Abstract
The etiology and pathogenesis of endometriosis are complex with both genetic and environmental factors contributing to disease risk. Genome-wide association studies (GWAS) have identified multiple signals in the estrogen receptor 1 (ESR1) region associated with endometriosis and other reproductive traits and diseases. In addition, candidate gene association studies identified signals in the ESR1 region associated with endometriosis risk suggesting genetic regulation of genes in this region may be important for reproductive health. This study aimed to investigate hormonal and genetic regulation of genes in the ESR1 region in human endometrium. Changes in serum oestradiol and progesterone concentrations and expression of hormone receptors ESR1 and progesterone receptor (PGR) were assessed in endometrial samples from 135 women collected at various stages of the menstrual cycle. Correlation between hormone concentrations, receptor expression and expression of genes in the ESR1 locus was investigated. The effect of endometriosis risk variants on expression of genes in the region was analyzed to identify gene targets. Hormone concentrations and receptor expression varied significantly across the menstrual cycle. Expression of genes in the ESR1 region correlated with progesterone concentration; however, they were more strongly correlated with expression of ESR1 and PGR suggesting coregulation of genes. There was no evidence that endometriosis risk variants directly regulated expression of genes in the region. Limited sample size and cellular heterogeneity in endometrial tissue may impact the ability to detect significant genetic effects on gene expression. Effects of these variants should be validated in a larger dataset and in relevant individual cell types.
Keywords: endometrium, oestradiol, progesterone, ESR1, gene expression
Introduction
Endometriosis, defined as the growth of endometrial-like tissue (glands/stroma) outside the uterine cavity (Giudice and Kao, 2004) affects 6–10% of reproductive age women and 35–50% of women with pain, infertility or both (Giudice and Kao, 2004). Endometriotic lesions are believed to stem from cells in the endometrium that are refluxed into the peritoneal cavity during retrograde menstruation. Once in the peritoneal cavity these cells attach, establish lesions and proliferate. Inherent variation in endometrial cells is believed to predispose some women to developing the disease. What these differences are is under investigation.
Endometriosis pathogenesis has a clear genetic component. Genome-wide association studies (GWAS) have to date, identified 27 genomic regions strongly associated with endometriosis risk (Nilufer et al., 2018). All regions, however, are intronic or intergenic, not specifically linked to protein coding of individual genes and their likely mechanistic contribution to endometriosis risk is subtle. In depth analysis is required to understand the influence of genetic variants on disease risk.
Genetic variants are major regulators of gene expression (Wright et al., 2014), accounting for 15–100% of variation in gene expression through their interaction with promoters of proximal genes (Powell et al., 2012). This interaction is most likely to occur in genes within 1 megabase (Mb) of the causal variant (Fung et al., 2017). Identifying the causal variants and understanding their influence on proximal gene regulation will be vital in determining genetic influence on disease.
The most recently published meta-analysis of endometriosis GWAS (Sapkota et al., 2017) identified four independent genetic risk variants within the chromosome 6q25.1–6q25.2 region, all of which flank the estrogen receptor 1 (ESR1) gene. Endometriosis is an oestrogen-dependent disease (Bulun et al., 2000). Biological effects of oestrogen are mediated by oestrogen receptors encoded by ESR1 and estrogen receptor 2 (ESR2). While both receptors are expressed in endometrium oestogenic activity is dominated by ESR1 activation (Matsuzaki et al., 2001). In addition variants in this region have also been independently associated with other reproductive traits and diseases including age at menarche (Perry et al., 2014), age at first birth (Barban et al., 2016), breast cancer (Michailidou et al., 2017) and uterine leiomyomata (Gallagher et al., 2019) suggesting genetic regulation in this region significantly influences endometrial function and reproductive health.
The endometrium is a complex tissue that undergoes cyclical breakdown and regeneration across the menstrual cycle. Changes in endometrial composition occur in response to oestrogen and progesterone concentrations and are accompanied by significant changes in genes expression (Ponnampalam et al., 2004; Fung et al., 2018; Mortlock et al., 2020). As the endometrium is the likely source of initiating cells of endometriotic lesions understanding the hormonal and genetic risk factors on gene transcription in endometrium and any differernces in endometriosis patients will be important in understanding potential mechanisms of disease pathogenesis.
Therefore, given the importance of oestrogen activity in endometrial function and reproductive traits and the identification of multiple, independent signals of genetic association in the region of ESR1, we analyzed genes expression in the ESR1 region, as well as their relationships between hormones, their receptors and genetic variants. We identify the expression for a set of genes in this region strongly correlated with ESR1 expression suggestive of significant coregulation of these genes that may have an impact on endometrial function. We also found no direct association between the genetic variants in this region and an influence on gene expression, although we did find further support for the genetic overlap between endometriosis and uterine fibroids.
Materials and methods
Study participants and sample collection
A total of 151 women of European ancestry were recruited at the Royal Women’s Hospital, Melbourne with the study protocols reported in detail previously. Participants were reproductive-aged women who had not received hormone treatment in the 3 months prior to surgery and all gave written informed consent prior to participation. A total of 109 women were surgically diagnosed with endometriosis and the remaining 42 women had no evidence of endometriosis at surgery. Endometrial tissue samples, collected by uterine curettage at the time of surgery were divided into two portions and either stored in RNAlater (Life Technologies, Grand Island, NY, USA) at −80°C until RNA extraction, or formalin fixed for routine pathology assessment. Stage of the menstrual cycle was determined by an experienced pathologist from histological assessment of endometrial biopsy samples (Noyes et al., 1950) and assigned to one of the seven stages (1 M = menstrual, 2 EP = early-proliferative, 3 MP = mid-proliferative, 4 LP = late-proliferative, 5 ES = early-secretory, 6 MS = mid-secretory, 7 LS = late-secretory). Samples were excluded if the histologic stage of the menstrual cycle could not be determined. The study protocol was approved by the Human Research Ethics Committees of the Royal Women’s Hospital, Melbourne (#11/24 and 16/43), the QIMR Berghofer Medical Research Institute and The University of Queensland.
Oestrogen and progesterone hormone assays
Serum oestradiol concentrations were measured in duplicate by radioimmunoassay (Human Estradiol Coated Tube Radio Immuno Assay Kit, Catalog No. 07238102, MP Biomedicals LLC, Solon, OH, USA). The functional sensitivity of the assay was 10 pg/ml. The interassay and intraassay coefficients of variation were 7.2 and 6.6, respectively.
