SUMMARY
Neuronal alternative splicing is a key gene regulatory mechanism in brain. Yet the spliceosome machinery is insufficient to fully specify splicing complexity. In considering the role of the epigenome in activity-dependent alternative splicing, we and others find the histone modification, H3K36me3, to be a putative splicing regulator. In the current study, we found that mouse cocaine self-administration caused widespread differential alternative splicing, concomitant with enrichment of H3K36me3 at differentially spliced junctions. Importantly, only targeted epigenetic editing can distinguish between a direct role of H3K36me3 in splicing and an indirect role via regulation of splice factor expression elsewhere on the genome. We targeted Srsf11, which was both alternatively spliced and H3K36me3 enriched in brain following cocaine self-administration. Epigenetic editing of H3K36me3 at Srsf11 was sufficient to drive its alternative splicing and enhanced cocaine self-administration, establishing the direct causal relevance of H3K36me3 to alternative splicing of Srsf11 and to reward behavior.
Graphical Abstract
eTOC
Drugs of abuse, such as cocaine, regulate neuronal gene expression through changes in histone modifications. Xu et al apply epigenetic editing to define a novel role for the histone modification, H3K36me3, in alternative splicing of splice factor gene, Srsf11, and regulation of cocaine reward behavior.
INTRODUCTION
Alternative splicing is a key mechanism for gene regulation, with over 95% of eukaryotic genes expressing multiple isoforms. Dysregulation of alternative splicing is implicated in many neurological diseases (Lipscombe and Lopez Soto, 2019; Parikshak et al., 2016; Soto et al., 2019) but is understudied in neuropsychiatric diseases, including drug addiction. A recent mouse study shows that cocaine treatment leads to widespread differential splicing in a key brain reward region, the nucleus accumbens (NAc) (Feng et al., 2014). In fact, alternative isoform expression accounts for the vast majority of all cocaine-induced changes in mRNA expression in NAc (Feng et al., 2014).
Such activity-dependent alternative splicing is common in neurons, yet its high complexity exceeds the information capacity of the canonical splice machinery (Lipscombe, 2005). One hypothesis for how splice events are specified is that chromatin modifications provide additional information through interactions with the splicing and/or transcriptional machinery (Javier et al., 2019). Recent bioinformatic studies find that epigenetic features may be as important as gene sequence in differentiating splicing patterns (De Almeida et al., 2011; Bonev et al., 2017; Pajoro et al., 2017; Yuan et al., 2017; Zhou et al., 2012). Indeed, differential chromatin modifications correlate with alternatively spliced transcripts in mouse NAc after investigator-administered cocaine (Feng et al., 2014; Heller et al., 2014)
We recently applied an unbiased approach to examine the enrichment of various histone modifications at alternatively spliced exon junctions in neural tissues (Hu et al., 2017, 2020). We find that H3K36me3 and H3K4me1 at splice junctions vary by exon type, while H3K9me3 and H3K27me3 do not, even when controlling for confounding factors such as gene expression level, the number of exons of the gene, the size of the gene, the location of the exon in the gene, and length of the analyzed region (Hu et al., 2017). Furthermore, using machine learning, we find that H3K36me3 enrichment is the most informative and predictive for classifying alternative exon types, across tissues and development (Hu et al., 2017, 2020).
The H3K36me3 methyltransferase, SET domain containing protein 2, is conserved in yeast (Set2), human (SETD2) and mouse (Setd2) (McDaniel and Strahl, 2017; Strahl et al., 2002), and is known to regulate splicing. Overexpression of Set2 in human stem cells leads to splice-site enrichment of H3K36me3 and alternative splicing of fibroblast growth factor (FGF2) (Luco et al., 2010). Consistent with these findings, SETD2 interacts with Heterogeneous ribonucleoprotein L (hnRNPL), an RNA binding molecule that regulates alternative splicing and is necessary for SETD2 methyltransferase activity (Bhattacharya et al., 2021; Yuan et al., 2009). Despite this progress in identifying H3K36me3 as a splicing regulator, it has not yet been possible to distinguish between the mere presence and causal relevance of H3K36me3 to alternative splicing in brain or any other context. We sought to determine whether enrichment of H3K36me3 at a spliced gene is sufficient to regulate splicing of the associated transcript, or if the regulation of splicing by H3K36me3 is indirect, for example, by regulation of splice factor expression elsewhere on the genome.
To address this, we applied epigenetic editing to test the hypothesis that H3K36me3 was sufficient to regulate alternative splicing of an endogenous gene in brain in vivo. First, we quantified concomitant splice-site enrichment of H3K36me3 and differential isoform expression following mouse cocaine self-administration (SA). We found widespread alternative splicing in three brain reward regions, expanding on prior findings limited to NAc following investigator-administered cocaine. The recognition motif for serine and arginine rich splicing factor 11 (Srsf11, also known as SRp54) (Gonatopoulos-Pournatzis et al., 2018), was highly enriched amongst cocaine spliced transcripts. Cocaine SA regulated both alternative splicing and enrichment of H3K36me3 at Srsf11, with no change in mRNA expression level. We used dCas9-Set2 to enrich H3K36me3 specifically at Srsf11, which was sufficient to drive alternative splicing of this transcript. Interestingly, alternative splicing of Srsf11 partially recapitulated cocaine-induced alternative splicing, and enhanced cocaine-reward behavior.
RESULTS
Alternative splicing in brain reward regions following cocaine self-administration
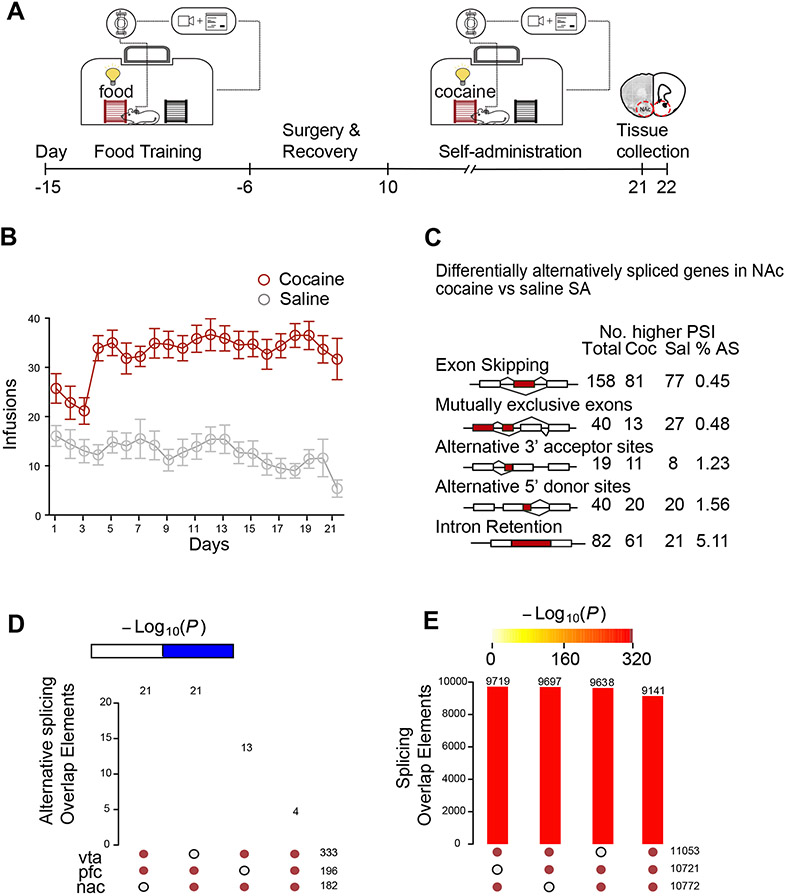
Cocaine SA is an operant paradigm modeling human addiction that induces neural mechanisms involved in drug seeking, reward, reinforcement, and withdrawal (Heilig et al., 2016). Mice are trained to associate an operant response (e.g., nose poke) with cocaine reward and reward cues (light, tone) and learn to discriminate between a cocaine-paired response and an unpaired response. Control animals are placed in an identical experimental apparatus but receive only saline and cues. This complex paradigm recapitulates drug motivation and saliency, as well as drug/cue-associations, all of which are key features of human addiction (Sadri-Vakili et al., 2010; Schmidt and Pierce, 2010; Vassoler et al., 2013). To investigate alternative splicing and global H3K36me3 enrichment in cocaine SA, we trained mice to self-administer cocaine or saline (control) (Figure 1A). Cocaine-treated mice volitionally infused cocaine during 21 daily two-hour sessions (Figure 1B) and discriminated between the cocaine and unpaired responses (Supplement figure 1A-C). For downstream biochemical analyses, we euthanized mice 24 hours after the last SA session and collected tissue from brain reward regions, NAc, ventral tegmental area (VTA) and prefrontal cortex (PFC) (Figure 1A).
Figure 1. Alternative splicing in brain reward regions following cocaine self-administration.
(A) Schematic showing cocaine self-administration (SA). Mice trained to self-administer food for 10 days prior to jugular catheterization surgery. Following recovery, mice (n=12/group) underwent 21 daily, two-hour SA sessions, in which one active response elicited one injection of either cocaine (0.7 mg/kg) or saline (control). Following the last day of SA, mice were subjected to forced (home cage) abstinence for 24 hours. NAc, VTA, and PFC tissues were then collected for downstream analysis.
(B) Average cocaine or saline infusions (n=12 per group) per session for SA treatments. Cocaine-administering mice showed a higher infusion rate than saline-administering mice (Repeated measures two-way ANOVA: Interaction effect F (20,440) =3.081, P<0.0001; drug effect F(1,22)=78.9, P<0.0001; session effect F (8.522,187.5) =2.298, P=0.0201).
(C) Summary of each type of alternative splicing event across cocaine and saline SA identified by rMATS. All events are significant (FDR < 0.05). Percent alternative splicing (%AS) was calculated as the percentage of each event across all identified splicing events.
(D-E) Comparison of skipped exon splicing events across NAc, PFC and VTA of (D) differentially spliced transcripts (FDR < 0.05, Δ PSI > 0.1) and (E) all spliced transcripts. Significance of overlap between brain regions was calculated by hypergeometric tests (P-value indicated in color legend bar).
To measure cocaine driven alternative splicing, we performed mRNA-sequencing and applied rMATS alternative splicing analysis (Shen et al., 2014) (Figure 1C) in NAc, PFC (Supplement figure 1D) and VTA (Supplement figure 1E), generating the first comprehensive dataset of differential isoform expression in three brain reward regions. The relative abundance of each isoform was quantified as percent spliced in (PSI); ΔPSI quantified the difference in relative isoform abundance between cocaine and saline SA. We identified 339, 369 and 799 transcripts that were differentially spliced in the NAc, PFC, and VTA, respectively, following cocaine SA (Figure 1C, Supplement figure 1D-E). We compared all saline and cocaine-induced changes in splicing in NAc, PFC and VTA (Figure 1C, Supplement figure 1D-E). We found negligible overlap of cocaine-driven alternative splicing events (Figure 1D, Supplement figure 1F), but similar isoform expression across all three brain regions (Figure 1E, Supplement figure 1G), suggesting a high degree of regional specificity of cocaine-regulated alternative splicing.
Cocaine SA regulated H3K36me3 enrichment at alternatively spliced exons.
