Abstract
The closely related species Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, and Haemophilus paraphrophilus are common findings in oral microbiota. The aims of this study were to evaluate the applicability of the Rapid NH and API ZYM kits and arbitrarily primed PCR (AP-PCR) in the identification and differentiation of the three species from each other. The material included 62 clinical isolates and three reference strains of A. actinomycetemcomitans representing the 5 serotypes and 18 AP-PCR genotypes. Haemophilus species included 12 clinical isolates and 11 reference strains of H. aphrophilus, H. paraphrophilus, and 5 other species. For the PCR amplification, the oligonucleotide 5′-CAGCACCCAC-3′ was used as a primer. Contrary to the consistent performance of API ZYM, the Rapid NH system was able to identify only 10 of 65 (15%) A. actinomycetemcomitans isolates, whereas all Haemophilus species were correctly identified. The API ZYM test differentiated A. actinomycetemcomitans from H. aphrophilus and H. paraphrophilus by negative β-galactosidase and α-glucosidase reactions and a positive esterase lipase reaction. However, the API ZYM test was unable to differentiate H. aphrophilus from H. paraphrophilus, it also could not differentiate A. actinomycetemcomitans serotypes from each other. Among the H. aphrophilus isolates three AP-PCR genotypes and among H. paraphrophilus isolates only one AP-PCR genotype, distinct from those of A. actinomycetemcomitans, were found. The Rapid NH test showed poor ability to identify clinical isolates of all A. actinomycetemcomitans serotypes. Moreover, AP-PCR genotyping proved to be a rapid method for the species differentiation of A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus.
The closely related Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, and Haemophilus paraphrophilus are gram-negative, facultatively anaerobic, and nonmotile coccobacilli that are frequently recovered from healthy and diseased oral cavities and may cause various extraoral infections, such as endocarditis, and abscesses in brain, neck, and lungs (1, 27, 32). During the past few decades A. actinomycetemcomitans has been implicated as a major periodontal pathogen, occurring in 30 to 90% of patients with adult and juvenile forms of periodontitis (10, 33). Based on differences in the carbohydrate moiety of cell surface lipopolysaccharide A. actinomycetemcomitans can be grouped into five serotypes (a to e) (11, 31). It has been suggested that certain serotypes are associated with specific forms of periodontitis, extraoral infections, and periodontal health (1, 11, 32). Although H. aphrophilus and H. paraphrophilus have also been isolated from periodontal pockets, they do not play a significant role in the etiology and pathogenesis of periodontal diseases (12). Therefore, the correct identification of A. actinomycetemcomitans in clinical samples is becoming increasingly important for screening, treatment planning, and monitoring of periodontal diseases (9). Although distinguishing H. aphrophilus from H. paraphrophilus may have limited clinical importance due to the infrequent occurrence of invasive infections, a species level identification is required for epidemiological characterization.
Differentiating between A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus by culture methods may be difficult due to the similarities in cell morphology, growth requirements, and colony morphology displayed on the commonly used selective culture medium for A. actinomycetemcomitans, tryptic soy-bacitracin-vancomycin agar (25). Additionally, identification problems may arise from inconsistent biochemical reactions: the presumptive differentiation of A. actinomycetemcomitans from Haemophilus species is generally based on the positive catalase test and the ability to ferment sucrose and lactose (25, 26). However, some A. actinomycetemcomitans strains are catalase negative, and some H. aphrophilus strains are catalase positive (3, 32, 28, 29). The V factor (NAD) requirement for the growth of H. paraphrophilus is the only criterion that differentiates this species from H. aphrophilus (27). However, this feature can be lost in individual strains or be falsely obviated by medium carryover effect (27). Therefore, in clinical laboratories A. actinomycetemcomitans may be overlooked, or H. aphrophilus and H. paraphrophilus be misidentified as A. actinomycetemcomitans. Additionally, intraspecies variation of A. actinomycetemcomitans may cause confusion in presumptive identification, e.g., there are limited data available on the biochemical characteristics of the five A. actinomycetemcomitans serotypes, especially of the recently found serotypes d and e (11, 21).
The aims of this study were to determine whether A. actinomycetemcomitans serotypes differ regarding biochemical reactions used for species identification and also to evaluate the applicability of the Rapid NH and API ZYM kits and of AP-PCR genotyping for the identification and species differentiation of A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus isolates.
MATERIALS AND METHODS
The study material comprised a total of 65 A. actinomycetemcomitans isolates, including 62 clinical isolates and three reference strains (ATCC 29523 serotype a, ATCC 43718 serotype b, and NCTC 9710 serotype c). The Haemophilus isolates included 12 clinical isolates and 11 reference strains (CCUG 3715, ATCC 13252, ATCC 19415, CCUG 1802, CCUG 23945, ATCC 49766, CCUG 12836, MQCD 1947, CCUG 12834, CCUG 3716, and CCUG 10787) of 7 Haemophilus species (H. aphrophilus, H. paraphrophilus, H. influenzae, H. parainfluenzae, H. haemolyticus, H. parahaemolyticus, and H. segnis).
