Abstract
S100 calcium binding protein A6 (S100A6) has been reported to involve in many kinds of cancers through regulating intracellular calcium homeostasis. Previous studies found that S100A6 increased in lung cancer patients' plasma and pleural effusion. This study focused on its function in Calu‐6 lung cancer cells. S100A6 gene was transferred into Calu‐6 lung cancer cell line by lentivirus vector, the empty vector transfected cells and the blank cells were set as control groups. MTT was evaluating cell proliferation. The transwell assay was reflecting cell migration and cell invasion. The flow cytometric analysis was detecting cell apoptosis and cell cycle of three groups (Calu‐6, Calu‐6/neo, Calu‐6/S100A6). Nude mouse tumorigenicity was then applied to evaluate S100A6's effect on cellular tumorigenicity. Compared with control groups, Calu‐6/S100A6 cells showed a weakening trend in the cell behaviours of proliferation, migration and invasiveness, while had an enhancement of cell apoptosis, with all P < .05. The cell cycle of Calu‐6/S100A6 cells had a reduction of S phase and an increase of G1 phase (P < .05). In animal study, after 5 weeks of cell injection, the tumour bulk of Calu‐6/S100A6 group was smaller than controls, with P < .05. Our results demonstrate S100A6 inhibits the growth of Calu‐6 lung cancer cells, as well as impairs Calu‐6's ability in tumorigenesis. At cellular level, S100A6 is supposed to act as a tumour suppressor gene in lung cancer.
Keywords: apoptosis, Calu‐6, migration, proliferation, S100A6
1. INTRODUCTION
Lung cancer is still the leading cause of cancer related mortality all over the world. As the global cancer statistics showed, lung cancer had an estimated 2.3 million new cases and caused an estimated 1.8 million deaths worldwide in 2020.1 For those air polluted countries, in particular, like India and China, lung cancer continues to be a dramatic threat to people's health, as well as a medical and financial burden.2 Histologically, lung cancer could be divided into non‐small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), the former one, accounting for around 85%, is the most common type and could be further classified into adenocarcinoma (38.5%), squamous carcinoma (30%) large cell carcinoma (2.9%), etc.3 Insidious onset of lung cancer, lack of typical symptoms and radiologically occult result in an advanced stage and poor prognosis when diagnosed. In recent years, the prognosis of some lung adenocarcinoma patients has been significantly improved due to widespread use of targeted therapies against diver mutations, for example, epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase rearrangement.4 However, for those patients without diver mutation genes who present with metastatic disease, including part of adenocarcinoma patients and a majority of squamous carcinoma patients, platinum‐based chemotherapy remains the first‐line treatment and the prognosis is still poor.5 Therefore, exploring the molecular mechanism involved in the occurrence of lung cancer is expected to determine the effective treatment target and improve the outcome of lung cancer.6
S100A6, is an important member of S100 protein family, has been confirmed to have a relationship with a variety of tumours.7 Currently, this calcium binding protein family is known to have 25 members, at least 16 of which are located in epidermal differentiation complex (EDC) on chromosome 1q21‐ an area prone to chromosomal rearrangements.8 This instability leads to the occurrence of many tumours. In lung cancer, S100A6 was found to have an abnormal expression in tumour cell lysates, plasma and pleural effusion samples of patients.9, 10 In addition, high S100 protein peak indicates a long median survival period. In this study, we plan to confirm the effects of S100A6 on the biological behaviours of lung cancer cell line, and explore the possible molecular mechanism of this malignancy.
2. MATERIALS AND METHODS
2.1. Cell culture
We purchased the lung cancer cell line Calu‐6 from Chinese Academy of Sciences, preserved them at 37°C and 5% CO2. Cells were cultured and expanded for 1 to 2 weeks in a medium containing high glucose DMEM (Gibco, USA) combined with 15% heat‐inactivated foetal bovine serum (FBS, Zhengbo, Beijing, China) and 100 U/mL streptomycin/penicillin (Gibco, USA).
2.2. pLVX‐AcGFP1‐N1‐S100A6 construction
The S100A6 gene originated from human, and we referred to the website of “https://www.ncbi.nlm.nih.gov/gene/6277” for its detailed information. HindIII/EcoRI was added as the restriction site, and then we designed the primers. The upstream primer information was 5‐CCCAAGCTTACCATGGCATGCCCCCTGGTCA‐3 and the sequence of downstream primer was 5‐CGGAATTCCGGTCAGCCCTTGAGGGCTTCATT‐3. The target gene was amplified by the S100A6 template. It was digested, linked and transformed with the carrier pcDNA3.1‐eGFP‐MCS, and the monoclonal plasmids were then selected. The size (273 bp + 900 bp = 1173 bp), which was identified by the method of PCR, was consistent with our expectation.
