Abstract
To monitor innate immune responses in the CNS, the 18 kDa Translocator protein (TSPO) is a frequently used target for PET imaging. The frequent assumption that increased TSPO expression in the human CNS reflects pro‐inflammatory activation of microglia has been extrapolated from rodent studies. However, TSPO expression does not increase in activated human microglia in vitro. Studies of multiple sclerosis (MS) lesions reveal that TSPO is not restricted to pro‐inflammatory microglia/macrophages, but also present in homeostatic or reparative microglia. Here, we investigated quantitative relationships between TSPO expression and microglia/macrophage phenotypes in white matter and lesions of brains with MS pathology. In white matter from brains with no disease pathology, normal appearing white matter (NAWM), active MS lesions and chronic active lesion rims, over 95% of TSPO+ cells are microglia/macrophages. Homeostatic microglial markers in NAWM and control tissue are lost/reduced in active lesions and chronic active lesion rims, reflecting cell activation. Nevertheless, pixel analysis of TSPO+ cells (n = 12,225) revealed that TSPO expression per cell is no higher in active lesions and chronic active lesion rims (where myeloid cells are activated) relative to NAWM and control. This data suggests that whilst almost all the TSPO signal in active lesions, chronic active lesion rims, NAWM and control is associated with microglia/macrophages, their TSPO expression predominantly reflects cell density and not activation phenotype. This finding has implications for the interpretation of TSPO PET signal in MS and other CNS diseases, and further demonstrates the limitation of extrapolating TSPO biology from rodents to humans.
Keywords: CD68, HLA‐DR, IBA‐1, microglia, multiple sclerosis, P2ry12, TSPO
MAIN POINTS
Increased TSPO in the MS brain is due to an increase in microglia/macrophage cell density rather than activation of these cells.
TSPO expression per microglia is similar in control and MS brain regardless of their activation state or morphology.
1. INTRODUCTION
Multiple sclerosis (MS) is a chronic demyelinating inflammatory disease of the central nervous system (CNS). While current anti‐inflammatory strategies targeting the adaptive immune response effectively reduce the frequency of relapses, they fail to limit the neurodegeneration driving disease progression and irreversible accumulation of disability. One potential explanation for this is that innate immune responses, particularly microglia activation linked to aging of the CNS and the immune system, may be a major contributing factor in neurodegeneration and irreversible disease progression (Labzin et al., 2018).
The 18 kDa translocator protein (TSPO) is widely used to monitor innate immune responses in the CNS in neuroinflammatory diseases using PET imaging in vivo (Banati et al., 2000; Colasanti et al., 2014; Venneti et al., 2006). While such expression is commonly considered to reflect activated pathogenic microglia, the early assumption that TSPO expression increases in human microglia as they become activated was inferred from animal studies using rodents (Bae et al., 2014; Gottfried‐Blackmore et al., 2008; Karlstetter et al., 2014; Owen et al., 2017; Wang et al., 2014). However, we recently demonstrated that TSPO expression in primary human macrophages and microglia does not increase with classical pro‐inflammatory or anti‐inflammatory activation in vitro (Owen et al., 2017). Similarly, TSPO mRNA was reported to be equal among all different microglia clusters in Alzheimer's disease (Olah et al., 2020). We subsequently showed that TSPO expression in microglia co‐localizes with both classical pro‐inflammatory and anti‐inflammatory phenotypic markers in brains from people with MS (Nutma et al., 2019). These studies also revealed expression of TSPO in a substantial number of brain glial cells which were negative for HLA‐DR, a classical marker for activated microglia. In inactive MS lesions and in the center of chronic active lesions, these TSPO+ cells were mainly astrocytes. However, in other regions, the identity of these TSPO+, HLA‐DR negative cells was not determined.
Here, we substantially extended the characterization of TSPO+ cells in the MS brain post mortem using a range of microglia and macrophage cell markers (IBA‐1, CD68, HLA‐DR) to test the hypothesis that the TSPO+, HLA‐DR negative cells were primarily HLA‐DR negative microglia. We also investigated quantitatively the relationship between TSPO expression and microglia/macrophage phenotypes across the MS brain. We show that previously unidentified TSPO+ cells are microglia and macrophages and that in control tissue and, in brains of people with MS, in normal appearing white matter (NAWM), active lesions and the rim of chronic active lesions nearly all TSPO+ cells (approximately 95%) are microglia and macrophages. As expected, the phenotype of these cells differs across regions. In control brain tissue and NAWM in MS, microglia and macrophages predominantly express IBA‐1 and CD68, while in active MS lesions HLA‐DR expression appears in addition. As previously documented, P2ry12 and TMEM119 are expressed in control tissue and NAWM microglia, but expression is lost or reduced in active lesions and in the rims of chronic active lesions. Nevertheless, despite the phenotypic change in microglial cells in control and NAWM relative to active and rim of chronic active lesions, the TSPO pixel count per cell is similar across regions. We conclude that the increased TSPO expression in the MS brain can be attributed to an increase in microglia and macrophage cell density predominantly and not due to activation of these cells. This is unlike the rodent brain, where both factors can contribute substantially in inflammatory pathology.
2. MATERIALS AND METHODS
2.1. Human brain tissue
Human brain tissues were obtained from 19 MS patients diagnosed by the McDonald criteria (Polman et al., 2005) and six age‐matched controls with non‐neurological conditions (Table 1). Controls were selected from a larger cohort based on the clinical and pathological profile and were excluded if they were recorded as having neurological disorders, cancer or other inflammatory CNS diseases. The rapid autopsy regime of the Netherlands Brain Bank in Amsterdam (coordinator, Dr. I. Huitinga) was followed to obtain donor tissues. Participants or next of kin had given informed consent for brain autopsy and for use of their tissue for research purposes.
TABLE 1.
