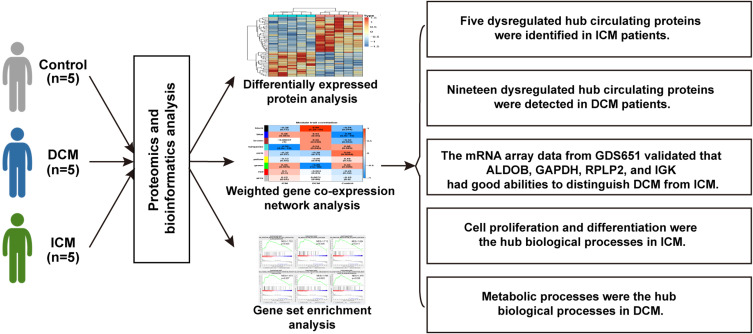
Abstract
Background
Ischemic cardiomyopathy (ICM) and nonischemic dilated cardiomyopathy (DCM) are the two most common causes of heart failure. However, our understanding of the specific proteins and biological processes distinguishing DCM from ICM remains insufficient.
Materials and Methods
The proteomics analyses were performed on serum samples from ICM (n=5), DCM (n=5), and control (n=5) groups. Proteomics and bioinformatics analyses, including weighted gene co-expression network analysis (WGCNA) and gene set enrichment analysis (GSEA), were performed to identify the hub circulating proteins and the hub biological processes in ICM and DCM.
Results
The analysis of differentially expressed proteins and WGCNA identified the hub circulating proteins in ICM (GAPDH, CLSTN1, VH3, CP, and ST13) and DCM (one downregulated protein, FGG; 18 upregulated proteins, including HEL-S-276, IGK, ALDOB, HIST1H2BJ, HEL-S-125m, RPLP2, EL52, NCAM1, P4HB, HEL-S-99n, HIST1H4L, HIST2H3PS2, F8, ERP70, SORD, PSMA3, PSMB6, and PSMA6). The mRNA expression of the heart specimens from GDS651 validated that ALDOB, GAPDH, RPLP2, and IGK had good abilities to distinguish DCM from ICM. In addition, GSEA results showed that cell proliferation and differentiation were the hub biological processes related to ICM, while metabolic processes and cell signaling transduction were the hub biological processes associated with DCM.
Conclusion
The present study identified five dysregulated hub circulating proteins among ICM patients and 19 dysregulated hub circulating proteins among DCM patients. Cell proliferation and differentiation were significantly enriched in ICM. Metabolic processes were strongly enhanced in DCM and may be used to distinguish DCM from ICM.
Keywords: proteomics, ischemic cardiomyopathy, dilated cardiomyopathy, weighted gene co-expression network analysis, gene set enrichment analysis
Graphical Abstract
Introduction
Heart failure (HF) is the final stage of diverse cardiovascular diseases, and it results in significant morbidity and mortality.1 HF is a complex multifactorial syndrome that results from myocardial injury and the consequent cardiac remodeling.2 The remodeling capability of the heart is important for its adaptation to the changing physiological circumstances. However, when the stimuli are strong enough to exceed the physiological adaptability of the heart, cardiac remodeling could induce pathological changes, such as hypertrophy, systolic and diastolic dysfunction, eventually resulting in HF.3
Ischemic cardiomyopathy (ICM) and nonischemic dilated cardiomyopathy (DCM) are the two most common outcomes of pathological cardiac remodeling, and also the two most common causes of HF. ICM mainly develops after a heart attack or coronary artery disease, while nonischemic DCM is associated with inherited genetic changes and viral infection. Recently, a large number of circulating proteins and biological processes have been reported to play important roles in the diagnosis and prognosis of HF.4–7 Some of them, such as ST2 and galectin-3, have been recognized by cardiovascular doctors worldwide, and have been included in clinical guidelines.8–10 However, our understanding of the specific proteins and biological processes that distinguish DCM from ICM remains insufficient.
In the present study, the proteomics analysis was performed with the serum samples from ICM patients, DCM patients, and healthy controls. Weighted gene co-expression network analysis (WGCNA) and differentially expressed protein analysis were performed to identify the hub circulating proteins that might distinguish DCM from ICM. In addition, gene set enrichment analysis (GSEA) was applied to detect the biological processes in DCM with the potential to distinguish DCM from ICM.
Materials and Methods
Specimen Collection
The blood samples were gathered from five typical ICM patients, five DCM patients, and five healthy controls. All the enrolled patients were diagnosed by two experienced physicians. All the procedures in the present study were approved by the Ethics Committee of the Liaocheng People’s Hospital (Liaocheng, China). The subjects provided written informed consent before enrollment. The ICM patients were those with previously documented myocardial infarction proved by an imaging study demonstrating coronary artery disease with the corresponding areas of akinesis, dyskinesis, or severe hypokinesis on maximal appropriate medical therapy with a confirmed decreased ejection fraction.11 Patients with primary valvular heart disease, known malignant tumor, acute infection, or acute coronary syndrome were excluded from the present study.11 The inclusion criteria for patients with DCM were as follows: i) Left ventricular end‑diastolic diameter (LVED) >50 mm (women) or >55 mm (men); and ii) left ventricular ejection fraction (LVEF) <45% and/or left ventricular fraction shortening <25%.12 Patients with clear causes of idiopathic DCM, including hypertension, coronary artery disease, valvular disease, congenital defect, alcoholic cardiomyopathy, tachycardia‑induced cardiomyopathy, and peripartum cardiomyopathy, were excluded from the present study.12 The clinical characteristics of the patients are presented in Table 1.
Table 1.
Clinical Characteristics of ICM Patients, DCM Patients and Healthy Controls
| Group | Controls | ICM | DCM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Gender | F | M | F | M | M | M | M | F | F | F | M | M | M | F | F |
| Age (years) | 34 | 42 | 31 | 40 | 45 | 69 | 57 | 63 | 63 | 69 | 56 | 43 | 38 | 64 | 65 |
| SBP (mmHg) | 118 | 120 | 122 | 118 | 124 | 90 | 124 | 110 | 106 | 128 | 112 | 104 | 108 | 110 | 112 |
| DBP (mmHg) | 74 | 76 | 72 | 70 | 74 | 64 | 62 | 60 | 70 | 76 | 70 | 64 | 70 | 60 | 72 |
| Heart rate (rpm) | 68 | 66 | 70 | 72 | 70 | 66 | 70 | 68 | 72 | 76 | 76 | 98 | 98 | 94 | 92 |
| LVED (mm) | 45 | 44 | 42 | 44 | 47 | 70 | 60 | 76 | 45 | 71 | 65 | 68 | 79 | 76 | 62 |
| LVEF (%) | 65 | 68 | 61 | 65 | 70 | 29 | 46 | 18 | 34 | 32 | 36 | 25 | 21 | 26 | 32 |
| NT-proBNP (pg/mL) | 90 | 82 | 84 | 70 | 96 | 7400 | 852 | 5670 | 2100 | 1350 | 1100 | 2240 | 1550 | 1670 | 3610 |
| NYHA class | / | / | / | / | / | III | II | III | III | III | III | III | III | III | III |
Abbreviations: ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; M, male; F, female; SBP, systolic blood pressure; DBP, diastolic blood pressure; LVED, left ventricular end-diastolic diameter; LVEF, left ventricular ejection fraction; NT-proBNP, N-terminal prohormone of brain natriuretic peptide; NYHA, New York Heart Association.