2.3. Production and concentration of lentivirus vector
Plvx‐acgfp1‐n1‐S100A6, pspax2 and pmd2.G were co‐transfected into 293 T cells in ratio 2:1:1. After 72 hours, we observed the fluorescence expression under microscope, collected the supernatant and filtered it by a PVDF (polyvinylidene fluoride) filter (0.45 μm, Millipore). Thirty‐fold concentrated stock was obtained using an ultracentrifugation under 4°C at 50 000 g for 150 minutes prior to be resuspended in PBS and stored at −80°C.
2.4. Cell's infection
Calu‐6 cells in logarithmic growth phase were collected, adjusted to a density of 1*106 mL by trypsin digestion and planted with a quantity of 2 mL per well in six‐well plates (JET BIOFIL, Guangzhou, China). Six well plates were then placed in the incubator for further culture. When cell convergence reached 80%‐90%, S100A6 over‐expressed lentivirus (MOI = 10), as well as empty lentivirus were separately added, and all the operations were strictly carried out following up kit instructions. We divided the cells into three groups, namely Calu‐6/S100A6, Calu‐6/neo and Calu‐6. After 48 hours, we used fluorescence microscopy to observe the infection efficiency, which was also detected accurately by QPCR and WB (western blot).
2.5. RNA extraction and QPCR
Sample processing: using TRIzol reagent (Invitrogen, USA) for cell lysis, after centrifugation, supernatant was collected. Chloroform, 2‐propanol and 75% ethanol were added into the supernatant sequentially to get purified RNA. After the extraction, we examined the purity and concentration of RNA by Nanodrop 2000 (Thermo, USA). cDNAs were synthesized from 2 mL RNA using an oligo dT primer and the RevertAid First Strand cDNA Synthesis Kit (Thermo, USA) as instructed by the manufacturer. PCR process was divided into four steps: incubation (95°C, 5 minutes), denaturation (94°C, 30 seconds for 44 circles), annealing (55°C, 30 seconds) and extension (72°C, 30 seconds). Primers' specific information was listed as follows: S1006 forward primer (5‐GGGAGGGTGACAAGCACAC‐3) and S1006 reversed primer (5‐AGCTTCGAGCCAATGGTGAG‐3); glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH, an endogenous control) forward primer (5‐GGAGCGAGATCCCTCCAAAAT‐3) and GAPDH reversed primer (5‐GGCTGTTGTCATACTTCTCATGG‐3).
2.6. Western blot
Western blot analysis was used to determine the expression of target protein. Cell extracts were acquired by a Total Protein Extraction Kit from infected cells and spun in a microfuge for 10 minutes at 14 000 rpm at 4°C. The whole cell lysates, which were first added to loading sample buffer, were then heated at 95°C for 5 minutes, separated by 10% sodium dodecyl sulphate‐polyacrylamide and terminally transferred to a nitrocellulose membrane. Membranes were pre‐incubated for 1 hour with 5% non‐fat milk, and then incubated with primary antibody against S100A6 (MAB769Hu22, USCN Life Sciences Inc., Wuhan, China, 1/500 dilution) at 4°C overnight. The control group, with the mouse GAPDH antibody, was set up at the same time. After washing three times at 25°C in TBST (tris buffered saline tween), the membranes were incubated with the horseradish peroxidase‐conjugated secondary antibodies (LAB769Hu71, USCN Life Sciences Inc., Wuhan, China, 1/5000 dilution). Immunoreactivity was visualized using enhanced chemiluminescence system.
2.7. MTT colorimetric assay
Cells of three groups, with a concentration of 5‐10 × 104/mL and a quantity of 100 μL per well, were seeded into every well of 96‐well plates. MTT assay was performed to analyse cell proliferation. In general, a total of 10 μL MTT stock solutions was added to every well and cells were incubated at 37°C for 4 hours before adding 110 μL Formazan solution. Next, the absorbance of each well was detected by ELISA (enzyme linked immunosorbent assay) at 490 nm. Proliferation rates were tested at 24, 36, 48, 60 and 72 hours, respectively, after transfection.