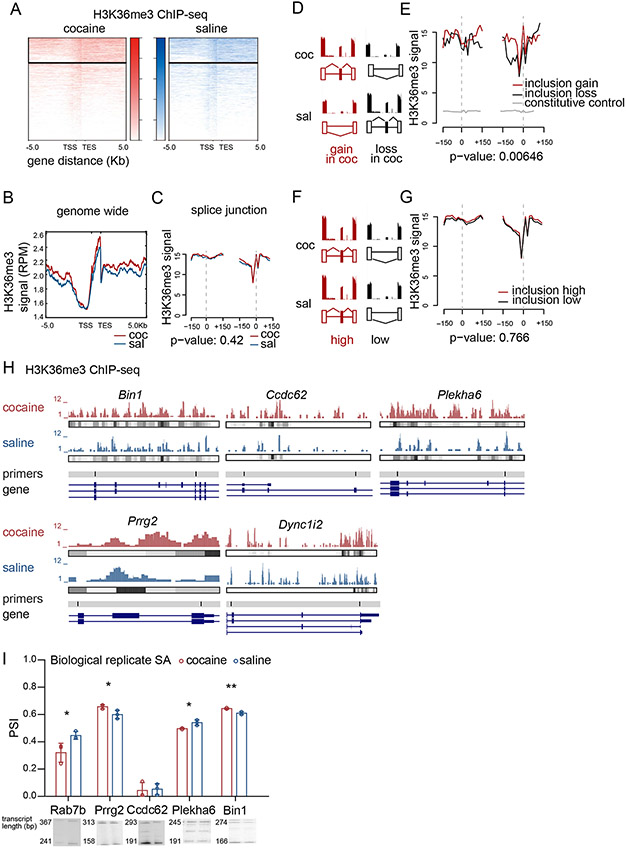
Prior work finds that global H3K36me3 enrichment predicts skipped exon selection in adult mouse NAc (Hu et al., 2017) and across mouse prenatal development (Hu et al., 2020; Xu et al., 2017). To further validate our published findings, we investigated global H3K36me3 enrichment at alternatively spliced exons in the NAc following cocaine SA by analyzing ChIP- and RNA-sequencing datasets collected from the same animal (Xu and Heller, 2018). We found cocaine SA caused concomitant changes in H3K36me3 enrichment and alternative splicing (Figure 2). Approximately 2% of H3K36me3 peaks were differentially enriched between saline and cocaine treated NAc (Figure 2A-B). The majority of differential H3K36me3 peaks (1060, 83.3%) were increased by cocaine SA. Cocaine SA did not change mRNA expression of endogenous H3K36me3 histone methyltransferase, Setd2 (not shown), which is consistent with the fact that cocaine did not grossly redistribute H3K36me3 across the genome or alter its genic distribution (Figure 2B), including at splice junctions of alternatively spliced transcripts (Figure 2C).
Figure 2. Cocaine regulation of H3K36me3 at alternatively spliced exons.
(A) Heatmap of H3K36me3 genic distribution after cocaine (red) or saline (blue) SA.
(B) Profile plots of H3K36me3 ChIP-seq peaks after cocaine (red) or saline (blue) SA.
(C) Distribution of H3K36me3 ChIP-seq reads after cocaine (red) and saline (blue) SA in the skipped exon flanking region (+/− 150bp). Dashed grey line shows exon-intron borders (ANOVA, P = 0.42).
(D) Schematic of gain and loss splicing events by cocaine (coc). Higher inclusion of skipped exon in cocaine (coc) relative to saline (sal) was defined as gain in coc (red, left), while lower inclusion of skipped exon in cocaine (coc) relative to saline (sal) was defined as loss in coc (black, right).
(E) Distribution of H3K36me3 ChIP-seq reads in the skipped exon flanking region (+/− 150bp) of cocaine-mediated gain (red) and loss (black) of skipped exon inclusion. Cocaine H3K36me3 ChIP-seq reads in the constitutive exon flanking region served as a control (+/− 150bp) (gray). Dashed grey line shows exon-intron borders (ANOVA, P=0.00646).
(F) Schematic of high (red, left) and low (black, right) splicing events, defined as the skipped exon inclusion isoforms in the upper (75%) and lower (25%) quantiles, respectively, with no difference in inclusion between cocaine and saline.
(G) Distribution of cocaine H3K36me3 ChIP-seq reads in the skipped exon flanking region (+/− 150bp) of high (red) and low (black) skipped exons. Dashed grey line shows exon-intron borders (ANOVA, P=0.766).
(H) Representative H3K36me3 ChIP-seq data and called peaks of alternatively spliced transcripts in NAc following cocaine and saline SA.
(I) Quantification (top) and representative blots (bottom) of PCR validation of selected cocaine regulated alternative splicing events identified by rMATS (Student’s t-test, *p<0.05, **p<0.01)
To define the correlation between alternative splicing events and global H3K36me3 enrichment, we quantified the mean H3K36me3 ChIP-seq signal distribution in a ±150 bp region flanking the splice sites of each skipped exon in both cocaine and saline SA samples (Figure 2C). We categorized two subtypes of skipped exons based on RNA-seq data, similarly to our published approach (Hu et al., 2020). Skipped exons were categorized as (i) ‘gain/loss’ if the isoform was differentially expressed following cocaine SA (Figure 2D-E) or (ii) ‘high/low’ if the isoform was in the 75% (high) and 25% (low) quantiles across both cocaine and saline treatments, and isoform expression was not different between saline and cocaine SA treatments (Figure 2F-G). We found that H3K36me3 was differentially enriched at splice junctions only in the cocaine gain/loss, but not high/low, category (Figures 2E & 2G). We found no difference in H3K36me3 enrichment at constitutive exon junctions, demonstrating the specific relevance of H3K36me3 junction enrichment to alternatively spliced exons (Figure 2E). H3K36me3 ChIP-seq data show differential enrichment of H3K36me3 at the relevant splice junction of representative genes (Figure 2H). We biochemically validated alternative splicing of a subset of these genes. Splicing PCR of NAc tissue from a biological replicate of cocaine treatment recapitulated NAc rMATS results for 4 out of 5 transcripts (Figure 2I). These findings indicate that cocaine SA regulates H3K36me3 enrichment specifically at differentially expressed isoforms, but does not grossly redistribute this modification genome-wide.
Alternative splicing and H3K36me3 enrichment of splice factor gene, Srsf11
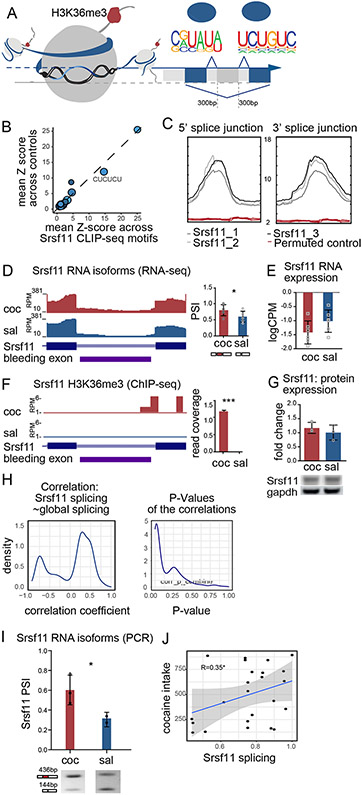
H3K36me3 may regulate cocaine-driven alternative splicing directly, via interactions with the splicing or transcriptional machinery at the spliced transcript, or indirectly via regulation of splice factor gene expression. In considering the latter case, we analyzed splice factor motif enrichment at cocaine alternative splice junctions located 150bps up- and down-stream of skipped exon splice site and identified 8 splice factor binding motifs. The corresponding splice factors were identified using MEME-TOMTOM (Gupta et al., 2007) and published iCLIP-sequencing data sets from mouse neurons (Chen et al., 2018; Rodor et al., 2017; Takeuchi et al., 2018; Vuong et al., 2016) (Figure 3A, Supplement figure 2). We focused on the motif for serine/arginine-rich splicing factor 11 (Srsf11), as the Srsf11 motifs identified in our dataset were also those most enriched by Srsf11 iCLIP-seq (CUCUCU) (Gonatopoulos-Pournatzis et al., 2018) (Figure 3B). We further validated the computational data by quantifying the enrichment distribution of Srsf11 at the junction regions. We identified robust Srsf11 binding motif enrichment at exon start and end sites, but a ‘flat-line’-like pattern of enrichment at the permuted junction controls (Figure 3C). Seven additional splice motifs were enriched in our dataset, including neuro-oncological ventral antigen 2 (Nova2) and polypyrimidine tract binding protein 2 (PTBP2) (Saito et al., 2019), but the enrichment patterns at splice junctions were highly similar to the permuted controls, indicating that those splice factors were not uniquely enriched at splice junctions of alternatively spliced transcripts in cocaine SA (Supplement figure 2).
Figure 3. Alternative splicing and H3K36me3 enrichment of splice factor gene, Srsf11.
(A) Unbiased motif analysis of alternative exons with higher inclusion in cocaine SA revealed enrichment motifs in the splice junctions (150bp up- and downstream of the splice site), including that of Srsf11.
(B) Scatter plot of averaged z-score of Srsf11 binding frequency on splicing motifs identified in cocaine SA splice junctions compared to saline SA control. Dot diameter corresponds to −log(p-value), and lighter blue corresponds to higher percentage of this motif found in the target sequence.
(C) Profile plot of Srsf11 iCLIP-seq signal at cocaine SA alternative splicing junctions (top, “bell-curve” like, gray) and permuted control (bottom, “flat-line” like, red).
(D) NAc RNA-seq visualization (left) and quantification (right) of RNA-seq at Srsf11 alternative splice junction after cocaine and saline SA.
(E) Quantification of Srsf11 NAc expression levels by RNA-seq (logCPM) after cocaine and saline SA.
(F) NAc H3K36me3 ChIP-seq visualization (left) and quantification (right) at alternative splice junction of Srsf11 after cocaine and saline SA.
(G) NAc western blot and quantification of Srsf11 protein expression after cocaine and saline SA.
(H) Density plot of correlation coefficient (left) and its corresponding P-value (right) between the PSI values of Srsf11 and those of its putative target genes following cocaine SA in all three brain regions.
(I) PCR quantification and representative gel of Srsf11 exon skipping event in biological replicate of cocaine and saline SA NAc (*p<0.05, Student’s t-test).
(J) Scatter plot of correlation between Srsf11 inclusion levels (PSI) and cocaine intake within each animal from cocaine SA (Pearson correlation R=0.35, *P<0.05) .
Having identified Srsf11 as a putative splice factor regulating cocaine-mediated alternative splicing, we next examined its differential expression and H3K36me3 enrichment. Cocaine SA increased Srsf11 alternative splicing relative to saline (Figure 3D) with no change in total Srsf11 mRNA expression (Figure 3E). Cocaine SA also increased enrichment of H3K36me3 at Srsf11, including at the spliced exon (Figure 3F). There was no change in total Srsf11 protein expression following cocaine SA relative to saline (Figure 3G). To examine the connection between alternative splicing of Srsf11 and global cocaine-mediated alternative splicing, we analyzed the correlation between the PSI values of Srsf11 and its putative target transcripts following cocaine SA in all three brain regions (Figure 3H). The significant correlation suggested a regulatory role of a skipped exon Srsf11 isoform in cocaine regulated alternative splicing. PCR validation of Srsf11 alternative splicing was accomplished in a biological replicate of cocaine SA mice, using a single pair of primers to amplify multiple isoforms; PSI was quantified as the relative abundance of each isoform in a single lane (Figure 3I) (Gonatopoulos-Pournatzis et al., 2018).
Given our prior data that H3K36me3 regulates alternative splicing in the context of cocaine reward (Hu et al., 2017), we hypothesized that the Srsf11 exon inclusion isoform was associated with cocaine reward behavior. Indeed, we measured a positive linear correlation between cocaine intake and Srsf11 exon inclusion levels (Figure 3J). Taken together, cocaine SA led to enrichment of H3K36me3 at Srsf11 splice junctions and alternative splicing of Srsf11 and its target transcripts, without changing Srsf11 gene expression.
Epigenetic editing of H3K36me3 was sufficient to drive alternative splicing of Srsf11
The previous experiments established a correlation between cocaine driven H3K36me3 and alternative splicing of Srsf11. To test whether H3K36me3 enrichment was sufficient to drive alternative splicing of Srsf11, it was necessary to experimentally manipulate H3K36me3 at Srsf11 alone. To accomplish this, we fused nuclease deficient Cas9 (dCas9) to histone methyltransferase, Set2, for targeted epigenetic editing of H3K36me3 in NAc neurons (Figure 4A). Although in higher eukaryotes multiple H3K36 methyltransferases catalyze H3K36me1 and H3K36me2, the yeast Set2 homolog is specific to H3K36me3 (McDaniel and Strahl, 2017; Sorenson et al., 2016; Strahl et al., 2002). Given that yeast Set2 had not previously been expressed in brain, we first confirmed that viral overexpression of Set2 in NAc increased global H3K36me3 by quantitative histone mass spectrometry (Supplement Figures 3A-B), western blot (Supplement Figure 3C), and ChIP-seq (Supplement Figures 3D-E), relative to catalytically dead control, HSV-Set2(R195G), HSV-GFP control, and virus-naïve animals. Co-infusion of HSV-Set2 with SET domain inhibitor, Bay598, restored H3K36me3 enrichment to baseline, confirming that the increased H3K36me3 was due to catalytic activity of Set2 (Supplement Figure 3C).