Proteomics
The proteomics analysis was performed by Novogene Co., Ltd (Beijing, China). The serum samples were labeled with a TMT kit (Thermo Fisher Scientific Inc, MA, USA) and fractionated by high-pH reverse-phase high-performance liquid chromatography with Thermo Q Exactive™ HF-X system. A total of 446 proteins were identified in at least 10 of the 15 serum samples of the ICM, DCM, and control groups (n=5 for each group). The data were standardized using the total sum intensity normalization.13,14
Weighted Gene Co-Expression Network Analysis (WGCNA)
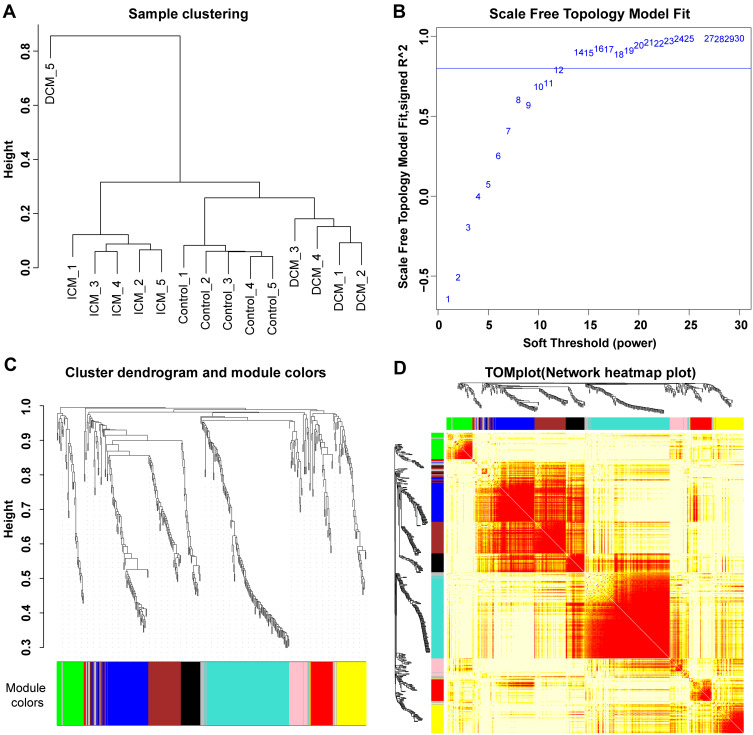
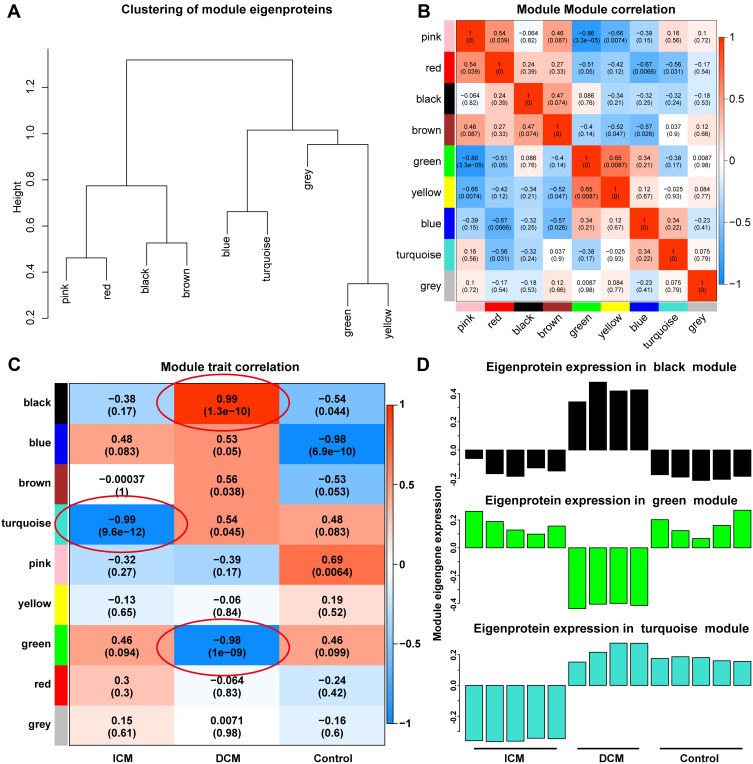
WGCNA is a handy way to find co-expressed gene modules and explore the relationship between different gene networks and clinical phenotypes, as well as core genes in the network. WGCNA was performed for the 446 proteins using the “WGCNA” package in R software (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/). The protein expression data from the DCM_5 sample were discarded in the subsequent analysis due to the heterogeneity of the data from other samples. Scale-free topology fitting index R2 and the soft threshold are of vital importance to construct the scale-free network and obtain the best-fit topology model. In the current study, the scale-free network was constructed with the scale-free R2=0.8,15,16 where the soft threshold was set at 12 to obtain the best-fit topology model. Finally, nine modules were detected, namely, black, blue, brown, green, grey, pink, red, turquoise, and yellow modules. The module–trait correlation plot was constructed to illustrate the correlation between the proteins and ICM or DCM.
Identification of Differentially Expressed Proteins in ICM and DCM Groups
Apart from the undetected proteins in the ICM group (73 undetected proteins) or the DCM group (16 undetected proteins), the data frame including the expression information of all of the other proteins was used to perform the differentially expressed protein analysis, compared with the expression data of the control group. The statistical significance was set at the mean cutoff value of 1.2-fold change and p<0.05. The hierarchical clustering plot was implemented to present the consistency of expression levels among different samples. The volcano plot was used to show the dysregulated proteins. The Venn diagram was applied to filter hub proteins associated with ICM and DCM. In addition, the RNA array data of the heart specimens from GDS651 (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS651, including left ventricles of ICM, DCM, and non-failing hearts (NF)) were used to verify the cardiac mRNA expression of hub proteins and draw the receiver operating characteristic (ROC) curve using Stata/SE 15.
Gene Set Enrichment Analysis (GSEA)
GSEA is a computational method to assess whether a set of genes associated with biological functions or disease phenotypes presents a statistical significance between biological samples. GSEA uses statistical approaches to identify significantly enriched or depleted classes or functions. Hub biological processes are analyzed with GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp).17,18 The gene sets of biological processes were obtained from the Gene Ontology (GO) gene sets annotated by the same GO terms. The significance criterion was set at nominal p<0.05.
Results
Sample Clustering and Modules Detected in WGCNA
A total of 446 proteins were detected in at least 10 of the 15 serum samples of ICM, DCM, and control groups (n=5 for each group). WGCNA was performed to detect different protein expression modules. The protein expression data from the DCM_5 sample were discarded in the subsequent analysis due to the heterogeneity of the data from other samples (Figure 1A). The scale-free network was constructed with the scale-free R2=0.8, where the soft threshold was set at 12 to obtain the best-fit topology model (Figure 1B). According to the hierarchical clustering dendrogram of proteins, nine modules were detected, namely, black, blue, brown, green, grey, pink, red, turquoise, and yellow modules (Figure 1C). The topological overlap map (TOM) plot of different modules is also presented in Figure 1D.
Figure 1.