2.8. Transwell migration and invasion assay
Transwell assay was applied to investigate S100A6's function in cell migration and invasion in vitro. Briefly, cells of three groups (each 100 μL), which were pre‐adjusted to a density of 1*106/mL, were placed in the upper chamber of six‐well transwell plates. Matrigel‐uncoated plates were used to evaluate cell migration, while matrigel‐coated plates were used for cell invasion detection, and all of them consisted of 8.0 μm pore size filters. Lower chamber was added 500 μL dulbecco's modified eagle medium (DMEM) containing 10% FBS, at the same time, bubbles should be avoided in the lower chamber. After incubation (37°C, 48 hours), lower chamber cells were fixed with 4% paraformaldehyde in PBS (room temperature, 10 minutes), and stained with 1% Crystal Violet (room temperature, 30 minutes). The migration and invasion cells were counted and took photography under the 100‐fold optical microscope. We calculated the cells' relative number in five separate fields.
2.9. Flow cytometry
Flow cytometry was performed to examine cell apoptosis and cell cycle. We collected those logarithmic phase cells, adjusted them to a density of 1*106/mL, seeded them in six‐well plates (2 mL per hole) and put the plates to incubator for 48 hours. Cells were then washed twice by cool PBS, centrifuged at 1000 rpm for 5 minutes at 4°C and 1‐5*105 of them were taken, mixed with 100 μL binding buffer after absorbing PBS. Next, cells were incubated at 4°C in the dark for 15 minutes at the presence of 10 μL Annexin V‐FITC and 5 μL of PI staining solution. The mixture was added to 400 μL binding buffer before being analysed by FACSCalibur system in 1 hour. In the detection of cell cycle, cells were fixed in 75% ethanol for 1 hour after centrifugation, then treated with 100 μL RNaseA solution, resuspended and bathed in water at 37°C for 30 minutes. Cell precipitation was then stained by PI dyeing solution (400 μL) in the dark at 4°C for 30 minutes, analysed by Cell Quest and MidFit software at an excitation wavelength of 488 nm. The cell cycle is mainly composed of three phases ‐ G1, S and G2.
2.10. Animal experiment
For the mouse experiment, we used 8‐ to 10‐weeks‐old Balb/c‐nude mice obtained from USCN Life Sciences Inc. Fifteen mice were grouped into Calu‐6 (four mice), Calu‐neo (four mice) and Calu‐6/S100A6 (seven mice), respectively. To analyse the influence of S100A6 to tumour formation, 2.5*106/L stably transfected Calu‐6/S100A6 cells (supplemented in 200 μL PBS) were injected into the mice's right flank. The same amount of Calu‐6 and Calu‐6/neo cells were injected to the controls simultaneously. We measured tumour volume in every mouse weekly, recorded the data. After 5 weeks, mice were sacrificed, and the tumour was removed, and photographed.
2.11. Statistical analysis
We selected GraphPad Prism 7.0 for making histogram, line chart and data analysis and used SPSS 22.0 to analyse the data. Every group of cells had two replicates and the final data were the average values of these three groups. The values in the histograms were showed with the style of average ± SE. In the migration figure (Figure 3) and invasion figure (Figure 4), the number of cells in the fourth quadrant of each field was counted in each group. The cell counts of three groups and their replicates were statistically analysed. The histogram was displayed in the form of mean ± SE using GraphPad Prism 7.0. In Figure 5, the proportion of apoptotic cells was counted as the sum of the proportion of the first quadrant and the fourth quadrant of each group and their repeated groups. The average value was taken, and the histogram was displayed in the form of average ± SE with GraphPad Prism 7.0. One way analysis of variance (ANOVA) was used to compare differences of the data among three groups after the confirmation of the normal distribution, and rank sum test was used to analyse the differences of the data that do not conform the normal distribution. We assessed the differences among three groups in MTT proliferation, cell cycle and animal experiment using two‐way ANOVA test. A two‐tailed P < .05 was considered statistically significance.
FIGURE 3.
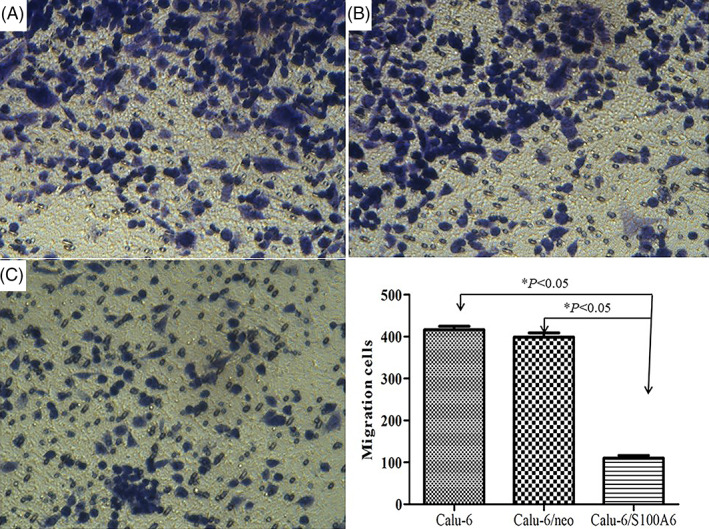
Transwell assay indicated the migration ability of Calu‐6/S100A6 cells decreased compared with Calu‐6 cells (418.6 ± 8.38) and Calu‐6/neo cells (395.7 ± 10.29)
FIGURE 4.
