Abstract
Background
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) circulates in the world and acquires mutations during evolution. To identify the new emergent variants, the surveillance of the variants of concern (VOC) and variants of interest (VOI) is ongoing. This study aimed to determine how the transition of viral lineage occurred by stationary genome analysis in Yamanashi, Japan.
Methods
We performed the whole genome sequencing using SARS-CoV-2 positive samples collected from February 2020 to the end of June 2021. Viral lineage was defined by the Phylogenetic Assignment of Named Global Outbreak (PANGO) lineages.
Results
We successfully obtained 325 viral genome sequences and the number of analyzed samples accounted for 15.4% of the total 2109 COVID-19 patients identified in our district. We identified 13 types of viral lineages including R.1, P.1, B.1.1.7 (Alpha) and B.1.617.2 (Delta). These virus lineages had distinct periods of expansion and decline. After the emerging of the R.1 lineage harboring E484K variant (designated VOI in Japan), the prevalent B.1.1.214 lineage were no longer identified. The R.1 lineages were temporarily prevalent afterwards, but the influx of B.1.1.7 lineage (designated VOC) led to a decline in R.1. Currently, B.1.1.7 has become dominant after mid-April 2021.
Conclusion
We clearly elucidated the transition and replacement of viral lineage by the community-based analysis. The virus completely replaced by more infectious lineages, therefore, it will be necessary to continue to monitor the VOC and VOI.
Keywords: SARS-CoV-2, Genome, B.1.1.7, VOC, VOI
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading worldwide and threatening human health. In countries where vaccination is widely available, the number of infected cases and deaths turned to be decreasing, giving a hope for the virus under the control. Meantime, various types of viral lineage have emerged during the virus evolution. In particular, mutations in receptor binding domain of spike protein are of interest for possible changes in the nature of the virus. The several viral lineages have been designated as variant of concern (VOC) or variant of interest (VOI).
On May 31, 2021, World Health Organization (WHO) proposed a new designation, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2) as the four VOCs and Epsilon (B.1.427/B.1.429), Zeta (P.2), Eta (B.1.525), Theta (B.1.525) Lota (B.1.526), Kappa (B.1.617.1) and Lambda (C.37) as the seven VOIs (World Health Organization, 2021). The U.S. Centers for Disease Control and Prevention (CDC) (CDC: SARS-CoV-2 Variant Classifications and Definitions, 2021), the European Centre for Disease Prevention and Control (ECDC) (European Centre for Disease Prevention and Control: SARS-CoV-2 Variants of Concern, 2021), the Public Health England (PHE) (Public Health England (PHE): COVID-19 (SARS-CoV-2) Variants, 2021) and National Institute of Infectious Diseases (NIID) in Japan also have their own designations for VOCs and VOIs (National Institute of Infectious Diseases (NIID), 2021). These lineages have hallmark mutations in spike protein, which was suggested to affect the transmissibility, immunity, and disease severity (Boehm et al., 2021). To prevent the spread of SARS-CoV-2, these VOCs and VOIs continue to be under surveillance in many countries.
In Japan, as of September 9, 2021, the cumulative number of COVID-19 cases is 1,603,112 and the number of deaths is 16,525. The number of people vaccinated with one or more doses of the vaccine is 77,749,452 (61.4% of the total population) and the number of people completing two doses of the vaccine is 62,546,331 (49.4%). In January 2021, we started to conduct genomic surveillance of SARS-CoV-2 (Hirotsu and Omata, 2021a; Hirotsu and Omata, 2021b; Hirotsu and Omata, 2021c). We previously reported on the SARS-CoV-2 R.1 lineage harboring spike W152L, E484K and G769V mutations (Hirotsu and Omata, 2021b). In Japan, the R.1 lineage was first registered in Global Initiative on Sharing All Influenza Data (GISAID) database in November 2020 and showed an increase around January 2021 (Hirotsu and Omata, 2021b; Sekizuka et al., 2021; Nagano et al., 2021). Although the NIID in Japan has designated R.1 lineage as a VOC, it is not clear there are associations with increased infectivity and transmissibility (National Institute of Infectious Diseases (NIID), 2021).
The B.1.1.7 lineage was first identified in the United Kingdom at September 2020 and detected at airport quarantine in Japan at December 2020. The B.1.1.7 was reported to be highly transmissible and increase the disease severity (Volz et al., 2021; Challen et al., 2021). Actually, the B.1.1.7 spreads rapidly and is identified in 150 countries as of June 30, 2021 (Hadfield et al., 2018; O'Toole et al., 2021). Although B.1.1.7 is reported to be highly transmissible, the transition of other virus lineage in Japan has not been fully elucidated after the influx of B.1.1.7.
