PXAs, we analyzed inflammatory, endothelial, and fibroblastic cell populations using methylCIBERSORT. In our study, PXAs were observed to have significantly increased CD8 T-cell epigenetic signatures compared with gangliogliomas. Both gangliogliomas and PXAs commonly harbor the BRAF V600E mutation (14). The methylation patterns illustrating increased infiltration of CD8 T-cells in PXAs suggests that tumor molecular profiles might be associated with increased immunogenicity and suggests a potential immunotherapy avenue involving cytotoxic T-cells in aggressive or recurrent tumors.
We confirm that CDKN2A/B deletion and BRAF V600E mutation are often present in PXA. In our cohort, CDKN2A/B deletion was not significantly associated with overall survival, but decreased survival appears to be observed in subjects lacking the BRAF V600E mutation. Our research suggests DNA methylation profiling could potentially aid in further prognostic characterization of PXAs. PXAs also showed significant upregulation of CD8 T-cell epigenetic signatures compared with gangliogliomas. This distinct characterization of immune cell-types in PXAs may have implications for future development of immunotherapy.
Research is in part supported by the Friedberg Charitable Foundation and the Making Headway Foundation and the NYU CTSA grant (TL1 TR001447) from the National Center for Advancing Translational Sciences, NIH.
The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1.Perkins SM, Mitra N, Fei W, et al. Patterns of care and outcomes of patients with pleomorphic xanthoastrocytoma: A SEER analysis. J Neurooncol 2012;110:99–104 [DOI] [PubMed] [Google Scholar]
- 2.Abid M, Haroon S, Memon AH, et al. Pleomorphic xanthoastrocytoma; clinicopathological spectrum of an intriguing neoplasm. Pak J Med Sci 2018;34:277–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56 [DOI] [PubMed] [Google Scholar]
- 4.Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol 2006;44:143–54 [PubMed] [Google Scholar]
- 5.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez R, Carmona FJ, Vizoso M, et al. DNA methylation alterations in grade II- and anaplastic pleomorphic xanthoastrocytoma. BMC Cancer 2014;14:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura T, Fukuoka K, Nakano Y, et al. Genome-wide DNA methylation profiling shows molecular heterogeneity of anaplastic pleomorphic xanthoastrocytoma. Cancer Sci 2019;110:828–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ida CM, Rodriguez FJ, Burger PC, et al. Pleomorphic xanthoastrocytoma: Natural history and long-term follow-up. Brain Pathol 2015;25:575–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaubel RA, Caron AA, Yamada S, et al. Recurrent copy number alterations in low-grade and anaplastic pleomorphic xanthoastrocytoma with and without BRAF V600E mutation. Brain Pathol 2018;28:172–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber RG, Hoischen A, Ehrler M, et al. Frequent loss of chromosome 9, homozygous CDKN2A/p14(ARF)/CDKN2B deletion and low TSC1 mRNA expression in pleomorphic xanthoastrocytomas. Oncogene 2007;26:1088–97 [DOI] [PubMed] [Google Scholar]
- 11.Mistry M, Zhukova N, Merico D, et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol 2015;33:1015–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korshunov A, Chavez L, Sharma T, et al. Epithelioid glioblastomas stratify into established diagnostic subsets upon integrated molecular analysis. Brain Pathol 2018;28:656–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korshunov A, Ryzhova M, Hovestadt V, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol 2015;129:669–78 [DOI] [PubMed] [Google Scholar]
- 14.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 2011;121:397–405 [DOI] [PubMed] [Google Scholar]
- 15.Serrano J, Snuderl M. Whole genome DNA methylation analysis of human glioblastoma using illumina BeadArrays. Methods Mol Biol 2018;1741:31–51 [DOI] [PubMed] [Google Scholar]
- 16.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425–37 [DOI] [PubMed] [Google Scholar]
- 17.Wiestler B, Capper D, Hovestadt V, et al. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro Oncol 2014;16:1630–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiestler B, Capper D, Sill M, et al. Integrated DNA methylation and copy-number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathol 2014;128:561–71 [DOI] [PubMed] [Google Scholar]
- 19.Orillac C, Thomas C, Dastagirzada Y, et al. Pilocytic astrocytoma and glioneuronal tumor with histone H3 K27M mutation. Acta Neuropathol Commun 2016;4:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huse JT, Snuderl M, Jones DT, et al. Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): An epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 2017;133:417–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson TE, Snuderl M, Serrano J, et al. Rapid progression to glioblastoma in a subset of IDH-mutated astrocytomas: A genome-wide analysis. J Neurooncol 2017;133:183–92 [DOI] [PubMed] [Google Scholar]
- 22.Chakravarthy A, Furness A, Joshi K, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun 2018;9:3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pradhan P, Dey B, Srinivas BH, et al. Clinico-histomorphological and immunohistochemical profile of anaplastic pleomorphic xanthoastrocytoma: Report of five cases and review of literature. Int J Hematol Oncol Stem Cell Res 2018;12:265–72 [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips JJ, Gong H, Chen K, et al. The genetic landscape of anaplastic pleomorphic xanthoastrocytoma. Brain Pathol 2019;29:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou H, Duan Y, Wei D, et al. Molecular features of pleomorphic xanthoastrocytoma. Hum Pathol 2019;86:38–48 [DOI] [PubMed] [Google Scholar]
- 26.Alexandrescu S, Korshunov A, Lai SH, et al. Epithelioid glioblastomas and anaplastic epithelioid pleomorphic xanthoastrocytomas—Same entity or first cousins? Brain Pathol 2016;26:215–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabouret E, Bequet C, Denicolai E, et al. BRAF mutation and anaplasia may be predictive factors of progression-free survival in adult pleomorphic xanthoastrocytoma. Eur J Surg Oncol 2015;41:1685–90 [DOI] [PubMed] [Google Scholar]
- 28.Wang SS, Bandopadhayay P, Jenkins MR. Towards immunotherapy for pediatric brain tumors. Trends Immunol 2019;40:748–61 [DOI] [PubMed] [Google Scholar]
- 29.Thomas AA, Ernstoff MS, Fadul CE. Immunotherapy for the treatment of glioblastoma. Cancer J 2012;18:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]