Clinical details of multiple sclerosis and control cases
| Case | Age | Sex | Diagnosis | Duration, years | PM delay (h:min) | Cause of death |
|---|---|---|---|---|---|---|
| 1 | 50 | F | SPMS | 17 | 7:35 | Euthanasia |
| 2 | 66 | F | PPMS | 13 | 9:35 | Euthanasia |
| 3 | 77 | F | PPMS | 24 | 10:00 | Euthanasia |
| 4 | 54 | F | Unknown | 27 | 9:25 | Respiratory failure |
| 5 | 63 | F | Unknown | Unknown | 10:50 | Unknown |
| 6 | 70 | M | SPMS | 38 | 9:25 | Euthanasia |
| 7 | 61 | M | SPMS | 18 | 9:15 | Euthanasia |
| 8 | 60 | F | SPMS | 10 | 10:40 | Euthanasia |
| 9 | 64 | F | SPMS | 31 | 10:10 | Urinary tract infection by MS |
| 10 | 56 | M | SPMS | 14 | 10:10 | Unknown |
| 11 | 71 | M | Unknown | Unknown | 7:15 | Urinary tract infection and pneumonia |
| 12 | 70 | M | SPMS | 38 | 9:25 | Euthanasia |
| 13 | 45 | M | PPMS | 10 | 7:45 | Pulmonary embolism or cardiac arrest |
| 14 | 63 | M | Unknown | Unknown | 10:00 | Aspiration pneumonia and sepsis |
| 15 | 63 | M | Unknown | Unknown | 11:00 | Aspiration pneumonia and sepsis |
| 16 | 67 | M | SPMS | 38 | 11:00 | Sudden death with MS, pneumonia |
| 17 | 77 | F | Unknown | Unknown | 9:45 | Aspiration pneumonia |
| 18 | 35 | F | SPMS | 10 | 10:20 | Euthanasia |
| 19 | 66 | F | Unknown | Unknown | 09:30 | Euthanasia |
| Controls | ||||||
| 1 | 49 | M | Control | N/A | 6:15 | Euthanasia/Hodgkin's lymphoma |
| 2 | 56 | M | Control | N/A | 14:00 | Heart failure |
| 3 | 81 | M | Control | N/A | 5:30 | Metastasis prostate carcinoma |
| 4 | 92 | F | Control | N/A | 7:00 | Acute death/probably pulmonary embolism |
| 5 | 62 | M | Control | N/A | 7:20 | Unknown |
| 6 | 71 | M | Control | N/A | 8:55 | Pancreatic and rectal carcinoma with hepatic metastases |
Abbreviations: F, female; M, male; MS, multiple sclerosis; N/A, not applicable; PM, post‐mortem; PP, primary progressive; SP, secondary progressive.
Tissues were fixed in 4% paraformaldehyde, processed, and embedded in paraffin. Tissues were selected and classified based on the size and type of lesion for quantitative analyses. Identification of the lesions was acquired by immunohistochemistry for myelin proteolipid protein (PLP) to detect myelin loss and HLA‐DR to detect microglia and macrophage activation (van der Valk & De Groot, 2000).
2.2. Immunofluorescence
Paraffin sections were immersed in xylene for 5 min for de‐paraffinization and rehydrated in descending concentrations of ethanol and washed in phosphate buffered saline (PBS). To reduce autofluorescence sections were incubated in 0.1% glycine for 10 min. Antigen retrieval was performed with citrate or TRIS buffer in the microwave (10 min, 180 Watt). Sections then were incubated with primary antibodies diluted in antibody diluent (Sigma, U3510) overnight, washed and afterwards incubated with Alexa Fluor®‐labeled secondary antibodies for 1 h at room temperature (Table 2). Autofluorescent background signal was reduced with Sudan black (0.1% in 70% EtOH) for 10 min after which sections were thoroughly rinsed. Nuclei were stained with 4,6‐diami‐dino‐2‐phenylindole (DAPI) and mounted with Fluormount (Invitrogen; #00‐4959‐52). For all fluorescent staining procedures, negative controls in which either the primary or secondary antibodies were omitted were used to control the presence of background signal.
TABLE 2.
Antibodies for immunohistochemistry
| Antigen | Host | Source | Antibody | Dilution |
|---|---|---|---|---|
| CD68 | Mouse | DAKO | M0814 | 1:500 |
| CD68 | Rabbit | Atlas Antibody | HPA048982 | 1:4000 |
| HLA‐DP,‐DQ,‐DR | Mouse | DAKO | M0775 | 1:400 |
| HLA‐DR (LN3) | Mouse | Biolegend | 327,011 | 1:750 |
| IBA‐1 | Rabbit | Wako | 1,919,741 | 1:10.000 |
| TSPO (PBR) | Goat | Novus Biologicals | NB100‐41398 | 1:750 |
| P2RY12 | Rabbit | ANASPEC | AS55042A | 1:200 |
| TMEM119 | Rabbit | Merck | HPA051870 | 1:250 |
2.3. Imaging and quantitative analyses
Images of each lesion type were collected using a Leica DM6000 microscope (Leica Microsystems, Heidelberg, Germany) for fluorescent images. Images were obtained randomly in the white matter of controls, and from MS in normal appearing white matter (NAWM) and white matter lesions. Pictures were analyzed using ImageJ software and stained cells were counted manually using the cell counter plugin (de Vos, University of Sheffield, UK). Cells were counted as single, double, or triple positive based, and co‐localization of markers was based on observation of overlapping fluorescent signal. To justify the accuracy of the counting, 18 pictures were manually counted by three independent observers (EN, EG, MM). The inter‐observer consistency was evaluated, resulting in a correlation coefficient of >0.90. For the Sholl analysis, microglia were manually traced using ImageJ software. Afterwards, the traced microglial masks were analyzed using the Sholl Analysis Plugin (Ferreira et al., 2014). Data was analyzed using GraphPad Prism 8.2.1 software. All data was tested for normal distribution, using the Shapiro–Wilk normality test (Ghasemi & Zahediasl, 2012). Significant differences between the lesions were tested using the one‐way analysis of variance test (ANOVA), (Kim, 2017) or the Kruskal‐Wallis test, in case of non‐normally distributed data. When positive, Dunnett's post‐hoc analysis was performed to analyze which groups differ significantly from their respective control or NAWM in MS tissue (Lee & Lee, 2018). Data was considered significant when p < .05.
3. RESULTS
3.1. Expression of microglia and macrophage markers in multiple sclerosis lesions
The LN3 antibody (Table 2) was used to denote the expression of HLA‐DR in human tissues (Peferoen et al., 2015). To investigate whether the lack of expression of HLA‐DR by some microglia or macrophages is due to the antibody specificity we used another antibody (CR3/43) targeting HLA‐DR, DP and DQ. We compared expression in the white matter of controls (Figure 1(a)) with non‐neurological diseases, and in NAWM, active (Figure 1(b)), chronic active (Figure 1(c)), and inactive (Figure 1(d)) MS lesions in post mortem tissue. Immunostaining of HLA‐DR, DP, DQ, and HLA‐DR showed a complete overlap with HLA‐DR in all MS and control cases. The number of cells positive for both HLA‐DR and HLA‐DR, DP, DQ was increased four‐fold in active lesions compared to control (p < .0001) or NAWM (p < .0001, Figure 1(e)).
FIGURE 1.