Sample clustering and modules detected in weighted gene co-expression network analysis. (A) The sample clustering plot. The protein expression data from the DCM_5 sample were discarded in the subsequent analysis due to the heterogeneity of the data from other samples. (B) The scale-free network was constructed with the scale-free R2=0.8, where the soft threshold was set at twelve to get the best-fit topology model. (C) According to the hierarchical clustering dendrogram of proteins, nine modules were detected, namely black, blue, brown, green, grey, pink, red, turquoise, and yellow modules. (D) The topological overlap map (TOM) plot of distinct modules detected. The heatmap showed the topological overlap matrix (TOM) among genes in the analysis. Different colors on the x-axis and y-axis represented different modules. The yellow brightness of the middle part represented the strength of connections between modules.
Modules Highly Associated with ICM and DCM
Different modules were well distinguished based on the clustering plot of module eigengenes (Figure 2A) and the module–module correlation plot (Figure 2B). The module–trait correlation plot (Figure 2C) showed that the turquoise module correlated well with ICM (Pearson correlation coefficient, PCC=−0.99, p=9.6×10−12). Additionally, the black module (PCC=0.99, p=1.3×10−10) and the green module (PCC=−0.98, p=1.0×10−9) were strongly associated with DCM. As presented in Figure 2D, proteins in the black module were expressed at a high level, and proteins in the green module were expressed at a low level in the DCM group. In addition, proteins in the turquoise module were significantly downregulated in the ICM group. Furthermore, hub proteins of the turquoise, black, and green modules are presented in the Supplementary Figures 1 and 2.
Figure 2.
Modules highly associated with ischemic and DCM. Different modules are well distinguished according to the clustering plot of module eigengenes (A) and the module-module correlation plot (B). The module-trait correlation plot (C) showed that the turquoise module was well correlated with ICM. The black module and the green module were strongly associated with DCM. The proteins in the black module were expressed at a high level and the proteins in the green module were at a low level in the DCM group. In addition, the proteins in the turquoise module were significantly down-regulated in the ICM group (D).
Hub Proteins Enriched in the ICM Patients
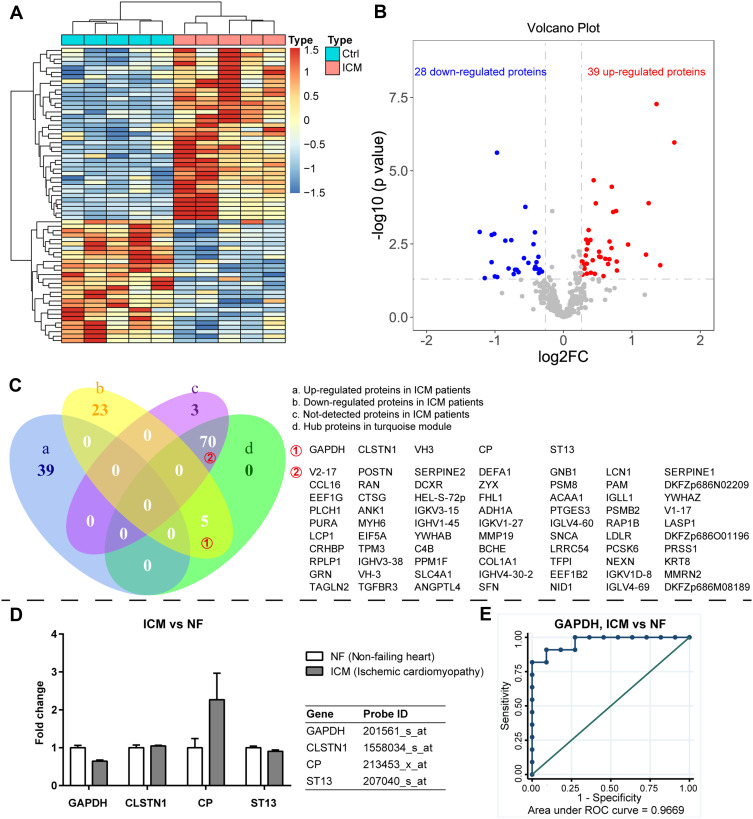
Among the 446 proteins, 73 proteins were undetected, 39 proteins were upregulated, and 28 proteins were downregulated in the ICM group. The hierarchical clustering plot (Figure 3A) and a volcano plot (Figure 3B) were constructed to present the dysregulated proteins in the ICM group. The Venn plot (Figure 3C) showed that five downregulated proteins (GAPDH, CLSTN1, VH3, CP, and ST13) overlapped with the hub proteins in the turquoise module, which was negatively associated with ICM in the module–trait correlation analysis of WGCNA. However, other 70 hub proteins in the turquoise module were detected only in the DCM group, but not in the ICM group.
Figure 3.
Hub proteins enriched in the ICM patients. The hierarchical clustering plot (A) and a volcano plot (B) were constructed to present the dysregulated proteins in the ICM group. The Venn plot (C) showed that five down-regulated proteins were overlapped with the hub proteins in the turquoise module, which was negatively associated with ICM in the module-trait correlation analysis of WGCNA. In addition, the other 70 hub proteins in the turquoise module could be detected only in the DCM group, but not in the ICM group. In addition, the validation results showed that mRNAs of GAPDH, CLSTN1, CP, and ST13 were detected in the heart specimens from GDS651, among which the protein expression change of GAPDH in the serum is consistent with its mRNA expression change in the heart specimens of ICM patients (D). The ROC curve illustrated that GAPDH could distinguish ICM from NF well (E).
In addition, the RNA array data of heart specimens of ICM and NF from GDS651 were used to verify the expression of the mRNA corresponding to hub proteins in the heart. The results showed that mRNAs of GAPDH, CLSTN1, CP, and ST13 were detected in the heart specimens, among which the protein expression change of GAPDH in the serum is consistent with its mRNA expression change in the heart specimens of ICM patients (Figure 3D). The ROC curve illustrated that GAPDH could distinguish ICM from NF well (Area under ROC curve (AUC) =0.9669, Figure 3E).
Hub Proteins Enriched in the DCM Patients
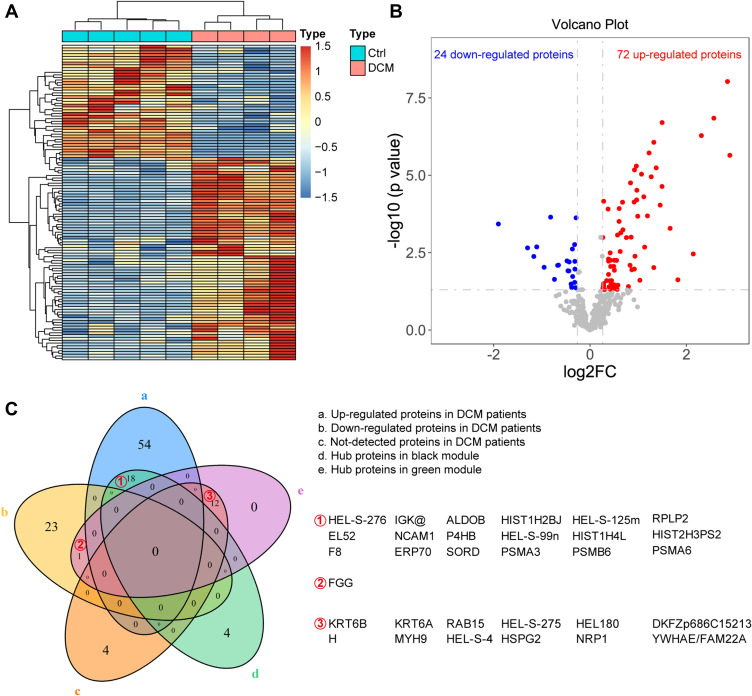
Among the 446 proteins, 16 proteins were undetected, 72 proteins were upregulated, and 24 proteins were downregulated in the DCM group. The hierarchical clustering plot (Figure 4A) and a volcano plot (Figure 4B) were constructed to present the dysregulated proteins in the DCM group. The Venn plot (Figure 4C) showed that 18 upregulated proteins, namely, HEL-S-276, IGK, ALDOB, HIST1H2BJ, HEL-S-125m, RPLP2, EL52, NCAM1, P4HB, HEL-S-99n, HIST1H4L, HIST2H3PS2, F8, ERP70, SORD, PSMA3, PSMB6, and PSMA6, overlapped with the hub proteins in the black module, which was positively associated with DCM in the module–trait correlation analysis of WGCNA. FGG was the only protein overlapping with the hub proteins of the green module; other 12 hub proteins of the green module were detected only in the ICM group, but not in the DCM group.