The identification of clinical isolates of A. actinomycetemcomitans, H. aphrophilus, H. paraphrophilus, H. influenzae, and H. parainfluenzae were carried out by established methods (4, 21). The A. actinomycetemcomitans isolates were serotyped as previously described (21) by using an immunodiffusion assay. Polyclonal serotype-specific rabbit antisera raised against the five serotypes (a to e) served as antibody, and the antigen extracts were prepared by autoclaving harvested A. actinomycetemcomitans cells. The 65 A. actinomycetemcomitans isolates represented the 5 serotypes (6 serotype a, 10 serotype b, 7 serotype c, 3 serotype d, and 38 serotype e isolates and 1 nonserotypeable isolate) and 18 AP-PCR genotypes (2, 7).
The isolates were revived from cultures frozen at −70°C in skim milk and subcultured three times on enriched blood agar and chocolate agar medium prior to biochemical testing.
Rapid NH test.
All A. actinomycetemcomitans isolates (n = 65) and all Haemophilus isolates (n = 23) were tested by the Rapid NH system. The Rapid NH system (Innovative Diagnostic Systems, Inc., Norcross, Ga.) is a qualitative micromethod based on the detection of preformed bacterial enzymes. Dehydrated reactants are included for the following tests: hydrolysis of amide substrates proline p-nitroanilide and γ-glutamyl p- nitroanilide; fermentation of glucose and sucrose; hydrolysis of o-nitrophenyl-β-d-galactoside; hydrolysis of fatty acid ester, resazurin, urea, and p-nitrophenyl phosphate; utilization of ornithine; utilization of tryptophane to form indole; and reduction of nitrate to nitrite and of nitrate to nitrogen. The Rapid NH panels were inoculated, incubated, and interpreted according to the manufacturer’s instructions. Bacterial cells from fresh cultures were suspended in 1 ml of Rapid NH inoculation fluid to the recommended turbidity (McFarland no. 3 standard), and the entire contents of the tube were transferred to the panel. After incubation in air at 37°C for 4 h, the color reactions were scored before and after reagent addition.
Nitrate broth test.
The indole-nitrate test was performed for the A. actinomycetemcomitans serotypes a, b, and e isolates (n = 15) which were nitrate negative in the Rapid NH test system. Indole-nitrate medium was prepared as previously described (15). The fresh A. actinomycetemcomitans isolates from enriched blood agar and Haemophilus isolates from chocolate agar were inoculated into indole-nitrate medium and incubated in air at 37°C for 2 days. The test was carried out as previously described (15).
Serum sugar fermentation.
Sucrose fermentation was carried out in 2 ml of phenol red broth (13) supplemented with 10% rabbit serum and 1% sucrose for the A. actinomycetemcomitans serotype d and e isolates (n = 18) which were sucrose-positive in the Rapid NH test system. Heavy suspension (corresponding to a McFarland no. 6 turbidity standard) was prepared from test strains, and 160 μl of the specimen was inoculated into the phenol red broth tube. The tubes were incubated in air at 35°C up to 9 days. A yellow color was interpreted as a positive fermentation reaction, and a reddish-pink color was considered a negative fermentation reaction.
API ZYM test.
A total of 34 A. actinomycetemcomitans isolates (6 serotype a, 10 serotype b, 7 serotype c, 1 serotype d, 12 serotype e, and 1 nonserotypeable) and all isolates (n = 23) of the seven Haemophilus species were tested by using the API ZYM system. The API ZYM system (bioMérieux Vitex, Inc., St. Louis, Mo.) is a micromethod that allows semiquantitative and rapid determination of 19 enzymatic reactions (alkaline phosphatase, esterase [C4], esterase lipase [C8], lipase, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase). The API ZYM strips were inoculated, incubated, and interpreted according to the manufacturer’s instructions. After inoculation each cupule of strips with 65 μl of a dense suspension (McFarland no. 5 or 6 standards) in water, the panel was incubated in air at 37°C for 4 h. Then, the reagents were added and reactions were interpreted. If poor reactivity was scored after 4 hours in order to confirm scoring the color intensity was checked again after an overnight incubation. Color reactions were scored from 0 to 5 according to a reference color chart supplied by the manufacturer. Color reaction grade 0 was interpreted to correspond to a negative reaction; grades 1 and 2 corresponded to a weak reaction (5 to <20 nmol), and grades 3, 4, and 5 corresponded to a strong reaction (>20 nmol). To confirm the reproducibility of the enzymatic reactions, 12 serotype e isolates and reference strains of serotypes a, b, and c were examined twice on different days from different subcultures.
AP-PCR genotyping.
