Abstract
The number of cholera cases and the mortality rates reported from different regions of Vietnam varied considerably in the period from 1979 to 1996, with between 2,500 and 6,000 cases reported annually from 1992 to 1995. Annual mortality rates ranged from 2.0 to 9.6% from 1979 to 1983 to less than 1.8% after 1983. Major cholera outbreaks were reported from the High Plateau region for the first time in 1994 and 1995; this is an area with limited access to health services and safe drinking-water supplies. All cases were associated with Vibrio cholerae O1. Using ribotyping, cholera toxin (CT) genotyping, and characterization of antibiotic susceptibility patterns and antibiotic resistance genes by PCR, we show that strains isolated after 1990 were clearly different from strains isolated before 1991. In contrast to strains isolated before 1991, 94% of 104 strains isolated after 1990 showed an identical ribotype R1, were resistant to sulfamethoxazole and streptomycin, and showed a different CT genotype. Furthermore, PCR analysis revealed that sulfamethoxazole-resistant strains harbored class I integrons containing a gene cassette ant(3")-1a encoding resistance to streptomycin and spectinomycin. This is, to our knowledge, the first report of class I integrons in V. cholerae. The development of cholera and the changes in the phenotypic and genotypic properties of V. cholerae O1 shown in the present study highlight the importance of monitoring V. cholerae O1 in Vietnam as in other parts of the world. In particular, the emergence of the new ribotype R1 strain containing class I integrons should be further studied.
Cholera has been an important cause of diarrhea in Vietnam for more than a century, with two million cases reported in 1850. In 1885, an outbreak of cholera was associated with mortality rates of up to 50% among French soldiers, and from 1910 to 1930 between 5,000 to 30,000 cholera cases were reported annually (27). After the emergence of the seventh pandemic in Indonesia in 1961, Vibrio cholerae O1 biotype El Tor entered South Vietnam in 1964 with a total of 20,009 cases and 821 deaths. Historically, the majority of cholera outbreaks have occurred in the central coastal areas and in South Vietnam. V. cholerae O1 El Tor was rarely reported in North Vietnam until 1976, where it caused outbreaks in the city of Hai Phong and in the Quang Ninh province.
During the last decade, remarkable changes have been reported in the incidence and characteristics of V. cholerae, including the entry of V. cholerae O1 into Latin America in 1991 (21, 33), the emergence in 1992 and rapid spread of O139 in Southeast Asia (39), recent major cholera outbreaks in Africa, and the recognition of non-O1, non-O139 serotypes of V. cholerae as important causes of diarrhea (24, 38). Studies of the molecular evolution of V. cholerae O1 strains isolated in Peru from 1991 to 1995 (11), in India (37, 45), and in Thailand (10) indicate that V. cholerae O1 undergoes genetic changes at relatively high rates. Although the significance of the genetic changes within V. cholerae remains uncertain, these changes are important in understanding the epidemiology and evolution of V. cholerae.
Despite the fact that V. cholerae continues to be an important cause of diarrhea in Vietnam, little information on cholera has been provided in the national and international scientific literature. Testing of an oral, killed whole-cell vaccine based on V. cholerae strains isolated outside of Vietnam has recently been initiated (43). However, V. cholerae O1 strains associated with cholera outbreaks in the different regions of Vietnam have not been characterized and possible changes in their phenotypic and genotypic traits are unknown.
In the present study, we report the number of cholera cases reported to the National Institute of Hygiene and Epidemiology (NIHE) from the different regions of Vietnam between 1979 and 1996. Taking advantage of several molecular-biology-based typing techniques and traditional phenotypic characterization, we selected and characterized 135 V. cholerae O1 biotype El Tor strains not only to describe possible geographic and time-dependent differences but also to investigate a possible epidemiological relationship among strains and V. cholerae O1 strains isolated in other Southeast Asian countries. Furthermore, strains were tested for their antibiotic susceptibilities, plasmid contents, and antibiotic resistance genes to evaluate if the current recommendations of antibiotic treatment of cholera are adequate and to determine if antibiotic-resistant V. cholerae O1 strains, which are increasingly being reported worldwide, are associated with cholera in Vietnam.
MATERIALS AND METHODS
Cholera surveillance.
Surveillance of cholera constitutes an important part of the continuing national surveillance program of notifiable infectious diseases carried out by the NIHE. At the district level, health centers collect surveillance data from each commune and report them monthly to 1 of 61 Provincial Preventive Medicine Centers, which then report the data to one of four regional Pasteur Institutes under the direction of the NIHE. The district centers report cases by residence and are often defined clinically without formal case definitions or culture of specimens. However, each provincial center does conduct standard bacteriological analysis of stool samples for Vibrio spp. by using alkaline-peptone-water, followed by plating onto thiosulfate-citrate-bile salt-sucrose (TCBS) agar.
Bacteriology.
Among a collection of more than 350 V. cholerae O1 biotype El Tor strains recovered from cholera patients from 1979 to 1996, 135 strains were included in the present study for further analysis (Tables 1 and 2). Strains were selected to represent cholera outbreaks reported in provinces throughout the country, although a few strains were also studied from single cholera cases. V. cholerae O1 strains isolated in 1984, 1987, 1988, 1989, 1991, 1992, and 1993 were not available in the strain collection at the NIHE.
TABLE 1.
The regional number of cholera cases in Vietnam reported by the provinces to the NIHE between 1979 and 1996
| Yr of isolation | No. of cholera cases by region
|
||||
|---|---|---|---|---|---|
| North | Center | South | High plateau | No. of deaths/total no. of cases (%) | |
| 1979 | 0 | 0 | 2,017 | 0 | 194/2,017 (9.6) |
| 1980 | 1,685 | 0 | 6,501 | 0 | 427/8,186 (5.2) |
| 1981 | 442 | 1,613 | 708 | 0 | 89/2,763 (3.2) |
| 1982 | 0 | 126 | 1,686 | 0 | 56/1,812 (3.1) |
| 1983 | 78 | 3,571 | 3,750 | 0 | 151/7,399 (2.0) |
| 1984 | 0 | 114 | 149 | 0 | 3/263 (1.1) |
| 1985 | 381 | 3,271 | 702 | 0 | 77/4,354 (1.8) |
| 1986 | 1,622 | 3,147 | 832 | 0 | 66/5,601 (1.2) |
| 1987 | 1,018 | 218 | 833 | 0 | 23/2,069 (1.1) |
| 1988 | 1,389 | 916 | 224 | 12 | 41/2,541 (1.6) |
| 1989 | 1 | 0 | 129 | 0 | 0/130 (0.0) |
| 1990 | 0 | 798 | 1,161 | 0 | 15/1,959 (0.8) |
| 1991 | 3 | 142 | 0 | 0 | 3/145 (2.1) |
| 1992 | 12 | 1,849 | 649 | 0 | 12/2,510 (0.5) |
| 1993 | 0 | 2,684 | 776 | 0 | 11/3,460 (0.3) |
| 1994 | 216 | 1,822 | 626 | 1,459 | 58/4,123 (1.4) |
| 1995 | 814 | 3,494 | 1,327 | 453 | 45/6,088 (0.7) |
| 1996 | 149 | 324 | 149 | 8 | 1/630 (0.2) |
TABLE 2.