Figure 4.
Hub proteins enriched in the DCM patients. The hierarchical clustering plot (A) and a volcano plot (B) were constructed to present the dysregulated proteins in the DCM group. The Venn plot (C) showed that 18 up-regulated proteins were overlapped with the hub proteins in the black module, which was positively associated with DCM in the module-trait correlation analysis of WGCNA. FGG was the only protein overlapped with the hub proteins in the green module. In addition, the other 12 hub proteins in the green module could be detected only in the ICM group, but not the DCM group.
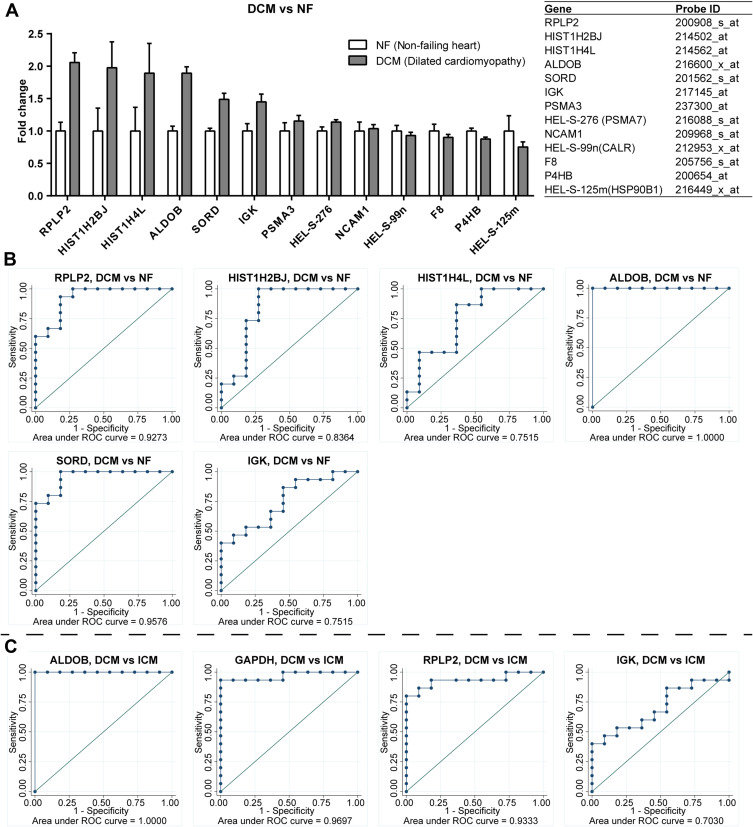
In addition, the RNA array data of heart specimens of DCM and NF from GDS651 were used to verify the expression of the mRNA corresponding to hub proteins in the heart. The results showed that mRNAs of RPLP2, HIST1H2BJ, HIST1H4L, ALDOB, SORD, IGK, PSMA3, HEL-S-276, NCAM1, HEL-S-99n, F8, P4HB, and HEL-S-125m were detected in the heart specimens, among which the protein expression changes of RPLP2, HIST1H2BJ, HIST1H4L, ALDOB, SORD, and IGK in the serum are consistent with its mRNA expression change in the heart specimens of DCM patients (Figure 5A). The ROC curve illustrated that RPLP2 (AUC=0.9273), HIST1H2BJ (AUC=0.8364), HIST1H4L (AUC=0.7515), ALDOB (AUC=1.0000), SORD (AUC=0.9576), and IGK (AUC=0.7515) could distinguish DCM from NF well (Figure 5B). Furthermore, ALDOB (AUC=1.0000), GAPDH (AUC=0.9697), RPLP2 (AUC=0.9333), and IGK (AUC=0.7030) could distinguish DCM from ICM well (Figure 5C).
Figure 5.
ALDOB, GAPDH, RPLP2, and IGK could distinguish DCM from ICM well. The RNA array data of heart specimens of DCM and NF from GDS651 were used to verify the expression of the mRNA corresponding to hub proteins in the heart. The results showed that mRNAs of RPLP2, HIST1H2BJ, HIST1H4L, ALDOB, SORD, IGK, PSMA3, HEL-S-276, NCAM1, HEL-S-99n, F8, P4HB, and HEL-S-125m were detected in the heart specimens, among which the protein expression changes of RPLP2, HIST1H2BJ, HIST1H4L, ALDOB, SORD, and IGK in the serum are consistent with its mRNA expression change in the heart specimens of DCM patients (A). The ROC curve illustrated that RPLP2, HIST1H2BJ, HIST1H4L, ALDOB, SORD, and IGK could distinguish DCM from NF well (B). Furthermore, ALDOB, GAPDH, RPLP2, and IGK could distinguish DCM from ICM well (C).