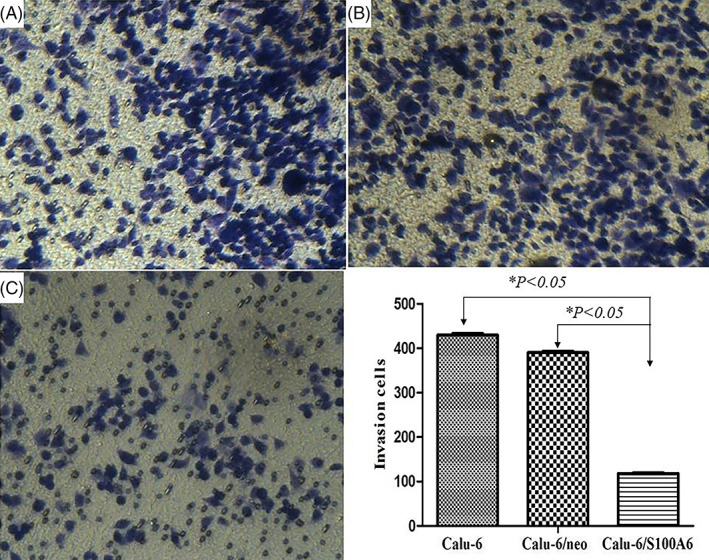
Transwell assay showed that the invasion capacity of Calu‐6/S100A6 cells reduced in contrast with control groups, with 118.2 ± 2.99 (Calu‐6/S100A6), 430.8 ± 7.12 (Calu‐6) and 390.5 ± 5.83 (Calu‐6/neo)
FIGURE 5.
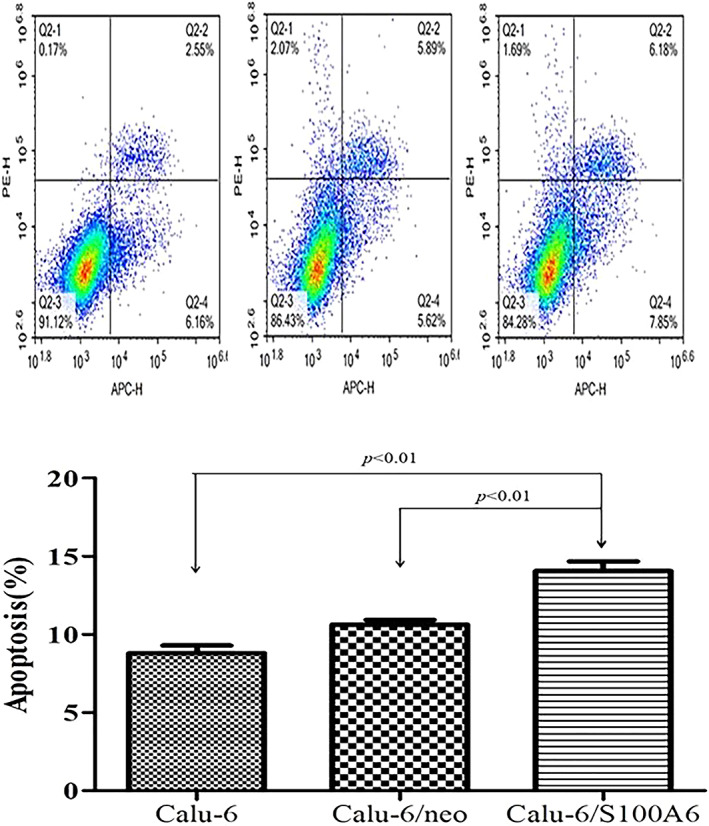
Flow cytometric analysis demonstrated the percentages of apoptotic cells. Calu‐6/S100A6 group had a higher apoptotic cells mean proportion of (13.88 ± 0.71)% than its counterparts ‐(8.86 ± 0.49)% and (10.11 ± 0.36)%, respectively
3. RESULTS
3.1. S100A6's expression at mRNA and protein levels after transfection
The infection efficiency was illustrated by fluorescence microscope, RT‐PCR method and western blot. As shown in Figure 1, S100A6 had an obviously elevated expression at mRNA level (Figure 1A) and protein level (Figure 1B) in the Calu‐6/S100A6 group, but not in the control groups, indicating a high transfection efficiency. In addition to that, Calu‐6 cells were observed having a weak green fluorescence after the transfection, as listed in Figure 1C, also suggesting a successful transfection.