Origin, antibiogram, and ribotype(s) of 133 clinical V. cholerae O1 strains isolated in Vietnam from 1979 to 1996
| Date of isolation (no. of isolates) | Province/region of isolation | Antibiograma | Ribotype |
|---|---|---|---|
| Mar.–Dec. 1979 (2) | Nghia Binh/center | Cl;SmI | R6 |
| Nov. 1979 (1) | Khanh Hoa/center | Cl;SmI (1) | R6 |
| Dec. 1979 (1) | Thuan Hai/center | Cl;SmI (1) | R6 |
| Feb. 1980 (1) | Khanh Hoa/center | Cl;SmI | R6 |
| Mar. 1980 (1) | Khanh Hoa/center | Cl;SmI | R1 |
| Apr. 1981 (1)b | Khanh Hoa/center | Cl;SmI | R6 |
| Aug. 1981 (2)c | Khanh Hoa/center | Cl;SmI (2) | R6 |
| Feb.–Mar. 1982 (3) | Khanh Hoa/center | Cl;SmI (1) | R6 |
| June 1983 (1)b | Hue/center | Cl;SmI | R4 |
| June 1983 (2) | Hue/center | Cl;SmI | R4, R6 |
| Aug. 1985 (2) | Ham Tan-Thuan Hai/center | Cl;SmI (1) | R5 |
| June 1986 (2) | Da Nang/center | Cl;SmI (1) | R5 |
| Aug. 1990 (7) | Hue/center | Cl;SmI (2) | R4(6), R6(1) |
| Aug. 1990 (3) | Khanh Hoa/center | Cl;SmI | R4 |
| Mar. 1994–June 1995 (13) | Ben Tre/south | Cl;Sm;Su (1) | R1 |
| Apr.–June 1994 (5) | Ho Chi Minh City/south | Cl;Sm;Su (2) | R1 |
| May–June 1995 (3) | Dong Thap/south | Cl;SmI;Su (2) | R1 |
| May 1994–June 1995 (7) | Dong Thap/south | Cl;Sm;Su (2) | R1 |
| May–June 1994 (4) | Dak Lak/high plateau | Cl;Sm;Su (1) | R1 |
| July 1994 (1) | Gia Lai/high plateau | Cl;Sm (1) | R1 |
| May–July 1994 (6) | Gia Lai/high plateau | Cl;Sm;Su (2) | R1 |
| June–July 1994 (3) | Da Lat/high plateau | Cl;Sm;Su | R1 |
| July 1994 (1) | Dak Lak/high plateau | Cl;Sm (1) | R1 |
| July 1994 (1) | Dak Lak/high plateau | Cl;Sm;Su (1) | R1 |
| July 1994–June 1995 (2) | Vinh Long/south | Cl;Sm;Su | R1 |
| Nov. 1994 (1) | Can Tho/south | Cl;Sm;Su | R1 |
| Nov. 1994 (1) | Minh Hai/south | Cl;SmI (1) | R4 |
| Nov. 1994 (1) | Ca Mau/south | Cl;SmI (1) | R3 |
| Nov. 1994–May 1995 (3) | Minh Hai/south | Cl;Sm;Su (2) | R1 |
| Dec. 1994 (3) | Dong Nai/south | Cl;Sm;Su | R1 |
| 1995 (1) | Gia Lai/high plateau | Cl;Sm;Su (1) | R2 |
| 1995 (1) | Dak Lak/high plateau | Cl;Sm;Su (1) | R2 |
| Mar. 1995 (1) | Dong Thap/south | Cl;Sm;Su (1) | R8 |
| Apr. 1995 (1) | Long An/south | Cl;Sm;Su | R1 |
| Apr. 1995 (2) | Soc Trang/south | Cl;Sm;Su | R1 |
| May 1995 (1) | Hue/center | Cl;Sm | R1 |
| Apr. 1995–Jan. 1996 (19) | Hue/center | Cl;Sm;Su (3) | R1 |
| May 1995 (1)b | Hue/center | Cl;Sm | R7 |
| June 1995 (1) | Soc Trang/south | Cl;Sm;Su | R1 |
| Sept. 1995–July 1996 (19) | Hai Phong/north | Cl;Sm;Su (2) | R1 |
| Oct. 1995 (1) | Gia Lai/high plateau | Cl;SmI (1) | R1 |
| June 1996 (1) | Hai Hung/north | Cl;Sm;Su (1) | R1 |
The numbers of strains studied by PCR analysis for the presence of integrons are shown in brackets. Cl, colistin; Sm, streptomycin; Su, sulfamethoxazole; subscript “I,” intermediate resistance.
Strains did not hybridize with the CT probe.
In addition to V. cholerae O1, V. mimicus, which contained CT genes and showed an antibiogram and ribotype R6 identical to that of the V. cholerae O1 strains, was isolated from both patients.
Fecal specimens were analyzed by the Provincial Preventive Medicine Centers, and the NIHE for V. cholerae by enrichment for 3 to 6 h in alkaline-peptone-water (pH 9.0) and were then plated onto TCBS agar (Difco, Detroit, Mich.). Presumptive identification was performed on the basis of standard biochemical reactions, including oxidase activity, Gram stain, growth in nutrient broth containing 0% NaCl, and acid production from glucose, inositol, mannitol, and sucrose. All suspected V. cholerae isolates were tested by agglutination tests with polyvalent O1 and monospecific Ogawa and Inaba antisera raised at the NIHE and O139 antisera (kindly provided by Yoshifumi Takeda, Research Institute, International Research Center of Japan). The O139 antiserum has been used for agglutination of V. cholerae isolates at the regional Pasteur Institutes and the NIHE since 1994. Further analysis and studies of strains were carried out at the Royal Veterinary and Agricultural University, Frederiksberg, Denmark.
V. cholerae isolates were tested for DNA sequences encoding cholera toxin (CT) in hybridization studies with an alkaline phosphatase-labeled 23-base oligonucleotide probe (44). The CT-producing V. cholerae strains 889 (3) and MS371 (15) were used as positive controls on the hybridization filters. An Escherichia coli K-12 strain was used as a negative control.