Serum concentrations of progesterone were determined in duplicate by the IMMULITE 2000 immunoassay for human progesterone (Catalogue No. L2KPW2/10381181, Siemens Healthcare Diagnostics, Los Angeles, CA, USA). The minimum detectable serum concentration was 0.1 ng/ml. The interassay coefficient of variation was 3.9 and the intraassay coefficient of variation was 6.8. These tests were conducted at the Ligand Assay and Analysis Core Laboratory at the University of Virginia Center for Research in Reproduction.
Raw progesterone concentrations (ng/ml) were compared with histological dating to confirm menstrual cycle stage (proliferative = 1.3 ng/ml, secretory = >3.03 ng/ml, etc.). Samples with progesterone concentrations outside the expected range for each menstrual stage, identified by histological dating, were removed from the analysis (n = 16). We retained 135 samples with cycle stages classified into M (n = 9), EP (n = 5), MP (n = 51), LP (n = 9), ES (n = 14), MS (n = 27) and LS (n = 20) phases. The participants were aged between 18 and 39 years (median = 31) with BMI between 15 and 45 (median = 24). Clinical data for the 135 women are listed in Supplementary Table SI. Prior to statistical analysis hormone concentrations were log transformed for normality.
Gene expression measurement and normalization
Total RNA was extracted from endometrial tissues using RNA lysis solution (RLT buffer) and AllPrep DNA/RNA mini kit (QIAGEN, Valencia, CA, USA). RNA integrity was assessed using an Agilent Bioanlayzer 2100 (Agilent Technologies, Santa Clara, CA, USA). All samples had RNA Integrity Number (RIN) > 8. RNA concentrations were measured using the NanoDropND-6000 as described previously (Fung et al., 2018). Total RNA was amplified using an Ambion Illumina TotalPrep RNA amplification kit (Ambion) and RNA was converted to biotinylated cRNA. Expression profiles were obtained by hybridizing 750 ng of cRNA to Illumina Human HT-12 v4.0 Beadchips (Illumina Inc., San Diego, USA), as described previously (Fung et al., 2017). Samples were scanned (Illumina iScan Reader) and randomized across arrays and array positions.
Gene expression normalization was performed as described by Fung et al. (2018). To achieve a stabilized distribution across average expression levels, preprocessed expression levels were transformed using a quantile adjustment across individuals, followed by scaling to log2. Further normalization was performed to allow expression levels to be compared across chips and genes.
Statistical analysis of serum hormone and endometrial gene expression
Comparison across menstrual cycle stage
Statistical analysis of the data was carried out using R software (Khan, 2013). A two-way ANOVA was performed to test for differences in mean oestradiol and progesterone serum levels between women at different menstrual cycle stages. Differences in ESR1 and PGR mRNA expression in endometrial tissue across the menstrual cycle were also assessed using a two-way ANOVA. A P-value <0.05 was considered statistically significant. Subsequent pair-wise t-tests were performed between menstrual cycle stages.
Correlation between target genes in the ESR1 region and reproductive hormones
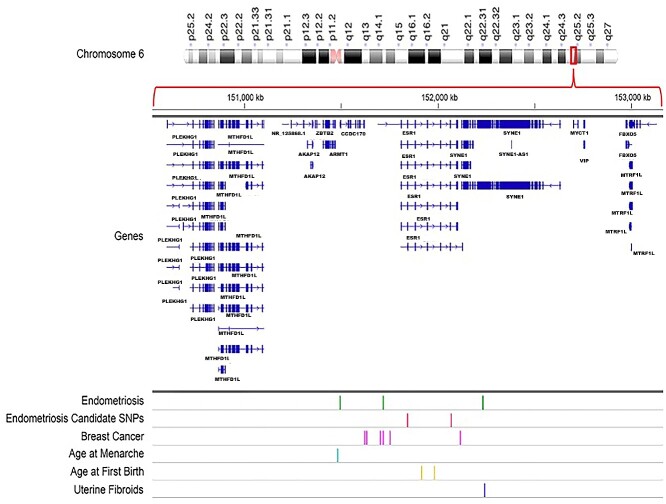
We defined target genes of the ESR1 region as genes within ±1 Mb of the independent genomic loci associated with endometriosis risk on chromosome 6 (Table I) and that showed positive expression in endometrial tissue. This range spanned from 1 Mb upstream from the most proximal signal to 1 Mb downstream from the most distal signal (range: 150816011–153554014) (Fig. 1).
Table I.
Four independent single nucleotide polymorphisms (SNPs) on chromosome 6 associated with endometriosis risk identified by GWAS (Sapkota et al., 2017).
| SNPs | Position (hg19) | GWAS P-value |
|---|---|---|
| rs1971256 | 151816011 | 3.74 × 10−08 |
| rs2206949 | 152037556 | 2.73 × 10−7 |
| rs17803970 | 152553718 | 7.04 × 10−8 |
| rs71575922 | 152554014 | 2.02 × 10−8 |
GWAS, genome-wide association studies.
Figure 1.
Genes surrounding the estrogen receptor 1 (ESR1) region on chromosome 6 and relative positions of independent signals from GWAS with reported significant associations for age at first birth (Barban et al., 2016), age at menarche (Perry et al., 2014), breast cancer (Zheng et al., 2009; Cai et al., 2011; Michailidou et al., 2017), uterine fibroids (Gallagher et al., 2019) and endometriosis (Sapkota et al., 2017). Endometriosis candidate SNPs refers to coding variants in ESR1 reported to be associated with endometriosis risk (Georgiou et al., 1999; Choi et al., 2001; Kitawaki et al., 2001; Hsieh et al., 2007; Govindan et al., 2009; Xie et al., 2009; Wang et al., 2013).
To investigate possible hormonal regulation of target genes, we performed a Spearman’s rank correlation test with both oestrogen and progesterone concentration and target gene mRNA expression. A correlation analysis was also performed between the respective hormone receptors ESR1 and PGR. Pair-wise correlations were calculated along with the correlation between each gene and oestrogen and progesterone concentrations. The resulting P-values were corrected for multiple testing using the Bonferroni method and P-value <0.05 was considered statistically significant. The significance of correlations was also considered controlling for genome-wide false discovery rate (FDR) using the Benjamini–Hochberg method, an FDR < 0.05 was considered significant.