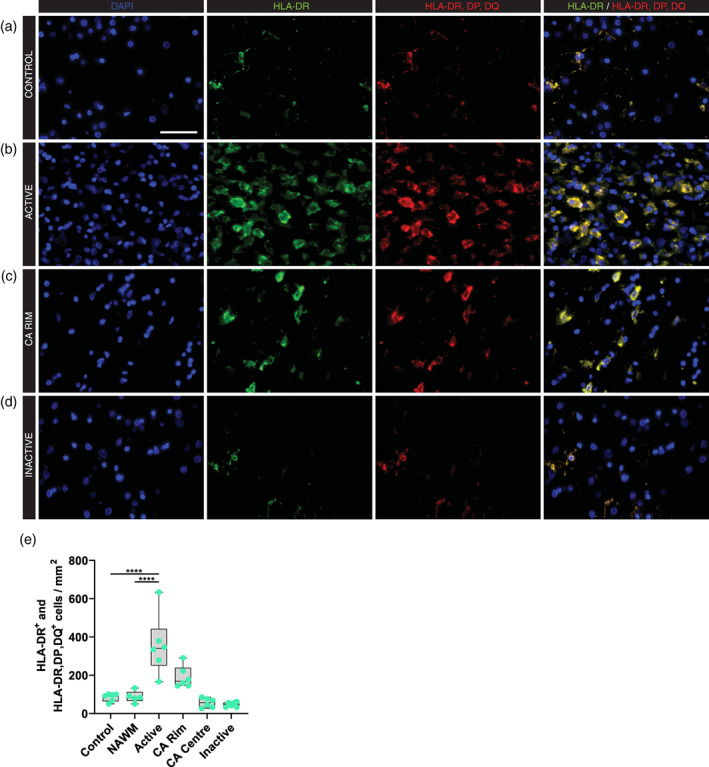
Expression of HLA‐DR/DP/DQ in white matter lesions in MS. Representative images of HLA‐DR/DP/DQ expression in control (a) and multiple sclerosis lesions (b–d); Active (b), chronic active (CA) rim (c) and inactive (d) lesions. (e) Quantitative analysis of the number of HLA‐DR and HLA‐DR/DP/DQ positive cells showed an increase in active lesions compared to control and NAWM tissues. (f) HLA‐DR positive cells and HLA‐DP/DQ/DR positive cells from single immunostainings show an overlap in the number of positive stained cells. ****p < .0001. Scale bar = 50 μm. NAWM = normal appearing white matter, CA = chronic active
To characterize the distribution of the markers IBA‐1, CD68 and HLA‐DR, a triple staining was performed in control (Figure 2(a)), and in NAWM, active (Figure 2(b)), chronic active (Figure 2(c)), and inactive (Figure 2(d)) MS lesions. In control and NAWM, most microglia and macrophages (64%, SEM 6%) were positive for IBA‐1 and CD68 but negative for HLA‐DR (Figure 2(e)). The few remaining cells were triple positive. However, in active white matter lesions and the rim of chronic active lesions, most microglia and macrophages (76%, SEM 19%) were triple positive, and the few remaining cells were double (either IBA‐1/CD68, IBA‐1/HLA‐DR, or HLA‐DR/CD68) or single positive (Figure 2(e)).
FIGURE 2.
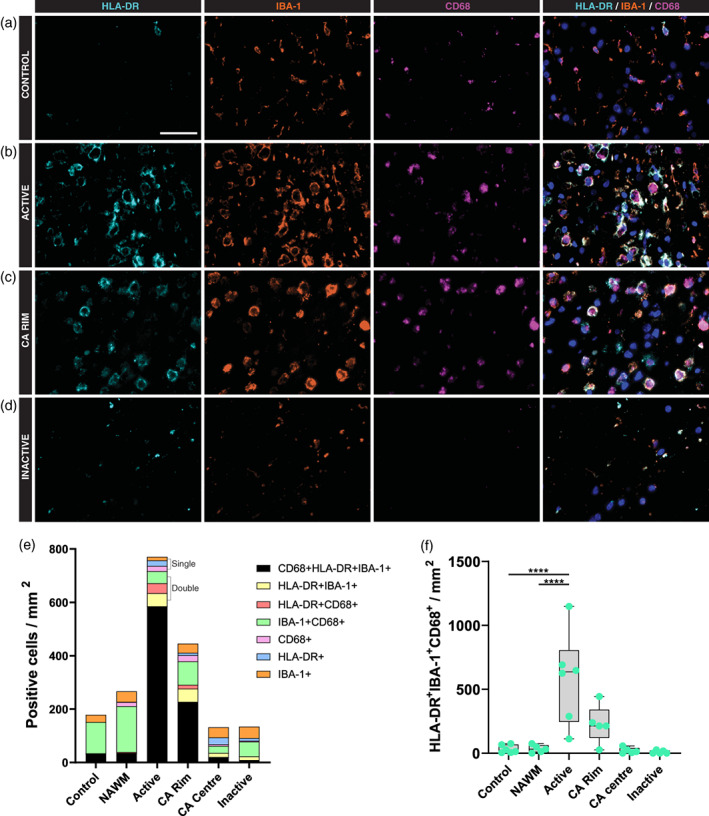
Expression of microglia/macrophage markers in white matter lesions in MS. Representative images of HLA‐DR, IBA‐1 and CD68 expression in control (a) and multiple sclerosis lesions (b–d); Active (b), chronic active (CA) rim (c) and inactive (d) lesions. (e) Quantitative analysis of the number of positive cells for the three microglia/macrophage markers showing an increase in microglia/macrophage cell density in the active lesion areas (f). ****p < .0001. Scale bar = 50 μm. CA, chronic active; NAWM, normal appearing white matter
The number of microglia and macrophages expressing all three markers was approximately 18‐fold greater in active lesions compared to control (p < .0001) or to NAWM (p < .0001) (Figure 2(f)). This large increase is a combination of (a) an increase in the percentage of microglia and macrophages which are triple positive, as described above, and (b) an increase in microglia and macrophage density in active lesions of approximately 4‐fold. In inactive lesions, the microglia and macrophage population was substantially smaller than in control and NAWM regions, and the percentage of triple positive cells was lower, too, contributing only approximately 15% of the microglia and macrophage population. In summary, most microglia and macrophages express IBA‐1 and CD68 in control and NAWM, while in active MS lesions, many microglia and macrophages also express HLA‐DR.
3.2. Identity of TSPO+ cells in MS lesions
As we reported previously (Nutma et al., 2019) TSPO + HLA− cells were observed in control, NAWM and lesional white matter (Figure 3(a)). TSPO + HLA− cells are most abundant in the centers of chronic active lesions and in inactive lesions. We have previously shown that the TSPO + HLA− cells are astrocytes in these regions (Nutma et al., 2019). In other regions, the TSPO + HLA− population is approximately 30% lower, but, and the identity of these cells has not been made clear. Morphologically these TSPO + HLA− cells resemble microglia or macrophages. To test the hypothesis that these are indeed macrophages and/or microglia, we performed triple‐immunostaining for TSPO and different pairs of three microglial and macrophage markers; TSPO/HLA‐DR/IBA (Figure 3(b)), TSPO/HLA‐DR/CD68 (Figure 3(c)), and TSPO/IBA‐1/CD68 (Figure 3(d)). All three triple stains (Figure 3(e)–(g)) showed an increase of triple positive cells in active lesion areas compared to control and NAWM. The triple stain for TSPO, IBA‐1 and CD68 (Figure 3(j)) showed that in control and NAWM and in the rims of chronic active lesions, almost all TSPO+ cells were microglia and macrophages. In control tissue, every TSPO+ cell was also double positive for IBA‐1 and CD68 (n = 36 cells/mm2) (Figure S1). In NAWM, every TSPO+ cell (n = 35 cells/mm2) expressed at least one of IBA‐1 or CD68, and most (88%) were double positive (Figure 3(j)). In the rim of chronic active lesions, 98% of TSPO+ cells were double positive for IBA‐1 and CD68. In active lesions, only 88% of TSPO+ cells expressed at least one of IBA‐1 or CD68. However, the majority of the remaining 12% of cells which were IBA‐1 and CD68 negative are likely to be microglia/macrophages, because the triple stains which included HLA‐DR, instead of CD68, showed that only 6% of TSPO+ cells were HLA‐DR and IBA‐1 negative (Figure 3(h),(i)). As expected, the percentage of TSPO+ cells which were negative for microglia/macrophage markers was high in inactive lesions (75%) and in the center of chronic active lesions (25%). These percentages are consistent with our previous work which identified these cells as astrocytes (Nutma et al., 2019).