For the AP-PCR amplification, A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus chromosomal DNA was extracted as previously described (22). The 50-μl PCR reaction volume consisted of 0.2 mM deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 2.5 U of AmpliTaq (Perkin-Elmer Cetus, Norwalk, Conn.), 5 μl of template DNA (1:100 to 1:500 dilution), and 0.4 μM primer OPA-13 (5′-CAGCACCCAC-3′). The amplification was carried out in a thermocycler (Perkin-Elmer) as previously described (2). Amplification products were analyzed electrophoretically in 1% (wt/vol) agarose gel stained with ethidium bromide (0.5 μg/ml) and photographed under UV light. The banding patterns were analyzed visually.
RESULTS
Rapid NH test.
The Rapid NH test results of 65 A. actinomycetemcomitans isolates of all serotypes and genotypes are shown in Table 1. For comparisons, the Rapid NH differential chart obtained for A. actinomycetemcomitans is included. The Rapid NH system was unable to identify 55 of 65 (85%) A. actinomycetemcomitans isolates due to false reactions to proline p-nitroanilide (n = 51), sucrose (n = 18), fatty acid ester (n = 42), resazurin (n = 38), p-nitrophenyl phosphate (n = 20), and nitrate reduction (n = 15). In other words, the Rapid NH system was unable to identify 5 of 6 (83%) A. actinomycetemcomitans serotype a, 1 of 3 (33%) serotype d, 31 of 38 (82%) serotype e, and any of the serotype b (n = 10), c (n = 7), and nonserotypeable (n = 1) isolates. The false reactions among serotypes can be seen in Table 1 in detail. Although the Rapid NH system was unable to differentiate H. aphrophilus from H. paraphrophilus or H. parainfluenzae from H. parahaemolyticus, the other Haemophilus species were correctly identified to the species level.
TABLE 1.
Comparison of Rapid NH test reaction results of 65 A. actinomycetemcomitans isolates representing five serotypes with those in the Rapid NH system differential chart for the identification of A. actinomycetemcomitans
| Substrate | Positive identification of A. actinomycetemcomitans according to Rapid NH system differential chart
|
Rapid NH system results for tested A. actinomycetemcomitans isolates (no. [%] of isolates) for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Reaction | %a | Serotype a (n = 6)b | Serotype b (n = 10) | Serotype c (n = 7) | Serotype d (n = 3) | Serotype e (n = 38) | Nonserotypeable (n = 1) | Total (n = 65) | |
| Proline p-nitroanilide | − | 100 | 1 (17) | 1 (10) | 2 (29) | 0 (0) | 10 (26) | 0 (0) | 14 (22) |
| γ-Glutamyl p-nitroanilide | + | 90 | 5 (83) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 64 (98) |
| o-Nitrophenyl-β-d-galactosidase | − | 100 | 6 (100) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 65 (100) |
| Glucose | + | 83 | 6 (100) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 65 (100) |
| Sucrose | − | 100 | 6 (100) | 10 (100) | 7 (100) | 2 (67) | 21 (55) | 1 (100) | 47 (72) |
| Fatty acid ester | − | 100 | 3 (50) | 2 (20) | 0 (0) | 2 (67) | 16 (42) | 0 (0) | 23 (35) |
| Resazurin | + | 92 | 2 (33) | 4 (40) | 5 (71) | 3 (100) | 13 (34) | 0 (0) | 27 (42) |
| p-Nitrophenyl phosphate | + | 92 | 6 (100) | 0 (0) | 3 (43) | 3 (100) | 35 (92) | 1 (100) | 45 (70) |
| Ornithine | − | 100 | 6 (100) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 65 (100) |
| Urea | − | 100 | 6 (100) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 65 (100) |
| Nitrate | + | 96 | 5 (83) | 9 (90) | 7 (100) | 3 (100) | 25 (66) | 1 (100) | 50 (77) |
| Tryptophane | − | 100 | 6 (100) | 10 (100) | 7 (100) | 3 (100) | 38 (100) | 1 (100) | 65 (100) |
| No identification | 5 (83) | 10 (100) | 7 (100) | 1 (33) | 31 (82) | 1 (100) | 55 (85) | ||
Proportion of strains.
Number of isolates tested.
The database codes of the Rapid NH test panel obtained from all A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus isolates are shown in Table 2. The predominant codes among nonidentified A. actinomycetemcomitans serotypes a, b, c, d, and e and the nonserotypeable isolates were 3122, 3502, 3512, 3722, 3732, and 3522, respectively. The false A. actinomycetemcomitans codes were not overlapping with other identifications. In the Rapid NH system, o-nitrophenyl-β-d-galactosidase was the only reaction negative for all A. actinomycetemcomitans isolates but positive for all H. aphrophilus and H. paraphrophilus isolates.
TABLE 2.