The following strains were included in the ribotyping studies for comparison purposes: V. cholerae O1 strain 33/97 isolated in 1997 in Bangkok, Thailand; V. cholerae O1 strain 697/38 isolated in 1994 in Samut Sakorn, Thailand (16); V. cholerae strain O1 VC20 isolated in 1992 in Calcutta, India (37); and V. cholerae O1 strain 1076/25 isolated in 1982 in Samut Sakorn, Thailand (41). V. cholerae strains contained in the oral vaccine currently being tested in Vietnam were included in the ribotyping studies, the antibiotic susceptibility testing, the plasmid analysis, the PCR analysis, and the CT genotyping. The vaccine strains included were V. cholerae O139 strain 4260B; the classic biotype V. cholerae O1 serotype Inaba strain, Cairo 48; the classic biotype V. cholerae O1 serotype Ogawa strain, Cairo 50; the El Tor biotype V. cholerae O1 serotype Inaba strain, Phil 6973; and the classic biotype V. cholerae O1 serotype Inaba strain, 569B (43).
Antibiotic susceptibility testing.
Each isolate was tested for susceptibility to 12 antibacterial agents by the disk diffusion method as recommended by the Swedish Reference Group for Antibiotics (SRGA) (29) by using Iso-Sensitest agar (Oxoid, Ltd., Basingstoke, United Kingdom). Testing was carried out according to the instructions of the disk manufacturer (Oxoid, Ltd.) and the SRGA with the following concentrations of antibiotics and inhibition zone diameter breakpoints (resistant and sensitive, respectively, in millimeters): ampicillin at 10 μg/disk (≤13 and ≥16), chloramphenicol at 30 μg/disk (≤19 and ≥24), ciprofloxacin at 5 μg/disk (≤17 and ≥20), colistin at 10 μg/disk (≤14 and 18≥), gentamicin at 10 μg/disk (≤12 and ≥16), kanamycin at 30 μg/disk (≤11 and ≥16), nalidixic acid at 30 μg/disk (≤16 and ≥21), neomycin at 30 μg/disk (≤11 and ≥15), streptomycin at 10 μg/disk (≤10 and ≥14), sulfamethoxazole at 100 μg/disk (≤19 and ≥24), tetracycline at 30 μg/disk (≤19 and ≥24), and trimethoprim-sulfamethoxazole (1.25:23.75) (≤19 and ≥24) at 25 μg/disk. Isolates were recorded as susceptible, intermediate, or resistant. The National Committee for Clinical Laboratory Standards has proposed breakpoint values for V. cholerae (26).
Isolation of plasmid DNA.
Plasmid preparation was carried out by the method of Kado and Liu (19), which was modified by incubating the cells at an elevated pH (12.54) for 30 min at 56°C during the lysis step. After lysis, phenol-chloroform-isoamylalcohol (25:24:1) was added, and the samples were mixed well. After centrifugation, a sample of the supernatant was electrophoresed, and plasmids were visualized essentially as described previously (28). V. cholerae O1 strain V 1075/25 containing an approximately 150-kb plasmid was used as the control strain (41). Plasmid sizes were estimated from the migration distance in the agarose gels relative to the migration distance of reference plasmids in E. coli V517 and 39R861 (22, 42) by the method of Rochelle et al. (34).
Ribotyping.
Total bacterial DNA was extracted by the method of Murray and Thompson (25). On the basis of previous studies (9, 31), the restriction enzyme BglI was used to digest chromosomal DNA. Ribotyping was performed by the procedure described by Dalsgaard et al. (9) with digoxigenin-labeled 16S and 23S rRNA probes. A 1-kb DNA molecular size standard (GIBCO-BRL, Gaithersburg, Md.) was used as a size marker. Ribotype patterns were considered to be different when there was a difference of one or more bands between isolates. Each ribotype was designated R, followed by an arbitrary number.
PCR.
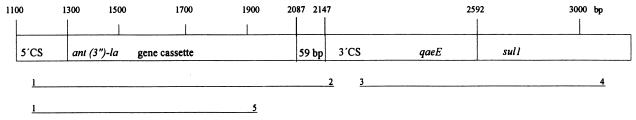
Since resistance to sulfonamides may be associated with the presence of class I integrons, we investigated whether a subset of the strains contained class I integrons (30, 32). Integrons comprise two conserved segments, the 5′ conserved segment (5′-CS) and the 3′ conserved segment (3′-CS) and an internal variable region which may contain gene cassettes encoding antibiotic resistance. Figure 1 was modified from the structure of class 1 integrons reported by Sandvang et al. (36). We initially selected a specific class I primer set sul1 and qacEΔ1 directed at the 3′ conserved segments of the integrons (Fig. 1, Table 3) (30, 32). A total of 34 V. cholerae O1 representative strains were selected based on their antibiogram, ribotypes, and year of isolation. The number of strains studied are marked in brackets in Table 2. The PCR results of a subset of the strains tested are shown in Fig. 3 and 4. PCR was carried out by suspension of one bacterial colony in 100 μl of distilled H2O, which was then boiled for 15 min at 95°C. After lysis, the suspension was centrifuged and stored at −20°C. For each reaction, 5 μl of the lysis suspension was used for PCR. The PCR amplification was performed according to the method of Aarestrup et al. (1) and optimized to detect amplicons in the range of 300 to 1,500 bp. The annealing temperature was set at 58°C. Strains yielding a PCR product with the class I primers were further amplified with the primers int1 F and int1 B, which amplify from each conserved segment products of variable sizes depending on the number and length of inserted gene cassettes (Fig. 1). Primers int1 F and ant(3")-Ia B were used to determine if the integron contained a gene cassette encoding resistance to streptomycin and spectinomycin (20). All PCR primers used are listed in Table 3. Salmonella serotype Typhimurium strain 9720921 and Acinetobacter spp. strain R4-96 were included as positive and negative controls, respectively (36). A 100-kb molecular mass standard (GIBCO-BRL) was used as a size marker during electrophoresis of the PCR products.
FIG. 1.
Association between integron structure and PCR products modified from Sandvang et al. (36). The lines below the integron structure represent amplicons. The numbers above the PCR products refer to the primers used on V. cholerae O1 (see Table 3). 5′-CS and 3′-CS represent the 5′ and 3′ conserved segments of the integron; qacE and sul1 encode resistance to disinfectant and sulfonamide, respectively. Numbers correspond to the sequence positions in GenBank accession number D43625 (20).
TABLE 3.