FIGURE 3.
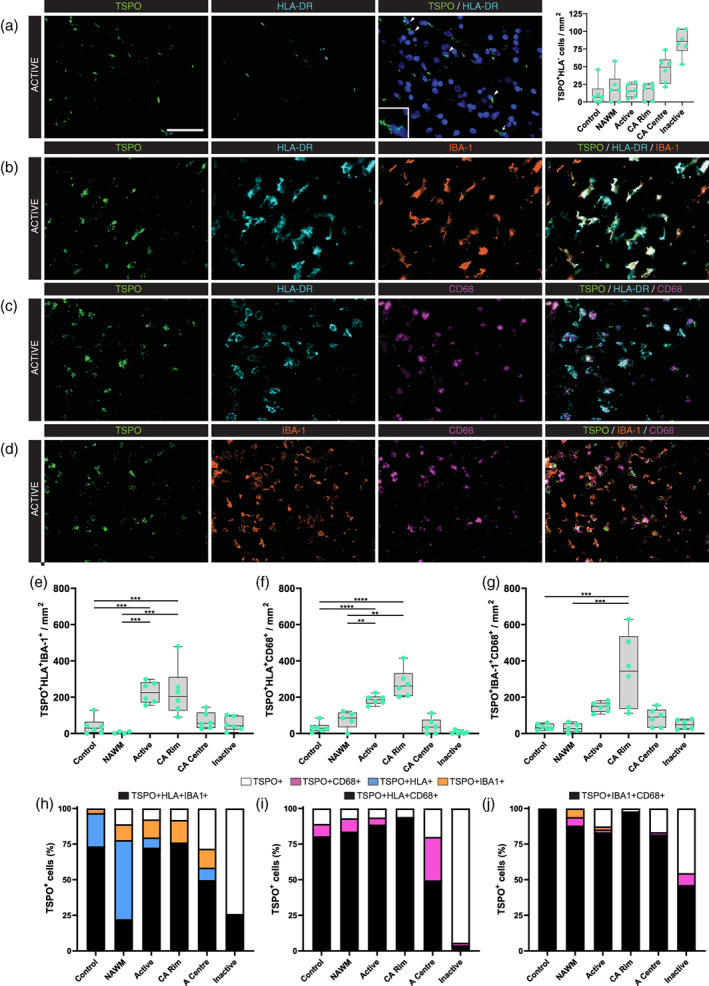
Co‐localization of TSPO with microglia/macrophage markers in white matter lesions in MS. Representative images of TSPO + HLA‐DR− cells in active lesion areas in MS (a), and representative images showing co‐localization between TSPO and HLA‐DR/IBA‐1 (b), HLA‐DR/CD68 (c), IBA‐1/CD68 (d). Quantitative analysis showed an increase in microglia markers in active lesion areas for all different staining combinations (e–g). Percentages showing that nearly all TSPO+ cells in white matter MS lesions co‐localize with at least one or two microglia/macrophage markers (h–j). **p < .01, ***p < .001, ****p < .0001. Scale bar = 50 μm. CA, chronic active; NAWM, normal appearing white matter
In summary, almost all TSPO+ cells in control, NAWM, active lesions, and the rims of chronic active lesions are microglia and/or macrophages, although, in the centers of chronic active lesions and inactive lesions, there is a substantial proportion of TSPO+ cells which are astrocytes.
3.3. Expression of homeostatic markers TMEM119 and P2ry12 by TSPO + HLA+ cells
Expression of the homeostatic markers TMEM119 and P2ry12 is reported to be mostly limited to NAWM and inactive lesions (Satoh et al., 2016; Zrzavy et al., 2017). To further characterize the relationship between TSPO expression and phenotypic markers, we performed triple staining of P2ry12 or TMEM119 in combination with HLA‐DR and TSPO to identify whether TSPO+ cells expressed markers of both activated (HLA‐DR) and homeostatic (TMEM119 and P2ry12) microglial cells. The triple stains showed that in control (Figure 4(a),(g)), NAWM, active (Figure 4(d),(j)), chronic active and inactive lesions P2ry12 + TSPO+ cells as well as TMEM119 + TSPO+ cells are found with (Figure 4(c),(f),(i),(l)) and without (Figure 4(b),(e),(h),(k)) HLA‐DR expression. As expected, all TSPO+ cells in control and NAWM, which we demonstrated above are microglia or macrophages, express both P2ry12 and TMEM119. Also as expected, TSPO+ cells in active lesions and the rim of chronic active lesions, which again we have demonstrated above are microglia and macrophages, showed evidence of altered phenotype compared to control and NAWM. In these lesion areas, P2ry12 expression is lost in almost all cells (Figure 4(m)), TMEM119 expression is lost in approximately 50% of cells, and HLA‐DR expression increases so that it appears in almost all cells (Figure 4(n)).
FIGURE 4.