FIGURE 1.

S100A6's expression in three groups after transfection. A, mRNA level of S100A6 was detected positive in Calu‐6/S100A6 cells by RT‐PCR. B, The protein level of S100A6 was detected positive in Calu‐6/S100A6 cells using Western‐blot. GAPDH expression was used as a control in the same panel. C, Calu‐6 cells were observed having a weak green fluorescence after the transfection
3.2. S100A6 inhibited cells' proliferation, migration and invasion
S100A6's effects on the proliferation of cells were measured by MTT method. The Figure 2 gave us the information that, Calu‐6/S100A6 cells had a downward trend of proliferating in comparison of control groups, with a P < .05.
FIGURE 2.
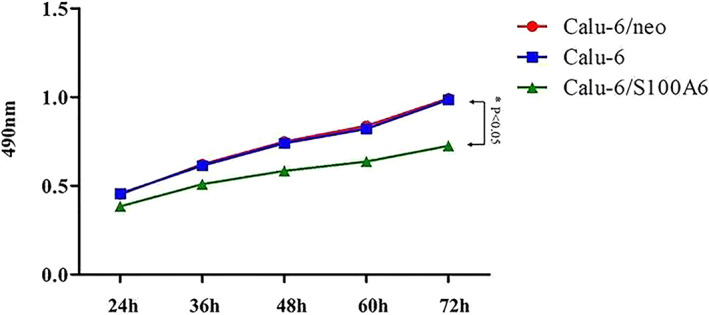
MTT assay determined Calu‐6/S100A6 cells had a downward trend of proliferating in comparison of control groups
Transwell assay was used to examine the functions of S100A6 on Calu‐6 cells' biological behaviours, including cell migration and invasion. As listed in Figure 3, with a mean cell count of 110.6 ± 6.36, the migration ability of Calu‐6/S100A6 cells decreased markedly compared with Calu‐6 cells (418.6 ± 8.38) and Calu‐6/neo cells (395.7 ± 10.29), the P‐value was less than .05. Meanwhile, the invasion capacity of Calu‐6/S100A6 cells reduced in contrast with control groups, with 118.2 ± 2.99 (Calu‐6/S100A6), 430.8 ± 7.12 (Calu‐6) and 390.5 ± 5.83 (Calu‐6/neo), respectively, the difference was statistically significant (P < .05), as showed in Figure 4.
3.3. S100A6 enhanced apoptosis and suppressed the cell cycle of Calu‐6 cells
We applied Flow cytometry assay to assess S100A6' effects on cell apoptosis and cell cycle. Results are listed in Figure 5, with a apoptotic cells mean proportion of (13.88 ± 0.71)%, the Calu‐6/S100A6 group had a stronger apoptotic ability than its counterparts ‐(8.86 ± 0.49)% and (10.11 ± 0.36)%, respectively (P < .05).
As for the changes of cell cycle after transfection, the proportions of Calu‐6/S100A6 cells in G1 phase and S phase accounted for (75.16 ± 0.78)% and (8.69 ± 0.84)%, respectively, which differed significantly from those in Calu‐6 cells (60.98 ± 0.64, 21.06 ± 0.70) and Calu‐6/neo cells (58.97 ± 0.18, 21.98 ± 1.80), indicating that S100A6 suppressed the cell cycle and the transition from G1 to S phase of Calu‐6 cells. Figure 6 listed the data, and the differences were dramatically significant (P < .05).
FIGURE 6.