PCR primers for identification and characterization of integrons and gene cassettes in V. cholerae O1 from Vietnam
| PCR primer
|
Accession no.b | Primer position | Reference | ||
|---|---|---|---|---|---|
| No. | Namea | Sequence | |||
| 1 | int1 Fa (5′–CS) | GGC ATC CAA GCA GCA AGC | U12338 | 1416→1433 | Collis and Hall (7) |
| 2 | int1 Ba (3′-CS) | AAG CAG ACT TGA CCT GAT | U12338 | 4831→4814 | Hall et al. (13) |
| 3 | qacEΔ1 F | ATC GCA ATA GTT GGC GAA GT | X15370 | 211→230 | Stokes and Hall (40) |
| 4 | sul1 B | GCA AGG CGG AAA CCC GCG CC | X12869 | 1360→1341 | Stokes and Hall (40) |
| 5 | ant(3")-Ia B | ATT GCC CAG TCG GCA GCG | M10241 | 1040→1023 | Hollingshead and Vapnek (17) |
F, nucleotide sequence forward 5′→3′; B, nucleotide sequence backward 3′←5′.
Accession numbers are from the published sequences in the GenBank database.
FIG. 3.
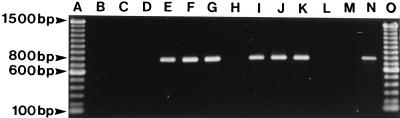
Examples of PCR for the presence of class I integrons among V. cholerae O1 strains isolated in Vietnam by using the primers qacEΔ1 F and sul1 B. Unless indicated otherwise, the following explanations for the contents of the lanes indicate province and year of isolation, antibiogram, and ribotype. Lanes: A, 100-bp molecular mass standard; B, Khanh Hoa, 1979, Cl;SmI, type R6; C, Ham Tan-Thuan Hai, 1985, Cl;SmI, type R5; D, Hue, 1990, Cl;SmI, type R4; E, Ben Tre, 1994, Cl;Sm;Su, type R1; F, Dak Lak, 1994, Cl;Sm,Su type R1; G, Hue, 1995, Cl;Sm;Su, type R1; H, Gia Lai, 1995, Cl;SmI, type R1; I, Dong Thap, 1995, Cl;Sm;Su, type R8; J, Dak Lak, 1995, Cl;Sm;Su, type R2; K, Hai Hung, 1996, Cl;Sm;Su, type R1; L, PCR mixture, negative control, M, R4-96, negative control; N, 9720921, positive control; and O, 100-bp molecular mass standard.
FIG. 4.
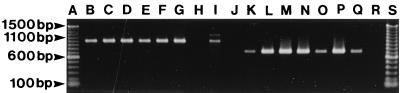
Examples of PCR products of V. cholerae O1 strains isolated in Vietnam by using the primers int1 F and int1 B (lanes B to G) and int1 F and ant(3")-Ia B (lanes K to P). Unless indicated otherwise, the following explanations for the contents of the lanes indicate the province and year of isolation, antibiogram, and ribotype. Lanes: A, 100-bp molecular mass standard; B, Ben Tre, 1994, Cl;Sm;Su, type R1; C, Dak Lak, 1994, Cl;Sm,Su, type R1; D, Hue, 1995, Cl;Sm;Su, type R1; E, Dong Thap, 1995, Cl;Sm;Su, type R8; F, Dak Lak, 1995, Cl;Sm;Su, type R2; G, Hai Hung, 1996, Cl;Sm;Su, type R1; H, R4-96, negative control; I, 9720921, positive control; J, PCR mixture, negative control; K, Ben Tre, 1994, Cl;Sm;Su, type R1; L, Dak Lak, 1994, Cl;Sm,Su, type R1; M, Hue, 1995, Cl;Sm;Su, type R1; N, Dong Thap, 1995, Cl;Sm;Su, type R8; O, Dak Lak, 1995, Cl;Sm;Su, type R2; P, Hai Hung, 1996, Cl;Sm;Su, type R1; Q, 9720921, positive control; R, R4-96, negative control; and S, 100-bp molecular mass standard.
CT genotyping.
Restriction fragment length polymorphism analysis of DNA sequences encoding CT genes (CT genotyping) was performed by hybridization of nylon membranes with BglI-digested DNA with a digoxigenin-labeled 23-base oligonucleotide CT probe (44). A total of 15 strains representing different ribotypes and the vaccine strains were typed. The hybridization procedures and detection of the probe were performed as described by the manufacturer (Boehringer GmbH, Mannheim, Germany). The restriction enzyme BglI was selected because it does not have any recognition sequence within the ctxA gene, but it has a single cleavage site located upstream and adjacent to the ctxA gene (23). Accordingly, the number of bands comprising each CT genotype pattern represent the possible number of copies of the CTX genetic element harbored by each strain.
RESULTS
Surveillance data.
The number of cases registered between 1979 and 1996 in the four regions of Vietnam are presented in Tables 1 and 2. A part of the data included in Table 1 was reported previously by Dalsgaard et al. (12). Several coastal areas of central Vietnam have experienced cholera outbreaks during the last decade, including repeated and recent outbreaks in the ancient city of Hue. As in the southern provinces, cholera appears to be endemic in certain central regions of the country (Tables 1 and 2). For the first time during the last decade, cholera outbreaks were reported in 1994 and 1995 in the provinces of Dac Lac and Gia Lai in the mountainous High Plateau area, including 1,459 and 453 cases, respectively. Hai Phong, an important port city in North Vietnam, experienced an outbreak of cholera in September 1995 which extended into July 1996 and included more than 250 cases (Tables 1 and 2). Although a number of cholera cases were reported in the northern provinces in the period from 1985 to 1988 and from 1994 to 1996, cholera is not recognized as endemic in North Vietnam. Information was not available to explain the low number of cases reported to the NIHE for certain regions in 1989, 1990, 1991, and 1993 (Table 1). From 1979 to 1983, the case fatality rate ranged from 2.0 to 9.6%. However, after 1983 the case fatality rate remained low, varying between 0.0 and 1.8%. The factor(s) responsible for the decrease in the case fatality rate were not identified. It should be noted that the strains studied from 1979 and 1980 originated from Central Vietnam, although the cases from which these strains were isolated were not reported to the NIHE (Tables 1 and 2). Bacteriological analysis of stool samples showed that all cases studied were associated with V. cholerae O1. Thus, V. cholerae O139 was not isolated from any cases. Furthermore, no outbreaks of diarrhea appeared to be associated with other non-O1 serogroups of V. cholerae which are increasingly being reported worldwide (8, 38).
Specimens, antibiotic susceptibility patterns, and plasmid analysis.