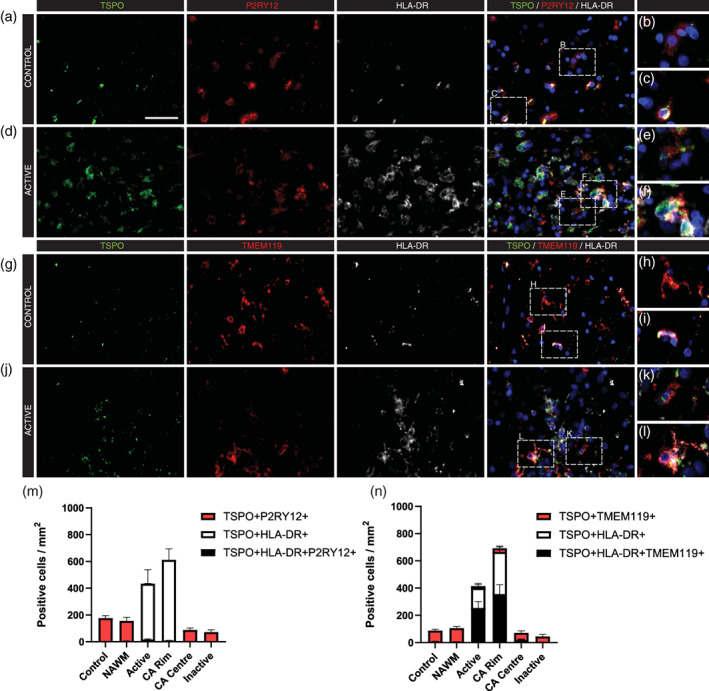
Microglia show a phenotypic shift in active lesions. Representative images of TSPO, P2ry12, and HLA‐DR (a–f) in control white matter (a–c) and an active MS white matter lesion (d–f) showing both TSPO + P2ry12 + HLA‐DR− cells (b,e) as well as TSPO + P2ry12 + HLA‐DR+ cells (c,f). Representative images of TSPO, TMEM119 and HLA‐DR (g–l) in control white matter (g–i) and an active white matter lesion (j–l) showing both TSPO + TMEM119 + HLA‐DR− cells (h,k) as well as TSPO + TMEM119 + HLA‐DR+ cells (i,l). Quantitative analysis showed a phenotypic shift and subsequent loss of P2ry12 expression in active lesion areas (m), as well as an increase in the cell density of microglia but a reduction in the percentage of TMEM119+ microglia/macrophages (n). Scale bar = 50 μm. Scale bar = 50 μm. CA, chronic active; NAWM, normal appearing white matter
3.4. TSPO pixels per cell do not change across regions with different microglial phenotypes and morphology
We have shown that almost all TSPO+ cells in control, NAWM, active lesions and rim of chronic active lesions are microglia and macrophages. We have also shown that, as expected, microglia and macrophages in the active lesions and rim of chronic active lesions express activation markers. Therefore, the TSPO expression per cell was compared across the different regions of white matter to determine whether it is increased when the microglial cells are activated compared to when the cells are homeostatic. To do this, the number of TSPO+ pixels across 12,225 TSPO+ cells were analyzed (Figure 5(a)–(c)). The percentages of pixels that were TSPO+ were similar across all of the regions of the white matter (control, 0.036% (±0.015), NAWM, 0.033% (±0.008), Active lesions; 0.041% (±0.014), CA rim; 0.038% (±0.014)) and were not statistically significant (Figure 5(a)). Furthermore, nearly all the cells in white matter lesions that express a microglia/macrophage marker are positive for TSPO (Figure 5(b)). To further investigate the amount of TSPO expression in microglia/macrophages on a per cell basis, the morphology was investigated using the Sholl analysis. Using this approach we found no correlation between the complex morphological changes that microglia/macrophages undertake, as shown by a loss in intersections of microglia/macrophages in active lesions (Figure 5(c)), and the amount of TSPO expression per cell (R 2 = 0.006278, p = .3521) (Figure 5(d)) when transitioning from an homeostatic to activated state.
FIGURE 5.
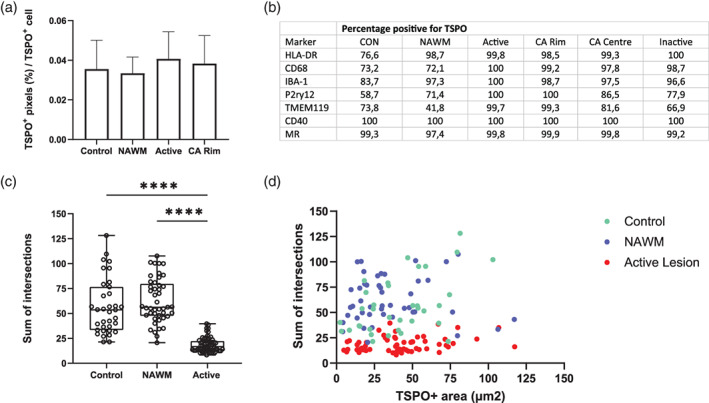
TSPO pixel count does not change across regions with different microglial morphology. The overall TSPO+ pixels per TSPO+ cell did not change in active lesion areas compared to control white matter and NAWM p > .05 (a). A table showing the percentages of microglia/macrophages that were positive for TSPO subdivided by lesion type (b). Microglia lose their complex morphology in active lesions, shown by a loss of intersections (c). No correlation was observed between the amount of TSPO+ area and the morphology of microglia in control, NAWM or active lesions (d)
In summary, although the microglia and macrophages in active and rim of chronic active lesions express activation markers, the expression of TSPO at protein level in these cells is no different to the microglia in control and NAWM regions, which express homeostatic microglia/macrophage markers.
4. DISCUSSION
TSPO is used widely as a target for PET imaging in studies of CNS inflammation. Many of these studies are interpreted assuming that TSPO is upregulated in activated glial cells such as microglia, macrophages and astrocytes with neuroinflammation (Beckers et al., 2018). This interpretation is based on studies of cellular expression of TSPO performed in rodents (Bae et al., 2014; Gottfried‐Blackmore et al., 2008; Karlstetter et al., 2014; Owen et al., 2017; Wang et al., 2014). However, recent investigations have shown that TSPO expression does not increase in human microglia and macrophages in vitro after classical pro‐inflammatory stimulation (Owen et al., 2017), and that in the TSPO expression in microglia in the MS brain post mortem co‐localizes with both classical pro‐inflammatory and anti‐inflammatory phenotypic markers (Nutma et al., 2019). Furthermore, an earlier study found that transformation of microglia to ameboid phagocytes is not necessary for maximal PK11195 binding (Banati et al., 1997).
In the current study, the expression of macrophage/microglia markers was examined in post mortem MS tissue to characterize the phenotype of cells expressing TSPO. We initially examined a large population of TSPO+ but HLA‐DR− cells, presumed to be microglia based on morphology and the lack of GFAP expression (Nutma et al., 2019). Examination of IBA‐1 and CD68 staining established that almost all cells were IBA‐1+ or CD68+, and hence this population must be microglia/macrophages.
To further characterize TSPO expression in microglia and macrophages, MS lesions were stained for combinations of microglia/macrophage markers with TSPO. Cells expressing TSPO and microglia/macrophage markers were most abundant in active lesion areas, as expected from PET data showing increased TSPO signal in lesions (Colasanti et al., 2014; Datta et al., 2017; Debruyne et al., 2003; Unterrainer et al., 2018; Vomacka et al., 2017). In control tissue, NAWM, active lesions and the rim of chronic active lesions very few TSPO+ cells were negative for the markers IBA‐1, CD68 or HLA‐DR indicating that the majority of the TSPO+ cells in these areas are likely to be microglia. In the center of chronic active lesions and in inactive lesion areas there were substantial numbers of TSPO+ cells that lacked expression of any microglial or macrophage marker. Indeed, up to 70% of TSPO+ cells do not co‐localize with IBA‐1, CD68, or HLA‐DR in inactive lesions. This is consistent with our previous findings, showing that approximately 65% of the total population of TSPO+ cells in inactive lesions are astrocytes (Dickens et al., 2014; Kaunzner et al., 2019; Nutma et al., 2019). Although TSPO is also expressed in endothelial cells an can be found at low levels in oligodendrocytes and neurons, our data suggests that in the MS brain the largest proportion of TSPO PET signal in the white matter originates from microglia, except in the proportionally smaller volumes of inactive lesions, in which most of the signal must originate from astrocytes (Banati, 2003; Cosenza‐Nashat et al., 2009; Nutma et al., 2019; Veronese et al., 2018; Wimberley et al., 2018).