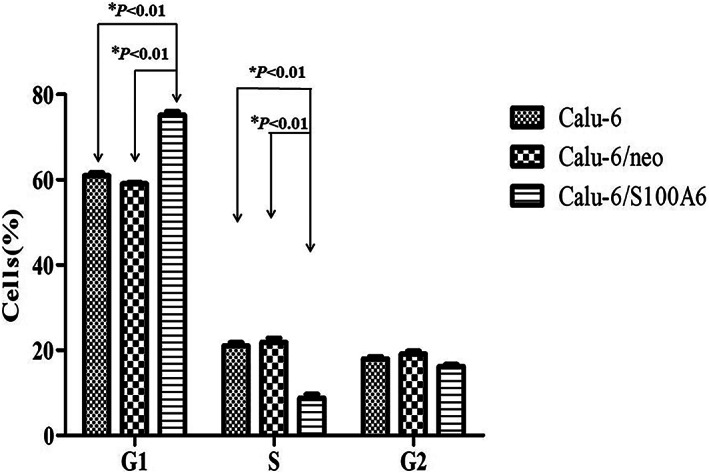
Flow cytometric analysis showed change in cell cycle. The proportions of Calu‐6/S100A6 cells in G1 phase and S phase accounted for (75.16 ± 0.78)% and (8.69 ± 0.84)%, respectively, differing from those in Calu‐6 cells (60.98 ± 0.64, 21.06 ± 0.70) and Calu‐6/neo cells (58.97 ± 0.18, 21.98 ± 1.80)
3.4. S100A6 suppressed tumour formation of Calu‐6 cells
The results of animal test were showed in Figure 7. After 2 weeks of injection, over time, the gap between Calu‐6/S100A6 group and control groups tumour volumes was widening. Specifically, at the end of the third week, the tumour volume in three groups was 32.86 ± 1.87 mm3 (Calu‐6), 31.71 ± 3.91 mm3 (Calu‐6/neo) and 7.97 ± 2.97 mm3 (Calu‐6/S100A6), respectively. Until the last day of the fourth week, compared with the groups of Calu‐6 (147.36 ± 17.48 mm3) and Calu‐6/neo (153.84 ± 16.93 mm3), the tumour size of Calu‐6/S100A6 group was obviously small, being 56.08 ± 8.95 mm3. After 5 weeks, the differences among three groups further expanded; in particular, the tumour bulk of Calu‐6/S100A6 group was 112.53 ± 15.19 mm3, which was smaller than that of Calu‐6 group (318.21 ± 53.27 mm3) and Calu‐6/neo group (332.49 ± 52.95 mm3), with P < .05.
FIGURE 7.
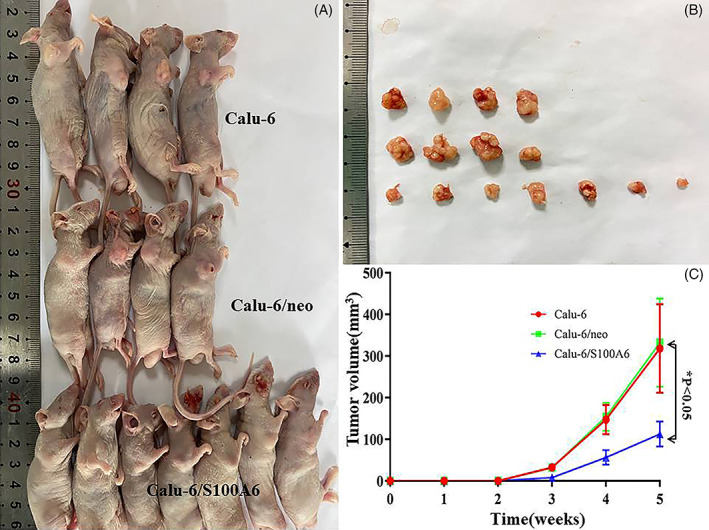
Animal experiments showed different tumour volume in three groups. After 5 weeks, the tumour bulk of Calu‐6/S100A6 group was 112.53 ± 15.19 mm3; it was smaller than that of Calu‐6 group (318.21 ± 53.27 mm3) and Calu‐6/neo group (332.49 ± 52.95 mm3)
All the results at the cellular level had three replications and were displayed as mean ± SD.
4. DISCUSSION
In this study, we investigated the role of S100A6 played in lung cancer cell line by transfecting Caulu‐6 cells with S100A6 overexpression lentivirus vector and injecting them to nude mice. Through comparing cells' behaviours and mice's tumour volume among three groups with different interventions, we believed overexpressed S100A6 has a close relationship with decreased cell proliferation, migration, invasion, cell cycle, tumorigenicity and an enhanced apoptosis of lung cancer cell, suggesting this protein may inhibit the genesis and development of this malignancy.