Cell Proliferation and Differentiation are the Hub Biological Processes Related to ICM
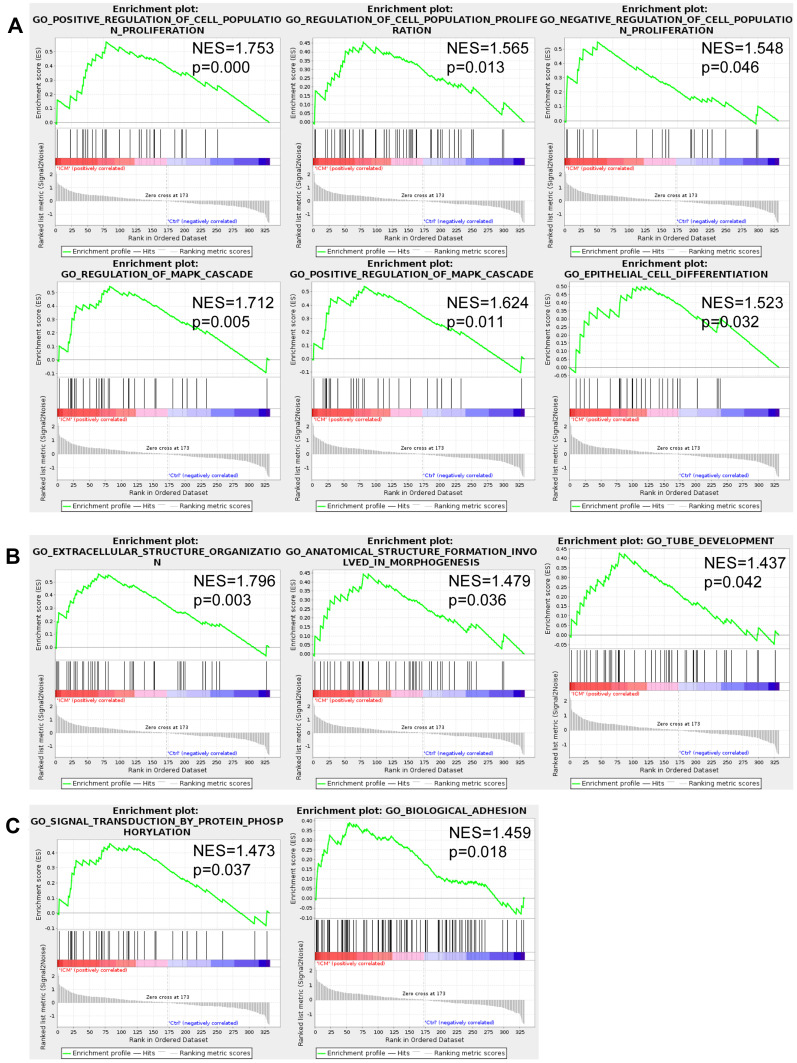
GSEA was performed to detect the hub biological processes in the ICM patients, which were significantly enriched in the ICM group but not significantly enriched in the DCM group (Table 2 and Figure 6). The results showed that 11 biological processes positively correlated with ICM, among which six biological processes were associated with cell proliferation and differentiation, including the regulation of the MAPK cascade. In addition, three biological processes were related to the structure organization, namely, extracellular structure organization, anatomical structure formation involved in morphogenesis, and tube development. The remaining two enriched biological processes were signal transduction by protein phosphorylation and biological adhesion.
Table 2.
Positive Biological Processes Only Enriched in ICM Patients with Gene Set Enrichment Analysis (GSEA)
| GO: Biological Process | Enriched in ICM | Compared with DCM | Only Significant in ICM | ||||
|---|---|---|---|---|---|---|---|
| GS | NES | p | GS | NES | p | ||
| Cell proliferation and differentiation | |||||||
| Positive regulation of cell population proliferation | 27 | 1.753 | 0.000 | 29 | 1.338 | 0.087 | Yes |
| Regulation of cell population proliferation | 46 | 1.565 | 0.013 | 51 | 0.998 | 0.487 | Yes |
| Negative regulation of cell population proliferation | 21 | 1.548 | 0.046 | 25 | 0.664 | 0.904 | Yes |
| Regulation of MAPK cascade | 29 | 1.712 | 0.005 | 30 | 0.803 | 0.756 | Yes |
| Positive regulation of MAPK cascade | 25 | 1.624 | 0.011 | 26 | 0.744 | 0.816 | Yes |
| Epithelial cell differentiation | 26 | 1.523 | 0.032 | 29 | 0.959 | 0.548 | Yes |
| Structure organization | |||||||
| Extracellular structure organization | 37 | 1.796 | 0.003 | 44 | 1.281 | 0.113 | Yes |
| Anatomical structure formation involved in morphogenesis | 41 | 1.479 | 0.036 | 47 | 0.832 | 0.755 | Yes |
| Tube development | 44 | 1.437 | 0.042 | 49 | 0.958 | 0.572 | Yes |
| Others | |||||||
| Signal transduction by protein phosphorylation | 34 | 1.473 | 0.037 | 37 | 1.364 | 0.084 | Yes |
| Biological adhesion | 82 | 1.459 | 0.018 | 89 | 1.147 | 0.231 | Yes |
Abbreviations: ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; GO, gene ontology; GS, gene size; NES, normalized enrichment score.
Figure 6.
Cell proliferation and differentiation were the hub biological processes related to ICM. The gene set enrichment analysis (GSEA) results showed that 11 biological processes were positively correlated to ICM (but not correlated to DCM), among which six biological processes were associated with cell proliferation and differentiation (A), three with the structure organization (B). The additional 2 enriched biological processes were the signal transduction by protein phosphorylation and biological adhesion (C).
Metabolic Processes are the Hub Biological Processes Associated with DCM
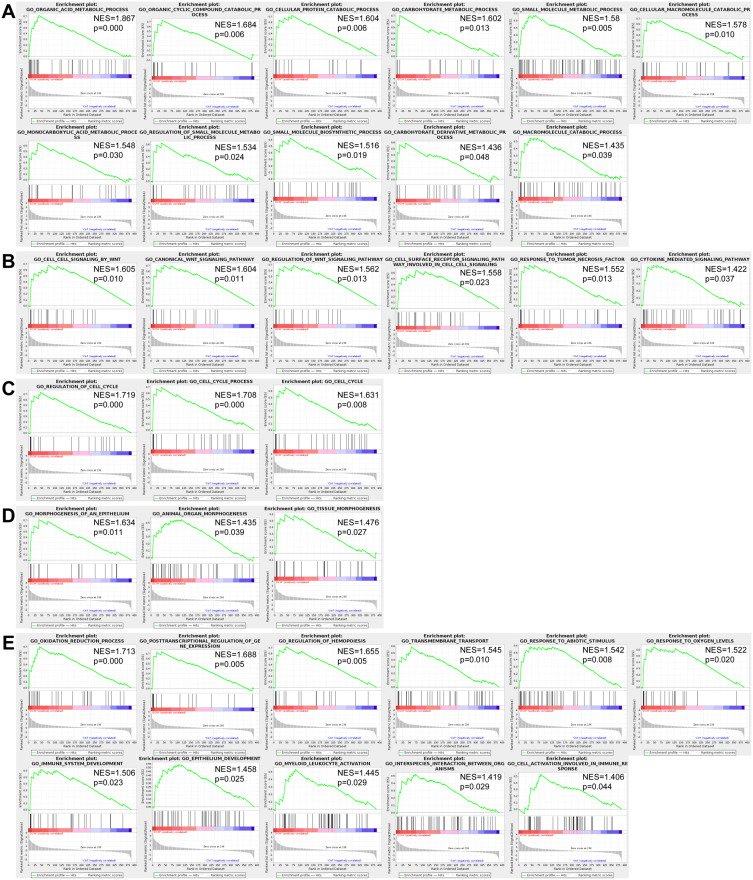
Based on GSEA, a total of 34 biological processes were significantly enriched in DCM patients but not significantly enriched in ICM patients (Table 3 and Figure 7). Eleven biological processes are involved in the metabolic processes, suggesting that the metabolic processes are the hub biological processes associated with DCM. In addition, there were six biological processes enriched in the cell signaling transduction, three in the cell cycle and three in the morphogenesis. The remaining biological processes included the oxidation reduction process, the post-transcriptional regulation of gene expression, the regulation of hemopoiesis, the transmembrane transport, the response to abiotic stimulus, the response to oxygen levels, the immune system development, the epithelium development, the myeloid leukocyte activation, the interspecies interaction between organisms, and the cell activation involved in the immune response.
Table 3.