In this study, we conducted whole genome analysis of SARS-CoV-2 in 325 SARS-CoV-2 positive samples collected from February 2020 to June 2021 in Kofu, Japan. The lineages other than VOC and VOI were observed until mid-January 2021, afterward, the R.1 lineage was dominant. However, the subsequent influx of B.1.1.7 increased rapidly, suggesting a rapid replacement of R.1 with B.1.1.7. We elucidated that the two major lineages have different infectivity based on genomic surveillance.
2. Materials and methods
2.1. Ethics statement
The Institutional Review Board of the Clinical Research and Genome Research Committee at Yamanashi Central Hospital approved this study and the use of an opt-out consent method (Approval No. C2019–30). The requirement for written informed consent was waived owing to it being an observational study and the urgent need to collect COVID-19 data.
2.2. Sample collection and nucleic acid extraction
From February 15, 2020 to June 30, 2021, a total of 2109 COVID-19 patients were confirmed in Yamanashi Prefecture, Japan. Nasopharyngeal swab samples were collected by using cotton swabs and placed in 3 ml of viral transport media (VTM) purchased from Copan Diagnostics (Murrieta, CA, United States). We used 200 μl of VTM for nucleic acid extraction, performed within 2 h of sample collection. Total nucleic acid was isolated using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific; Waltham, MA, United States) as previously described (Hirotsu et al., 2020a). Of the 2109 COVID-19 patients in Yamanashi Prefecture, 335 patients visited our hospital and tested positive by RT-qPCR and/or quantitative antigen test as previously described (Hirotsu et al., 2020a; Hirotsu et al., 2020b; Hirotsu et al., 2020c; Hirotsu et al., 2021a). All 335 samples of nucleic acids were stored in the freezer.
2.3. Whole-genome sequencing
We subjected 335 samples collected from COVID-19 patients to whole genome analysis and successfully obtained 325 sequence data, except for 10 samples with very low viral load. Whole genome sequencing analysis was performed as we previously described (Hirotsu and Omata, 2021b). In brief, SARS-CoV-2 genomic RNA was reverse transcribed into cDNA and amplified by using the Ion AmpliSeq SARS-CoV-2 Research Panel (Thermo Fisher Scientific) on the Ion Torrent Genexus System in accordance with the manufacturer's instructions (Hirotsu and Omata, 2021b; Hirotsu and Omata, 2021c). Sequencing reads were processed, and their quality was assessed by using Genexus Software with SARS-CoV-2 plugins. The sequencing reads were mapped and aligned by using the torrent mapping alignment program. After initial mapping, a variant call was performed by using the Torrent Variant Caller. The COVID19AnnotateSnpEff plugin was used for the annotation of variants. Assembly was performed with the Iterative Refinement Meta-Assembler (Shepard et al., 2016).
2.4. Clade and lineage classification
The viral clade and lineage classifications were conducted by using Nextstrain (Hadfield et al., 2018), and Phylogenetic Assignment of Named Global Outbreak (PANGO) Lineages (Rambaut et al., 2020). The sequences data was deposited in the Global Initiative on Sharing Avian Influenza Data (GISAID) EpiCoV database (Shu and McCauley, 2017).
3. Results
From February 2020 to the end of June 2021, we collected 335 SARS-CoV-2 positive samples determined by RT-qPCR and/or quantitative antigen tests (Hirotsu et al., 2020a; Hirotsu et al., 2020b; Hirotsu et al., 2020c; Hirotsu et al., 2021a). We subjected these samples to the whole genome analysis. As a result, sequencing analysis could successfully determine viral sequence from 325 individuals, excluding 10 individuals due to the very low viral load. As of June 30, this represented 15.4% of the 2109 infected individuals identified in our district.
To characterize the viral lineage, the yielded sequence data were analyzed by PANGO lineage (O'Toole et al., 2021; Rambaut et al., 2020). The result showed the 325 samples were classified into 13 types of lineage (B, B.1, B.1.1, B.1.149, B.1.1.284, B.1.1.214, B.1.411, B.1.346, R.1, P.1, B.1.1.220 B.1.1.7 and B.1.617.2) (Table 1 ).
Table 1.