Influence of genetic variants on target gene expression
Identification of genetic variants within the ESR1 region
In addition to the large-scale GWAS studies, other targeted candidate gene studies have assessed genetic variants in the ESR1 region and their relationship to endometriosis. To include these variants in the analysis of target gene expression, we conducted a literature search for scientific articles reporting genetic association between endometriosis risk and polymorphisms in the ESR1 gene. The search was performed in electronic databases PubMed, EMBASE and Web of Science and used the following keywords ‘endometriosis’, ‘polymorphism’, ‘ESR1’, ‘estrogen receptor alpha’, ‘ER alpha’. No restrictions were set on sample size or ethnicity. Additional relevant manuscripts were obtained from the reference lists. Studies with the following criteria were included: (i) full-text articles; (ii) studies that evaluated the relationship between the ESR1 polymorphisms and the risk of endometriosis; (iii) case–control studies, where patients were diagnosed with endometriosis, while controls were patients not affected by endometriosis. The exclusion criteria were: (i) meta-analyses, reviews, editorial articles or publications where the number of cases and/or controls in the analysis was not clearly stated; (ii) publications that did not report allele frequencies separately; (iii) studies where no statistically significant associations with endometriosis risk were reported.
Genetic variants in the ESR1 regions and endometrial eQTLs
Expression quantitative trait loci (eQTLs) are genetic variants that influence gene expression. We assessed the overlap between genetic variants associated with endometriosis in the ESR1 region and endometrial eQTL data generated by Fung et al. (2018). For this analysis, we restricted the data to probes/genes expressed in >90% of samples which included a total of 45 923 cis-eQTLs for 417 unique genes and 2968 trans-eQTLs for 82 unique genes. Genetic variants associated with endometriosis were classified as overlapping an eQTL if the risk variant, or variants in high linkage disequilibrium (LD) (r2>0.8) with the risk variant, corresponded to the eQTL SNP (eSNP) significantly associated with gene expression.
Summary data-based Mendelian randomization assessment of association
A summary data-based Mendelian randomiszation (SMR) analysis can assess if any variants in the ESR1 region have a causal or pleiotropic association with the expression of nearby genes and endometriosis risk. Using previously generated SMR analysis data available from Fung et al. (2018) we extracted information on the predefined target genes. The SMR was conducted using GWA meta-analysis summary data from Sapkota et al. (2017) consisting of 7 899 416 SNPs genotyped across >12 000 cases together with endometrial eQTL data from Fung et al. (2018).
Association with other reproductive traits
Variants in the ESR1 locus associated with other reproductive traits, including breast cancer (Dunning et al., 2016; Michailidou et al., 2017), age at menarche (Perry et al., 2014), age at first birth (Barban et al., 2016) and uterine fibroids (Gallagher et al., 2019), were also tested for correlation with endometriosis risk variants to assess if any variants were shared between traits. Correlation was assessed by LD between genetic variants, estimated using LDlink tools (DOI: 10.1093/bioinformatics/btv402) and reference haplotypes from Phase 3 of the 1000 Genomes Project European population. We analyzed pair-wise LD between these variants and the independent association signals for endometriosis risk. A threshold of r2<0.8 was applied to classify variants in strong LD with the endometriosis associated SNP.
Results
Hormone concentrations across the menstrual cycle
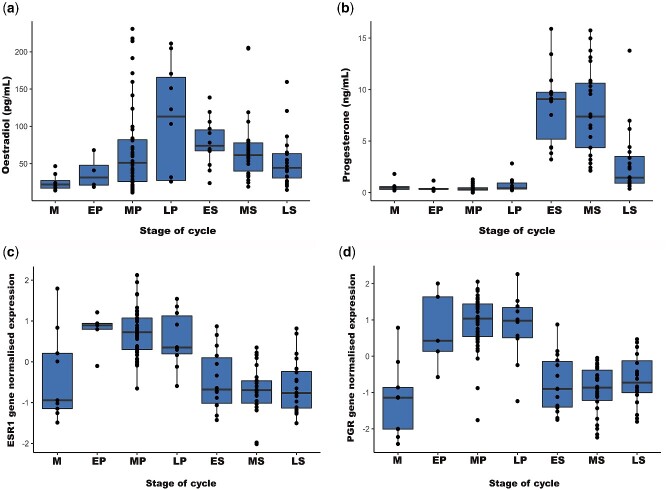
Oestradiol concentrations across the menstrual cycle ranged from 10 to 300 pg/ml (Fig. 2a and Supplementary Fig. S1a log concentrations). Oestradiol concentrations increased during the proliferative phase of the cycle followed by a decline in the secretory phase (Fig. 2a). Differences in oestradiol concentrations across the menstrual cycle were consistent in women with and without endometriosis and samples were analyzed together to increase power in subsequent analyses (Supplementary Fig. S2). There was a significant difference in the oestradiol concentration between menstrual cycle stages (P < 2.5 × 10−3). Pair-wise analysis identified significant differences between menstrual stage and an increase in oestradiol concentration in late-proliferative (P = 0.009), early-secretory (P = 0.008) and mid-secretory (P = 0.023) stages (Supplementary Table SII).
Figure 2.
Raw concentrations of (a) oestradiol and (b) progesterone across the menstrual cycle in serum samples from 135 women and gene expression changes of (c) ESR1 and (d) progesterone receptor (PGR). X-axis represents stages of menstrual cycle (M, menstrual; EP, early-proliferative; MP, mid-proliferative; LP, late-proliferative; ES, early-secretory; MS, mid-secretory; LS, late-secretory) and Y-axis represents raw concentrations of oestradiol, progesterone and normalized gene expression of ESR1 and PGR. Boxplots represents two central quartiles with the mean shown by the horizontal line. Dots represent the individual data points.
Progesterone concentrations ranged from 0.1 to 21.3 ng/ml across the menstrual cycle (Fig. 2b and Supplementary Fig. S1b, log concentrations). Progesterone concentrations were low during the proliferative phase and increased in the early-secretory phase, followed by decline in the late-secretory phase of the cycle (Fig. 2b). These differences in progesterone concentrations between stages of the menstrual cycle were highly significant (P < 2 × 10−16). Pair-wise analysis results showed significantly higher progesterone concentrations in the secretory stages compared with proliferative stages (Supplementary Table SIII).
Hormone receptor mRNA expression across the menstrual cycle
There were no significant differences in hormone receptor mRNA expression between women with and without endometriosis and differences in expression across the menstrual cycle were consistent regardless of endometriosis diagnosis. Samples were analyzed together to increase power in subsequent analyses (Supplementary Fig. S3). ESR1 mRNA expression was significantly higher (P < 2.2 × 10−16) in the proliferative stage than in the secretory stage of the menstrual cycle (Fig. 2c). Subsequent pair-wise analysis indicated significant differences between all proliferative stages with menstrual and secretory stages (Supplementary Table SIV). In the secretory stage, oestrogen receptor gene expression was inversely related with progesterone concentration across the menstrual cycle (Fig. 2c and b).