Positive Biological Processes Only Enriched in DCM Patients with Gene Set Enrichment Analysis (GSEA)
| GO: Biological Process | Enriched in DCM | Compared with ICM | Only Significant in DCM | ||||
|---|---|---|---|---|---|---|---|
| GS | NES | p | GS | NES | p | ||
| Metabolic Processes | |||||||
| Organic acid metabolic process | 31 | 1.867 | 0.000 | Not enriched | Yes | ||
| Organic cyclic compound catabolic process | 16 | 1.684 | 0.006 | Not enriched | Yes | ||
| Cellular protein catabolic process | 15 | 1.604 | 0.006 | Not enriched | Yes | ||
| Carbohydrate metabolic process | 15 | 1.602 | 0.013 | Not enriched | Yes | ||
| Small molecule metabolic process | 53 | 1.580 | 0.005 | Not enriched | Yes | ||
| Cellular macromolecule catabolic process | 20 | 1.578 | 0.010 | Not enriched | Yes | ||
| Monocarboxylic acid metabolic process | 19 | 1.548 | 0.030 | Not enriched | Yes | ||
| Regulation of small molecule metabolic process | 21 | 1.534 | 0.024 | Not enriched | Yes | ||
| Small molecule biosynthetic process | 21 | 1.516 | 0.019 | Not enriched | Yes | ||
| Carbohydrate derivative metabolic process | 21 | 1.436 | 0.048 | Not enriched | Yes | ||
| Macromolecule catabolic process | 34 | 1.435 | 0.039 | Not enriched | Yes | ||
| Cell signaling transduction | |||||||
| Cell cell signaling by Wnt | 18 | 1.605 | 0.010 | Not enriched | Yes | ||
| Canonical Wnt signaling pathway | 15 | 1.604 | 0.011 | Not enriched | Yes | ||
| Regulation of Wnt signaling pathway | 15 | 1.562 | 0.013 | Not enriched | Yes | ||
| Cell surface receptor signaling pathway involved in cell-cell signaling | 21 | 1.558 | 0.023 | Not enriched | Yes | ||
| Response to tumor necrosis factor | 19 | 1.552 | 0.013 | Not enriched | Yes | ||
| Cytokine mediated signaling pathway | 31 | 1.422 | 0.037 | 24 | 0.536 | 0.988 | Yes |
| Cell cycle | |||||||
| Regulation of cell cycle | 18 | 1.719 | 0.000 | 15 | 0.919 | 0.571 | Yes |
| Cell cycle process | 21 | 1.708 | 0.000 | Not enriched | Yes | ||
| Cell cycle | 24 | 1.631 | 0.008 | 21 | 0.817 | 0.704 | Yes |
| Morphogenesis | |||||||
| Morphogenesis of an epithelium | 16 | 1.634 | 0.011 | 17 | 0.556 | 0.979 | Yes |
| Animal organ morphogenesis | 36 | 1.435 | 0.039 | 31 | 1.336 | 0.095 | Yes |
| Tissue morphogenesis | 24 | 1.476 | 0.027 | 23 | 0.918 | 0.584 | Yes |
| Others | |||||||
| Oxidation reduction process | 25 | 1.713 | 0.000 | Not enriched | Yes | ||
| Posttranscriptional regulation of gene expression | 15 | 1.688 | 0.005 | Not enriched | Yes | ||
| Regulation of hemopoiesis | 17 | 1.655 | 0.005 | 16 | 1.104 | 0.321 | Yes |
| Transmembrane transport | 34 | 1.545 | 0.010 | 27 | 0.634 | 0.943 | Yes |
| Response to abiotic stimulus | 39 | 1.542 | 0.008 | 29 | 1.056 | 0.391 | Yes |
| Response to oxygen levels | 20 | 1.522 | 0.020 | Not enriched | Yes | ||
| Immune system development | 23 | 1.506 | 0.023 | 21 | 1.083 | 0.333 | Yes |
| Epithelium development | 44 | 1.458 | 0.025 | 39 | 1.211 | 0.189 | Yes |
| Myeloid leukocyte activation | 46 | 1.445 | 0.029 | 38 | 1.321 | 0.096 | Yes |
| Interspecies interaction between organisms | 51 | 1.419 | 0.029 | Not enriched | Yes | ||
| Cell activation involved in immune response | 44 | 1.406 | 0.044 | 37 | 1.221 | 0.189 | Yes |
Abbreviations: ICM, ischemic cardiomyopathy; DCM, dilated cardiomyopathy; GO, gene ontology; GS, gene size; NES, normalized enrichment score.
Figure 7.
Metabolic processes were the hub biological processes associated with DCM. A total of thirty-four biological processes were significantly enriched in the DCM patients (but not significantly enriched in the ICM patients) via performing the gene set enrichment analysis (GSEA). The results showed that 11 biological processes were associated with the metabolic processes (A). In addition, there were six biological processes enriched in the cell signaling transduction (B), three in the cell cycle (C), and three in the morphogenesis (D). The other biological processes included the oxidation reduction process, the post-transcriptional regulation of gene expression, the regulation of hemopoiesis, the transmembrane transport, the response to abiotic stimulus, the response to oxygen levels, the immune system development, the epithelium development, the myeloid leukocyte activation, the interspecies interaction between organisms and the cell activation involved in immune response (E).
Discussion
In the present study, proteomics and bioinformatics analyses were performed to identify the hub proteins and key biological processes in ICM and DCM. Identification of hub proteins is of great importance to understand the development of diseases and identify proteins relevant for disease diagnosis and prognosis. The combination of WGCNA and differentially expressed protein analysis showed that five downregulated proteins (GAPDH, CLSTN1, VH3, CP, and ST13) were identified as the hub circulating proteins in ICM. As for DCM, one downregulated protein (FGG) and 18 upregulated proteins (HEL-S-276, IGK, ALDOB, HIST1H2BJ, HEL-S-125m, RPLP2, EL52, NCAM1, P4HB, HEL-S-99n, HIST1H4L, HIST2H3PS2, F8, ERP70, SORD, PSMA3, PSMB6, and PSMA6) were identified as the hub circulating proteins. The mRNA expression of the heart specimens from GDS651 validated that ALDOB, GAPDH, RPLP2, and IGK had good abilities to distinguish DCM from ICM. In addition, GSEA results showed that the metabolic processes were the hub biological processes in DCM, while the cell proliferation and differentiation were the hub biological processes in ICM. Metabolic processes are potential biological processes distinguishing nonischemic DCM from ICM.