Identified viral lineages in the community of Japan.
| Lineage | Designation by WHO | Designation by CDC (USA) | Designation by ECDC (Europe) | Designation by NIID (Japan) | Number of samples | Identified period |
|---|---|---|---|---|---|---|
| B | – | – | – | – | 1 | February 2020 |
| B.1 | – | – | – | – | 2 | March 2020 |
| B.1.1 | – | – | – | – | 8 | March to November 2020 |
| B.1.149 | – | – | – | – | 1 | March 2020 |
| B.1.1.284 | – | – | – | – | 20 | July 2020 to January 2021 |
| B.1.1.214 | – | – | – | – | 96 | August 2020 to February 2021 |
| B.1.411 | – | – | – | – | 4 | December 2020 |
| B.1.346 | – | – | – | – | 3 | January 2021 |
| R.1 | – | – | – | VOI | 20 | January to May 2021 |
| P.1 | VOC | VOC | VOC | VOC | 1 | February 2021 |
| B.1.1.220a | – | – | – | – | 3 | April 2021 |
| B.1.1.7 | VOC | VOC | VOC | VOC | 165 | April to June 2021 |
| B.1.617.2 | VOC | VOC | VOC | VOC | 1 | June 2021 |
| Total | 325 |
VOI, variant of interest; VOC, variant of concern; WHO, World Health Organization; CDC, Centers for Disease Control and Prevention; ECDC, European Centre for Disease Prevention and Control; NIID, National Institute of Infectious Diseases.
Harboring E484K mutation.
During the first wave (March to May 2020) in Japan, the B.1 and B.1.1 lineages with spike D614G mutation were predominant (Yurkovetskiy et al., 2020) (Table 1). At the second (from July to September 2020) and third waves (from October 2020 to February 2021), the B.1.1.214 (a total of 96 samples) and B.1.1.284 (20 samples) lineages were prevailed (Table 1). Almost of the B.1.1.214 and B.1.1.284 lineages were identified only in Japan and did not spread to other countries (Julia et al., 2020).
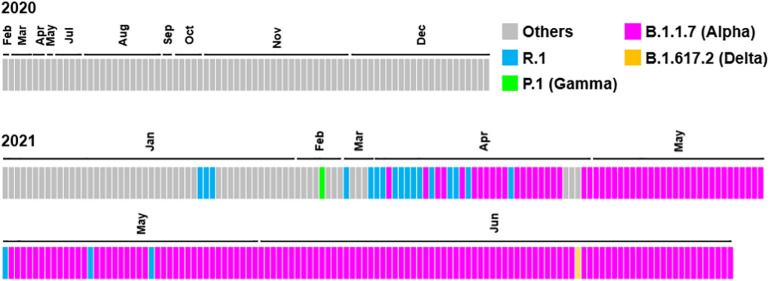
Afterward, several types of VOC or VOI were identified in our district (Fig. 1 and Table 1). The R.1 was first detected in January 2021 and a total of 20 were identified so far (Table 1). Although P.1 was detected in a patient at February 2021 (Fig. 1) (Hirotsu and Omata, 2021c), there was no further spread of P.1 thereafter, suggesting early containment was successful. In the early part of the fourth wave (March to mid-April 2021), the R.1 and B.1.1.7 lineages coexisted, but after the late part of the fourth wave (after mid-April 2021), B.1.1.7 became dominant (Fig. 1). The first case of B.1.617.2 was found in Yamanashi, Japan in June, but no further spread of the infection has occurred at this point. The results show that the once potentially dominant R.1 lineage declined rapidly along with the influx of B.1.1.7 lineage.
Fig. 1.
Transition of virus lineages.
The data showed the virus lineages identified from February 2020 to the end of June 2020 in a community of Japan. The lineages were analyzed by PANGO lineage. Each box represents a detected lineage and is indicated by R.1 (light blue), P.1 (light green) B.1.1.7 (pink), B.1.617.2 (orange), and others (gray). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The B.1.1.7 (Alpha) has N501Y mutation in receptor binding domain of spike protein, binds to the angiotensin-converting enzyme 2 with high affinity and acquires a high transmission rate (Ramanathan et al., 2021). This study revealed that the B.1.1.7 rather than the R.1 expanded in Japan during the April to June 2021. These two lineages were mixed temporally from March to mid-April 2021, but eventually B.1.1.7 became dominant. Our epidemiological analysis clearly showed the R.1 and B.1.1.7 competed in a community with human migration and that the higher propagating B.1.1.7 lineage survived.