Similarly, PGR mRNA expression was higher in the proliferative compared with the secretory stage of the menstrual cycle. PGR expression increased from the proliferative to early-secretory stages of the menstrual cycle (Fig. 2d) in the opposite direction to progesterone concentrations (Fig. 2b). Variation in PGR expression across the menstrual cycle was highly significant (P < 2.2 × 10−16). Subsequent pair-wise analysis indicated significant differences between all proliferative stages and secretory stages (Supplementary Table SV).
Correlation between target genes in the ESR1 risk region
Given the multiple independent signals of genetic association in the region of ESR1 with many reproductive traits and diseases and the potential these variants lead to a coordinated influence on gene expression in the ESR1 region we analyzed the relationship between these genes. From the expression arrays, we identified 22 probes representing 15 genes within 1 Mb of genetic markers significantly associated with endometriosis risk (Fig. 1). Following Bonferroni correction for multiple testing, 16 out of the 22 probes showed significant pair-wise correlations of mRNA expression in several groups (Fig. 3). The expression of RMND1, ARMT1, CCDC170, ZBTB2 and FBXO5 were all positively correlated with ESR1 expression (r > 0.4) (Table II). Expression of MTHFD1L (r = −0.47), AKAP12 (r = −0.40) and MTRF1L (r = −0.32) were negatively correlated with ESR1 expression (Table II). There were no data for SYNE1, the gene immediately downstream of ESR1, as the probes for SYNE1 on the HT12 chips did not detect expression of this gene.
Figure 3.
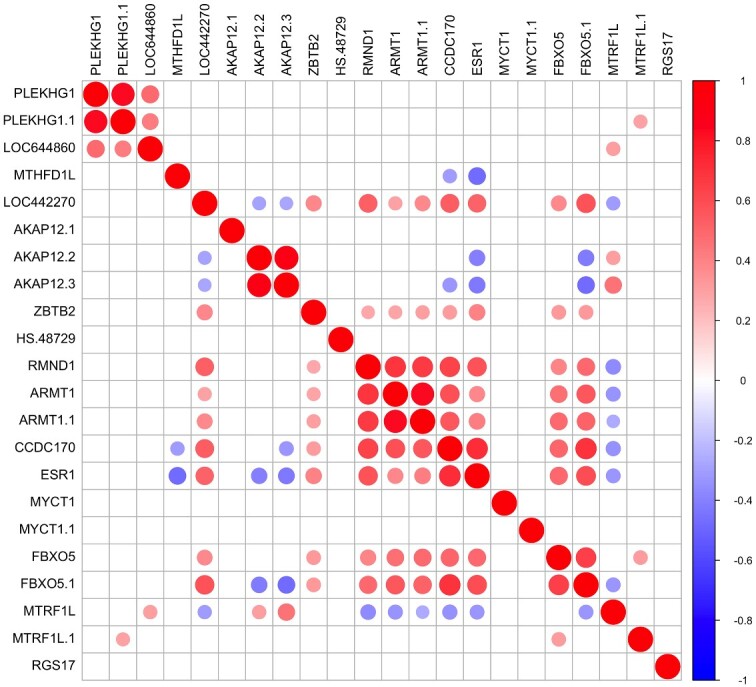
Correlation matrix plot showing the correlation coefficients for the level of expression of pairs of genes as they appear on chromosome 6q25.1–6q25.2 within the ESR1 region. Gene expression was measured in endometrial samples from women collected across the menstrual cycle. Red color denotes genes showing positive correlations and blue denotes genes showing negative correlations.
Table II.
Genes significantly correlated with ESR1 expression in endometrium and their correlation with serum oestradiol and progesterone concentrations and expression of PGR in samples from 135 women.
| Genes | Gene names | ESR1 | PGR | Oestradiol | Progesterone |
|---|---|---|---|---|---|
| MTHFD1L | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like | −0.47* | −0.45* | −0.17 | 0.16 |
| AKAP12 | A-kinase anchoring protein 12 | −0.40* | −0.49* | −0.17 | 0.24 |
| ZBTB2 | Zinc finger and BTB domain containing 2 | 0.40* | 0.23 | −0.26* | −0.31* |
| RMND1 | Required for meiotic nuclear division 1 homolog | 0.57* | 0.59* | −0.24 | −0.39* |
| ARMT1 | Acidic residue methyltransferase 1 | 0.38* | 0.51* | −0.21 | −0.33* |
| CCDC170 | Coiled-coil domain containing 170 | 0.71* | 0.80* | −0.07 | −0.52* |
| ESR1 | Estrogen receptor 1 | 1.00* | 0.83* | −0.15 | −0.59* |
| FBXO5 | F-box protein 5 | 0.59* | 0.76* | 0.07 | −0.41* |
| MTRF1L | Mitochondrial translational release factor 1 like | −0.32* | −0.36* | 0.08 | −0.17 |
Correlation is significant following Bonferroni correction.
We considered a set of eight genes which correlated significantly with ESR1 mRNA expression to further investigate their pattern of expression relative to serum progesterone and oestrogen concentrations and their receptors (Fig. 3 and Table II). Correlations with oestradiol concentrations were low and not significant while correlations with progesterone were high and significant for only RMND1, ARMT1, CCDC170, ESR1, FBXO5. Expression of these genes had a higher correlation with the changes in the endometrial expression of receptor genes (ESR1 and PGR) (Supplementary Tables SVI and SVII) than with hormone concentrations (oestradiol and progesterone) (Supplementary Tables SVIII and SIX), suggesting likely coregulation between this set of genes and ESR1.
Analysis of genetic influence on gene expression
Association between genetic variants in the ESR1 region
Endometriosis GWAS report significant associations between endometriosis risk and four independent SNPs in the ESR1 region (Table I). Two independent signals were identified in the genes CCDC170 (rs1971256, P = 3.74 × 10−8) and SYNE1 (rs71575922, P = 2.20 × 10−8). Further analysis by Sapkota et al. (2017), conditioning on these original signals, identified two further independent signals in the ESR1 region (Table I and Supplementary Fig. S4).