Microglia adopt intricate and complicated phenotypes that cannot be well defined by single markers (Holtman et al., 2017; Vogel et al., 2014; Zrzavy et al., 2017). As expected, using a range of markers, our data revealed evidence of a phenotypic shift across lesion type. In active MS lesions microglia and macrophages predominantly express all three commonly used markers CD68, IBA‐1 and HLA‐DR suggesting that these cells have the capacity to scavenge, present antigens as well as phagocytose debris from their environment (Hendrickx et al., 2017; Ito et al., 1998; Ohsawa et al., 2004; Sasaki et al., 2001). In control and NAWM tissue, microglia and macrophages predominantly expressed IBA‐1 and CD68 but not HLA‐DR, indicating that HLA‐DR expression arises during activation of microglia in active MS lesions, corroborating previous studies (Zrzavy et al., 2017). Recently, P2ry12 and TMEM119 have been identified as markers of homeostatic microglia (Bennett et al., 2016; Butovsky et al., 2014; Satoh et al., 2016), some of which express TSPO (Nutma et al., 2019). Microglia expressing TMEM119 also express CD68 in control tissue as well as in early MS lesions (Zrzavy et al., 2017) indicating that resident microglia (expressing P2ry12 and TMEM119) express activation markers the balance of which is likely regulated by inflammatory cytokines in their environment (van Wageningen et al., 2019). As expected, we show that TMEM119 and P2ry12 expression is constitutive in regions where microglia are homeostatic (control, NAWM) and lost in areas where microglia are activated (active lesions and the rim of chronic active lesions). We also show the co‐localization of P2ry12 and HLA‐DR, suggesting that these cells are either transitioning from their homeostatic status toward a more inflammatory profile or vice versa, from active to homeostatic cells.
That almost all TSPO+ cells across a range of white matter regions were microglia, and that – as expected ‐ the phenotype in control and NAWM was homeostatic, and the phenotype in active lesions and the rim of chronic active lesions was activated, allowed for examination of the relationship between activation and TSPO expression. We analyzed the percentage of TSPO+ pixels in TSPO+ cells (n = 12,225 cells) and compared regions containing homeostatic microglia with regions containing activated microglia. There was no difference in pixel count per cell between these regions. This implies that although TSPO+ cells in active lesion areas are activated microglia/macrophages, their TSPO expression does not increase on a per cell basis relative to microglia/macrophages from homeostatic regions. Additionally, there was no correlation between the morphology and the amount of TSPO expression in microglia and macrophages. These findings are consistent with previous reports from our own group and others showing a lack of increase in TSPO expression in activated myeloid cells (Harberts et al., 2013; Owen et al., 2017), and co‐localization of TSPO with markers of both pro‐inflammatory and reparative phenotypes in the MS brain (Nutma et al., 2019). In support of our studies, a recent study showed that single cell RNA‐seq of microglia in Alzheimer's disease reported comparable TSPO mRNA levels across all microglial sub‐types suggesting TSPO PET to be a good proxy for total microglia count (Olah et al., 2020). Taken together, these findings support generalization of the hypothesis that when human myeloid cells adopt a pro‐inflammatory phenotype, TSPO expression itself does not increase, and any change in signal likely reflects an increase in myeloid cell density rather than a phenotypic shift. The data also highlights the limitations of extrapolating from rodents to humans when examining TSPO biology.
AUTHOR CONTRIBUTIONS
Erik Nutma, Emeline Gebro, and Manuel C. Marzin: Performed experiments. Erik Nutma and Emeline Gebro: Analysed the data and wrote the manuscript. Erik Nutma, Paul van der Valk, Paul M. Matthews, David R. Owen, and Sandra Amor: Conceived the study and edited the final manuscript. All authors discussed the results and contributed to the study.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Supporting information
Figure S1 Number of microglia/macrophage markers in white matter lesions in MS. Numbers of positive cells per mm2 showing that nearly all TSPO+ cells in white matter MS lesions co‐localize with at least one or two microglia/macrophage markers (a–c). Scale bar = 50 μm. NAWM = normal appearing white matter, CA = chronic active.
ACKNOWLEDGMENTS
We thank the Multiple Sclerosis Society of GB and NI for financial support.
Nutma, E., Gebro, E., Marzin, M. C., van der Valk, P., Matthews, P. M., Owen, D. R., & Amor, S. (2021). Activated microglia do not increase 18 kDa translocator protein (TSPO) expression in the multiple sclerosis brain. Glia, 69(10), 2447–2458. 10.1002/glia.24052
David R. Owen and Sandra Amor share senior authorship.
Funding information Multiple Sclerosis Society, Grant/Award Number: C008‐16.1
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bae, K. R., Shim, H. J., Balu, D., Kim, S. R., & Yu, S. W. (2014). Translocator protein 18 kDa negatively regulates inflammation in microglia. Journal of Neuroimmune Pharmacology, 9(3), 424–437. 10.1007/s11481-014-9540-6 [DOI] [PubMed] [Google Scholar]
- Banati, R. B. (2003). Neuropathological imaging: In vivo detection of glial activation as a measure of disease and adaptive change in the brain. British Medical Bulletin, 65(1), 121–131. 10.1093/bmb/65.1.121 [DOI] [PubMed] [Google Scholar]
- Banati, R. B., Myers, R., & Kreutzberg, G. W. (1997). PK (‘peripheral benzodiazepine’)—binding sites in the CNS indicate early and discrete brain lesions: Microautoradiographic detection of [3H]PK11195 binding to activated microglia. Journal of Neurocytology, 26(2), 77–82. 10.1023/a:1018567510105 [DOI] [PubMed] [Google Scholar]
- Banati, R. B., Newcombe, J., Gunn, R. N., Cagnin, A., Turkheimer, F., Heppner, F., Price, G., Wegner, F., Giovannoni, G., Miller, D. H., Perkin, G. D., Smith, T., Hewson, A. K., Bydder, G., Kreutzberg, G. W., Jones, T., Cuzner, M. L., & Myers, R. (2000). The peripheral benzodiazepine binding site in the brain in multiple sclerosis: Quantitative in vivo imaging of microglia as a measure of disease activity. Brain, 123(Pt 11), 2321–2337. 10.1093/brain/123.11.2321 [DOI] [PubMed] [Google Scholar]