The S100 protein family, which is the largest subfamily of calcium‐binding proteins, has been proved to get involved in a variety of biological processes of multiple tumours.11 In this family, S100A6 is the first one to be found to have a close association with cellular proliferation, and has been identified to participate in tumour progression and invasion.12 For example, in cervical cancer, investigators demonstrated that S100A6 overexpression promoted the metastatic ability and epithelial‐mesenchymal transition of HeLa and CaSki cells, whereas had no significant effect on apoptosis.13 In gastric cancer, Wang et al found S100A6 had a remarkable increased expression in patients' tissues, and this overexpression had a clear correlation with high invasive cell behaviours, including positive lymph node involvement as well as vascular invasion.14 Similar conclusions were also reached in pancreatic cancer and human nasopharyngeal carcinoma.15, 16 However, S100A6, along with another member‐S100A1, was considered to be associated with better prognosis in breast cancer patients.17
In lung cancer, the action mechanism of S100A6 is suggested to be complex and it has an altered expression in different subtypes. It was reported that S100A6 has a higher expression in lung adenocarcinoma mixed subtype with bronchioloalveolar carcinoma (BAC) component than in pure BAC. Ishii et al assessed S100A6 expression in tumour cells' nucleus and cytoplasm of 92 lung adenocarcinoma using immunohistochemistry, and data analysis showed invasive tumour, compared with normal lung tissue and non‐advanced lesions, had significantly higher cytoplasmic S100A6 expression, but have no obvious nuclear S100A6 immunoreactivity. This trend was more obvious in adenocarcinoma with a BAC component.18 In lung squamous cell cancer (SCC), investigators detected the expression of S100A6 in 177 patients and further divided the cohort into two groups according to S1006's level. Data showed that S100A6 was an independent risk factor for the prognosis of SCC, predicting unfavourable prognosis.19 However, a separate investigation came to opposite conclusion. In the study of De Petris et al, the presence of post‐translationally modified S100A6 was confirmed in tumour cell lysates, as well as pleural effusion and plasma samples. In addition, high S100A6 peak intensity was demonstrated to be related with longer median survival (35 months for high peak intensity vs 18 months for low peak intensity).20 Moreover, S100A6 tended to have a lower expression in squamous histology than other subtypes. As to the SCLC, S100A6 was identified to be under expressed as compared with NSCLC and normal lung tissue.21 These contradictory research results suggested S100A6 may take on various functions in different pathological subtypes of lung cancer. At the cellular level, compared with previous research, our study drew an opposite conclusion. Investigators analysed the S100A6 expression in the human bronchial epithelioid cell lines (HBE) and NSCLC cell lines (A549, H441 and H1975), finding that S100A6 had the highest expression in A549 cells. Moreover, subsequent experiment indicated that S100A6 overexpression could promote the invasion, migration, proliferation and angiogenesis of A549 lung cancer cells by inhibiting the P53 acetylation.22 The main reason may be that A549 belongs to lung adenocarcinoma cell line, while Calu‐6 belongs to undifferentiated lung cancer cell line. This also suggested that S100A6 may play a different role in different subtypes of lung cancer. This is not the first contradiction in S100 protein family. S100A2, as another important member in S100 protein family, has been proved to play inconsistent roles in lung cancer.23, 24, 25 We will demonstrate the exact effect on diverse lung cancer cell types using multiple SCC cell lines and adenocarcinoma cell strains in our future experiments.
There is still lack of researches on the mechanism of S100A6 in lung cancer, only a small amount of experiments confirmed that S100A6 has an interaction with P53, promotes its transcriptional activity, consequently facilitating the apoptotic activity of this antioncogene.26, 27 In future, we need more in‐depth researches to clarify the exact mechanism of S100A6 in lung cancer.