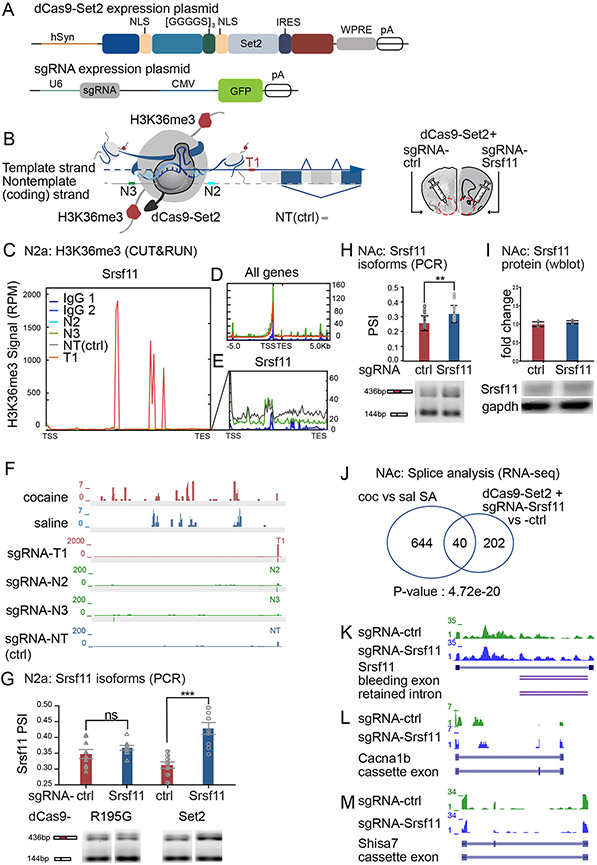
Figure 4. Epigenetic editing of H3K36me3 was sufficient to drive alternative splicing of Srsf11.
(A) Schematics of dCas9-Set2 and sgRNA expression plasmids.
(B) Schematic of Srsf11 and associated H3K36me3-enriched nucleosome shows binding sites of Srsf11-sgRNAs relative to other features of Srsf11. Srsf11-sgRNAs target the endogenous H3K36me3 enriched region of Srsf11 following cocaine SA. sgRNAs: T=template strand, N=non-template strand, NT=non-targeting, Ctrl = control.
(C-E) H3K36me3 enrichment at Srsf11 (C,E) and genome wide (D) in N2a cells transfected with dCas9-Set2 and Srsf11-sgRNAs. H3K36me3 enrichment following transfection with Srsf11-sgRNAs N2, N3 and NT was not above background (C). Note difference in RPM scale relative to panel D.
(F) H3K36me3 ChIP-seq of NAc following HSV-Set2 or -R195G treatment, and H3K36me3 CUT&RUN in N2a cells of dCas9-Set2 with Srsf11-sgRNAs T1, N2, N3 and NT control. Gray bar: 20bp protospacer motif. Note difference in RPM scale for Srsf11-sgRNA T1.
(G) Quantification of Srsf11 alternative isoform expression in N2a cells transfected with dCas9-Set2 or dCas9-Set2(R195G) and sgRNA T1 or NT (ns, non-significant, *** p<0.001. (One-way ANOVA with Tukey’s multiple comparison, P(dCas9-Set2, NT-sgRNA vs. Srsf11-T1-sgRNA)=0.0007; P(dCas9-Set2(R195G), NT-sgRNA vs. Srsf11-T1-sgRNA)=0.18).
(H) PCR quantification and representative gel of Srsf11 exon skipping event in biological replicate of dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA in NAc (Student’s t-test, **p<0.01).
(I) Srsf11 protein quantification and representative western blots following dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA in NAc (Student’s t-test).
(J) Comparison of alternatively spliced transcripts across dCas9-Set2 + Srsf11-sgRNA vs NT-sgRNA and cocaine vs saline SA (Fisher’s Exact Test, P<0.00001).
(K-M) NAc RNA-seq visualization and splicing event of Set2 (J), Cacna1b (K), and Shisa7 (L) following treatment with dCas9-Set2 + ctrl-sgRNA and dCas9-Set2 + Srsf11-sgRNA.
We designed Srsf11 sgRNAs to target dCas9-Set2 to the endogenous site of H3K36me3 enrichment by cocaine SA (Figure 4B). We noted previous data that intragenic targeting of dCas9 to the non-template (coding) DNA strand causes expression silencing, while targeting to the template DNA strand does not (Qi et al., 2013). We thus designed the Srsf11 sgRNAs to target either the template (T1) or non-template (N2, N3) strands, with minimal predicted off-target mismatch (see STAR methods) (Figure 4B). A control (ctrl), non-targeting (NT)-sgRNA does not align to any genomic sequence (Lorsch et al., 2019).
Next, we measured H3K36me3 enrichment using ‘Cleavage Under Targets and Release Using Nuclease’ (CUT&RUN) (Skene and Henikoff, 2017; Skene et al., 2018) in N2a cells transfected with dCas9-Set2 and sgRNAs T1, N2, or N3 (Figure 4C-E, Supplement figure 3F). dCas9-Set2 plus Srsf11-sgRNA-T1 led to a 200-fold increase of H3K36me3 enrichment compared to control, non-targeting (NT)-sgRNA, while sgRNAs N2 and N3 showed similar enrichment to NT (Figures 4C-E, Supplement figures 3F-G). sgRNAs T1, N2, N3 and NT all showed similar global H3K36me3 distributions, suggesting negligible off-target effects (Figures 4D-E; note change in RPM scale compared to Figure 4C). Track coverage confirmed that Srsf11-sgRNA-T1 enriched H3K36me3 at precisely the Srsf11-sgRNA-T1 photospacer motif, which was not observed for the control NT-sgRNA , or N2 and N3 (Figure 4F, note y-axis scales). Differential binding analysis found Srsf11 was the locus of greatest H3K36me3 enrichment (Supplement figure 3G), while off-target enrichment largely mapped to intergenic regions (Supplement figure 3H). We validated that N2a cell transfection of dCas9-Set2 with Srsf11-sgRNA-T1 increased Srsf11 exon inclusion compared to sgRNA-NT-control (Figure 4G, right), with no difference in mRNA expression measured by qPCR (not shown). There was no change in isoform abundance or total mRNA expression when N2a cells were transfected with dCas9-Set2(R195G), which lacks methyltransferase activity (Strahl et al., 2002) (Figure 4G, left). All following studies utilized the Srsf11-sgRNA-T1, referred to as Srsf11-sgRNA, and the non-targeting (NT)-sgRNA, referred to as ctrl-sgRNA.
We next expressed dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA in mouse NAc (Figure 4H). Neuron-specific expression was conferred by the human synapsin 1 gene promoter (Lorsch et al., 2019). We used within-animal controls wherein each NAc was injected unilaterally with dCas9-Set2 and either ctrl-sgRNA or Srsf11-sgRNA (Figure 4B). We found that expression of dCas9-Set2 with Srsf11-sgRNA increased Srsf11 skipped exon inclusion compared to control-NT-sgRNA (Figure 4H). There was no change in mRNA expression (not shown) or Srsf11 protein (Figure 4I). We confirmed Cas9 immunoreactivity localized specifically to NeuN+ neurons, such that 100% of dCas9+ cells were NeuN+ (Carpenter et al., 2020). Overall, we found that in mouse NAc neurons, intragenic H3K36me3 enrichment was sufficient to drive alternative isoform expression of Srsf11.
To determine whether alternative splicing of Srsf11 was relevant to cocaine-mediated alternative splicing, we performed RNA-sequencing and splice analysis of NAc expressing dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA (Figure 4 J-M). We found a common set of NAc transcripts that were alternatively spliced following Srsf11 splicing and cocaine treatment (Figure 4J, Table 1), including Srsf11 (Figure 4K). Many of the spliced transcripts function in neurobiology and substance use disorder (Table 1), such as Calcium Voltage-Gated Channel Subunit Alpha1 B (Cacna1b, Figure 4L), and Shisa Family Member 7 (Shisa7, Figure 4M), discussed below. Taken together, these data support a model in which cocaine-regulated splicing of Srsf11 leads to downstream splicing of neurobiologically-relevant Srsf11 target transcripts.
Table 1: Overlap of genes alternatively spliced in both cocaine and dCas9-Set2 contexts.
Key word searches were performed to assess how these genes might contribute to the underlying biology of alternative splicing in response to cocaine. A global interrogation of the literature suggests that most of the genes have known involvement in alternative splicing and addiction phenotypes. All overlapping genes have a Srsf11 DNA binding motif within the splice junction at which alternative splicing occurs, suggesting the role that Srsf11 might play as a critical regulator of splicing in response to cocaine. Several genes had limited literature hits, suggesting that these might be novel targets. Key words: Cocaine, Addiction, Splicing, Neuronal Context.