SARS-CoV-2 R.1 lineage emerged rapidly in Japan at January 2021 (Fig. 1). The R.1 carries the W152L N-terminal domain and E484K in receptor binding domain (Hirotsu and Omata, 2021b), which have been shown to be of concern for immune escape (Liu et al., 2021a), while there are no reports on its transmission potential. Of note, the fact that the B.1.1.214 lineage was replaced by the R.1 lineage suggests that R.1 possibly has higher transmissibility. This situation is observed by the data of genomic epidemiological study throughout Japanese dataset and in Tokyo (Sekizuka et al., 2021; Nagano et al., 2021; Tokumasu et al., 2021), indicating current study shows that the situation is not limited to one district.
Recent study showed the relative instantaneous reproduction numbers of the R.1, B.1.1.7, and B.1.617.2 compared to other strains were estimated at 1.256 (range: 1.198–1.335), 1.449 (range: 1.342–1.596), and 1.776 (range: 1.557–2.00), respectively (Ito et al., 2021). This data is consistent with the situation observed in this study, suggesting that a virus lineage will be replaced by another lineage with high transmissibility. As of June 30, B.1.1.7 was the predominant in Yamanashi, Japan, while the first B.1.617.2 strain was identified on June 21. The B.1.617.2 is completely replacing the B.1.1.7 that was rampant and spreading in the United Kingdom (Hadfield et al., 2018). Ito et al. estimated that B.1.617.2 lineage will replace others including the B.1.1.7 around mid-July 2021 in Japan (Ito et al., 2021). In fact, as of September 2021, our genomic surveillance data show that B.1.617.2 is dominant over B.1.1.7, indicating that a replacement is occurring (data not shown). The COVID-19 mRNA vaccines were shown to induce robust immune response and be effective against these circulating VOCs (Liu et al., 2021b; Choi et al., 2021; Hirotsu et al., 2021b), however, it is important to continue the genomic surveillance and monitor trends for signs of an increase in the number of infected individuals (Kustin et al., 2021) and breakthrough infection after vaccination (Teran et al., 2021; Duerr et al., 2021).
In summary, we present a community-based stationary observation of the viral linage replacement. Genomic epidemiological investigations revealed that the whole picture of shifting of VOI and VOC in the community. Prospective whole genome analysis could reveal the linage transitions in real time and the key viral lineage in the infection explosion (Chrysostomou et al., 2021). Real-time monitoring can provide suggestive insights for public health containment of the virus.
Financial disclosure
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to Y.H. and M.O.), Grant-in-Aid for Early-Career Scientists 18K16292 (to Y.H.) and Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.) from the Japan Society for the Promotion of Science (JSPS) KAKENHI, a Research Grant for Young Scholars (to Y.H.) funded by Yamanashi Prefecture, the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank Makoto Maejima, Masahiro Shibusawa and clinical laboratory technicians at the Division of Microbiology in Clinical Laboratory for sample collection and COVID-19 test, and Masato Kondo, Ryota Tanaka, Kazuo Sakai, Manami Nagano, Takuhito Fukami, and Ryo Kitamura (Thermo Fisher Scientific) for technical help. We also thank all of the medical and ancillary hospital staff for their support, and the patients for their participation. We thank all researchers who share genome data on GISAID (http://www.gisaid.org).