All Rapid NH numerical codes obtained from 65 A. actinomycetemcomitans isolates and 13 H. aphrophilus and H. paraphrophilus isolatesa
| Identified A. actinomycetemcomitans isolates (n = 10) (%) | Identified H. aphrophilus or paraphrophilus isolates (n = 13) (%) | Unidentified A. actinomycetemcomitans isolates
|
|||||
|---|---|---|---|---|---|---|---|
| Serotype a isolates (n = 5) (%) | Serotype b isolates (n = 10) (%) | Serotype c isolates (n = 7) (%) | Serotype d isolates (n = 1) (%) | Serotype e isolates (n = 31) (%) | Nonserotypeable isolate (n = 1) (%) | ||
| 2122 (2) (20) | 6722 (1) (8) | 3532 (1) (20) | 2100 (1) (10) | 3512 (3) (43) | 3722 (1) (100) | 2720 (1) (3) | 3522 (1) (100) |
| 2110 (1) (10) | 6332 (10) (77) | 1520 (1) (20) | 3112 (1) (10) | 2512 (1) (14) | 2520 (2) (6) | ||
| 3132 (2) (20) | 6330 (2) (15) | 3502 (1) (20) | 3502 (5) (50) | 3522 (1) (14) | 3122 (4) (13) | ||
| 2120 (1) (10) | 3122 (2) (40) | 3512 (3) (30) | 3532 (1) (14) | 3732 (7) (23) | |||
| 2132 (2) (20) | 2522 (1) (14) | 2712 (1) (3) | |||||
| 2130 (2) (20) | 3712 (1) (3) | ||||||
| 3722 (3) (10) | |||||||
| 3522 (3) (10) | |||||||
| 3120 (3) (10) | |||||||
| 3100 (1) (3) | |||||||
| 3322 (1) (3) | |||||||
| 3720 (3) (10) | |||||||
| 3520 (1) (3) | |||||||
The number of isolates and the proportion of isolates are given in the first and second sets of parentheses, respectively.
Serum sugar fermentation and nitrate broth tests.
All tested A. actinomycetemcomitans isolates (n = 18) were negative to sucrose in the serum sugar test. Moreover, all A. actinomycetemcomitans isolates (n = 15) which were negative for nitrate reduction in the Rapid NH test showed a positive nitrate reduction result in the nitrate broth test.
API ZYM test.
All A. actinomycetemcomitans isolates, representing 5 serotypes and 1 nonserotypeable isolate and 18 AP-PCR genotypes showed an indistinguishable biochemical profile in the API ZYM test. Additionally, the A. actinomycetemcomitans isolates examined in duplicate showed no variation in their enzyme-producing capacity. All of the test isolates gave negative reactions to 9 enzymes: lipase, trypsin, chymotrypsin, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. Table 3 shows the results for the remaining 10 enzymatic reactions of the API ZYM test panel concerning the 34 A. actinomycetemcomitans isolates representing all serotypes and genotypes and all isolates (n = 23) of the 7 Haemophilus species. All Haemophilus and A. actinomycetemcomitans test isolates exhibited a strong reaction to acid and alkaline phosphatases and to leucine arylamidase. None of the Haemophilus species produced esterase lipase, but A. actinomycetemcomitans isolates were rather weakly positive for this enzyme. In contrast to A. actinomycetemcomitans, H. aphrophilus and H. paraphrophilus produced β-galactosidase and α-glucosidase. The API ZYM system was unable to differentiate between H. aphrophilus and H. paraphrophilus.
TABLE 3.
Results of API ZYM characterization of clinical A. actinomycetemcomitans isolates representing all serotypes and various Haemophilus speciesa
| Enzyme activity |
Haemophilus spp. (S/W/VW)
|
A. actinomycetemcomitans (S/W/VW)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H. aphrophilus(n = 11) | H. paraphrophilus(n = 2) | H. influenzae(n = 4) | H. parain-fluenzae(n = 3) | H. haemolyticus(n = 1) | H. parahae-molyticus(n = 1) | H. segnis(n = 1) | Serotype a (n = 6) | Serotype b (n = 10) | Serotype c (n = 7) | Serotype d (n = 2) | Serotype e (n = 11) | Nonsero-typeable (n = 1) | |