Recent studies have reported various potential proteins for the diagnosis and prognosis of HF.5–7,19 However, specific circulating proteins of ICM and DCM, the two most common causes of HF,20,21 remain obscure. In our previous study, small RNA-sequencing has been employed to identify dysregulated circulating miRNAs in DCM.12 The present study presented five downregulated circulating proteins that might be potential biomarkers of ICM. CLSTN1 is an essential regulator of axon branching and endosomal trafficking during sensory neuron development and is associated with Alzheimer’s disease.22,23 However, the nature of calcium handling (Ca2+ induces Ca2+ release) in cardiomyocytes is quite different from the Ca2+ signaling in postsynaptic neurons. CLSTN1 has been reported to have a cytoplasmic calcium-binding domain,24 suggesting that CLSTN1 might be associated with the regulation of calcium ions in cardiomyocytes in patients with ICM. Solid basic studies are still needed to confirm the potential role of CLSTN1 in ICM. ST13 may mediate the intrinsic apoptotic pathway and inhibit tumor growth,25,26 indicating its potential effects in the regulation of cardiomyocytes’ apoptosis in ICM. Furthermore, FGG, which is associated with left ventricular diastolic dysfunction induced by the depletion of the β3-adrenergic receptor,27 was downregulated in DCM. Other 18 upregulated circulating proteins, including NCAM1 (a protein associated with left ventricular wall thickness28) and components of the 20S core proteasome complex (such as PSMA3, PSMB6, and PSMA629–31), were also identified as hub proteins in DCM. We noticed that the expression changes of some of these hub proteins are not consistent with the expression changes of the mRNAs in the heart specimens from GDS651. However, it is not enough to conclude that these proteins are not valuable for the identification of ICM or DCM, considering that there would be some adaptive changes in tissues and cells other than the hearts of these patients. In addition, several studies also detected the serum proteins of patients with HF and performed bioinformatic analyses. For example, Milting et al analyzed the plasma biomarkers for myocardial remodeling from end stage heart failure patients with the need for mechanical circulatory support.32 Chan MY et al evaluated the prioritizing candidates of post–myocardial infarction heart failure using plasma proteomics and single-cell transcriptomics.33 However, they have different research populations and purposes from ours.
Cell proliferation capability of the cardiac fibroblasts and endothelial cells plays crucial roles in cardiac remodeling.34,35 The regenerative capacity of intrinsic stem cells also has important effects on tissue repair after cardiac injury.36 In the present study, six of 11 significant biological processes were associated with cell proliferation and differentiation, including the regulation of the MAPK cascade. This indicated that cell proliferation and differentiation were the hub biological processes related to ICM. The hearts of mammals need a lot of energy to keep them beating in the best condition. The high-energy phosphate storage in cardiomyocytes is far from enough to keep the heart beating for a long time. The heart has the ability to use fatty acids, glucose, ketones, and amino acids to obtain enough energy through specific metabolism pathways. Cardiac metabolism plays crucial role in maintaining the normal structure and function of the heart. Improving cardiac metabolism is of great significance to improve cardiac function in DCM.37,38 The GSEA results showed that 11 of 34 significant biological processes were related to the metabolic processes, indicating that the metabolic processes were the hub biological processes associated with DCM. Metabolic processes are also potential biological processes distinguishing nonischemic DCM from ICM.
There are several limitations to the present study. First, the sample size was small because of logistic difficulties to collect enough specimens to perform extensive proteomics analysis within a short time. It is due to the consideration of minimizing the storage time of the collected specimens to avoid protein degradation and increase the credibility of the test data. Second, even though both WGCNA and differentially expressed protein analysis were performed to increase the credibility of the selected hub proteins in the present study, basic research is still necessary to validate the results, and we are planning to carry out related research. Third, the cell sources of the proteins, which are not discussed in this study, are quite important for their potential roles in DCM and ICM. However, basic research studies, such as immunofluorescence co-localization, flow cytometry, and even single-cell sequencing, are needed to identify the specific cell sources of these hub proteins, which will be the focus of our future research.
Conclusions
The proteomics and bioinformatics analyses identified the hub circulating proteins in ICM and DCM. In addition, GSEA results showed that cell proliferation and differentiation were the hub biological processes related to ICM, while the metabolic processes and cell signaling transduction were the hub biological processes associated with DCM. Metabolic processes are potential biological processes to distinguish nonischemic DCM from ICM.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81573095, 82100386).
Abbreviations
AUC, Area under ROC curve; DCM, Dilated cardiomyopathy; GO, Gene Ontology; GS, gene size; GSEA, Gene set enrichment analysis; F, Heart failure; ICM, Ischemic cardiomyopathy; NES, normalized enrichment score; NF, Non-failing hearts; PCC, Pearson correlation coefficient; ROC curve, Receiver operating characteristic curve; TOM, Topological overlap map; WGCNA, Weighted gene co-expression network analysis.
Data Sharing Statement
The datasets supporting the results of this article are included within the article. In addition, the comparative data have been uploaded to the integrated proteome resources database (PXD028101, iProX database, https://www.iprox.org//page/project.html?id=IPX0002777000).
Ethics Approval and Consent to Participate
This study was conducted in accordance with the Declaration of Helsinki. All procedures in the present study were subject to approval by the Ethics Committee of the Liaocheng People’s Hospital (Liaocheng, China). All subjects were taken and provided written informed consent before enrollment.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Roth GA, Johnson C, Abajobir A, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70(1):1–25. doi: 10.1016/j.jacc.2017.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W, Zhang A, Han Y, et al. Cyclin-Dependent Kinase Inhibitor 2b Controls Fibrosis and Functional Changes in Ischemia-Induced Heart Failure via the BMI1-p15-Rb Signalling Pathway. Can J Cardiol. 2021;37(4):655–664. doi: 10.1016/j.cjca.2020.05.016 [DOI] [PubMed] [Google Scholar]
- 3.Kelkar AA, Butler J, Schelbert EB, et al. Mechanisms Contributing to the Progression of Ischemic and Nonischemic Dilated Cardiomyopathy. J Am Coll Cardiol. 2015;66(18):2038–2047. doi: 10.1016/j.jacc.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 4.Lubrano V, Balzan S. Role of oxidative stress-related biomarkers in heart failure: galectin 3, alpha1-antitrypsin and LOX-1: new therapeutic perspective? Mol Cell Biochem. 2020;464(1–2):143–152. doi: 10.1007/s11010-019-03656-y [DOI] [PubMed] [Google Scholar]
- 5.Mallick A, Januzzi JL. Biomarkers in Acute Heart Failure. Revista Española de Cardiología. 2015;68(6):514–525. doi: 10.1016/j.rec.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 6.Tromp J, Khan MAF, Mentz RJ, et al. Biomarker Profiles of Acute Heart Failure Patients With a Mid-Range Ejection Fraction. JACC Heart Fail. 2017;5(7):507–517. doi: 10.1016/j.jchf.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 7.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. 2016;18(5):457–468. doi: 10.1002/ejhf.495 [DOI] [PubMed] [Google Scholar]