| ENSEMBL | SYMBOL | SRSF11 Motif ? |
Key word search (Cocaine, Addiction) |
Key word search (splicing, neuronal context) |
Citations | |
|---|---|---|---|---|---|---|
| 1 | ENSMUSG00000025085 | Ablim1 | Y | (1) Polymorphisms associated with novelty seeking, personality traits and alcohol dependency | (2) SNP in neuronal tissue involved in ADHD and BPD (3) Alternative splicing involved in ALS linked neurodegeneration |
(1) Wang KS, Liu X, Aragam N, Mullersman JE, Jian X, Pan Y, Liu Y. Polymorphisms in ABLIM1 are associated with personality traits and alcohol dependence. J Mol Neurosci. 2012 Feb;46(2):265-71. doi: 10.1007/s12031-011-9530-6. (2) Kimm J.E. van Hulzen, Claus J. Scholz, Barbara Franke, Stephan Ripke, Marieke Klein, Andrew McQuillin, Edmund J. Sonuga-Barke, John R. Kelsoe, Mikael Landén, Ole A. Andreassen, Klaus-Peter Lesch, Heike Weber, Stephen V. Faraone, Alejandro Arias-Vasquez, Andreas Reif. Genetic Overlap Between Attention-Deficit/Hyperactivity Disorder and Bipolar Disorder: Evidence From Genome-wide Association Study Meta-analysis. Biological Psychiatry. 2017 Nov;82(9):634-641. doi:10.1016/j.biopsych.2016.08.040 (3) Orozco, D., Edbauer, D. FUS-mediated alternative splicing in the nervous system: consequences for ALS and FTLD. J Mol Med 91, 1343–1354 (2013). https://doi.org/10.1007/s00109-013-1077-2 |
| 2 | ENSMUSG00000040537 | Adam22 | Y | NA | (1) ADAM22 and ADAM family genes found alternatively spliced in human brain tissue and glioma (2) Major neuronal receptor involved in Schwann cell signaling |
(1) Gödde NJ, D'Abaco GM, Paradiso L, Novak U. Differential coding potential of ADAM22 mRNAs. Gene. 2007 Nov 15;403(1-2):80-8. doi: 10.1016/j.gene.2007.07.033. (2) Ozkaynak E, Abello G, Jaegle M, van Berge L, Hamer D, Kegel L, Driegen S, Sagane K, Bermingham JR Jr, Meijer D. Adam22 is a major neuronal receptor for Lgi4-mediated Schwann cell signaling. J Neurosci. 2010 Mar 10;30(10):3857-64. doi: 10.1523/JNEUROSCI.6287-09.2010. |
| 3 | ENSMUSG00000019986 | Ahi1 | Y | (1) Loss promotes depression via tyrosine hydroxylase down-regulation and circadian clock pathway in mice | (2) Reduced serum levels associated with Alheimer's disease (3) Abberent splicing results in oncogene status |
(1) Guo D, Zhang S, Sun H, Xu X, Hao Z, Mu C, Xu X, Wang G, Ren H. Tyrosine hydroxylase down-regulation after loss of Abelson helper integration site 1 (AHI1) promotes depression via the circadian clock pathway in mice. J Biol Chem. 2018 Apr 6;293(14):5090-5101. doi: 10.1074/jbc.RA117.000618. (2) Ting LL, Lu HT, Yen SF, Ngo TH, Tu FY, Tsai IS, Tsai YH, Chang FY, Li XJ, Li S, Lee CK, Kao SH, Kuo YM, Lin YF. Expression of AHI1 Rescues Amyloidogenic Pathology in Alzheimer's Disease Model Cells. Mol Neurobiol. 2019 Nov;56(11):7572-7582. doi: 10.1007/s12035-019-1587-1. (3) Kennah E, Ringrose A, Zhou LL, Esmailzadeh S, Qian H, Su MW, Zhou Y, Jiang X. Identification of tyrosine kinase, HCK, and tumor suppressor, BIN1, as potential mediators of AHI-1 oncogene in primary and transformed CTCL cells. Blood. 2009 May 7;113(19):4646-55. doi: 10.1182/blood-2008-08-174037. |
| 4 | ENSMUSG00000004113 | Cacna1b | Y | (1) Cocaine seeking reinstatement (2) Altered expression after methanphetamine treatment (3) Reversal of Peripheral Neuropathic Pain by small molecule inhibition |
(4) Alternative splicing impacts excitatory neurotransmission and is linked to behavioral responses to aversive stimuli (5) Cell-specific alternative splicing modulates opioid sensitivity (6) Alternative splicing of Cacna1b pre-mRNA modifies CaV2.2 channel properties across different neurons |
(1) Buchta WC, Moutal A, Hines B, Garcia-Keller C, Smith ACW, Kalivas P, Khanna R, Riegel AC. Dynamic CRMP2 Regulation of CaV2.2 in the Prefrontal Cortex Contributes to the Reinstatement of Cocaine Seeking. Mol Neurobiol. 2020 Jan;57(1):346-357. doi: 10.1007/s12035-019-01711-9. (2) González B, Rivero-Echeto C, Muñiz JA, Cadet JL, García-Rill E, Urbano FJ, Bisagno V. Methamphetamine blunts Ca(2+) currents and excitatory synaptic transmission through D1/5 receptor-mediated mechanisms in the mouse medial prefrontal cortex. Addict Biol. 2016 May;21(3):589-602. doi: 10.1111/adb.12249. (3) Shan Z, Cai S, Yu J, Zhang Z, Vallecillo TGM, Serafini MJ, Thomas AM, Pham NYN, Bellampalli SS, Moutal A, Zhou Y, Xu GB, Xu YM, Luo S, Patek M, Streicher JM, Gunatilaka AAL, Khanna R. Reversal of Peripheral Neuropathic Pain by the Small-Molecule Natural Product Physalin F via Block of CaV2.3 (R-Type) and CaV2.2 (N-Type) Voltage-Gated Calcium Channels. ACS Chem Neurosci. 2019 Jun 19;10(6):2939-2955. doi: 10.1021/acschemneuro.9b00166. (4) Bunda, A., LaCarubba, B., Bertolino, M. et al. Cacna1b alternative splicing impacts excitatory neurotransmission and is linked to behavioral responses to aversive stimuli. Mol Brain 12, 81 (2019). https://doi.org/10.1186/s13041-019-0500-1 (5) López Soto, EJ, Lipscombe, D. Cell-specific exon methylation and CTCF binding in neurons regulate calcium ion channel splicing and function. eLife 2020;9:e54879. DOI: 10.7554/eLife.54879 (6) Lopez Soto EJ, Gandal MJ, Gonatopoulos-Pournatzis T, Heller EA, Luo D, Zheng S. Mechanisms of Neuronal Alternative Splicing and Strategies for Therapeutic Interventions. J Neurosci. 2019;39(42):8193-8199. doi:10.1523/JNEUROSCI.1149-19.2019 |
| 5 | ENSMUSG00000027829 | Ccnl1 (Ania-6) | Y | (1) RNA polymerase II-associated cyclin that is induced by dopamine, cocaine, and glutamate. | (2) Splice variants regulated distinct signaling pathways in striatal neurons | (1) Nairn AC, Greengard P. A novel cyclin provides a link between dopamine and RNA processing. Neuron. 2001 Oct 25;32(2):174-6. doi: 10.1016/s0896-6273(01)00469-x. PMID: 11683987. (2) Sgambato V, Minassian R, Nairn AC, Hyman SE. Regulation of ania-6 splice variants by distinct signaling pathways in striatal neurons. J Neurochem. 2003 Jul;86(1):153-64. doi: 10.1046/j.1471-4159.2003.01816.x. |
| 6 | ENSMUSG00000055447 | Cd47 | Y | (1) Cell-surface signaling molecule that has decreased expression (mPFC) at 1-day and 10-days but not 100 days of abstinence. | (2) SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-κB-CD47 axis. (3) Plasma membrane protein with four known isoforms that encode variation in its intracytoplasmic tail. Isoforms containing exon 10 are associated with memory retention in rats. CD47-deficient neurons have impaired axon and dendrite formation in development. |
(1) Freeman WM, Lull ME, Patel KM, et al. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29. Published 2010 Feb 26. doi:10.1186/1471-2202-11-29 (2) Liu F, Dai M, Xu Q, et al. SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-κB-CD47 axis. Oncogene. 2018;37(18):2394-2409. doi:10.1038/s41388-017-0119-6 (3) Begg, B.E., Jens, M., Wang, P.Y. et al. Concentration-dependent splicing is enabled by Rbfox motifs of intermediate affinity. Nat Struct Mol Biol 27, 901–912 (2020). https://doi.org/10.1038/s41594-020-0475-8 |
| 7 | ENSMUSG00000112964 | Riken | Y | NA | (1) Alternatively spliced into LncRNAs that modulate neural development by regulating the Sox6 through sequestering miRNAs | (1) Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019 May;52(3):e12573. doi: 10.1111/cpr.12573. |
| 8 | ENSMUSG00000031819 | Emc8 | Y | NA | NA | |
| 9 | ENSMUSG00000019978 | Epb41l2 | Y | (1) Four SNPs in EPB41 have significant male-specific association with smoking cessation and abstinence. | (2) Alternartively spliced in a tissue specifica manner with two homologues that are strongly expressed in the brain (4.1N, EPB41L1; and 4.1B, EPB41L3). (3) High level, focal expression of splice variant (4.1B mRNA) in select neuronal populations within the mouse brain, including Purkinje cells of the cerebellum, pyramidal cells in hippocampal regions CA1-3, thalamic nuclei, and olfactory bulb. |
(1) Lee W, Bergen AW, Swan GE, et al. Gender-stratified gene and gene-treatment interactions in smoking cessation. Pharmacogenomics J. 2012;12(6):521-532. doi:10.1038/tpj.2011.30 (2) Peters LL, Weier HU, Walensky LD, Snyder SH, Parra M, Mohandas N, Conboy JG. Four paralogous protein 4.1 genes map to distinct chromosomes in mouse and human. Genomics. 1998 Dec 1;54(2):348-50. doi: 10.1006/geno.1998.5537. (3) Parra M, Gascard P, Walensky LD, Gimm JA, Blackshaw S, Chan N, Takakuwa Y, Berger T, Lee G, Chasis JA, Snyder SH, Mohandas N, Conboy JG. Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J Biol Chem. 2000 Feb 4;275(5):3247-55. doi: 10.1074/jbc.275.5.3247. |
| 10 | ENSMUSG00000075415 | Fnbp1 | Y | (1) Upregulated in the rat medial prefrontal cortex following a history of alcohol dependence. | (2) Multiple splice variants, one of which is predominately expressed in the brain whereas others expressed ubiquitously. Functionally different splicing variants have different responses to Rnd2 in association with N-WASP to induces neurite branching. (3) Indication in ALS and limb morphogenesis |
(1) Tapocik JD, Solomon M, Flanigan M, et al. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J. 2013;13(3):286-296. doi:10.1038/tpj.2012.17 (2) Kakimoto T, Katoh H, Negishi M. Identification of splicing variants of Rapostlin, a novel RND2 effector that interacts with neural Wiskott-Aldrich syndrome protein and induces neurite branching. J Biol Chem. 2004 Apr 2;279(14):14104-10. doi: 10.1074/jbc.M312763200. Epub 2004 Jan 19. (3) Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N, Hayashi N, Yokoi D, Watanabe H, Watanabe H, Katsuno M, Izumi Y, Kanai K, Hattori N, Morita M, Taniguchi A, Kano O, Oda M, Shibuya K, Kuwabara S, Suzuki N, Aoki M, Ohta Y, Yamashita T, Abe K, Hashimoto R, Aiba I, Okamoto K, Mizoguchi K, Hasegawa K, Okada Y, Ishihara T, Onodera O, Nakashima K, Kaji R, Kamatani Y, Ikegawa S, Momozawa Y, Kubo M, Ishida N, Minegishi N, Nagasaki M, Sobue G. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol. 2020 Sep 23;3(1):526. doi: 10.1038/s42003-020-01251-2. |
| 11 | ENSMUSG00000038766 | Gabpb2 | Y | NA | (1) p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. | (1) Mardaryev AN, Gdula MR, Yarker JL, et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development. 2014;141(1):101-111. doi:10.1242/dev.103200 |
| 12 | ENSMUSG00000021552 | Gkap1 | Y | NA | NA | |
| 13 | ENSMUSG00000056073 | Grik2 | Y | (1) Upregulated in the striatum after adolescent administration of methylphenidate in rats. | (2) Evidence for several different splicing variants with variable expression in NT2 cells and in human hippocampus. | (1) Adriani W, Leo D, Greco D, Rea M, di Porzio U, Laviola G, Perrone-Capano C. Methylphenidate administration to adolescent rats determines plastic changes on reward-related behavior and striatal gene expression. Neuropsychopharmacology. 2006 Sep;31(9):1946-56. doi: 10.1038/sj.npp.1300962. (2) Barbon A, Vallini I, Barlati S. Genomic organization of the human GRIK2 gene and evidence for multiple splicing variants. Gene. 2001;274(1–2):187-197. doi:10.1016/S0378-1119(01)00611-4. |