| Alkaline phosphatase | 11/0/0 | 2/0/0 | 4/0/0 | 3/0/0 | 1/0/0 | 1/0/0 | 1/0/0 | 6/0/0 | 10/0/0 | 7/0/0 | 2/0/0 | 11/0/0 | 1/0/0 |
| Esterase (C4) | 0/6/5 | 0/2/0 | 0/0/2 | 0/0/3 | 0/0/0 | 0/1/0 | 0/0/1 | 0/6/0 | 0/9/0 | 0/7/0 | 0/2/0 | 0/11/0 | 0/1/0 |
| Esterase lipase (C8) | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/5/1 | 0/7/3 | 0/3/3 | 0/1/1 | 0/7/3 | 0/0/1 |
| Leucine arylamidase | 11/0/0 | 2/0/0 | 4/0/0 | 3/0/0 | 1/0/0 | 1/0/0 | 1/0/0 | 6/0/0 | 10/0/0 | 7/0/0 | 2/0/0 | 11/0/0 | 1/0/0 |
| Valine arylamidase | 1/2/8 | 0/1/1 | 0/4/0 | 0/0/3 | 0/0/0 | 0/0/0 | 0/0/1 | 0/4/2 | 0/3/7 | 0/4/2 | 0/1/1 | 0/9/2 | 0/1/0 |
| Cystine arylamidase | 0/2/6 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/4 | 0/4/3 | 0/1/4 | 0/0/1 | 0/4/1 | 0/0/1 |
| Acid phosphatase | 11/0/0 | 2/0/0 | 4/0/0 | 3/0/0 | 1/0/0 | 1/0/0 | 1/0/0 | 6/0/0 | 10/0/0 | 7/0/0 | 2/0/0 | 11/0/0 | 1/0/0 |
| Naphthol-AS-BI-phosphohydrolase | 3/8/0 | 2/0/0 | 1/3/0 | 3/0/0 | 1/0/0 | 1/0/0 | 0/1/0 | 0/0/6 | 0/2/8 | 0/1/6 | 0/1/1 | 0/6/4 | 0/0/1 |
| β-Galactosidase | 11/0/0 | 2/0/0 | 0/0/0 | 0/2/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 |
| α-Glucosidase | 11/0/0 | 2/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 | 0/0/0 |
Sign: S, number of strains with strong reaction (grades 3, 4, and 5 according to color chart: ≥20 nmol); W, number of strains with weak reaction (grades 1 and 2: 5 to <20 nmol); VW, number of strains with very weak reaction (<5 nmols). n, number of isolates tested.
Genetic diversity of A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus isolates as determined by AP-PCR genotyping.
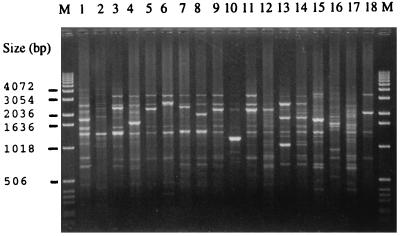
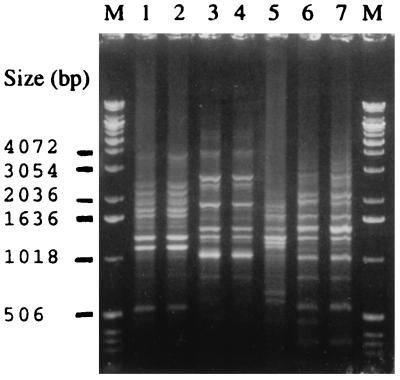
The oligonucleotide 5′-CAGCACCCAC-3′ primer distinguished 3 AP-PCR genotypes among the H. aphrophilus isolates and 1 AP-PCR genotype among the H. paraphrophilus isolates that were distinct from the 18 AP-PCR genotypes of the present A. actinomycetemcomitans isolates (Fig. 1). Of 11 H. aphrophilus isolates, 5 (45%) belonged to the AP-PCR genotype 1, 5 (45%) belonged to the AP-PCR genotype 2, and 1 belonged to the AP-PCR genotype 3. Two H. paraphrophilus isolates showed identical banding patterns with each other, and the patterns were different from those of 11 H. aphrophilus isolates (Fig. 2). Of the 18 AP-PCR genotypes of the A. actinomycetemcomitans isolates, 2 belonged to serotype a, 6 to serotype b, 4 to serotype c, 2 to serotype d, and 3 to serotype e and 1 was nonserotypeable.
FIG. 1.
AP-PCR banding patterns of 18 A. actinomycetemcomitans isolates representing 18 AP-PCR genotypes and 5 serotypes. Lanes: 1 and 2, serotype a; 3 to 8, serotype b; 9 to 12 serotype c; 13 and 14, serotype d; 15 to 17 serotype e; 18, nonserotypeable isolate; M, molecular size marker.
FIG. 2.
AP-PCR banding patterns of five H. aphrophilus and two H. paraphrophilus isolates. Lanes: 1 and 2, H. aphrophilus AP-PCR genotype 1; 3 and 4, H. aphrophilus AP-PCR genotype 2; 5, H. aphrophilus AP-PCR genotype 3; 6 and 7, H. paraphrophilus AP-PCR genotype 1; M, molecular size marker.
DISCUSSION
The Rapid NH identification kit has a database for Haemophilus, Neisseria, and some other genera that include fastidious gram-negative bacilli, such as A. actinomycetemcomitans. Although the identification performance of the Rapid NH has been extensively studied for Haemophilus and Neisseria species (6, 8, 20), no data were available for the five A. actinomycetemcomitans serotypes.