In conclusion, our experiments identify S100A6 played an inhibitive role in the carcinogenesis and progress of Calu‐6 cell line via inhibiting proliferation, migration, invasion and enhancing apoptosis as well as suppressing cell division. In future, we will illustrate its function using more kinds of lung cancer cell lines.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Wang T, Han S, Du G. S100A6 represses Calu‐6 lung cancer cells growth via inhibiting cell proliferation, migration, invasion and enhancing apoptosis. Cell Biochem Funct. 2021;39(6):771–779. 10.1002/cbf.3639
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM, Xu R‐H. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond). 2019;39:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez‐Canales J, Parra‐Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Treat Res. 2016;170:25‐46. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz‐Patiño A, Castro CD, Ricaurte LM, et al; Latin‐American Consortium for the Investigation of Lung Cancer (CLICaP). EGFR amplification and sensitizing mutations correlate with survival in lung adenocarcinoma patients treated with erlotinib (MutP‐CLICaP). Target Oncol. 2018;13:621‐629. [DOI] [PubMed] [Google Scholar]
- 5.Tagliamento M, Genova C, Rijavec E, et al. Afatinib and Erlotinib in the treatment of squamous‐cell lung cancer. Expert Opin Pharmacother. 2018;19:2055‐2062. [DOI] [PubMed] [Google Scholar]
- 6.Aguiar PN Jr, De Mello RA, Hall P, Tadokoro H, de Lima Lopes G. PD‐L1 expression as a predictive biomarker in advanced non‐small‐cell lung cancer: updated survival data. Immunotherapy. 2017;9:499‐506. [DOI] [PubMed] [Google Scholar]
- 7.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357‐364. [DOI] [PubMed] [Google Scholar]
- 8.Ma N, Zhu L, Yang L, Cui Y, Zhan Y. Prognostic values of S100 family mRNA expression in ovarian cancer. Cancer Biomark. 2019;25:67‐78. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Huo X, Chong Z, et al. A review of S100 protein family in lung cancer. Clin Chim Acta. 2018;476:54‐59. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Liang Y, Thakur A, et al. Diagnostic significance of S100A2 and S100A6 levels in sera of patients with non‐small cell lung cancer. Tumour Biol. 2016;37:2299‐2304. [DOI] [PubMed] [Google Scholar]
- 11.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15:96‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donato R, Sorci G, Giambanco I. S100A6 protein: functional roles. Cell Mol Life Sci. 2017;74:2749‐2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A, Gu Y, Li X, et al. S100A6 promotes the proliferation and migration of cervical cancer cells via the PI3K/Akt signaling pathway. Oncol Lett. 2018;15:5685‐5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XH, Du H, Li L, et al. Increased expression of S100A6 promotes cell proliferation in gastric cancer cells. Oncol Lett. 2017;13:222‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohuchida K, Mizumoto K, Ishikawa N, et al. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11:7785‐7793. [DOI] [PubMed] [Google Scholar]
- 16.Li A, Shi D, Xu B, et al. S100A6 promotes cell proliferation in human nasopharyngeal carcinoma via the p38/MAPK signaling pathway. Mol Carcinog. 2017;56:972‐984. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Wang Z, Liu W, et al. Distinct prognostic values of S100 mRNA expression in breast cancer. Sci Rep. 2017;7:39786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii A, Suzuki M, Satomi K, et al. Increased cytoplasmic S100A6 expression is associated with pulmonary adenocarcinoma progression. Pathol Int. 2009;59:623‐630. [DOI] [PubMed] [Google Scholar]
- 19.He X, Xu X, Khan AQ, Ling W. High expression of S100A6 predicts unfavorable prognosis of lung squamous cell cancer. Med Sci Monit. 2017;23:5011‐5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Petris L, Orre LM, Kanter L, et al. Tumor expression of S100A6 correlates with survival of patients with stage I non‐small‐cell lung cancer. Lung Cancer. 2009;63:410‐417. [DOI] [PubMed] [Google Scholar]
- 21.Lee HS, Park JW, Chertov O, et al. Matrix‐assisted laser desorption/ionization mass spectrometry reveals decreased calcylcin expression in small cell lung cancer. Pathol Int. 2012;62:28‐35. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Lv X, Zhang Z, Xie S. S100A6/miR193a regulates the proliferation, invasion, migration and angiogenesis of lung cancer cells through the P53 acetylation. Am J Transl Res. 2019;11:4634‐4649. [PMC free article] [PubMed] [Google Scholar]
- 23.Feng G, Xu X, Youssef EM, Lotan R. Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis. Cancer Res. 2001;61:7999‐8004. [PubMed] [Google Scholar]
- 24.Heighway J, Knapp T, Boyce L, et al. Expression profiling of primary non‐small cell lung cancer for target identification. Oncogene. 2002;21:7749‐7763. [DOI] [PubMed] [Google Scholar]
- 25.Smith SL, Gugger M, Hoban P, et al. S100A2 is strongly expressed in airway basal cells, preneoplastic bronchial lesions and primary non‐small cell lung carcinomas. Br J Cancer. 2004;91:1515‐1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Króliczak W, Pietrzak M, Puzianowska‐Kuznicka M. P53‐dependent suppression of the human calcyclin gene (S100A6): the role of Sp1 and of NFkappaB. Acta Biochim Pol. 2008;55:559‐570. [PubMed] [Google Scholar]
- 27.Słomnicki ŁP, Nawrot B, Leśniak W. S100A6 binds p53 and affects its activity. Int J Biochem Cell Biol. 2009;41:784‐790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