| 14 | ENSMUSG00000000568 | Hnrnpd | Y | (1) Quantitative trait gene underlying differential behavioral sensitivity to methamphetamine. (2) Central Node Protein Networks in D1 medium spiny neurons. |
(1) Alternative splicing in C- and N-terminal domains in a cell-type specific manner. D1 receptor activation increases avalibility of hnRNP H C-terminal epitope as evidenced by decrease in Hnmph1 transcript but no change in H protein levels. (3) Differences in splicing variants of Hnmpd alters predicted splicing program based on its RNA binding motifs. |
(1) Ruan QT, Yazdani N, Beierle JA, et al. Changes in neuronal immunofluorescence in the C- versus N-terminal domains of hnRNP H following D1 dopamine receptor activation. Neurosci Lett. 2018;684:109-114. doi:10.1016/j.neulet.2018.07.015 (2) Mansuri MS, Peng G, Wilson RS, et al. Differential Protein Expression in Striatal D1- and D2-Dopamine Receptor-Expressing Medium Spiny Neurons. Proteomes. 2020;8(4):27. Published 2020 Oct 13. doi:10.3390/proteomes8040027 (3) Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Human molecular genetics. 2012;21:4171–4186. |
| 15 | ENSMUSG00000022556 | Hsf1 | Y | (1) Alcohol activates HSF1 and other heat shock factor genes. HSF1 may be necessary to confer Sp1 regulation of ethanol sensitivity. (2) HSF1 transcriptional activity mediates alcohol induction of Vamp2 expression and GABA release. |
(3) Long noncoding RNAs regulate Hsf1's expression level and splicing isoforms which can cause changes in oligomerization, subcellular compartmentalization, PTMs, target gene activation and protein stability in normal and disease conditions. Protein misfolding associated with Huntington disease and Parkinson disease. | (1) Pignataro L, Varodayan FP, Tannenholz LE, Harrison NL. The regulation of neuronal gene expression by alcohol. Pharmacol Ther. 2009;124(3):324-335. doi:10.1016/j.pharmthera.2009.09.002 (2) Varodayan FP, Harrison NL. HSF1 transcriptional activity mediates alcohol induction of Vamp2 expression and GABA release. Front Integr Neurosci. 2013;7:89. Published 2013 Dec 11. doi:10.3389/fnint.2013.00089 (3) Gomez-Pastor, R., Burchfiel, E. & Thiele, D. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19, 4–19 (2018). https://doi.org/10.1038/nrm.2017.73 |
| 16 | ENSMUSG00000030556 | Lrrc28 | Y | NA | NA | |
| 17 | ENSMUSG00000031683 | Lsm6 | Y | (1) Associated with smoking-related phenotypes. | (2) Sm-like proteins are thought to be important for pre-mRNA splicing. (3) LSMs play a critical role in modulating development-related gene expression through the regulation of mRNA splicing and decay in Arabidopsis. |
(1) Drgon, T., Montoya, I., Johnson, C. et al. Genome-Wide Association for Nicotine Dependence and Smoking Cessation Success in NIH Research Volunteers. Mol Med 15, 21–27 (2009). https://doi.org/10.2119/molmed.2008.00096 (2) Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005 Jun;33(Pt 3):433-8. doi: 10.1042/BST0330433. (3) Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012 Dec;24(12):4930-47. doi: 10.1105/tpc.112.103697. |
| 18 | ENSMUSG00000007411 | Mark3 | Y | NA | NA | |
| 19 | ENSMUSG00000000631 | Myo18a | Y | (1) Downregulated in prelimbic cortex after chronic cocaine exposure. | (2) Alternative splicing of the Myo18A results in two isoforms with differential expression, priming and activating functions in tissue resident macrophages. (3) Involved in cell polarity during neurogenesis. |
(1) Pignataro L, Varodayan FP, Tannenholz LE, Harrison NL. The regulation of neuronal gene expression by alcohol. Pharmacol Ther. 2009;124(3):324-335. doi:10.1016/j.pharmthera.2009.09.002 (2) Yang L, Carrillo M, Wu YM, DiAngelo SL, Silveyra P, Umstead TM, et al. (2015) SP-R210 (Myo18A) Isoforms as Intrinsic Modulators of Macrophage Priming and Activation. PLoS ONE 10(5): e0126576. https://doi.org/10.1371/journal.pone.0126576 (3) Xie Z, Hur SK, Zhao L, Abrams CS, Bankaitis VA. A Golgi Lipid Signaling Pathway Controls Apical Golgi Distribution and Cell Polarity during Neurogenesis. Dev Cell. 2018;44(6):725-740.e4. doi:10.1016/j.devcel.2018.02.025 |
| 20 | ENSMUSG00000055053 | Nfic | Y | (1) Identified as a CREB Binding Protein (CBP)-Interacting protein in NAc and Striatum after cocaine injections in mice. | (2) Tissue- and cell-specific alternative splicing resulting in different biological functions in CNS versus other tissues. | (1) Funahashi Y, Ariza A, Emi R, Xu Y, Shan W, Suzuki K, Kozawa S, Ahammad RU, Wu M, Takano T, Yura Y, Kuroda K, Nagai T, Amano M, Yamada K, Kaibuchi K. Phosphorylation of Npas4 by MAPK Regulates Reward-Related Gene Expression and Behaviors. Cell Rep. 2019 Dec 3;29(10):3235-3252.e9. doi: 10.1016/j.celrep.2019.10.116. (2) Lamani E, Wu Y, Dong J, Litaker MS, Acevedo AC, MacDougall M. Tissue- and cell-specific alternative splicing of NFIC. Cells Tissues Organs. 2009;189(1-4):105-10. doi: 10.1159/000152912. |
| 21 | ENSMUSG00000021068 | Nin | Y | NA | (1) RNA splicing via exon switching regulates neural stem cell fate. (2) Rbfox proteins promote neuronal differentiation by switching Ninein from a centrosomal splice form in NPCs to a non-centrosomal isoform in neurons. |
(1) Hayakawa-Yano Y, Yano M. An RNA Switch of a Large Exon of Ninein Is Regulated by the Neural Stem Cell Specific-RNA Binding Protein, Qki5. Int J Mol Sci. 2019 Feb 26;20(5):1010. doi: 10.3390/ijms20051010. (2) Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, Black DL, Kharchenko PV, Sharp PA, Walsh CA. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016 Aug 25;166(5):1147-1162.e15. doi: 10.1016/j.cell.2016.07.025. |
| 22 | ENSMUSG00000021772 | Nkiras1 | Y | NA | (1) Spliced into two variants that differentially interact with NF-kB | (1) Sarais F, Rebl H, Verleih M, Ostermann S, Krasnov A, Köllner B, Goldammer T, Rebl A. Characterisation of the teleostean κB-Ras family: The two members NKIRAS1 and NKIRAS2 from rainbow trout influence the activity of NF-κB in opposite ways. Fish Shellfish Immunol. 2020 Nov;106:1004-1013. doi: 10.1016/j.fsi.2020.08.052. Epub 2020 Sep 2. PMID: 32890762. |
| 23 | ENSMUSG00000037514 | Pank2 | Y | (1) Mutations involved in disruptions of iron regulation in the brain and periphery in cocaine addiction (2) Hypomethylated after thirty days of cocaine withdrawal relative to one day. |
(3) Mutations involvted in neurodegenerative disorders. | (1) Ersche, K., Acosta-Cabronero, J., Jones, P. et al. Disrupted iron regulation in the brain and periphery in cocaine addiction. Transl Psychiatry 7, e1040 (2017). https://doi.org/10.1038/tp.2016.271 (2) Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, Kennedy P, Nestler EJ, Szyf M, Yadid G. Role of DNA Methylation in the Nucleus Accumbens in Incubation of Cocaine Craving. Journal of Neuroscience. May 2015, 35 (21) 8042-8058; DOI: 10.1523/JNEUROSCI.3053-14.2015 (3) Dastsooz, H., Nemati, H., Fard, M.A.F. et al. Novel mutations in PANK2 and PLA2G6 genes in patients with neurodegenerative disorders: two case reports. BMC Med Genet 18, 87 (2017). https://doi.org/10.1186/s12881-017-0439-y |
| 24 | ENSMUSG00000056851 | Pcbp2 | Y | (1) PCBP2 can bind to the Mu Opior Receptor promoter to activate transcription. | (1) Polycytosine-binding proteins (PCBPs) bind to poly(C) DNA and RNA sequences to regulate transcription and post-transcriptional processing. PCBs regulate axonogenesis in neurodevelopment. | (1) Bryant CD, Yazdani N. RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 2016;15(1):169-186. doi:10.1111/gbb.12273 |
| 25 | ENSMUSG00000027881 | Prpf38b | Y | (1) Increased expression in adolescent bindge drinking. | (2) Pre-mRNA splicing factor predictive of responsiveness to trastuzumab treatment in breast cancer. | (1) McBride WJ, Kimpel MW, McClintick JN, et al. Changes in gene expression within the extended amygdala following binge-like alcohol drinking by adolescent alcohol-preferring (P) rats. Pharmacol Biochem Behav. 2014;117:52-60. doi:10.1016/j.pbb.2013.12.009 (2) Abdel-Fatah TMA, Rees RC, Pockley AG, et al. The localization of pre mRNA splicing factor PRPF38B is a novel prognostic biomarker that may predict survival benefit of trastuzumab in patients with breast cancer overexpressing HER2. Oncotarget. 2017;8(68):112245-112257. Published 2017 Nov 18. doi:10.18632/oncotarget.22496 |
| 26 | ENSMUSG00000040225 | Prrc2c | Y | (1) Differentially expressed in hippocampus after cocaine exposure. | (2) SRSF1 downregulation leds to alternative splicing of Prrc2c via the skipping of an exon in primary lung tumors. | (1) Webb, A., Papp, A.C., Curtis, A. et al. RNA sequencing of transcriptomes in human brain regions: protein-coding and non-coding RNAs, isoforms and alleles. BMC Genomics 16, 990 (2015). https://doi.org/10.1186/s12864-015-2207-8 (2) de Miguel FJ, Sharma RD, Pajares MJ, Montuenga LM, Rubio A, Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014 Feb 15;74(4):1105-15. doi: 10.1158/0008-5472.CAN-13-1481. |
| 27 | ENSMUSG00000063179 | Pstk | Y | NA | NA | |
| 28 | ENSMUSG00000010608 | Rbm25 | Y | (1) Involved in pre-mRNA splicing of the Mu Opiod Receptor gene OPRM1 through interactions with SRSF proteins and U1 snRNP. (2) Regulator of spliceosome assembly and alternative mRNA splicing activity that is downregulated in prenatal alcohol exposure in females and upregulated in chronic alcohol exposure in males (3). |
(4) Global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. | (1) Xu J, Lu Z, Xu M, et al. A heroin addiction severity-associated intronic single nucleotide polymorphism modulates alternative pre-mRNA splicing of the μ opioid receptor gene OPRM1 via hnRNPH interactions. J Neurosci. 2014;34(33):11048-11066. doi:10.1523/JNEUROSCI.3986-13.2014 (2) Salem NA, Mahnke AH, Konganti K, Hillhouse AE, Miranda RC. Cell-type and fetal-sex-specific targets of prenatal alcohol exposure in developing mouse cerebral cortex. iScience. 2021;24(5):102439. Published 2021 Apr 20. doi:10.1016/j.isci.2021.102439 (3) Wilhelm CJ, Hashimoto JG, Roberts ML, Sonmez MK, Wiren KM. Understanding the addiction cycle: a complex biology with distinct contributions of genotype vs. sex at each stage. Neuroscience. 2014;279:168-186. doi:10.1016/j.neuroscience.2014.08.041 (4) Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, Gozani O. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J Biol Chem. 2017 Aug 11;292(32):13381-13390. doi: 10.1074/jbc.M117.784371. |