The present study examined the ability of the Rapid NH test in the identification of A. actinomycetemcomitans isolates representing all five currently known serotypes and all AP-PCR genotypes so far distinguished by the oligonucleotide 5′-CAGCACCCAC-3′ primer among A. actinomycetemcomitans isolates in our culture collection (2, 7). The results showed that 55 of 65 (85%) A. actinomycetemcomitans isolates were not identified with the Rapid NH system due to false-positive reactions for proline p-nitroanilide, sucrose, and fatty acid ester and false-negative reactions for resazurin, p-nitrophenyl phosphate, and nitrate reduction. Several studies have reported negative sucrose and lactose fermentation reactions and positive nitrate reduction of A. actinomycetemcomitans (3, 18, 23, 26, 27, 31). In these studies either the serotype distribution of the test material was not mentioned or the recently designated serotypes d and e were not included. Interestingly, in the Rapid NH test the positive reaction for sucrose was only observed in some isolates of the recently found, rare A. actinomycetemcomitans serotypes d and e (11, 21). In order to be sure that the sucrose fermentation and nitrate reduction by certain isolates of these new serotypes are different from the literature description of this species, all of the A. actinomycetemcomitans isolates which were positive for sucrose and/or negative for nitrate reduction in the Rapid NH test were tested by other testing methods, i.e., the serum sugar test and the nitrate-broth test. The results of these tests showed that the sucrose-positive and nitrate reduction-negative reactions obtained from the Rapid NH test system were false-positive and false-negative reactions. The false-negative test reaction in the Rapid NH test system may be due to the fact that some of our A. actinomycetemcomitans isolates may have been unable to react with the substrates in the Rapid NH test, but we found no answer for the false-positive reactions. As no information was obtained from the manufacturer about the A. actinomycetemcomitans strains used to set up the database of the Rapid NH identification system, it is difficult to explain the discrepancy between the results on our A. actinomycetemcomitans strains and those of the manufacturer. However, it may be related to serotype, the number of strains, or genetic or environmental differences among isolates. Our test A. actinomycetemcomitans isolates showed several codes which were not present in the database of the kit. New codes obtained from our test isolates, if added to the Rapid NH database, may help to identify clinical A. actinomycetemcomitans strains.
Although the Rapid NH system cannot differentiate between H. aphrophilus and H. paraphrophilus and between H. parainfluenzae and H. parahaemolyticus without a supplemental growth factor dependence test and the ability to grow on blood agar, the Haemophilus isolates belonging to the seven tested species were correctly identified. The key reaction differentiating H. aphrophilus and H. paraphrophilus from A. actinomycetemcomitans was the hydrolysis of o-nitrophenyl-β-d-galactoside. It was positive among all H. aphrophilus and H. paraphrophilus isolates tested but negative among all A. actinomycetemcomitans isolates, as previously shown by Slots (26).
In the present study, the enzymatic activity of A. actinomycetemcomitans and Haemophilus species was also evaluated by using the API ZYM rapid micromethod. All serotypes and nonserotypeable isolates of A. actinomycetemcomitans showed indistinguishable enzymatic profiles in the API ZYM test. However, the API ZYM system was unable to differentiate between H. aphrophilus and H. paraphrophilus isolates. The kit differentiates A. actinomycetemcomitans from H. aphrophilus and H. paraphrophilus on the basis of β-galactosidase and α-glucosidase reactions. Our results confirm the previous results published by Slots (24, 26) and Sakazaki et al. (23), but differ from those by Myhrvold et al. (16). Slots (24, 26) reported that 70 to 86% of A. actinomycetemcomitans isolates produced weak leucine aminopeptidase but all of our test A. actinomycetemcomitans isolates strongly produced leucine aminopeptidase. This discrepancy may be related to diversity in the origin of the isolates.
Conventional phenotypic tests, including fermentation of sucrose and lactose, used for differentiating A. actinomycetemcomitans from H. aphrophilus and H. paraphrophilus are time-consuming and labor-intensive. The conventional fermentation tests take 5 or 10 days depending on the method (18, 26). Moreover, phenotypical tests can sometimes be unreliable, since, for example, some catalase-negative strains of A. actinomycetemcomitans and catalase-positive strains of H. aphrophilus or H. paraphrophilus have been reported (28, 29). In such cases, these species would be misidentified due to these phenotypically variable strains. Molecular methods such as hybridization to species-specific oligonucleotide probes (5) or cloned DNA probes (28) or analysis of the 23S rRNA gene (17) or the 16S rRNA gene (19) have been developed to discriminate between A. actinomycetemcomitans and H. aphrophilus or H. paraphrophilus. However, sufficient differentiation between H. aphrophilus and H. paraphrophilus could not be achieved in most of these studies (5, 17, 28). AP-PCR, which has proven to be a rapid and useful technique for detecting DNA polymorphism in various prokaryotic organisms (30), applies short oligonucleotide primers of random sequence for the amplification of certain DNA fragments. Although isolates of many bacterial species, such as A. actinomycetemcomitans and Porphyromonas gingivalis, can be differentiated by using AP-PCR (2, 7, 14), to our knowledge this technique has not previously been applied for H. aphrophilus and H. paraphrophilus isolates.