- Beckers, L., Ory, D., Geric, I., Declercq, L., Koole, M., Kassiou, M., Bormans, G., & Baes, M. (2018). Increased expression of translocator protein (TSPO) Marks pro‐inflammatory microglia but does not predict neurodegeneration. Molecular Imaging and Biology, 20(1), 94–102. 10.1007/s11307-017-1099-1 [DOI] [PubMed] [Google Scholar]
- Bennett, M. L., Bennett, F. C., Liddelow, S. A., Ajami, B., Zamanian, J. L., Fernhoff, N. B., Mulinyawe, S. B., Bohlen, C. J., Adil, A., Tucker, A., Weissman, I. L., Chang, E. F., Li, G., Grant, G. A., Hayden Gephart, M. G., & Barres, B. A. (2016). New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences of the United States of America, 113(12), E1738–E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky, O., Jedrychowski, M. P., Moore, C. S., Cialic, R., Lanser, A. J., Gabriely, G., Koeglsperger, T., Dake, B., Wu, P. M., Doykan, C. E., Fanek, Z., Liu, L. P., Chen, Z., Rothstein, J. D., Ransohoff, R. M., Gygi, S. P., Antel, J. P., & Weiner, H. L. (2014). Identification of a unique TGF‐beta‐dependent molecular and functional signature in microglia. Nature Neuroscience, 17(1), 131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, A., Guo, Q., Muhlert, N., Giannetti, P., Onega, M., Newbould, R. D., Ciccarelli, O., Rison, S., Thomas, C., Nicholas, R., Muraro, P. A., Malik, O., Owen, D. R., Piccini, P., Gunn, R. N., Rabiner, E. A., & Matthews, P. M. (2014). In vivo assessment of brain white matter inflammation in multiple sclerosis with (18)F‐PBR111 PET. Journal of Nuclear Medicine, 55(7), 1112–1118. 10.2967/jnumed.113.135129 [DOI] [PubMed] [Google Scholar]
- Cosenza‐Nashat, M., Zhao, M. L., Suh, H. S., Morgan, J., Natividad, R., Morgello, S., & Lee, S. C. (2009). Expression of the translocator protein of 18 kDa by microglia, macrophages and astrocytes based on immunohistochemical localization in abnormal human brain. Neuropathology and Applied Neurobiology, 35(3), 306–328. 10.1111/j.1365-2990.2008.01006.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, G., Colasanti, A., Kalk, N., Owen, D., Scott, G., Rabiner, E. A., Gunn, R. N., Lingford‐Hughes, A., Malik, O., Ciccarelli, O., Nicholas, R., Nei, L., Battaglini, M., Stefano, N. D., & Matthews, P. M. (2017). (11)C‐PBR28 and (18)F‐PBR111 detect white matter inflammatory heterogeneity in multiple sclerosis. Journal of Nuclear Medicine, 58(9), 1477–1482. 10.2967/jnumed.116.187161 [DOI] [PubMed] [Google Scholar]
- Debruyne, J. C., Versijpt, J., van Laere, K., de Vos, F., Keppens, J., Strijckmans, K., Achten, E., Slegers, G., Dierckx, R. A., Korf, J., & de Reuck, J. L. (2003). PET visualization of microglia in multiple sclerosis patients using [11C]PK11195. European Journal of Neurology, 10(3), 257–264. 10.1046/j.1468-1331.2003.00571.x [DOI] [PubMed] [Google Scholar]
- Dickens, A. M., Vainio, S., Marjamäki, P., Johansson, J., Lehtiniemi, P., Rokka, J., Rinne, J., Solin, O., Haaparanta‐Solin, M., Jones, P. A., Trigg, W., Anthony, D. C., & Airas, L. (2014). Detection of microglial activation in an acute model of neuroinflammation using PET and radiotracers 11C‐(R)‐PK11195 and 18F‐GE‐180. Journal of Nuclear Medicine, 55(3), 466–472. 10.2967/jnumed.113.125625 [DOI] [PubMed] [Google Scholar]
- Ferreira, T. A., Blackman, A. V., Oyrer, J., Jayabal, S., Chung, A. J., Watt, A. J., Sjöström, P. J., & van Meyel, D. J. (2014). Neuronal morphometry directly from bitmap images. Nature Methods, 11(10), 982–984. 10.1038/nmeth.3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi, A., & Zahediasl, S. (2012). Normality tests for statistical analysis: A guide for non‐statisticians. International Journal of Endocrinology and Metabolism, 10(2), 486–489. 10.5812/ijem.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried‐Blackmore, A., Sierra, A., Jellinck, P. H., McEwen, B. S., & Bulloch, K. (2008). Brain microglia express steroid‐converting enzymes in the mouse. The Journal of Steroid Biochemistry and Molecular Biology, 109(1–2), 96–107. 10.1016/j.jsbmb.2007.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberts, E., Datta, D., Chen, S., Wohler, J. E., Oh, U., & Jacobson, S. (2013). Translocator protein 18 kDa (TSPO) expression in multiple sclerosis patients. Journal of Neuroimmune Pharmacology, 8(1), 51–57. 10.1007/s11481-012-9397-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx, D. A. E., van Eden, C. G., Schuurman, K. G., Hamann, J., & Huitinga, I. (2017). Staining of HLA‐DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. Journal of Neuroimmunology, 309, 12–22. 10.1016/j.jneuroim.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Holtman, I. R., Skola, D., & Glass, C. K. (2017). Transcriptional control of microglia phenotypes in health and disease. The Journal of Clinical Investigation, 127(9), 3220–3229. 10.1172/JCI90604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, D., Imai, Y., Ohsawa, K., Nakajima, K., Fukuuchi, Y., & Kohsaka, S. (1998). Microglia‐specific localisation of a novel calcium binding protein, Iba1. Brain Research. Molecular Brain Research, 57(1), 1–9. 10.1016/s0169-328x(98)00040-0 [DOI] [PubMed] [Google Scholar]
- Karlstetter, M., Nothdurfter, C., Aslanidis, A., Moeller, K., Horn, F., Scholz, R., Neumann, H., Weber, B. H. F., Rupprecht, R., & Langmann, T. (2014). Translocator protein (18 kDa) (TSPO) is expressed in reactive retinal microglia and modulates microglial inflammation and phagocytosis. Journal of Neuroinflammation, 11(1), 1–13. 10.1186/1742-2094-11-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunzner, U. W., Kang, Y., Zhang, S., Morris, E., Yao, Y., Pandya, S., Hurtado Rua, S. M., Park, C., Gillen, K. M., Nguyen, T. D., Wang, Y., Pitt, D., & Gauthier, S. A. (2019). Quantitative susceptibility mapping identifies inflammation in a subset of chronic multiple sclerosis lesions. Brain, 142(1), 133–145. 10.1093/brain/awy296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. K. (2017). Understanding one‐way ANOVA using conceptual figures. Korean Journal of Anesthesiology, 70(1), 22–26. 10.4097/kjae.2017.70.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin, L. I., Heneka, M. T., & Latz, E. (2018). Innate immunity and neurodegeneration. Annual Review of Medicine, 69, 437–449. 10.1146/annurev-med-050715-104343 [DOI] [PubMed] [Google Scholar]