| 29 | ENSMUSG00000035325 | Sec31a | Y | NA | (1) Protein transport protein, involved in COPII SEC31A mutation that affects ER homeostasis, causing a neurological syndromes. | (1) Halperin D, Kadir R, Perez Y, Drabkin M, Yogev Y, Wormser O, Berman EM, Eremenko E, Rotblat B, Shorer Z, Gradstein L, Shelef I, Birk R, Abdu U, Flusser H, Birk OS. SEC31A mutation affects ER homeostasis, causing a neurological syndrome. J Med Genet. 2019 Mar;56(3):139-148. doi: 10.1136/jmedgenet-2018-105503. |
| 31 | ENSMUSG00000021870 | Slmap | Y | NA | (1) Undergoes developmentally regulated and tissue specific alternative splicing that results in two mutually exclusive isoforms with different biological functions. | (1) Wielowieyski PA, Sevinc S, Guzzo R, Salih M, Wigle JT, Tuana BS. Alternative splicing, expression, and genomic structure of the 3' region of the gene encoding the sarcolemmal-associated proteins (SLAPs) defines a novel class of coiled-coil tail-anchored membrane proteins. J Biol Chem. 2000 Dec 8;275(49):38474-81. doi: 10.1074/jbc.M007682200. |
| 32 | ENSMUSG00000020439 | Smtn | Y | NA | NA | |
| 33 | ENSMUSG00000025006 | Sorbs1 | Y | (1) Methamphetamineinduced methylation of promoter region. | (2) Splice variants associated with COPD and obesity. | (1) Itzhak, Y., Ergui, I. & Young, J. Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol Psychiatry 20, 232–239 (2015). https://doi.org/10.1038/mp.2014.7 (2) Brandsma CA, Guryev V, Timens W, Ciconelle A, Postma DS, Bischoff R, Johansson M, Ovchinnikova ES, Malm J, Marko-Varga G, Fehniger TE, van den Berge M, Horvatovich P. Integrated proteogenomic approach identifying a protein signature of COPD and a new splice variant of SORBS1. Thorax. 2020 Feb;75(2):180-183. doi: 10.1136/thoraxjnl-2019-213200. |
| 34 | ENSMUSG00000031626 | Sorbs2 | Y | (1) Associated with nicotine dependency and latency to smoke first cigarette in the morning. | (2) Impaired dendritic development and memory in Sorbs2 knock-out mice. | (1) Chen J, Loukola A, Gillespie NA, Peterson R, Jia P, Riley B, Maes H, Dick DM, Kendler KS, Damaj MI, Miles MF, Zhao Z, Li MD, Vink JM, Minica CC, Willemsen G, Boomsma DI, Qaiser B, Madden PAF, Korhonen T, Jousilahti P, Hällfors J, Gelernter J, Kranzler HR, Sherva R, Farrer L, Maher B, Vanyukov M, Taylor M, Ware JJ, Munafò MR, Lutz SM, Hokanson JE, Gu F, Landi MT, Caporaso NE, Hancock DB, Gaddis NC, Baker TB, Bierut LJ, Johnson EO, Chenoweth M, Lerman C, Tyndale R, Kaprio J, Chen X. Genome-Wide Meta-Analyses of FTND and TTFC Phenotypes. Nicotine Tob Res. 2020 May 26;22(6):900-909. doi: 10.1093/ntr/ntz099. (2) Zhang Q, Gao X, Li C, Feliciano C, Wang D, Zhou D, Mei Y, Monteiro P, Anand M, Itohara S, Dong X, Fu Z, Feng G. Impaired Dendritic Development and Memory in Sorbs2 Knock-Out Mice. J Neurosci. 2016 Feb 17;36(7):2247-60. doi: 10.1523/JNEUROSCI.2528-15.2016. |
| 35 | ENSMUSG00000021007 | Spata7 | Y | NA | NA | |
| 36 | ENSMUSG00000055436 | Srsf11 | Y | (1) Implicated as hub gene in methamphetamine intake in mice. | (2) Involved in early splicing complex assembly in neuronal microexon usage. (3) Coactivates neuronal microexons disrupted in autism. (4) Inhibits exon 7 inclusion in SMN2, associated with worse spinal muscular atrophy. |
(1) Hitzemann R, Iancu OD, Reed C, Baba H, Lockwood DR, Phillips TJ. Regional Analysis of the Brain Transcriptome in Mice Bred for High and Low Methamphetamine Consumption. Brain Sci. 2019;9(7):155. Published 2019 Jun 30. doi:10.3390/brainsci9070155 (2) Lopez Soto EJ, Gandal MJ, Gonatopoulos-Pournatzis T, Heller EA, Luo D, Zheng S. Mechanisms of Neuronal Alternative Splicing and Strategies for Therapeutic Interventions. J Neurosci. 2019;39(42):8193-8199. doi:10.1523/JNEUROSCI.1149-19.2019 (3) Gonatopoulos-Pournatzis T, Wu M, Braunschweig U, Roth J, Han H, Best AJ, Raj B, Aregger M, O'Hanlon D, Ellis JD, Calarco JA, Moffat J, Gingras AC, Blencowe BJ. Genome-wide CRISPR-Cas9 Interrogation of Splicing Networks Reveals a Mechanism for Recognition of Autism-Misregulated Neuronal Microexons. Mol Cell. 2018 Nov 1;72(3):510-524.e12. doi: 10.1016/j.molcel.2018.10.008. (4) Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS One. 2014 Dec 15;9(12):e115205. doi: 10.1371/journal.pone.0115205. |
| 37 | ENSMUSG00000004626 | Stxbp2 | Y | (1) (2) Genetic biomarker for alcoholism and alcohol-responsiveness in human brain. (3) Syntaxin binding proteins associated with cocaine self-administration. |
(4) Splice variant is a biomarker in Schizophrenia. | (1) Manzardo AM, McGuire A, Butler MG. Clinically relevant genetic biomarkers from the brain in alcoholism with representation on high resolution chromosome ideograms. Gene. 2015 Apr 15;560(2):184-94. doi: 10.1016/j.gene.2015.01.064. (2) Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002 May;81(4):802-13. doi: 10.1046/j.1471-4159.2002.00860.x. (3) Tannu, N., Howell, L. & Hemby, S. Integrative proteomic analysis of the nucleus accumbens in rhesus monkeys following cocaine self-administration. Mol Psychiatry 15, 185–203 (2010). https://doi.org/10.1038/mp.2008.53 (4) Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(4):382-392. doi:10.1002/ajmg.b.31181 |
| 38 | ENSMUSG00000029452 | Tmem116 | Y | (1) (2) Positively associated with binge drinking. | (3) Identified target for alternative pre-mRNA polyadenylation and splicing by Herpes simplex virus ICP27. | (1) Saba LM, Flink SC, Vanderlinden LA, et al. The sequenced rat brain transcriptome--its use in identifying networks predisposing alcohol consumption. FEBS J. 2015;282(18):3556-3578. doi:10.1111/febs.13358 (2) Chen IC, Kuo PH, Yang AC, et al. CUX2, BRAP and ALDH2 are associated with metabolic traits in people with excessive alcohol consumption. Sci Rep. 2020;10(1):18118. Published 2020 Oct 22. doi:10.1038/s41598-020-75199-y (3) Tang S, Patel A, Krause PR. Herpes simplex virus ICP27 regulates alternative pre-mRNA polyadenylation and splicing in a sequence-dependent manner. Proc Natl Acad Sci U S A. 2016;113(43):12256-12261. doi:10.1073/pnas.1609695113 |
| 39 | ENSMUSG00000052707 | Tnrc6a | Y | NA | (1) TNRC6 proteins are important factors for RNA-mediated modulation of transcription. | (1) Kalantari R, Chiang CM, Corey DR. Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res. 2016;44(2):524-537. doi:10.1093/nar/gkv1305 |
| 40 | ENSMUSG00000045648 | Vwc2l | Y | (1) Associated with opioid use disorder in humans. | (2) Increased in CSF of patients with frontotemporal dementia. (3) (4) Implicated in neurogenesis in neural precursor cells. |
(1) Alblooshi H, Al Safar H, El Kashef A, et al. Stratified analyses of genome wide association study data reveal haplotypes for a candidate gene on chromosome 2 (KIAA1211L) is associated with opioid use in patients of Arabian descent. BMC Psychiatry. 2020;20(1):41. Published 2020 Jan 31. doi:10.1186/s12888-019-2425-8 (2) Remnestål J, Öijerstedt L, Ullgren A, et al. Altered levels of CSF proteins in patients with FTD, presymptomatic mutation carriers and non-carriers. Transl Neurodegener. 2020;9(1):27. Published 2020 Jun 23. doi:10.1186/s40035-020-00198-y (3) Kolson DR, Wan J, Wu J, et al. Temporal patterns of gene expression during calyx of held development. Dev Neurobiol. 2016;76(2):166-189. doi:10.1002/dneu.22306 (4)Chang C. Agonists and Antagonists of TGF-β Family Ligands. Cold Spring Harb Perspect Biol. 2016;8(8):a021923. Published 2016 Aug 1. doi:10.1101/cshperspect.a021923 |
Epigenetic editing of Srsf11 increased cocaine reward behavior
Having determined that the Srsf11 motif was highly enriched among cocaine-regulated alternative splicing events (Figure 2) and that cocaine and Srsf11 regulate alternative splicing of a common set of transcripts (Figure 4J), we next tested the hypothesis that alternative splicing of Srsf11 was sufficient to increase cocaine reward behavior. We first modeled cocaine reward using conditioned place preference (CPP), which measured the strength of the association between investigator administered reward and associated contextual cues. In this paradigm, mice freely explore a two-sided chamber, to confirm a lack of innate preference for either side (pre-test). Mice are then trained to associate investigator-administered cocaine or saline each with exploration of one side of the chamber. After training, mice are again allowed to freely explore the two chambers (post-test). The difference in percentage of total time spent on the cocaine-paired side before (pre-test) and after (post-test) training quantifies CPP. Manipulations that enhance cocaine reward lead to increased time spent in the cocaine-paired side (Heller et al., 2016; McClung, 2007).
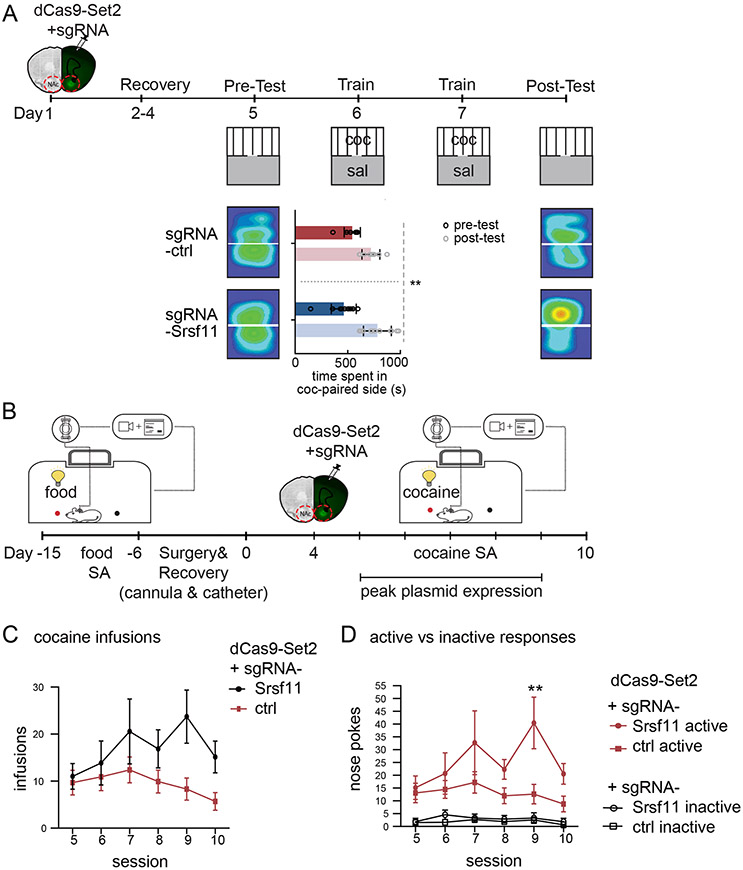
We transfected dCas9-Set2 with Srsf11-sgRNA or control-NT-sgRNA into mouse NAc and subjected animals to cocaine CPP (Figure 5A). dCas9-Set2 plus Srsf11-sgRNA-T1 caused an increase in cocaine CPP, as measured by time spent on the cocaine-paired side, compared to control-NT-sgRNA (Figure 5A). There were no observed locomotor differences measured as total distance traveled (Supplement figure 4A). Taken together, targeted enrichment of H3K36me3 at Srsf11 was sufficient to enhance cocaine CPP.
Figure 5. Epigenetic editing of H3K36me3 at Srsf11 increased cocaine reward behavior.
(A) Epigenetic editing in NAc with dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA, followed by CPP. Time spent in cocaine-paired chamber following each treatment (Repeated measures, two-way ANOVA: session effect F(1,19) =139.4, P<0.001; treatment effect F(1,19) = 0.0454, P=0.8335; interaction effect F(1,19) = 11.39; P=0.0032).
(B) Timeline and schematic of mouse cocaine SA with epigenetic editing in NAc with dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA (n=12 mice per group). Plasmid infusion through cannulas implanted at time of jugular catheterization allows cocaine SA to proceed the following day, during plasmid expression.
(C) Average cocaine infusions per session for cocaine SA show an increase upon treatment with dCas9-Set2 with Srsf11-sgRNA relative to ctrl-sgRNA (Repeated measures, two-way ANOVA: session x sgRNA F(5,90) = 2.152, P=0.0664; sgRNA F(1,18)=3.874, P=0.0647; subject F(18.90)=7.174, P<0.0001; session F(2.691,48.44)=2.403, P=0.0852).
(D) Mice treated with dCas9-Set2 and Srsf11-sgRNA show greater operant responses (nose pokes) of the active vs inactive holes relative to ctrl-sgRNA (Three-way ANOVA: session x response x sgRNA F(5,90) = 2.785, *P=0.0219; response x sgRNA F(1,18)=3.287, P=0.0866; session x sgRNA F(5,90)=2.319, *P=0.0497; session x response F(3.231,58.15)=2.999, *P=0.0345; sgRNA F(1,18)=6.307, *P=0.0218; Lever F(0.6938,12.49)=29.77, **P=0.0003; Session F(5.0,90)=3.287, **P=0.0017).