In this study, the inter- and intraspecies genetic heterogeneity of A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus isolates was determined by AP-PCR genotyping by using oligonucleotide 5′-CAGCACCCAC-3′ as a primer. The primer distinguished 3 AP-PCR genotypes among 11 H. aphrophilus and 18 AP-PCR genotypes among a total of 65 A. actinomycetemcomitans isolates. It is obvious that more AP-PCR genotypes can be distinguished when more H. aphrophilus isolates are analyzed. Although a limited number of H. aphrophilus and H. paraphrophilus isolates were analyzed in the present study, it is worth noticing that AP-PCR genotypes of these three species were distinct from each other. We suggest that the AP-PCR method may prove to be a useful supplement to conventional methods in the differentiation of the three species, A. actinomycetemcomitans, H. aphrophilus, and H. paraphrophilus.
In conclusion, we showed that the present Rapid NH system has a poor ability for identifying A. actinomycetemcomitans, whereas the API ZYM system consistently gave correct identifications and was able to differentiate A. actinomycetemcomitans from H. aphrophilus and H. paraphrophilus. However, the API ZYM system is unable to distinguish the A. actinomycetemcomitans serotypes from each other and H. aphrophilus from H. paraphrophilus. The performance of Rapid NH may be improved by including the present serotype-specific test results in a reconstructed database. To discriminate these three closely related species from each other we have developed a rapid-PCR-based method which can be alternatively used as an aid for identifying isolates that exhibit atypical phenotypic features.
ACKNOWLEDGMENTS
This work was supported by Ankara Gazi University, Ankara, Turkey, grant 2547-33; Center for International Mobility, grant 97-2908212; and The Academy of Finland, grant 1011575.
We thank Arja Kanevo for expert technical assistance.
REFERENCES
- 1.Asikainen S, Lai C-H, Alaluusua S, Slots S. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991;6:115–118. doi: 10.1111/j.1399-302x.1991.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 2.Asikainen S, Chen C, Slots J. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol Immunol. 1995;10:65–68. doi: 10.1111/j.1399-302x.1995.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 3.Boyce J M H, Frazer J, Zinnemann K. The growth requirements of Haemophilus aphrophilus. J Med Microbiol. 1969;2:55–62. doi: 10.1099/00222615-2-1-55. [DOI] [PubMed] [Google Scholar]
- 4.Campos J M. Haemophilus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual for clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 556–565. [Google Scholar]
- 5.Dix K, Watanabe S M, McArdle S, Lee D I, Randolph C, Moncla B, Schwartz D E. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J Clin Microbiol. 1990;28:319–323. doi: 10.1128/jcm.28.2.319-323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doern G V, Chapin K C. Laboratory identification of Haemophilus influenzae: basal media and the results of the satellitism test and evaluation of the RapID NH system. J Clin Microbiol. 1984;20:599–601. doi: 10.1128/jcm.20.3.599-601.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doğan, B., M. H. Saarela, H. Jousimies-Somer, S. Alaluusua, and S. Asikainen. Actinobacillus actinomycetemcomitans serotype e—biotypes, genetic diversity and distribution in relation to periodontal status. Oral Microbiol. Immunol., in press. [DOI] [PubMed]
- 8.Eriquez L A, Hodinka N E. Development of a test system for rapid differentiation of Neisseria and Haemophilus spp. J Clin Microbiol. 1983;18:1032–1039. doi: 10.1128/jcm.18.5.1032-1039.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemmig T F, Rüdiger S, Hofmann U, Schmidt H, Plaschke B, Strätz A, Klaiber B, Karch H. Identification of Actinobacillus actinomycetemcomitans in subgingival plaque by PCR. J Clin Microbiol. 1995;33:3102–3105. doi: 10.1128/jcm.33.12.3102-3105.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fives-Taylor P, Meyer D, Mintz K. Virulence factors of the periodontopathogen Actinobacillus actinomycetemcomitans. J Periodontol. 1996;67(Suppl.):291–297. doi: 10.1902/jop.1996.67.3s.291. [DOI] [PubMed] [Google Scholar]
- 11.Gmür R, McNabb H, Van Steenbergen T J M, Baehni P, Mombelli A, Van Winkelhoff A J, Guggenheim B. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993;8:116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 12.Liljemark W F, Bloomquist C G, Uhl L A, Schaffer E M, Wolff L F, Pihlstrom B L, Bandt C L. Distribution of oral Haemophilus species in dental plaque from a large adult population. Infect Immun. 1984;46:778–786. doi: 10.1128/iai.46.3.778-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacFaddin J F. Media for isolation-cultivation-identification-maintenance of medical bacteria. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1985. pp. 620–628. [Google Scholar]