- 8.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2016;37(27):2129–2200. doi: 10.1093/eurheartj/ehw128 [DOI] [PubMed] [Google Scholar]
- 9.van der Meer P, Gaggin HK, Dec GW. ACC/AHA Versus ESC Guidelines on Heart Failure. J Am Coll Cardiol. 2019;73(21):2756–2768. doi: 10.1016/j.jacc.2019.03.478 [DOI] [PubMed] [Google Scholar]
- 10.Yancy CW, Jessup M, Bozkurt B, et al. 2016 ACC/AHA/HFSA Focused Update on New Pharmacological Therapy for Heart Failure: an Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure. J Am Coll Cardiol. 2016;68(13):1476–1488. doi: 10.1016/j.jacc.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 11.Wei X-M, Yang W-B, Su -X-X, Zhang A-D, Jin W, Fang Y-H. Plasma free fatty acid is associated with ischemic cardiomyopathy and cardiac dysfunction severity in systolic heart failure patients with diabetes. Chin Med J. 2021;134(4):472–474. doi: 10.1097/CM9.0000000000001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang G, Liu J, Yang C, et al. RNA sequencing discloses the genome wide profile of long noncoding RNAs in dilated cardiomyopathy. Mol Med Rep. 2019;19(4):2569–2580. doi: 10.3892/mmr.2019.9937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittenhafer-Reed KE, Richards AL, Fan J, et al. SIRT3 Mediates Multi-Tissue Coupling for Metabolic Fuel Switching. Cell Metab. 2015;21(4):637–646. doi: 10.1016/j.cmet.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sialana FJ, Wang A-L, Fazari B, et al. Quantitative Proteomics of Synaptosomal Fractions in a Rat Overexpressing Human DISC1 Gene Indicates Profound Synaptic Dysregulation in the Dorsal Striatum. Front Mol Neurosci. 2018;11:26. doi: 10.3389/fnmol.2018.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Xu M, Cui Z, et al. Genome-wide identification of FHL1 as a powerful prognostic candidate and potential therapeutic target in acute myeloid leukaemia. EBioMedicine. 2020;52:102664. doi: 10.1016/j.ebiom.2020.102664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rangaraju S, Dammer EB, Raza SA, et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer s disease. Mol Neurodegener. 2018;13(1):24. doi: 10.1186/s13024-018-0254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1± -responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc National Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger AL, Stojkovic S, Diedrich A, Demyanets S, Wojta J, Pezawas T. Elevated plasma levels of asymmetric dimethylarginine and the risk for arrhythmic death in ischemic and non-ischemic, dilated cardiomyopathy A prospective, controlled long-term study. Clin Biochem. 2020;83:37–42. doi: 10.1016/j.clinbiochem.2020.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters S, Kumar S, Elliott P, Kalman JM, Fatkin D. Arrhythmic Genotypes in Familial Dilated Cardiomyopathy: implications for Genetic Testing and Clinical Management. Heart Lung Circ. 2019;28(1):31–38. doi: 10.1016/j.hlc.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 21.Rady M, Ulbrich S, Heidrich F, et al. Left Ventricular Torsion A New Echocardiographic Prognosticator in Patients With Non-Ischemic Dilated Cardiomyopathy. Cir J. 2019;83(3):595–603. doi: 10.1253/circj.CJ-18-0986 [DOI] [PubMed] [Google Scholar]
- 22.Ponomareva OY, Holmen IC, Sperry AJ, Eliceiri KW, Halloran MC. Calsyntenin-1 Regulates Axon Branching and Endosomal Trafficking during Sensory Neuron Development In Vivo. J Neurosci. 2014;34(28):9235–9248. doi: 10.1523/JNEUROSCI.0561-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagnoni A, Perkinton MS, Gray EH, Francis PT, Noble W, Miller CCJ. Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates A production. Hum Mol Genet. 2012;21(13):2845–2854. doi: 10.1093/hmg/dds109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt L, Schrimpf SP, Meskenaite V, et al. Calsyntenin-1, a Proteolytically Processed Postsynaptic Membrane Protein with a Cytoplasmic Calcium-Binding Domain. Mol Cell Neurosci. 2001;17(1):151–166. doi: 10.1006/mcne.2000.0937 [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Yu M, Guan D, et al. ASK1-JNK signaling cascade mediates Ad-ST13-induced apoptosis in colorectal HCT116 cells. J Cell Biochem. 2010;110(3):581–588. doi: 10.1002/jcb.22551 [DOI] [PubMed] [Google Scholar]
- 26.Yang M, Cao X, Yu MC, et al. Potent Antitumor Efficacy of ST13 for Colorectal Cancer Mediated by Oncolytic Adenovirus via Mitochondrial Apoptotic Cell Death. Hum Gene Ther. 2008;19(4):343–353. doi: 10.1089/hum.2007.0137 [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Wei X, Su X, Shen Y, Jin W, Fang Y. Depletion of beta3-adrenergic receptor induces left ventricular diastolic dysfunction via potential regulation of energy metabolism and cardiac contraction. Gene. 2019;697:1–10. doi: 10.1016/j.gene.2019.02.038 [DOI] [PubMed] [Google Scholar]
- 28.Arnett DK, Meyers KJ, Devereux RB, et al. Genetic Variation in NCAM1 Contributes to Left Ventricular Wall Thickness in Hypertensive Families. Circ Res. 2011;108(3):279–283. doi: 10.1161/CIRCRESAHA.110.239210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barac YD, Emrich F, Krutzwakd-Josefson E, et al. The ubiquitin-proteasome system: a potential therapeutic target for heart failure. J Heart Lung Transplantation. 2017;36(7):708–714. doi: 10.1016/j.healun.2017.02.012 [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Su H, Wang X. The COP9 signalosome coerces autophagy and the ubiquitin-proteasome system to police the heart. Autophagy. 2016;12(3):601–602. doi: 10.1080/15548627.2015.1136773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagan J, Seto T, Pagano M, Cittadini A. Role of the Ubiquitin Proteasome System in the Heart. Circ Res. 2013;112(7):1046–1058. doi: 10.1161/CIRCRESAHA.112.300521 [DOI] [PubMed] [Google Scholar]
- 32.Milting H, Ellinghaus P, Seewald M, et al. Plasma biomarkers of myocardial fibrosis and remodeling in terminal heart failure patients supported by mechanical circulatory support devices. J Heart Lung Transplant. 2008;27(6):589–596. doi: 10.1016/j.healun.2008.02.018 [DOI] [PubMed] [Google Scholar]
- 33.Chan MY, Efthymios M, Tan SH, et al. Prioritizing Candidates of Post-Myocardial Infarction Heart Failure Using Plasma Proteomics and Single-Cell Transcriptomics. Circulation. 2020;142(15):1408–1421. doi: 10.1161/CIRCULATIONAHA.119.045158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Xu Q, Yang Q, et al. Vascular peroxidase 1 is a novel regulator of cardiac fibrosis after myocardial infarction. Redox Biol. 2019;22:101151. doi: 10.1016/j.redox.2019.101151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang J, Gao Z-Y, Zhang L-Y, Wang C-Y. Over-expression of JAZF1 promotes cardiac microvascular endothelial cell proliferation and angiogenesis via activation of the Akt signaling pathway in rats with myocardial ischemia-reperfusion. Cell Cycle. 2019;18(14):1619–1634. doi: 10.1080/15384101.2019.1629774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du G-Q, Shao Z-B, Wu J, et al. Targeted myocardial delivery of GDF11 gene rejuvenates the aged mouse heart and enhances myocardial regeneration after ischemia reperfusion injury. Basic Res Cardiol. 2017;112(1):7. doi: 10.1007/s00395-016-0593-y [DOI] [PubMed] [Google Scholar]
- 37.Beadle RM, Williams LK, Kuehl M, et al. Improvement in Cardiac Energetics by Perhexiline in Heart Failure Due to Dilated Cardiomyopathy. JACC Heart Fail. 2015;3(3):202–211. doi: 10.1016/j.jchf.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 38.Tuunanen H, Engblom E, Naum A, et al. Trimetazidine, a Metabolic Modulator, Has Cardiac and Extracardiac Benefits in Idiopathic Dilated Cardiomyopathy. Circulation. 2008;118(12):1250–1258. doi: 10.1161/CIRCULATIONAHA.108.778019 [DOI] [PubMed] [Google Scholar]