A literature search for candidate gene studies identified a further three SNPs with reported association with endometriosis risk (Table III; smallest P-value rs2234693, P < 0.005). Two variants, rs2234693 and rs9340799, are located in intron 1 and the variant rs3798573 is located in intron 6 of the ESR1 gene. We found no correlation between these three candidate ESR1 variants with the variants identified in the GWAS studies (LD estimates r2 < 0.1). Using the large-scale data from the GWAS meta-analysis, we found no evidence of an association with endometriosis for these three candidate SNPs (rs2234693, P = 0.38; rs9346799, P = 0.92; rs3798573, P = 0.2).
Table III.
List of studies that evaluated the relationship between ESR1 gene polymorphisms and endometriosis risk.
| Variant | Alleles | Ethnicity | Cases | Controls | P-value | References |
|---|---|---|---|---|---|---|
| rs2234693 | C > T | Asian (Indian) | 110 | 117 | <0.05 | Govindan et al. (2009) |
| Greek | 57 | 57 | <0.05 | Georgiou et al. (1999) | ||
| Asian (Japanese) | 109 | 27 | <0.0005 | Kitawaki et al. (2001) | ||
| Asian (Taiwanese) | 112 | 110 | <0.005 | Hsieh et al. (2007) | ||
| Asian (South Korean) | 77 | 67 | <0.05 | Choi et al. (2001) | ||
| rs9340799 | A > T | Asian (Taiwanese) | 112 | 110 | <0.005 | Hsieh et al. (2007) |
| Asian (Chinese) | 214 | 160 | <0.002 | Xie et al. (2009) | ||
| rs3798573 | A > G | Asian (Chinese) | 312 | 357 | <0.01 | Wang et al. (2013) |
In addition to the four independent, GWAS-identified, variants for endometriosis risk there are also additional variants associated with breast cancer (Zheng et al., 2009; Cai et al., 2011; Dunning et al., 2016), age at first birth (Barban et al., 2016), age at menarche (Perry et al., 2014) and uterine fibroids (Gallagher et al., 2019) (Fig. 1) within the ESR1 region. We found that none of the SNPs associated with age at first birth, age at menarche or breast cancer were in strong LD with the endometriosis associated SNP (r2 < 0.7). The uterine fibroid SNP (rs58415480), however, was in strong LD (r2 = 0.9) with the endometriosis SNP (rs71575922), located in the SYNE1 region.
Genetic regulation of gene expression in the ESR1 region and their relationship with endometriosis risk
We investigated previously published endometrial eQTLs (Fung et al., 2018) to determine whether they influence expression of the 15 target genes in the region. There were significant cis-eQTLs for three genes (ZBTB2, ARMT1, MTHFD1L) and trans-eQTLs for 10 genes (RMND1, CCDC170, ARMT1, FBXO5, PLEKHG1, MTHFD1L, AKAP12, LOC442270, MTRF1L, RGS17). We restricted our analysis to consider only cis-eQTLs, resulting in 38 SNPs that influence ARMT1 expression, 16 SNPs that influence MTHFD1L expression and 22 SNPs that influence ZBTB2 expression. Signals for genetic regulation of MTHFD1L and ZBTB2 expression did not overlap with the GWAS signals for genetic association with endometriosis. Analysis of LD between the endometrial cis-eQTLs and the lead SNPs from the four independent ESR1 GWAS association signals (listed in Table I) identified 20 cis-eQTLs for ARMT1 in LD (r2 > 0.8) with the rs2206949 GWAS-identified risk SNP in the ESR1 region.
Overlap between the genetic regions that regulate ARMT1 expression and the endometriosis risk SNPs in the ESR1 region was tested using the SMR analysis. The SMR test employees statistical modeling that can be used to infer causal relationships (Zhu et al., 2016). Results of the SMR analysis showed no evidence of a causal or pleiotropic association between the variants regulating ARMT1 expression and endometriosis risk, suggesting the endometrial eQTL for ARMT1 and endometriosis risk SNPs in the ESR1 region are independent. We also tested for an association between the eQTLs found to influence other genes in the ESR1 region and endometriosis risk and found no evidence for a causal relationship between these genetic variants.
We also tested whether the lead SNPs associated with age at menarche, age at first birth, breast cancer and uterine leiomyomata in the ESR1 region were in LD (r2 > 0.8) with endometrial eQTLs. None of the trait associated SNPs in this region showed evidence of regulation of gene expression in endometrium therefore no further analysis of causality with the SMR test was performed.
Discussion
In recent years, GWAS studies have made large inroads into identifying the genetic basis of many diseases. The mechanistic relationship between genetic risk factors and disease, however, is proving difficult to elucidate, as the gene or genes they influence is not clear. Understanding the regulation of genes in risk regions will help prioritize gene targets for future functional studies. GWAS studies have reported multiple independent risk variants for endometriosis, breast cancer and other reproductive traits near the ESR1 locus on chromosome 6. An influence on ESR1, or coregulated genes could contribute to disease risk. Endometrium, the likely source of cells initiating endometriotic lesions, is significantly regulated by hormone concentrations. We analyzed potential target genes around the ESR1 loci, expressed in endometrium and determined their relationship to both hormones and receptor concentrations and identified a set of genes coregulated with ESR1. We also assessed the association between relevant genetic variants in this region from other reproductive traits or candidate gene studies and found no association with endometriosis or expression of these genes.
Sex hormones have a significant role in the regulation of biological systems. Concentrations of oestrogen and progesterone (Draper et al., 2018) and expression of genes in the endometrium vary across the menstrual cycle in association with the changing histology and structure of the endometrium (Ponnampalam et al., 2004; Talbi et al., 2006; Fung et al., 2018; Mortlock et al., 2020). The protein expression patterns of steroid receptors ESR and PGR also vary across the cycle (Lessey et al., 2006; Critchley and Saunders, 2009). Results of our studies showed that ESR1 and PGR in endometrium gradually decreased from the proliferative to secretory phases. These changes in expression pattern reflect cyclic changes in oestradiol and progesterone concentrations in the circulation. Our results showed that oestradiol concentrations increased throughout the proliferative phase followed by a decline in the secretory phase whereas, the progesterone concentrations remained low during the proliferative phase and increased in the early-secretory phase, followed by decline in the late-secretory phase of the cycle.
Oestrogenic effects act via the oestrogen receptors (ER) localized in epithelial, stromal and vascular cells. Of the two isoforms, the expression of ESR1 mRNA (Parl et al., 1987; Koji and Brenner, 1993) and protein (ERα) (Press and Greene, 1984; Bergeron et al., 1988; Lessey et al., 1988) has been the most extensively studied in the uterus. ERα protein is expressed in the epithelial glands and in the stroma of both the functional and basal layers of the endometrium. In response to progesterone, the expression of ERα is downregulated in the stromal and epithelial cells of endometrium during the secretory phase (Lessey et al., 1988; Young, 2013).