References
- Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., Kaiser L. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin. Microbiol. Infect. 2021;27(8):1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC: SARS-CoV-2 Variant Classifications and Definitions. 2021. https://wwwcdcgov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-infohtml
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A., Koch M., Wu K., Dixon G., Oestreicher J., Legault H., GBE Stewart Jones, Colpitts T., Pajon R., Bennett H., et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.06.28.449914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysostomou A.C., Vrancken B., Koumbaris G., Themistokleous G., Aristokleous A., Masia C., Eleftheriou C., Iomicronannou C., Stylianou D.C., Ioannides M., et al. A comprehensive molecular epidemiological analysis of SARS-CoV-2 infection in cyprus from april 2020 to january 2021: evidence of a highly polyphyletic and evolving epidemic. Viruses. 2021;13(6) doi: 10.3390/v13061098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr R., Dimartino D., Marier C., Zappile P., Wang G., Lighter J., Elbel B., Troxel A.B., Heguy A. Dominance of alpha and iota variants in SARS-CoV-2 vaccine breakthrough infections in new York City. J. Clin. Invest. 2021;131(18):e152702.. doi: 10.1172/JCI152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control: SARS-CoV-2 Variants of Concern. 2021. https://wwwecdceuropaeu/en/covid-19/variants-concern
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Omata M. Household transmission of SARS-CoV-2 R.1 lineage with spike E484K mutation in Japan. medRxiv. 2021 doi: 10.1101/2021.03.16.21253248. (2021.2003.2016.21253248) [DOI] [Google Scholar]
- Hirotsu Y., Omata M. Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLoS Pathog. 2021;17(6) doi: 10.1371/journal.ppat.1009619. (e1009619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Omata M. Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Inf. 2021;82(6):276–316. doi: 10.1016/j.jinf.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci. Rep. 2020;10(1):18899. doi: 10.1038/s41598-020-76043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Mochizuki H., Omata M. Double-quencher probes improve detection sensitivity toward severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J. Virol. Methods. 2020;284:113926. doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. Int. J. Infect. Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Amemiya K., Sugiura H., Shinohara M., Takatori M., Mochizuki H., Omata M. Robust antibody responses to the BNT162b2 mRNA vaccine occur within a week after the first dose in previously infected individuals and after the second dose in uninfected individuals. Front. Immunol. 2021;12:722766. doi: 10.3389/fimmu.2021.722766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Piantham C., Nishiura H. Predicted domination of variant Delta of SARS-CoV-2 before Tokyo Olympic games, Japan. Euro Surveill. 2021;26(27):2100570. doi: 10.2807/1560-7917.ES.2021.26.27.2100570. (2021.2006.2012.21258835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julia L.M., Ginger T., Alaa A.L., Manar A., Marco C., Emily H., Jerry Z., Mark Z., Nate M., Kristian G.A., et al. 2020. Outbreak.Info. [Google Scholar]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A., Cai H., Cutler M., Cooper D., Muik A., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- Nagano K., Tani-Sassa C., Iwasaki Y., Takatsuki Y., Yuasa S., Takahashi Y., Nakajima J., Sonobe K., Ichimura N., Nukui Y., et al. SARS-CoV-2 R.1 lineage variants prevailed in Tokyo in March 2021. J Med Virol. 2021 doi: 10.1002/jmv.27240. (2021.2005.2011.21257004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Infectious Diseases (NIID) 2021. https://wwwniidgojp/niid/ja/diseases/ka/corona-virus/2019-ncov/10280-covid19-41html
- O’Toole Á., Hill V., Pybus O., Watts A., Bogoch I., Khan K., Messina J., consortium TC-GUC-U, Network for Genomic Surveillance in South Africa (NGS-SA) B-UCGN, Tegally H., et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 2021;6(121) doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (PHE): COVID-19 (SARS-CoV-2) Variants. 2021. https://wwwgovuk/government/collections/new-sars-cov-2-variant
- Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 2021;21:1070. doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekizuka T., Itokawa K., Hashino M., Okubo K., Ohnishi A., Goto K., Tsukagoshi H., Ehara H., Nomoto R., Ohnishi M., et al. A discernable increase in the severe acute respiratory syndrome coronavirus 2 R.1 lineage carrying an E484K spike protein mutation in Japan. Infect. Genet. Evol. 2021;5:105013. doi: 10.1016/j.meegid.2021.105013. (2021.2004.2004.21254749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard S.S., Meno S., Bahl J., Wilson M.M., Barnes J., Neuhaus E. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics. 2016;17:708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Eur. Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teran R.A., Walblay K.A., Shane E.L., Xydis S., Gretsch S., Gagner A., Samala U., Choi H., Zelinski C., Black S.R. Postvaccination SARS-CoV-2 infections among skilled nursing facility residents and staff members — Chicago, Illinois, December 2020–march 2021. Am. J. Transplant. 2021;21(6):2290–2297. doi: 10.1111/ajt.16634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumasu R., Weeraratne D., Snowdon J., Parida L., Kudo M., Koyama T. Introductions and evolutions of SARS-CoV-2 strains in Japan. medRxiv. 2021 doi: 10.1101/2021.02.26.21252555. (2021.2002.2026.21252555) [DOI] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization Tracking SARS-CoV-2 Variants. 2021. https://wwwwhoint/en/activities/tracking-SARS-CoV-2-variants/
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751. doi: 10.1016/j.cell.2020.09.032. (e738) [DOI] [PMC free article] [PubMed] [Google Scholar]