We next applied the cocaine SA paradigm to evaluate the effect of H3K36me3 enrichment and splicing of Srsf11 on animals’ motivation, cue discrimination, and drug intake (Carpenter et al., 2020; Sadri-Vakili et al., 2010; Vassoler et al., 2013; Walker et al., 2018). Mice were trained to self-administer food for 10 days before undergoing intracranial cannulation and intravenous catheterization followed by a recovery period (Figure 5B). Mice were then trained to self-administer either cocaine or saline. For both groups, a single nose-poke into the appropriate ‘active’ hole evoked one infusion (cocaine for cocaine treatment group, saline for saline treatment group) and associated tone/light cues (Supplement figure 4B-F). Poking in the ‘inactive’ hole elicited no response for either group. Cocaine-treated animals acquired stable responding (Supplement figure 4B-F). dCas9-Set2 and Srsf11-sgRNA or ctrl-sgRNA were then injected into bilateral NAc following the day 4 SA session. Cocaine SA was continued for the subsequent days 5-10 (Figure 5B-D, Supplement figure 4B-H). We found that infusion of dCas9-Set2 plus Srsf11-sgRNA increased total drug intake relative to control-NT-sgRNA (Figure 5C, Supplement figure 4B) and increased cocaine-paired responding (active hole) relative to non-paired responding (inactive hole) in Srsf11-sgRNA but not NT-sgRNA controls (Figure 5D, Supplement figures 4B-E). Epigenetic editing of control animals receiving saline produced no effect on intake or response discrimination (Supplement figures 4G-H). Taken together, these data revealed that targeted enrichment of H3K36me3 at Srsf11 was sufficient to enhance cocaine self-administration and CPP in mice.
Global H3K36me3 enrichment by Set2 overexpression regulates alternative splicing in NAc.
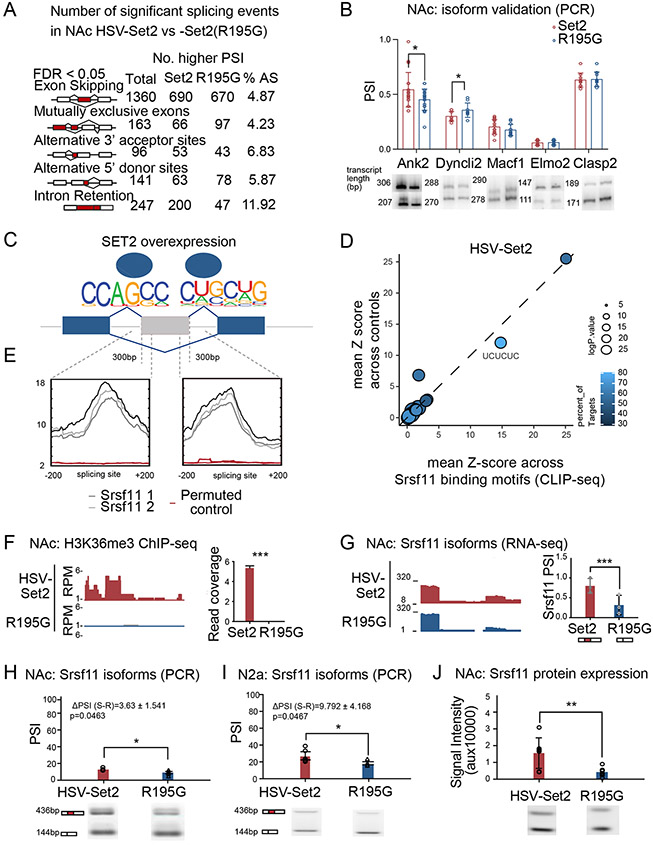
We next considered whether genome-wide H3K36me3 enrichment by Set2 overexpression would regulate alternative splicing, and the similarity of this splicing to that of cocaine SA. We performed RNA-sequencing and splicing analysis using rMATS on NAc with viral overexpression of Set2 or Set2(R195G) (Figure 6A). We identified 1360 exon skipping events between Set2 and Set2(R195G) treated NAc. Out of the five events we tested, two were biochemically validated in a replicate biological cohort of Set2 overexpression in NAc (Figure 6B). We then applied motif analysis at Set2-mediated alternative splicing junctions (150bps up- and down-stream of skipped exon site) and found enrichment of the Srsf11 splice site motif, and six other motifs, enriched in both cocaine SA and Set2 overexpression conditions, relative to respective Set2 (R195G) or saline SA controls (Figure 6C). Overlapping Srsf11 iCLIP-seq motifs with Set2 alternative splicing motifs confirmed Srsf11 as a potential splice factor (UCUCUC) regulating Set2 mediated alternative splicing (Figure 6D). Similarly to cocaine SA, we found a ‘bell’-like Srsf11 iCLIP-seq distribution at Set2 alternative splice junctions and a ‘flat-line’-like pattern of enrichment at the permuted junction controls (Figure 6E).
Figure 6. Global H3K36me3 enrichment by Set2 overexpression regulates alternative splicing in NAc , including at Srsf11.
(A) Summary of differential alternative splicing events between HSV-Set2 and HSV-Set2(R195G) identified by rMATS. All events are significant (FDR < 0.05). Percent alternative splicing was calculated as the percentage of total events in all identified splicing events.
(B) Splicing PCR quantification and representative gel image for validating exon skipping events in biological replicate of HSV-Set2 and -Set2(R195G) injected NAc (Student’s t-test, *p<0.05).
(C) Unbiased motif analysis of alternative exons with higher inclusion following HSV-Set2 vs HSV-Set2(R195G) identified Srsf11 enrichment motifs in the splice junctions (150bp upstream and downstream of the splice site).
(D) Profile plot of Srsf11 iCLIP-seq signal at differential alternative splicing junctions (top, “bell-curve” like, gray) and permuted control (bottom, “flat-line” like, red) following HSV-Set2 vs HSV-Set2(R195G).
(E) Scatter plot of averaged z-score of Srsf11 binding frequency compared to control on splicing motifs identified Set2 splice junctions. Dot circumference corresponds to −log(p-value), and lighter blue corresponds to higher percentage of this motif found in the target sequence.
(F) H3K36me3 ChIP-seq and (G) RNA-seq visualization (left) and quantification (right) at Srsf11 splice junction following NAc injection of HSV-Set2 or -Set2(R195G).
(H) Quantification and representative PCR gel for validating Srsf11 exon skipping event in NAc after NAc injection of HSV-Set2 or -Set2(R195G) (Student’s t-test, *p<0.05).
(I) Splicing PCR quantification and representative gel to validate Srsf11 exon skipping event in Set2 or Set2(R195G) transfected N2a cells (Student’s t-test, *p<0.05).
(J) Quantification and representative western blot showing increased Srsf11 protein following NAc injection of HSV-Set2 relative to HSV-Set2(R195G) (Student’s t-test, *p<0.05).
We compared changes in mRNA expression and splicing to H3K36me3 global enrichment by HSV-Set2 in NAc. Like cocaine SA, Set2 overexpression caused enrichment of H3K36me3 at Srsf11 (Figure 6F), but there was no change in Srsf11 gene expression by RNA-seq or qPCR in a biological replicate (Supplement figures 5A-B). Similarly to cocaine SA, Set2 overexpression increased abundance of the Srsf11 inclusion isoform, relative to control Set2(R195G) (Figure 6G, Supplement figure 5C). Srsf11 alternative splicing following Set2 overexpression was biochemically validated in a biological replicate of NAc treatment (Figure 6H) and transfected N2a cells (Figure 6I). Srsf11 protein expression was greater following global enrichment of H3K36me3 (Figure 6J). There was little correlation between global alternative splicing (ΔPSI) and gene expression (fold change) following Set2 overexpression (R=-0.01420054) (Supplement figure 5D). The Srsf11 splice motif was among those highly represented in an overlapping set of cocaine and Set2 regulated splice events (Figure 6E). Taken together, these data suggested a model whereby cocaine SA or Set2 overexpression led to H3K36me3 enrichment and alternative splicing of Srsf11, and its downstream target transcripts.
Set2 overexpression also regulated gene expression in NAc (Supplement Figure 5F-L), consistent with the role of H3K36me3 in marking transcriptionally active loci (Strahl et al., 2002; Wilhelm et al., 2011) and abundance at genic regions (Supplement figure 5F-H). We validated H3K36me3-mediated downregulation of transmembrane protein 25 (Tmem25) and upregulation of plexin A1 (Plxna1) in NAc and N2a biological replicates (Supplement figures 5I-K). We also found a significant overlap of differentially expressed genes (DEG) in NAc following Set2 overexpression and cocaine SA (Supplement figure 5L). These results showed that Set2 overexpression increased global H3K36me3 enrichment in NAc affecting both steady state mRNA levels and alternative splicing.
Set2 overexpression increased cocaine reward behavior.
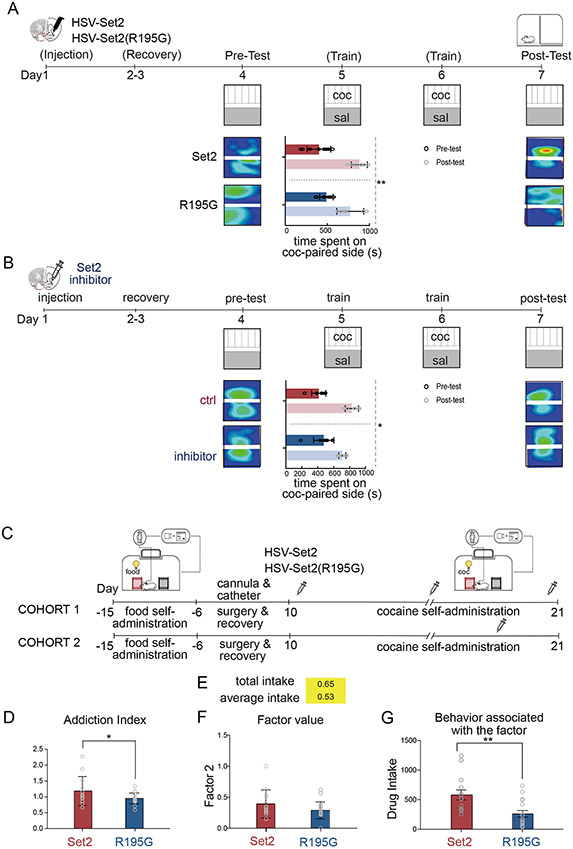
Having established a convergence in the genes regulated by Set2 overexpression and cocaine SA, we next tested whether Set2 was sufficient to regulate cocaine reward behavior (Figure 7, Supplement figure 6). Mice were injected intra-NAc with either HSV-Set2, HSV-Set2(R195G), or were virus naive (mock surgery) and subjected to cocaine CPP (Figure 7A, Supplement figure 6A) (Carpenter et al., 2020). Amongst cocaine treated mice, Set2 overexpression enhanced CPP compared to Set2(R195G) control (Figure 7A) and virus naïve mice (Supplement figure 6A). There was no difference in the total distance traveled between HSV-Set2, HSV-Set2(R195G) injected, or virus naive animals, indicating that the Set2-enhanced cocaine CPP was not due to an effect on locomotion. All mice that received cocaine showed an increase in time spent in the cocaine-paired chamber, indicating that virus treatment did not wholly prevent formation of a drug-context association (Figure 7A). Small molecule inhibition of Set2 enzymatic activity (Bay598) rescued the effect of Set2 overexpression on cocaine CPP (Supplement figure 6B) and had no effect on locomotor activity (not shown). Finally, to determine if endogenous Set2 methyltransferase activity was necessary for cocaine CPP, we performed CPP after intra-NAc injection of Set2 inhibitor. Set2 inhibition reduced CPP compared to vehicle control (Figure 7B), suggesting that decreased H3K36me3 enrichment reduces the rewarding effects of cocaine. Further, we found a positive linear correlation between global enrichment of H3K36me3 and cocaine CPP, underscoring the potential importance of Set2 in cocaine reward (Supplement figure 6C). These data are especially significant given the need to identify novel targets that reverse the effects of drug exposure in order to treat addiction.
Figure 7. Set2 overexpression increased mouse cocaine reward behavior.
(A) Schematic and results of cocaine CPP following HSV-Set2 or HSV-Set2(R195G) NAc injection. HSV-Set2 increased time spent in cocaine-paired chamber compared to HSV-Set2(R195G) (Repeated measures, 2-way ANOVA, session effect F(1,16) =67.98, P<0.0001; treatment effect F(1,16) = 0.094, P=0.7620; interaction effect F(1,16) = 4.497, P=0.04).
(B) Schematic and results of cocaine CPP following treatment with Set2 inhibitor (Bay598, 4 μM) or control (DMSO). Inhibition of Set2 methyltransferase activity attenuated cocaine CPP (Repeated measures, 2-way ANOVA, session effect F(1,26) =93.71, P<0.0001; treatment effect F(1,26) = 0.7578, P=0.3920; interaction effect F(1,26) = 4.497, P=0.011).