- Lee, S., & Lee, D. K. (2018). What is the proper way to apply the multiple comparison test? Korean Journal of Anesthesiology, 71(5), 353–360. 10.4097/kja.d.18.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutma, E., Stephenson, J. A., Gorter, R. P., de Bruin, J., Boucherie, D. M., Donat, C. K., Breur, M., van der Valk, P., Matthews, P. M., Owen, D. R., & Amor, S. (2019). A quantitative neuropathological assessment of translocator protein expression in multiple sclerosis. Brain, 142(11), 3440–3455. 10.1093/brain/awz287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa, K., Imai, Y., Sasaki, Y., & Kohsaka, S. (2004). Microglia/macrophage‐specific protein Iba1 binds to fimbrin and enhances its actin‐bundling activity. Journal of Neurochemistry, 88(4), 844–856. 10.1046/j.1471-4159.2003.02213.x [DOI] [PubMed] [Google Scholar]
- Olah, M., Menon, V., Habib, N., Taga, M. F., Ma, Y., Yung, C. J., Cimpean, M., Khairallah, A., Coronas‐Samano, G., Sankowski, R., Grün, D., Kroshilina, A. A., Dionne, D., Sarkis, R. A., Cosgrove, G. R., Helgager, J., Golden, J. A., Pennell, P. B., Prinz, M., … de Jager, P. L. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer's disease. Nature Communications, 11(1), 6129. 10.1038/s41467-020-19737-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, D. R., Narayan, N., Wells, L., Healy, L., Smyth, E., Rabiner, E. A., Galloway, D., Williams, J. B., Lehr, J., Mandhair, H., Peferoen, L. A., Taylor, P. C., Amor, S., Antel, J. P., Matthews, P. M., & Moore, C. S. (2017). Pro‐inflammatory activation of primary microglia and macrophages increases 18 kDa translocator protein expression in rodents but not humans. Journal of Cerebral Blood Flow & Metabolism, 37(8), 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peferoen, L. A., Vogel, D. Y., Ummenthum, K., Breur, M., Heijnen, P. D., Gerritsen, W. H., Peferoen‐Baert, R. M., van der Valk, P., Dijkstra, C. D., & Amor, S. (2015). Activation status of human microglia is dependent on lesion formation stage and remyelination in multiple sclerosis. Journal of Neuropathology and Experimental Neurology, 74(1), 48–63. 10.1097/NEN.0000000000000149 [DOI] [PubMed] [Google Scholar]
- Polman, C. H., Reingold, S. C., Edan, G., Filippi, M., Hartung, H. P., Kappos, L., Lublin, F. D., Metz, L. M., McFarland, H. F., O'Connor, P. W., Sandberg‐Wollheim, M., Thompson, A. J., Weinshenker, B. G., & Wolinsky, J. S. (2005). Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Annals of Neurology, 58(6), 840–846. 10.1002/ana.20703 [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Ohsawa, K., Kanazawa, H., Kohsaka, S., & Imai, Y. (2001). Iba1 Is an Actin‐cross‐linking protein in macrophages/microglia. Biochemical and Biophysical Research Communications, 286(2), 292–297. 10.1006/bbrc.2001.5388 [DOI] [PubMed] [Google Scholar]
- Satoh, J., Kino, Y., Asahina, N., Takitani, M., Miyoshi, J., Ishida, T., & Saito, Y. (2016). TMEM119 marks a subset of microglia in the human brain. Neuropathology, 36(1), 39–49. 10.1111/neup.12235 [DOI] [PubMed] [Google Scholar]
- Unterrainer, M., Mahler, C., Vomacka, L., Lindner, S., Havla, J., Brendel, M., Böning, G., Ertl‐Wagner, B., Kümpfel, T., Milenkovic, V. M., Rupprecht, R., Kerschensteiner, M., Bartenstein, P., & Albert, N. L. (2018). TSPO PET with [(18)F]GE‐180 sensitively detects focal neuroinflammation in patients with relapsing‐remitting multiple sclerosis. European Journal of Nuclear Medicine and Molecular Imaging, 45(8), 1423–1431. 10.1007/s00259-018-3974-7 [DOI] [PubMed] [Google Scholar]
- van der Valk, P., & De Groot, C. J. (2000). Staging of multiple sclerosis (MS) lesions: Pathology of the time frame of MS. Neuropathology and Applied Neurobiology, 26(1), 2–10. 10.1046/j.1365-2990.2000.00217.x [DOI] [PubMed] [Google Scholar]
- van Wageningen, T. A., Vlaar, E., Kooij, G., Jongenelen, C. A. M., Geurts, J. J. G., & van Dam, A. M. (2019). Regulation of microglial TMEM119 and P2RY12 immunoreactivity in multiple sclerosis white and grey matter lesions is dependent on their inflammatory environment. Acta Neuropathologica Communications, 7(1), 206. 10.1186/s40478-019-0850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti, S., Lopresti, B. J., & Wiley, C. A. (2006). The peripheral benzodiazepine receptor (translocator protein 18kDa) in microglia: From pathology to imaging. Progress in Neurobiology, 80(6), 308–322. 10.1016/j.pneurobio.2006.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese, M., Reis Marques, T., Bloomfield, P. S., Rizzo, G., Singh, N., Jones, D., Agushi, E., Mosses, D., Bertoldo, A., Howes, O., Roncaroli, F., & Turkheimer, F. E. (2018). Kinetic modelling of [(11)C]PBR28 for 18 kDa translocator protein PET data: A validation study of vascular modelling in the brain using XBD173 and tissue analysis. Journal of Cerebral Blood Flow and Metabolism, 38(7), 1227–1242. 10.1177/0271678X17712388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, D. Y., Glim, J. E., Stavenuiter, A. W., Breur, M., Heijnen, P., Amor, S., Dijkstra, C. D., & Beelen, R. H. (2014). Human macrophage polarization in vitro: Maturation and activation methods compared. Immunobiology, 219(9), 695–703. 10.1016/j.imbio.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Vomacka, L., Albert, N. L., Lindner, S., Unterrainer, M., Mahler, C., Brendel, M., Ermoschkin, L., Gosewisch, A., Brunegraf, A., Buckley, C., Kümpfel, T., Rupprecht, R., Ziegler, S., Kerschensteiner, M., Bartenstein, P., & Böning, G. (2017). TSPO imaging using the novel PET ligand [(18)F]GE‐180: Quantification approaches in patients with multiple sclerosis. EJNMMI Research, 7(1), 89. 10.1186/s13550-017-0340-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Wang, X., Zhao, L., Ma, W., Rodriguez, I. R., Fariss, R. N., & Wong, W. T. (2014). Macroglia‐microglia interactions via TSPO signaling regulates microglial activation in the mouse retina. The Journal of Neuroscience, 34(10), 3793–3806. 10.1523/JNEUROSCI.3153-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberley, C., Lavisse, S., Brulon, V., Peyronneau, M. A., Leroy, C., Bodini, B., Remy, P., Stankoff, B., Buvat, I., & Bottlaender, M. (2018). Impact of endothelial 18‐kDa translocator protein on the quantification of (18)F‐DPA‐714. Journal of Nuclear Medicine, 59(2), 307–314. 10.2967/jnumed.117.195396 [DOI] [PubMed] [Google Scholar]
- Zrzavy, T., Hametner, S., Wimmer, I., Butovsky, O., Weiner, H. L., & Lassmann, H. (2017). Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain, 140(7), 1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Number of microglia/macrophage markers in white matter lesions in MS. Numbers of positive cells per mm2 showing that nearly all TSPO+ cells in white matter MS lesions co‐localize with at least one or two microglia/macrophage markers (a–c). Scale bar = 50 μm. NAWM = normal appearing white matter, CA = chronic active.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