Steroid receptor expression in the endometrium may influence gynecological pathologies. Uterine receptivity defects such as luteal phase defect, endometriosis and polycystic ovarian syndrome have been related to ERα downregulation in endometrial epithelial cells during the mid-secretory phase (Lessey et al., 2006). It was reported that ERβ mRNA was present in glandular epithelial cells and stromal cells throughout the menstrual cycle with predominant expression of ERβ in glandular epithelial cells (Matsuzaki et al., 1999). ERβ protein (Lecce et al., 2001) has been studied in human endometrial vascular, stromal and epithelial compartments with highest levels of ERβ expression observed during periovulatory period in epithelial cells. In our data, ESR2 was not detected on the array chip.
PGR is expressed as two functionally distinct isoforms, PR-A and PR-B acting as transcriptional regulators of progesterone responsive genes (Kastner et al., 1990). PGR expression in healthy cycling women exhibits well-described temporal and locational expression patterns (Lessey et al., 1988; Snijders et al., 1992). Studies on protein expression in human endometrium shows that the predominant form during the secretory phase is the PR-A isoform, whereas the PR-B isoform has been reported to decline in both stromal and glandular cells during the latter half of the menstrual cycle (Brosens et al., 1999; Mote et al., 2001). Reduced PGR expression in the endometrium of women with endometriosis has been proposed, however this evidence has limitations (McKinnon et al., 2018). Reduced expression has been shown in endometriotic lesions (Attia et al., 2000). In this study, we found no significant differences in ESR1 or PGR expression in eutopic endometrium of women with and without endometriosis, supporting our previously published analyses (Fung et al., 2018). It is possible subtle differences in the timing of changes in hormones, their receptors or cellular content between women with and without endometriosis may contribute to varied expression.
We assessed patterns of expression for 15 genes within 2 Mb of the ESR1 locus in endometrium. We identified a set of genes that show correlated changes in expression indicative of coregulation with ESR1. The set included genes immediately upstream of ESR1 (RMND1, ARMT1, CCDC170) and a gene (FBXO5) downstream of ESR1 whose expression was significantly positively correlated with ESR1 expression (r > 0.4). Two genes located upstream of ESR1 (MTHFD1L and AKAP12) and MTRF1, located downstream of ESR1 were significantly negatively correlated with ESR1. Patterns of expression for these genes across the menstrual cycle were more strongly correlated with receptor gene expression (ESR1 and PGR) than with hormone concentrations. Neighboring genes in close proximity in humans can be expressed concurrently (Hurst et al., 2002; Takai and Jones, 2004; Trinklein et al., 2004). There is also evidence that RMND1, CCDC170 and ARMT1 expression, immediately upstream of the ESR1 are correlated with ESR1 expression in tumor biopsies taken from postmenopausal women with stage I to IIIB ER+ breast cancer (Dunbier et al., 2011). Therefore, genes in the ESR1 region may be coregulated and not just menstrual cycle-dependent.
Genetic variants may also influence gene expression, contributing to gynecological pathologies. Four independent genetic risk variants for endometriosis around ESR1 have been identified using GWAS, although the causal variant, the variant that directly contributes to endometriosis risk, has not yet been confirmed. Candidate gene association studies based on selected genes with inferred biological function have also identified additional genetic variants in the ESR1 region that may be related to endometriosis risk. In the literature search we identified three candidate SNPs (Choi et al., 2001; Kitawaki et al., 2001; Hsieh et al., 2007; Govindan et al., 2009; Xie et al., 2009; Wang et al., 2013) that were located in intronic regions of ESR1 that have been proposed as causal variants. Investigating these SNPs in the endometriosis GWAS meta-analysis data (Sapkota et al., 2017), we found no evidence of an association with endometriosis (smallest P-value rs3798573, P = 0.2), nor was there a correlation with genome-wide significant GWAS signals in the ESR1 region. These results suggest the true signals in the region of ESR1 associated with endometriosis risk are noncoding variants located in intergenic regions which have subtle influences on gene regulation in the region.
The chromosome 6q25.1 region is a well replicated risk locus for breast cancer in Europeans and Asians (Cai et al., 2011). In our analysis, none of the breast cancer associated signals correlated with the endometriosis risk SNPs in the ESR1 region. Similarly, SNPs in the ESR1 region that were also reported to be associated with age at menarche (rs6933660) and age at first birth (rs726281, rs67229052) were not correlated with the endometriosis risk SNPs and were not associated with endometriosis. Thus, although there is a known relationship between age at menarche, age at first birth, breast cancer and endometriosis, these phenotypes do not appear to share the same candidate causal variants in the ESR1 region. Evidence of independent genetic risk factors for these related traits and diseases together with correlated expression of genes linked to ESR1 expression suggest complex regulation of genes in this region in different tissue types.
Women with endometriosis are two times more likely to have a uterine fibroid diagnosis (Gallagher et al., 2019). GWAS of uterine fibroids also identified one signal in the ESR1 region (Gallagher et al., 2019). The uterine fibroids associated signal (rs58415480) is in strong LD with one of the endometriosis signals (rs71575922). Biological overlap/coexistence between these two gynecological diseases has long been suspected due to the similarities in molecular mechanisms and progenitor cells (Uimari et al., 2011; Tanmahasamut et al., 2014; Gallagher et al., 2019). Gallagher et al. (2019) report a significant moderate genetic correlation between uterine fibroids and endometriosis (r = 0.39, P = 9.77 × 10−13) and evidence of a causal relationship. Our findings support the presence of genetic overlap between uterine leiomyomata and endometriosis. Further studies are necessary to discover the underlying biological relationships in development of these two diseases.
Annotation of risk SNPs identified by GWAS with gene expression data can expand our ability to understand the genetic basis of complex traits. We therefore annotated trait associated GWAS SNPs to endometrium specific eQTLs. We found that none of the trait associated lead SNPs, or correlated SNPs (estimates for LD r2 > 0.8) in the ESR1 region, showed evidence regulating gene expression for this set of target genes in endometrium. Variants located in regulatory elements might activate promoters affecting more than one gene. Variants associated with breast cancer have been reported to regulate ESR1 in reporter assays and may regulate other genes RMND1, CCDC170 and ARMT1 supporting coregulation of genes in this region (Dunbier et al., 2011; Dunning et al., 2016). Other approaches will be required to understand how independent genetic risk factors in the region of ESR1 increase risk for multiple diseases and traits.