Strains in the present study showed biochemical and serological reactions typical of those of V. cholerae and were identified as V. cholerae O1 biotype El Tor. However, in repeated testing, two strains were sucrose negative and did not agglutinate the O1 antisera. Each of the strains, which were identified as V. mimicus, were recovered from stool samples from young girls that also yielded V. cholerae O1 (Table 2). Strains isolated from 1979 to 1981 agglutinated Ogawa antisera, whereas strains isolated from 1982 to 1990 were all serotype Inaba. Strains isolated after 1990 were all serotype Ogawa. In colony hybridization studies with the CT probe, 132 of the 135 strains tested were deemed positive, yielding dark blue or purple colonies. The two V. mimicus strains hybridized with the CT probe in repeated studies, whereas the three CT-negative strains did not hybridize with the probe in repeated testing (Table 2).
All strains isolated in Vietnam showed resistance to colistin but were generally sensitive to most of the remaining antibacterial agents tested, including tetracycline. However, irrespective of the province of isolation and with few exceptions, each of the strains isolated after 1990 were found to be resistant to streptomycin and sulfamethoxazole. This was in contrast to strains isolated before 1991, which were susceptible to sulfamethoxazole and showed intermediate resistance to streptomycin. Among the vaccine strains, strain 4260B showed resistance to colistin, streptomycin, sulfamethoxazole, and trimethoprim, while the remaining vaccine strains were susceptible or showed intermediate resistance to colistin and streptomycin. Plasmid analysis revealed that only one strain isolated in Hai Phong in September 1995 carried plasmids (a single 70-kb plasmid). The vaccine strain Cairo 50 contained two plasmids that were 43 and 6.2 kb, respectively. The control strain showed an approximately 150-kb plasmid (results not shown).
Ribotyping.
Ribotyping of strains isolated in Vietnam produced two groups of rRNA restriction patterns (Fig. 2, Table 2). The DNA fragments containing rRNA genes ranged in size from 1.5 to 12.5 kb. All patient isolates showed common fragments of 2.3, 4.1, 4.6, 5.7, 6.0, 6.4, and 9.3 kb. The first group consisted of V. cholerae O1 strains isolated after 1990, of which the majority (98 of 104 strains) showed an identical ribotype, ribotype R1 (Fig. 2). Two strains isolated from outbreaks in Gia Lai and Dak Lak in the High Plateau region in 1995 showed ribotype R2, which differed from R1 by a single 12.4-kb fragment not seen among the R1 strains. One strain isolated in Dong Thap in 1995 showed a ribotype R8 that differed from R1 by a single fragment. A CT-negative V. cholerae O1 strain isolated in Hue in 1997 showed a unique ribotype R7 (Table 2).
FIG. 2.
Examples of BglI ribotypes of V. cholerae O1 strains associated with cholera outbreaks in Vietnam from 1979 to 1996, of clinical strains from Thailand and India, and of strains used in an oral vaccine. Unless otherwise stated, explanations for the contents of the lanes indicate the strain designation, the place and date of isolation, and the ribotype designation. Lanes: A, 1-kb molecular mass standard; B, Gia Lai, 1995, type R2; C, Ho Chi Minh City, June 1994, type R1; D, 33/97, Bangkok 1997, type R1; E, 697/38, Samut Sakorn 1994, type R1; F, VC20, Calcutta, 1992, type R1; G, Dong Thap, March 1995, type R8; H, Thuan Hai, December 1979, type R6; I, Minh Hai, November 1994, type R3; J, Ham Tan Thuan Hai, August 1985, type R5; K, Hue, June 1983, type R4; L, 1076/25, Samut Sakorn, Thailand, 1982, type R4; M, Hue, May 1995, type R7; N, 4260B, V. cholerae O139 vaccine strain; O, Phil 6973, vaccine strain, type R4; P, Cairo 48, vaccine strain; Q, Cairo 50, vaccine strain; R, 569B, vaccine strain; and S, 1-kb molecular mass standard.
The second group consisted of 29 strains isolated before 1991 which showed four closely related ribotypes, R3, R4, R5, and R6, which differed by one or two fragments only (Fig. 2, Table 2). Each of these ribotypes lacked a 10.7-kb fragment seen among isolates of the R1, R2, and R8 ribotypes. With one exception, ribotype R6 was shown only by V. cholerae O1 strains isolated from 1979 to 1983. The two V. mimicus strains also showed ribotype R6. Ribotype R5 was shown only by strains isolated during two major cholera outbreaks in central Vietnam in 1985 and 1986 (Fig. 2). Ribotypes R3 and R4 were also shown by two strains isolated in the Minh Hai province in South Vietnam in 1994 (Table 2).
Strains 33/97 and 697/38 isolated in Thailand in 1997 and 1994, respectively, and strain VC20 isolated in Calcutta in 1992 showed identical ribotypes that were similar to ribotype R1 (Fig. 2). Strain 1076/25 isolated in Thailand in 1982 showed ribotype R4. The vaccine strain V. cholerae O139 strain 4260B showed a unique ribotype, whereas V. cholerae O1 strain Phil 6973 showed ribotype R4. The three remaining vaccine strains showed different but closely related ribotypes demonstrating two unique large size fragments typical of classical strains (31). Strains Cairo 48 and Cairo 50 showed an identical ribotype. Repeated studies of the strains showed no variation in ribotype patterns, although differences in band intensity and the degree of background were observed for a few strains.
The division of strains into two groups based on ribotype patterns and date of isolation thus corroborates the results of the antibiotic susceptibility testing in which strains isolated before 1991 were found to be sensitive to sulfamethoxazole and intermediately resistant to streptomycin, whereas strains isolated after 1990 were resistant to both antibiotics.
PCR.
An approximately 800-bp class I integron PCR product was obtained from each of the 20 sulfamethoxazole-resistant V. cholerae O1 strains studied by using the primers qacEΔ1 F and sul1 B (Fig. 3, Table 2). Strains that contained integrons represented ribotypes R1, R2, and R8, whereas PCR of 14 sulfamethoxazole-sensitive strains did not yield any product, indicating that integrons were not present (Fig. 3, Table 2). The four sulfamethoxazole-sensitive ribotype R1 strains isolated after 1990 and the vaccine strains did not yield of PCR product (Table 2).
A PCR product of approximately 1,000 bp was obtained from each of the 20 class I integron strains by using the int1 F and int1 B primers (Fig. 4, Table 3). The control strain 9720921 showed two distinct integrons, which were earlier found to be 1,008 and 1,133 bp in size, respectively (Fig. 4, lane I) (36). An amplicon of approximately 750 bp was obtained from each of the 20 strains by using the int1 F and ant(3")-Ia B primers (17), thus confirming the presence of the aminoglycoside resistance gene cassette ant(3")-Ia, which is 752 bp and confers resistance to streptomycin and spectinomycin (Fig. 4, Table 3) (17, 20).