- 14.Ménard C, Brousseau R, Mouton C. Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification of Porphyromonas (Bacteroides) gingivalis. FEMS Microbiol Lett. 1992;95:163–168. doi: 10.1111/j.1574-6968.1992.tb05360.x. [DOI] [PubMed] [Google Scholar]
- 15.Moore L V H, Moore W E C. Supplement to the VPI anaerobe laboratory manual. 4th ed. (1977). Blacksburg, Va: Virginia Polytechnic Institute and State University; 1991. Anaerobe laboratory manual update; p. 146. [Google Scholar]
- 16.Myhrvold V, Brondz I, Olsen I. Application of multivariate analyses of enzymic data to classification of members of the Actinobacillus-Haemophilus-Pasteurella group. Int J Syst Bacteriol. 1992;42:12–18. doi: 10.1099/00207713-42-1-12. [DOI] [PubMed] [Google Scholar]
- 17.Preus H R, Sunday G J, Haraszthy V I, Zambon J J. Rapid identification of Actinobacillus actinomycetemcomitans based on analysis of 23S ribosomal RNA. Oral Microbiol Immunol. 1992;7:372–375. doi: 10.1111/j.1399-302x.1992.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 18.Pulverer G, Ko H L. Actinobacillus actinomycetemcomitans: fermentative capabilities of 140 strains. Appl Microbiol. 1970;20:693–695. doi: 10.1128/am.20.5.693-695.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggio M P, Lennon A. Rapid identification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, and Haemophilus paraphrophilus by restriction enzyme analysis of PCR-amplified 16S rRNA genes. J Clin Microbiol. 1997;35:1630–1632. doi: 10.1128/jcm.35.6.1630-1632.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson M J, Oberhofer T R. Identification of pathogenic Neisseria species with Rapid NH system. J Clin Microbiol. 1983;17:400–404. doi: 10.1128/jcm.17.3.400-404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saarela M, Asikainen S, Alaluusua S, Asikainen T, Pyhälä L, Lai C-H, Jousimies-Somer H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d, or e. Oral Microbiol Immunol. 1992;7:277–279. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 22.Saarela M, Asikainen S, Chen C, Alaluusua S, Slots J. Comparison of arbitrarily primed polymerase chain reaction and ribotyping for subtyping Actinobacillus actinomycetemcomitans. Anaerobe. 1995;1:97–102. doi: 10.1006/anae.1995.1004. [DOI] [PubMed] [Google Scholar]
- 23.Sakazaki R, Yoshizaki E, Tamura K, Kuramochi S. Increased frequency of isolation of Pasteurella and Actinobacillus species and related organisms. Eur J Clin Microbiol. 1984;3:244–248. doi: 10.1007/BF02014894. [DOI] [PubMed] [Google Scholar]
- 24.Slots J. Enzymatic characterization of some oral and nonoral gram-negative bacteria with the API ZYM system. J Clin Microbiol. 1981;14:288–294. doi: 10.1128/jcm.14.3.288-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slots J. Selective medium for isolation Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1982;15:606–609. doi: 10.1128/jcm.15.4.606-609.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slots J. Salient biochemical characteristics of Actinobacillus actinomycetemcomitans. Arch Microbiol. 1982;131:60–67. doi: 10.1007/BF00451500. [DOI] [PubMed] [Google Scholar]
- 27.Tanner A, Lai C-H, Maiden M. Characteristics of oral gram-negative species. In: Slots J, Taubman M, editors. Contemporary oral microbiology and immunology. St. Louis, Mo: Mosby-Year Book, Inc.; 1992. pp. 299–341. [Google Scholar]
- 28.Tønjum T, Bukholm G, Bøvre K. Identification of Haemophilus aphrophilus and Actinobacillus actinomycetemcomitans by DNA-DNA hybridization and genetic transformation. J Clin Microbiol. 1990;28:1994–1998. doi: 10.1128/jcm.28.9.1994-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tønjum T, Haas R. Identification of Actinobacillus actinomycetemcomitans by leukotoxin gene-specific hybridization and polymerase chain reaction assays. J Clin Microbiol. 1993;31:1856–1859. doi: 10.1128/jcm.31.7.1856-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zambon J J, Umemoto T, De Nardin E, Nakazawa F, Christersson L A, Genco R J. Actinobacillus actinomycetemcomitans in the pathogenesis of human periodontal disease. Adv Dent Res. 1988;2:269–274. doi: 10.1177/08959374880020021101. [DOI] [PubMed] [Google Scholar]
- 33.Zambon J J. Periodontal disease: microbial factors. Ann Periodontol. 1996;1:879–925. doi: 10.1902/annals.1996.1.1.879. [DOI] [PubMed] [Google Scholar]