A limitation in our study was that the power to detect tissue specific eQTLs was limited by the sample size. Other limitations include the use of endometrial tissue consisting of multiple cell types (including stroma and epithelium) undergoing changes in cellular composition and cell activity across the menstrual cycle. Genetic regulation of genes in the ESR1 region may be cell-type specific and the effects of risk variants in specific cell types remains to be investigated. Statistical methods developed to predict cell composition in whole blood without cell sorting require validated reference sets that are not yet available for endometrium. New techniques such as single-cell RNA-seq provide the expression profiles of individual cells and are a promising approach to provide additional insights into cell-type-specific gene expression. However, single-cell seq is expensive and can also introduce new practical, economic and computational challenges.
In summary, GWAS results show strong association between genetic risk variants and endometriosis risk on chromosome 6q25. The ESR1 gene in this region is a strong candidate, but the signals are intergenic and genetic studies show multiple independent genetic signals associated with endometriosis and other reproductive traits and diseases. ESR1 expression was correlated with the expression of seven other genes across the region with strongest correlated expression for CCDC170 providing strong evidence of coregulation of genes in this region. We found no evidence that genetic markers for endometriosis risk regulate the expression of ESR1 or other genes from this region in endometrium. These SNPs may have more subtle effects on gene expression or regulate genes in cell-specific manner. Other studies will be necessary to identify the functional effects of these variants on endometriosis risk. The roles of other genes on chromosome 6q25 that we show have correlated expression with ESR1 should also be considered.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary Material
Acknowledgments
We thank all the women who participated in the study and J. Donoghue, Ranita Charitra, Tracy Middleton and Irene Bell for support in patient recruitment and collection of samples and clinical data at the RWH, and the surgeons and anesthetists who collected tissue and blood samples.
Authors’ roles
Su.M., Sa.M. and G.W.M. designed the study with input from the other authors. Su.M., Sa.M., J.N.F., S.H., S.J.H. and J.G. coordinated data collection and quality control of data with support, input and oversight from S.H., P.A.W.R., L.C.G., B.M. and G.W.M. Data analysis was performed by Su.M. and Sa.M., which was interpreted by all authors. Su.M., Sa.M. and G.W.M. drafted the report with input from all other authors. The final manuscript has been critically revised and approved by all authors.
Funding
Research reported in this publication was supported by the National Health and Medical Research Council (NHMRC) project grants GNT1026033, GNT1049472, GNT1050208, GNT1105321 and GNT1147846. G.W.M. is supported by the NHMRC Fellowships (GNT1078399 and GNT1177194); L.C.G. and S.H. are supported by the National Institutes of Health, Eunice Kennedy Shriver National Institute for Child Health and Human Development R01 HD089511.
Conflict of interest
None declared.
References
- Attia GR, Zeitoun K, Edwards D, Johns A, Carr BR, Bulun SE. Progesterone receptor isoform A but not B is expressed in endometriosis. J Clin Endocrinol Metab 2000;85:2897–2902. [DOI] [PubMed] [Google Scholar]
- Barban N, Jansen R, de Vlaming R, Vaez A, Mandemakers JJ, Tropf FC, Shen X, Wilson JF, Chasman DI, Nolte IM, BIOS Consortium et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet 2016;48:1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron C, Ferenczy A, Shyamala G. Distribution of estrogen receptors in various cell types of normal, hyperplastic, and neoplastic human endometrial tissues. Lab Invest 1988;58:338–345. [PubMed] [Google Scholar]
- Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells1. Endocrinology 1999;140:4809–4820. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Zeitoun KM, Takayama K, Sasano H. Estrogen biosynthesis in endometriosis: molecular basis and clinical relevance. J Mol Endocrinol 2000;25:35–42. [DOI] [PubMed] [Google Scholar]
- Cai Q, Wen W, Qu S, Li G, Egan KM, Chen K, Deming SL, Shen H, Shen CY, Gammon MD et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res 2011;71:1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Lim Y, Ku S, Park S, Kim J, Moon S. Association of estrogen receptor gene polymorphism with endometriosis. Fertil Steril 2001;76:S152. [Google Scholar]
- Critchley HO, Saunders PT. Hormone receptor dynamics in a receptive human endometrium. Reprod Sci 2009;16:191–199. [DOI] [PubMed] [Google Scholar]
- Draper CF, Duisters K, Weger B, Chakrabarti A, Harms AC, Brennan L, Hankemeier T, Goulet L, Konz T, Martin FP et al. Menstrual cycle rhythmicity: metabolic patterns in healthy women. Sci Rep 2018;8:14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbier AK, Anderson H, Ghazoui Z, Lopez-Knowles E, Pancholi S, Ribas R, Drury S, Sidhu K, Leary A, Martin LA et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet 2011;7:e1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, Healey CS, Kar S, Pooley KA, Lopez-Knowles E, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet 2016;48:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JN, Girling JE, Lukowski SW, Sapkota Y, Wallace L, Holdsworth-Carson SJ, Henders AK, Healey M, Rogers PAW, Powell JE et al. The genetic regulation of transcription in human endometrial tissue. Hum Reprod 2017;32:893–904. [DOI] [PubMed] [Google Scholar]
- Fung JN, Mortlock S, Girling JE, Holdsworth-Carson SJ, Teh WT, Zhu Z, Lukowski SW, McKinnon BD, McRae A, Yang J et al. Genetic regulation of disease risk and endometrial gene expression highlights potential target genes for endometriosis and polycystic ovarian syndrome. Sci Rep 2018;8:11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CS, Mäkinen N, Harris HR, Rahmioglu N, Uimari O, Cook JP, Shigesi N, Ferreira T, Velez-Edwards DR, Edwards TL, the 23andMe Research Team et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat Commun 2019;10:4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeorgiouI, SyrrouM, BoubaI, DalkalitsisN, PaschopoulosM, NavrozoglouI, Lolis D. Association of estrogen receptor gene polymorphisms with endometriosis. Fertil Steril 1999;72:164–166. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC. Endometriosis. The Lancet 2004;364:1789–1799. [DOI] [PubMed] [Google Scholar]
- Govindan S, Shaik NA, Vedicherla B, Kodati V, Rao KP, Hasan Q. Estrogen receptor-alpha gene (T/C) Pvu II polymorphism in endometriosis and uterine fibroids. Dis Markers 2009;26:149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YY, Wang YK, Chang CC, Lin CS. Estrogen receptor α-351 XbaIG and -397 PvuIIC-related genotypes and alleles are associated with higher susceptibilities of endometriosis and leiomyoma. Mol Hum Reprod 2007;13:117–122. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Williams EJB, Pál C. Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet 2002;18:604–606. [DOI] [PubMed] [Google Scholar]
- Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J 1990;9:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AM. R-software: a newer tool in epidemiological data analysis. Indian J Community Med 2013;38:56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitawaki J, Obayashi H, Ishihara H, Koshiba H, Kusuki I, Kado N, Tsukamoto K, Hasegawa G, Nakamura N, Honjo H. Oestrogen receptor-alpha gene polymorphism is associated with endometriosis, adenomyosis and leiomyomata. Hum Reprod 2001;16:51–55. [DOI] [PubMed] [Google Scholar]
- Koji T, Brenner RM. Localization of estrogen receptor messenger ribonucleic acid in rhesus monkey uterus by nonradioactive in situ hybridization with digoxigenin-labeled oligodeoxynucleotides. Endocrinology 1993;132:382–392. [DOI] [PubMed] [Google Scholar]
- Lecce G, Meduri G, Ancelin M, Bergeron C, Perrot-Applanat M. Presence of estrogen receptor β in the human endometrium through the cycle: expression in glandular, stromal, and vascular cells. J Clin Endocrinol Metab 2001;86:1379–1386. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Killam AP, Metzger DA, Haney AF, Greene GL, McCarty KS Jr. Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J Clin Endocrinol Metab 1988;67:334–340. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Palomino WA, Apparao KB, Young SL, Lininger RA. Estrogen receptor-alpha (ER-alpha) and defects in uterine receptivity in women. Reprod Biol Endocrinol 2006;4:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki S, Fukaya T, Suzuki T, Murakami T, Sasano H, Yajima A. Oestrogen receptor α and β mRNA expression in human endometrium throughout the menstrual cycle. Mol Hum Reprod 1999;5:559–564. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Murakami T, Uehara S, Canis M, Sasano H, Okamura K. Expression of estrogen receptor alpha and beta in peritoneal and ovarian endometriosis. Fertil Steril 2001;75:1198–1205. [DOI] [PubMed] [Google Scholar]
- McKinnon B, Mueller M, Montgomery G. Progesterone resistance in endometriosis: an acquired property? Trends Endocrinol Metab 2018;29:535–548. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A, NBCS Collaborators et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017;551:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock S, Kendarsari RI, Fung JN, Gibson G, Yang F, Restuadi R, Girling JE, Holdsworth-Carson SJ, Teh WT, Lukowski SW et al. Tissue specific regulation of transcription in endometrium and association with disease. Hum Reprod 2020;35:377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mote PA, Johnston JF, Manninen T, Tuohimaa P, Clarke CL. Detection of progesterone receptor forms A and B by immunohistochemical analysis. J Clin Pathol 2001;54:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilufer R, Karina B, Paraskevi C, Rebecca D, Genevieve G, Ayush G, Stuart M, Sally M, Yadav S, Andrew SJ et al. Large-scale genome-wide association meta-analysis of endometriosis reveals 13 novel loci and genetically-associated comorbidity with other pain conditions. bioRxiv 2018;406967. [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Obst Gynecol Surv 1950;5:561–564. [DOI] [PubMed] [Google Scholar]
- Parl FF, Schonbaum CP, Cox DL, Cavener DR. Detection of estrogen receptor mRNA in human uterus. Mol Cell Endocrinol 1987;52:235–242. [DOI] [PubMed] [Google Scholar]
- Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Australian Ovarian Cancer Study et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014;514:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnampalam AP, Weston GC, Trajstman AC, Susil B, Rogers PAW. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol Hum Reprod 2004;10:879–893. [DOI] [PubMed] [Google Scholar]
- Powell JE, Henders AK, McRae AF, Caracella A, Smith S, Wright MJ, Whitfield JB, Dermitzakis ET, Martin NG, Visscher PM et al. The Brisbane Systems Genetics Study: genetical genomics meets complex trait genetics. PLoS One 2012;7:e35430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MF, Greene GL. An immunocytochemical method for demonstrating estrogen receptor in human uterus using monoclonal antibodies to human estrophilin. Lab Invest 1984;50:480–486. [PubMed] [Google Scholar]
- Sapkota Y, Steinthorsdottir V, Morris AP, Fassbender A, Rahmioglu N, De Vivo I, Buring JE, Zhang F, Edwards TL, Jones S, iPSYCH-SSI-Broad Group et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat Commun 2017;8:15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders MP, de Goeij AF, Debets-Te Baerts MJ, Rousch MJ, Koudstaal J, Bosman FT. Immunocytochemical analysis of oestrogen receptors and progesterone receptors in the human uterus throughout the menstrual cycle and after the menopause. J Reprod Fertil 1992;94:363–371. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Origins of bidirectional promoters: computational analyses of intergenic distance in the human genome. Mol Biol Evol 2004;21:463–467. [DOI] [PubMed] [Google Scholar]
- Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, Le Shay N, Nezhat CN, Kempson R, Lessey BA et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006;147:1097–1121. [DOI] [PubMed] [Google Scholar]
- Tanmahasamut P, Noothong S, Sanga-Areekul N, Silprasit K, Dangrat C. Prevalence of endometriosis in women undergoing surgery for benign gynecologic diseases. J Med Assoc Thai 2014;97:147–152. [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res 2004;14:62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uimari O, Jarvela I, Ryynanen M. Do symptomatic endometriosis and uterine fibroids appear together? J Hum Reprod Sci 2011;4:34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li Y, Maitituoheti M, Yang R, Wu Z, Wang T, Ma D, Wang S. Association of an oestrogen receptor gene polymorphism in Chinese Han women with endometriosis and endometriosis-related infertility. Reprod BioMed Online 2013;26:93–98. [DOI] [PubMed] [Google Scholar]
- Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, Madar V, Jansen R, Chung W, Zhou Y-H et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet 2014;46:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wang S, He B, Pan Y, Li Y, Zeng Q, Jiang H, Chen J. Association of estrogen receptor alpha and interleukin-10 gene polymorphisms with endometriosis in a Chinese population. Fertil Steril 2009;92:54–60. [DOI] [PubMed] [Google Scholar]
- Young SL. Oestrogen and progesterone action on endometrium: a translational approach to understanding endometrial receptivity. Reprod Biomed Online 2013;27:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, Wen W, Levy S, Deming SL, Haines JL et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet 2009;41:324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016;48:481–487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.