CT genotypes.
Southern blot hybridization with the CT probe of BglI-digested genomic DNAs of strains isolated after 1990 which belonged to ribotypes R1, R2, and R8 showed identical patterns demonstrating fragments of 3.5 and 7.0 kb (Fig. 5, lanes C to I). By contrast, the five V. cholerae O1 strains isolated before 1991 representing ribotypes R3, R4, R5, and R6 all showed an identical but different pattern which demonstrated three fragments (Fig. 5, lanes J to N; since the approximately 6.7-kb fragment appeared weakly in repeated testing and was thus difficult to visualize, the fragments are indicated by arrowheads). Strain VC-346 contained a single copy of the CTX element, strain VC-351 contained three copies and showed a unique CT genotype; the CT negative control strain VC-302, however, did not hybridize with the probe. The vaccine strains contained two to four copies of the CTX element (Fig. 5, lanes Q to U).
FIG. 5.
Southern hybridization analysis of genomic DNA from 15 V. cholerae strains isolated from patients in Vietnam and 5 vaccine strains digested with BglI and probed with a cholera toxin probe. Unless stated otherwise, explanations of the contents of the lanes indicate the strain designation, the province/region of isolation, and the ribotype. Lanes: A, HindIII digest of phage lambda as molecular size markers; B, VC-351, Khanh Hoa/Center ribotype R1; C, VC-320, Hue/Center, type R1; D, VC-233, Dong Thap/South, type R1; E, VC-288, Gia Lai/High Plateau, type R1; F, VC-378, Dak Lak/High Plateau, type R1; G, VC-281, Dong Thap/South, type R8; H, VC-181, Dak Lak/High Plateau, type R2; I, VC-182, Gia Lai/High Plateau, type R2; J, VC-203, Minh Hai/South, type R3; K, VC-367, Khanh Hoa/Center, type R4; L, VC-362, Ham Tan-Huan Hai/Center, type R5; M, VC-364, Da nang/Center, type R5; N, VC-336, Hue/Center, type R6; O, VC-346, Khanh Hoa/Center, type R6; P, VC-302, Hue/Center, type R7; Q, Cairo 48, vaccine strain; R, Cairo 50, vaccine strain; S, 569B, vaccine strain; T, 4260 B, vaccine strain; U, Phil 6973, vaccine strain; and V, HindIII digest of phage lambda as molecular size markers. Since the approximately 6.7-kb fragment appeared weakly (lanes J to N) in repeated testing and was difficult to visualize, the position of the fragment is indicated by arrowheads.
DISCUSSION
The present study describes the development of cholera in Vietnam in the period from 1979 to 1996. The number of cholera cases reported to the NIHE from the provinces in the four regions of the country showed that V. cholerae O1 remains an important cause of diarrhea, especially in the central and southern provinces where cholera appears to be endemic. The annual total number of cholera cases showed a high degree of variation, and no decreases in the number of cholera cases were registered in the study period with recent reports of between 2,500 and 6,000 cases reported annually from 1992 to 1995. Cholera was not reported from the provinces in the High Plateau region until two major outbreaks occurred in 1949 and 1995. However, data allowing identification of the causes of the two outbreaks were not available. The introduction of V. cholerae O1 into the High Plateau provinces is disturbing since this region is one of the Vietnam’s poorest, with the inhabitants having limited access to health services and safe drinking-water supplies. V. cholerae O139 has so far not been reported in Vietnam, in contrast to most of the neighboring countries; however, agglutination of V. cholerae in O139 antisera remains important in monitoring a possible introduction of the O139 serotype into Vietnam.
Based on the promising results of vaccine trials with orally administered cholera vaccines in Bangladesh, the NIHE decided that deployment of an effective cholera vaccine would be a useful preventive strategy to protect human health. Initial testing suggested that a two-dose regimen of an oral killed whole-cell vaccine can confer substantial protection against cholera (43). The appearance of V. cholerae O139 in India and Bangladesh in 1992 (2) and the subsequent spread to other Southeast Asian countries (5) was a cause for concern that O139 might spread into Vietnam. Since immunity to the O1 serogroup is not cross-protective against the O139 serogroup, V. cholerae O139 strain 4260B was included in a modified vaccine, for which testing began in central Vietnam in 1997. As no information was available about the immunogenic and genotypic properties of V. cholerae O1 strains isolated from patients in Vietnam, all of the strains included in the vaccine were isolated outside of Vietnam. However, we suggest based on the results of the present study that in the further development of an oral cholera vaccine, the new V. cholerae O1 ribotype R1 strain should be considered as a vaccine candidate strain.
The NIHE is aware that the current system for reporting cholera cases is inadequate and that the actual number of cases is higher than that shown in Table 1. Accordingly, the NIHE is promoting the definition of cholera cases according to the recommendations from the World Health Organization. In addition, bacteriological media and training of staff for the isolation of V. cholerae O1 from stool specimens are provided by the NIHE to provincial and regional laboratories. Despite some improvements in the reporting of cholera cases, it is likely also in the future that a significant proportion of cases will not be reported, partly because there does not exist a formal reporting system for the increasing number of private physicians and laboratories.
Based on ribotype patterns and the CT genotypes, the strains could be divided into two similar groups as described for the antibiotic susceptibility testing. The first and largest group contained 98 V. cholerae O1 strains isolated after 1990, of which 94% showed an identical ribotype R1. Ribotype R1 was shown by strains isolated in provinces in each of the four regions studied. It appears that this particular ribotype has established itself in Vietnam as the major genotype of V. cholerae O1 associated with cholera. As strains from 1991, 1992, and 1993 were not available for our study, it is unknown if the ribotype R1 strain was already present before 1994 as the major ribotype associated with cholera. The two large cholera outbreaks reported in the High Plateau region in 1995 showed a unique ribotype R2. However, ribotype R2 strains differed from R1 by a single fragment only and showed phenotypic characteristics identical to those of the ribotype R1 strains, including identical antibiotic susceptibility patterns.
Strains 33/97 and 697/38 isolated in Thailand and strain VC20 from India showed ribotype R1. Strain 697/38 was included in a recent study of phenotypic and molecular characteristics of V. cholerae O1 isolated in Samut Sakorn, Thailand, before, during, and after the emergence of V. cholerae O139 in August 1993 in Samut Sakorn (10). Among 43 strains isolated during and after the O139 epidemic, all strains showed ribotype R1 (10). Thus, the emergence and disappearance of the O139 serotype in Samut Sakorn was associated with the appearance of a V. cholerae O1 strain which showed a ribotype R1 identical to the type shown by O1 strains isolated in Vietnam after 1990. As indicated by the ribotype R1 shown by strain 697/38 isolated in 1997, this ribotype R1 seems to continue to be associated with cholera in Thailand. Although strain VC20 represents the major ribotype R1 associated with cholera in India before the emergence of O139 in 1992, a new but closely related ribotype seems to have displaced ribotype R1 in India (37).
Four closely related ribotypes were shown by strains isolated before 1990, whereas none of the strains isolated after 1990 showed similar ribotypes (Fig. 2, Table 2). Although closely related, certain ribotypes were only associated with cholera outbreaks in certain geographical regions. For example, ribotype R5 was associated only with two major outbreaks in the central region in 1985 and 1986 (Table 2).
It was interesting that each of the two V. mimicus strains recovered from two different cholera patients showed identical ribotypes R6 and antibiograms, as demonstrated by the V. cholerae O1 strains isolated from the patients. This is, to our knowledge, the first report of a simultaneous isolation of CT-positive V. cholerae O1 and V. mimicus strains showing identical ribotypes and antibiograms from cholera patients. Thus, our strains of the two species appear to differ only in the utilization of sucrose.
Except for four strains, ribotype R1 and R2 strains isolated in Vietnam after 1990 showed resistance to sulfamethoxazole, streptomycin, and colistin whereas strains isolated before 1991 were susceptible to sulfamethoxazole and showed resistance to colistin and intermediate resistance to streptomycin. Thus, the current recommendations for the treatment of cholera patients by the administration of tetracycline to adults remain valid (12). However, the resistance to sulfamethoxazole shown by strains isolated after 1990 is disturbing, since trimethoprim-sulfamethoxazole or ampicillin is recommended for the treatment of children and pregnant women (12).
Using PCR with a combination of primers, we showed that each of the sulfamethoxazole-resistant strains tested contained class I integrons and the aminoglycoside resistance gene cassette ant(3")-Ia, which encodes resistance to streptomycin and spectinomycin (Fig. 1, 3, and 4; Table 2) (20). In addition, two sulfamethoxazole-sensitive and streptomycin-resistant strains also contained integrons and the gene cassette indicating that the genetic elements encoding sulfamethoxazole were not expressed. This is, to our knowledge, the first report of V. cholerae O1 strains containing class I integrons and the resistance gene cassette ant(3")-Ia. Integrons are mobile DNA elements able to incorporate single or groups of antibiotic resistance genes by site-specific recombination within and between bacterial species (14). A scheme for the evolution of integrons, including the existence of a putative ancestor In0, has been proposed by Bissonnette and Roy (4). They further proposed that the streptomycin gene aadA2 was first inserted at the hot spot of In0, followed by the insertion of other resistance genes (4). Similarly, we found only one gene cassette encoding resistance to streptomycin and spectinomycin; however, the origin of the integrons found in V. cholerae O1 in Vietnam is unknown and remains to be determined.
Chromosomal repetitive sequence elements that conform to the definition of an integron were recently described for V. cholerae (6). However, these elements, which were designated VCR elements, seem to differ in that they contain the promoters for the 3′ genes and the terminators for the 5′ genes, a feature that is not common in the known integrons (6). Also, the VCR elements seem to contain more genes than integrons (6).
Isolates of Enterobacteriaceae often carry integrons, especially class I, encoding resistance to various antibacterial compounds (18, 35). Interestingly, similar changes in antibiotic susceptibility patterns, as seen in the present study, were reported in the Samut Sakorn study in Thailand, where strains isolated after 1990 showed ribotype R1, and were resistant to sulfamethoxazole, streptomycin, and colistin (10). Preliminary results indicate that strains isolated in Samut Sakorn also contain identical class I integrons, suggesting that the V. cholerae O1 ribotype R1 strain became established in both countries as the main cause of cholera and that it could have been transferred between the two countries (data not shown). Since antibiotic resistance gene cassettes can be inserted in integrons and integrons may also be carrying genes encoding other important properties, the emergence, distribution, and evolution of integrons should be closely monitored.
In conclusion, we have shown that cholera remains an important cause of diarrhea in Vietnam. The reports of cholera in the High Plateau region, which was previously free of the disease, underscores the need to monitor the epidemiology of V. cholerae. The phenotypic and genotypic characterization showed that V. cholerae O1 isolated before 1991 and after 1990 were clearly different, suggesting that V. cholerae O1 strains are continuously undergoing changes or that new strains of V. cholerae O1 strains may have been introduced. The emergence of a new ribotype R1 strain containing sul1 gene-associated class I integrons with a resistance gene cassette encoding resistance to streptomycin and spectinomycin is disturbing and needs to be further studied.
ACKNOWLEDGMENTS
We are grateful for the technical assistance provided by Nguyen Thi Gam and Bui Thu Hien at the National Institute of Hygiene and Epidemiology, Hanoi, and by Anne-Mette Petersen at the Royal Veterinary and Agricultural University in Denmark. We thank Andreas Petersen and Derek Brown of the Royal Veterinary and Agricultural University for help in planning the PCR studies and Dorthe Sandvang at the Danish Veterinary Laboratory for suggestions in the preparation of Fig. 1 and for the supply of control strains and the primer ant(3")-Ia used in the PCR analysis. We also thank Nikolaj Larsen and Kamilla Louise Mørk Pedersen for secretarial assistance in the preparation of Fig. 2 and James D. Oliver for comments on the manuscript. The Thai strains were provided by Peter Echeverria, Armed Forces Research Institute of Medical Sciences, Thailand, and the Indian strain was provided by Balakrish G. Nair, National Institute of Cholera and Enteric Diseases, India. We are grateful for the help provided by Dao Xuan Vinh, Buon Me Thuot Pasteur Institute, in collecting the data on cholera cases.
Anders Dalsgaard was supported by the Danish Council for Development Research, Danida grant 90810.
REFERENCES
- 1.Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;340:1938–1940. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert M J. Vibrio cholerae O139 Bengal. J Clin Microbiol. 1994;32:2345–2349. doi: 10.1128/jcm.32.10.2345-2349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagchi K, Echeverria P, Arthur J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissonnette L, Roy P H. Characterization of In0 of Pseudomonas aeruginosa plasmid pSV1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1991;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chongsa-nguan M, Chaicumpa W, Moolasart P, Kandhasingha P, Shimada T, Kurazono H, Takeda Y. Vibrio cholerae O139 Bengal in Bangkok. Lancet. 1993;342:430–431. doi: 10.1016/0140-6736(93)92841-g. [DOI] [PubMed] [Google Scholar]
- 6.Clark C A, Purins L, Kaewrakon P, Manning P A. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol Microbiol. 1997;26:1137–1138. doi: 10.1046/j.1365-2958.1997.d01-5533.x. [DOI] [PubMed] [Google Scholar]
- 7.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalsgaard A, Echeverria P, Larsen J L, Siebeling R, Serichantalergs O, Huss H H. Application of ribotyping for differentiating Vibrio cholerae non-O1 isolates from shrimp farms in Thailand. Appl Environ Microbiol. 1995;61:245–251. doi: 10.1128/aem.61.1.245-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalsgaard A, Serichantalergs O, Forslund A, Pitarangsi C, Echeverria P. Phenotypic and molecular characterization of Vibrio cholerae O1 isolated in Samutsakorn, Thailand before, during and after the emergence of V. cholerae O139. Epidemiol Infect. 1998;121:259–268. doi: 10.1017/s0950268898001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalsgaard A, Skov M N, Serichantalergs O, Echeverria P, Meza R, Taylor D N. Molecular evolution of Vibrio cholerae O1 isolated in Lima, Peru, from 1991–1995. J Clin Microbiol. 1997;35:1151–1156. doi: 10.1128/jcm.35.5.1151-1156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalsgaard A, Tam N V, Cam P D. cholera in Vietnam. Southeast Asian J Trop Med Public Health. 1997;28:69–72. [PubMed] [Google Scholar]
- 13.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 14.Hall R M, Stokes H W. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 15.Hanchalay S, Seriwatana J, Echeverria P, Holmgren J, Tirapat C, Moseley S L, Taylor D N. Non-O1 Vibrio cholerae in Thailand: homology with cloned cholera toxin genes. J Clin Microbiol. 1985;21:288–289. doi: 10.1128/jcm.21.2.288-289.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoge C W, Bodhidatta L, Echeverria P, Deesuwan M, Kitporka P. Epidemiologic study of Vibrio cholerae O1 and O139 in Thailand: at the advancing edge of the eighth pandemic. Am J Epidemiol. 1996;143:263–268. doi: 10.1093/oxfordjournals.aje.a008737. [DOI] [PubMed] [Google Scholar]
- 17.Hollingshead S K, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 18.Jones M E, Peters E, Weersink A-M, Fluit A, Verhoef J. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 19.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazama H, Kizu K, Iwasaki M, Hamashima H, Sasatsu M, Arai T. Isolation and structure of a new integron that includes a streptomycin resistance gene from the R plasmid of Pseudomonas aeroginosa. FEMS Microbiol Lett. 1995;134:137–141. doi: 10.1111/j.1574-6968.1995.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 21.Levine M M. South America: the nature of cholera. Lancet. 1991;338:45–46. [Google Scholar]
- 22.Macrina F L, Kopecko D J, Jones K R, Ayers D F, McCowen S K. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1987;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 23.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Rajendran K, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray M G, Thompson W F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S8. Vol. 18. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 27.Nguyen V A. Cholera in Vietnam. In: Nguyen VA, editor. Medical microbiology. Ho Chi Minh City, Vietnam: Saigon Pasteur Institute; 1962. pp. 436–444. [Google Scholar]
- 28.Olsen J E, Larsen J L. Restriction fragment length polymorphism of the Vibrio anguillarum serovar O1 virulence plasmid. Appl Environ Microbiol. 1990;56:3130–3132. doi: 10.1128/aem.56.10.3130-3132.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsson-Liljequist B, Larsson P, Walder M, Miörner H. Antimicrobial susceptibility testing in Sweden. III. Methodology for susceptibility testing. Scand J Infect Dis Suppl. 1997;105:13–23. [PubMed] [Google Scholar]
- 30.Paulsen I T, Littlejoh T G, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popovic T, Bopp C A, Olsvik Ø, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 33.Ries A A, Vugia D J, Beingolea L, Palacios A M, Vasquez E, Wells J G, Baca N G, Swerdlow D L, Pollack M, Bean N H, Seminario L, Tauxe R V. Cholera in Piura, Peru: a modern urban epidemic. J Infect Dis. 1992;166:1429–1433. doi: 10.1093/infdis/166.6.1429. [DOI] [PubMed] [Google Scholar]
- 34.Rochelle P A, Fry J C, Day M J, Bale M J. An accurate method for estimating sizes of small and large plasmids and DNA fragments by gel electrophoresis. J Gen Appl Microbiol. 1985;132:53–59. doi: 10.1099/00221287-132-1-53. [DOI] [PubMed] [Google Scholar]
- 35.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 36.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1997;157:177–181. doi: 10.1111/j.1574-6968.1997.tb12770.x. [DOI] [PubMed] [Google Scholar]
- 37.Sharma C, Ghosh A, Dalsgaard A, Forslund A, Ghosh R K, Bhattacharya S K, Nair G B. Molecular evidence that a distinct Vibrio cholerae O1 biotype El Tor strain in Calcutta may have spread to the African continent. J Clin Microbiol. 1998;36:843–844. doi: 10.1128/jcm.36.3.843-844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada T, Nair G B, Deb B C, Albert M J, Sack R B. Outbreaks of Vibrio cholerae non-O1 in India and Bangladesh. Lancet. 1993;341:1347. doi: 10.1016/0140-6736(93)90855-b. [DOI] [PubMed] [Google Scholar]
- 40.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 41.Tabtieng R, Wattanasri S, Echeverria P, Seriwatana J, Bodhidatta L, Chatkeomorakot A, Rowe B. An epidemic of V. cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid coding for type II dihydrofolate reductase in Thailand. Am J Trop Med Hyg. 1989;41:680–686. doi: 10.4269/ajtmh.1989.41.680. [DOI] [PubMed] [Google Scholar]
- 42.Threlfall E J, Rowe B, Ferguson J L, Ward L R. Characterization of plasmids conferring resistance to gentamycin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J Hyg (London) 1986;97:419–426. doi: 10.1017/s0022172400063609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trach D D, Clemens J D, Ke N T, Thuy H T, Son N D, Canh D G, Hang P V D, Rao M R. Field trial of a locally produced, killed, oral cholera vaccine in Vietnam. Lancet. 1997;349:231–235. doi: 10.1016/s0140-6736(96)06107-7. [DOI] [PubMed] [Google Scholar]
- 44.Wright A C, Guo Y, Johnson J A, Nataro J P, Morris J G., Jr Development and testing of a non-radioactive DNA oligonucleotide probe that is specific for Vibrio cholerae cholera toxin. J Clin Microbiol. 1992;30:2302–2306. doi: 10.1128/jcm.30.9.2302-2306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae serogroup O1 bioptype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]