(C) Schematic showing cocaine SA with Set2 viral injection. Mice were food trained for 10 days prior to jugular catheterization and NAc cannulation surgeries. Following recovery, mice underwent 21 daily 2hr SA sessions. HSV-Set2 or HSV-Set2(R195G) were injected at different time points between the two cohorts (indicated by syringes). Following the last day of SA, mice were subjected to forced (home cage) abstinence for 1 day. NAc tissue was then collected for downstream analysis.
(D-G) Addiction index (AI) of factors that are most strongly associated with addiction-like behaviors. Set2 treated group showed higher AI than Set2(R195G) group (Student’s t-test, * P<0.05).
(E) Factor loading of Factor 2 with SA behavior cocaine intake (yellow = positive).
(F) Individual transformed data from cocaine total intake during SA following Set2 or Set2(R195G) NAc injection (Student’s t-test, non-significant). The transformed value was calculated to generate AI for each individual.
(G) Individual cocaine total intake data presented for the behavior associated with factor 2 (Student’s t-test, ** P<0.01).
To link the effect of Set2 overexpression on CPP to its role in alternative splicing, we measured splicing of six transcripts (Plekha6, Rab7b, Prrg2, Bin1, Dyncli2 and Srsf11) that were enriched in H3K36me3 and alternatively spliced following either cocaine SA or Set2 overexpression (Supplement figure 6D-E). Similar to cocaine SA, Srsf11, Plekha6, Rab7b and Bin1 were alternatively spliced by cocaine CPP with Set2 overexpression (Supplement figure 6D-E) but not when Set2 inhibitor was co-infused (Supplement figure 6F). Taken together, these data indicate that Set2 methyltransferase activity is necessary to enrich H3K36me3 and regulate alternative splicing of certain genes in both cocaine CPP and SA paradigms.
Given our finding that Set2 overexpression enrichment enhanced cocaine CPP, we next tested the role of Set2 in cocaine SA. Cocaine SA models drug saliency, motivation and cue discrimination using volitional drug seeking. Similar to human addicts, these features of drug behavior are variable across animal subjects. We therefore used exploratory factor analysis to reduce multidimensional behavioral data to latent factors associated with interrelated behavioral traits (Figure 7C-D) (Walker et al., 2018). This approach discriminates between baseline individual differences in behavior and those driven by cocaine treatment (Supplement figure 6G). Using published criteria (Walker et al., 2018), we identified 3 factors that are associated with SA behavior and reflect important components of addiction: cocaine intake (Factor 2, Figures 7E-G), total paired responses (Factor 5, Supplement figures 6I-J), and discrimination between paired and unpaired responses (Factor 7, Supplement figures 6H-M). Based on these measures of addiction-related behaviors, we calculated a composite score, or Addiction Index (AI), for each animal (Walker et al., 2018). We found that animals with overexpression of Set2 in NAc had a higher Addiction Index (Figure 7D-G), compared to Set2(R195G) controls. We confirmed cocaine regulated alternative splicing of 5 transcripts (Srsf11, Plekha6, Bin1, Prrg2, Rab7b) following cocaine SA with Set2 overexpression (Supplement figure 6N-O). Taken together, these behavioral data demonstrated that global enrichment of H3K36me3 by Set2 overexpression in NAc enhanced cocaine reward behavior.
DISCUSSION
An outstanding question in splicing biology concerns the mechanism of splicing specificity and splice factor targeting. Recent studies, including our own, suggest a key function of histone modifications and other epigenetic features in comprehensively understanding splicing machinery (Gonatopoulos-Pournatzis et al., 2020; Hnilicová et al., 2011, 2013; Soto et al., 2019; Xu et al., 2017). It is well established that cocaine treatment leads to vast alternative splicing and epigenetically regulated differential gene expression, yet analysis of epigenetic regulation of alternative splicing is limited to only one prior study of cocaine treated mice (Feng et al., 2014).
To add to this nascent body of literature on chromatin-regulated alternative splicing in brain and other systems, we previously applied agnostic classical statistical and machine-learning methods to find that H3K36me3 is the most informative hPTM in predicting splicing events in the NAc (Hu et al., 2017) and other neural tissues at several developmental timepoints (Hu et al., 2020). Analysis of NAc enrichment of 8 hPTMs in the exon flanking regions reveals that only H3K36me3 and H3K4me1 are differentially enriched with respect to skipped exon category and best predict skipped exon events (Hu et al., 2017, 2020). A role of H3K36me3 in splicing is supported by its primary enrichment in exonic regions across many organisms (Ernst and Kellis, 2017; Hu et al., 2017; Leung et al., 2019; Luco et al., 2010; Meers et al., 2017; Xu et al., 2017). Importantly, the H3K36 methyltransferase, Setd2, interacts with hnRNPL, a key component of the canonical spliceosome, and this interaction appears to be necessary for histone methyltransferase activity (Bhattacharya et al., 2021; Yuan et al., 2009).
In the current study, we identified a target of H3K36me3/Set2-mediated alternative splicing in brain, the splice factor gene, Srsf11. Importantly, both cocaine treatment and Set2 overexpression enriched H3K36me3 and alternative splicing of Srsf11, with no change in Srsf11 gene expression, indicating that H3K36me3 regulated alternative splicing independently of gene expression. We found that targeted epigenetic editing of H3K36me3 using dCas9-Set2 recapitulated endogenous enrichment of H3K36me3 at Srsf11 and drove alternative splicing both in vitro and in vivo. Our results suggested that H3K36me3 enrichment need not be limited to the immediate splice junction, given that cocaine SA and Set2 both led to alternative splicing of Srsf11 at the same junction but enriched H3K36me3 at different distances from the alternatively spliced exon.
Srsf11 was recently implicated in microexon expression in neurons (Gonatopoulos-Pournatzis et al., 2018). It will be interesting to analyze microexon expression and splicing in our datasets, especially following dCas9-Set2 driven splicing of Srsf11. Furthermore, we expect that additional splice factors are involved in cocaine driven alternative splicing, such as A2BP1, the motif for which was identified in our datasets and is enriched in spliced transcripts following investigator administered cocaine (Feng et al., 2014). In identifying key splicing factors that regulate cocaine driven alternative splicing, we were limited to available database of splicing factor motifs and CLIP-seq data that matched our species and tissue type of interest. As more comprehensive CLIP-seq data become available, it will be possible to identify additional components of the splice machinery relevant in cocaine and chromatin-mediated alternative splicing.
Our finding that H3K36me3/Set2 drives splicing of Srsf11 aligns with similar findings at other transcripts. A recent report finds that H3K36me3 regulates alternative splicing and nonsense-mediated mRNA decay of BRCA1-associated RING domain protein 1 (BARD1) in human stem cells, which is crucial to activate the Ataxia telangiectasia mutated (ATM) and RAD3-related (ATR) pathway to maintain pluripotency (Xu et al., 2017). This alternative splicing related enrichment is highly evolutionarily conserved between humans and mice (Kolasinska-Zwierz et al., 2009; Wilhelm et al., 2011). A related study finds that histone modifications influence splice site selection. Specifically, HDAC inhibition or HDAC1 depletion reduce co-transcriptional association of the splicing regulator SRp40 (Srsf5) with the target fibronectin exon (Hnilicová et al., 2011). Chromatin modifications may directly regulate splicing via recruitment of splice factors/adapter or regulation of the kinetics of RNAPolII. Evidence for direct recruitment of an adapter system by H3K36me3 is demonstrated at the fibroblast growth factor (FGFR2) gene during cell differentiation (Luco et al., 2010). This adaptor system consisting of hPTM (H3K36me3), a chromatin-binding protein (mortality factor 4 like 1, MRG15) and a splicing regulator (polypyrimidine tract binding protein 1, PTB), regulates FGFR2 alternative splicing (Luco et al., 2010). However, in these prior studies it is difficult to distinguish between the direct action of histone modifications at the spliceosome and pleiotropic effects.
Given our research focus on neurobiology of substance use disorders, we investigated the relevance of Srsf11 splicing to cocaine reward behavior. We found that H3K36me3-mediated splicing of Srsf11 by dCas9-Set2 was sufficient to enhance cocaine reward behavior. Studies of splicing in other neurological conditions have highlighted the key role of isoform expression in mediating disease phenotypes (Soto et al., 2019). One important example is amyotrophic lateral sclerosis (ALS), a fatal adult-onset motor neuron disease characterized by progressive loss of upper and lower motor neurons, and frontotemporal dementia (FTD). Depletion of transactivation response element DNA-binding protein 43 (TDP-43, TARDBP) leads cryptic exons to be spliced into messenger RNAs, disrupting translation and promoting nonsense-mediated decay. These findings link TDP-43 to extensive splicing changes seen in ALS-FTD (Arnold et al., 2013; Ito et al., 2020; Ling et al., 2015). Another study in Alzheimer's disease (AD) postmortem brains revealed that reduction in the ratio of transmembrane to soluble neurexin isoforms promotes the neuronal AD phenotype (Hishimoto et al., 2019). Alternative splicing of presynaptic neurexin-1 at splice site 4 (SS4) enhances postsynaptic NMDA-receptor-mediated response, while alternative splicing of neurexin-3 at SS4 suppresses AMPA-receptor-mediated synaptic responses in hippocampus (Dai et al., 2019). There current study provided additional evidence that alternative splicing events were associated with the pathology of addiction, in that cocaine self-administration led to genome-wide alternative splicing and that alternative splicing of Srsf11 was sufficient to augment cocaine preference and self-administration. Interestingly, a recent study finds a novel function of Setd5 in trimethylating H3K36 in neurons (Sessa et al., 2019). Further investigation into the roles of both Setd2 and Setd5 in cocaine-mediated H3K36me3 enrichment will shed light onto the physiological relevance of our findings.
We identified a subset of transcripts that are alternatively spliced both after cocaine self-administration and chromatin-mediated splicing of Srsf11, many of which have previously been implicated in reward biology and/or alternative splicing in brain, and all of which contained an Srsf11 splice motif (Table 1). One such gene, Rbm25, encodes an RNA-binding protein that promotes inclusion of alternatively spliced exons (Carlson et al., 2017). Rbm25 complexes with Srsf protein family members and the U1 snRNP to regulate splicing of μ opioid receptor, OPRM1, which is associated with opioid dependence (Xu et al., 2014a). Alternative splicing of Cacna1b, which encodes the α1 subunit of Cav2.2 channels, was also found downstream of both cocaine SA and Srsf11 epigenetic editing. Cav2.2 is implicated in cocaine taking and seeking, through its expression in the medial PFC (Buchta et al., 2020). Furthermore, alternative splicing of Cacna1b functionally differentiates several protein isoforms. For example, splicing within the proximal region of the Cav2.2 C-terminus specifically impacts nociception and morphine analgesia (Javier et al., 2019; Soto and Lipscombe, 2020). We also characterized novel alternative splicing of Shisa7, a GABA(A) receptor auxiliary subunit that regulates inhibitory neural circuits and the sedative effects of benzodiazepines (Han et al., 2019).
Looking ahead, we are interested in cell type specificity of cocaine regulated alternative splicing. Although we did not observe Setd2 expression changes in total heterogenous NAc after cocaine treatment, published single cell data finds neuronal-subtype specific differential Setd2 expression following cocaine treatment (Supplement figure 3I) (Bhattacherjee et al., 2019, and personal communication, Eric Nestler). One approach to profile isoforms of different neuronal sub-types in the mouse brain applies tagged-ribosomal affinity purification (RiboTRAP). In addition, ‘isolation of nuclei tagged in specific cell types (INTACT)’ (Mo et al., 2015) allows purification of chromatin and mRNA from specific neuronal sub-types of genetically modified mice to investigate chromatin-regulated alternative splicing in a cell-type specific manner. To address the low input of mRNA from single cells or cell-types, we are seeking the potential of leveraging nanopore technology to sequence spliced RNA sequence from a single cell (Arzalluz-Luqueángeles and Conesa, 2018; Lebrigand et al., 2020), and the recently developed Paired-Tag for chromatin and splicing profiling in single neurons (Zhu et al., 2021) .
Conclusion
Epigenetic editing of H3K36me3 at Srsf11 was sufficient to drive alternative splicing of Srsf11 and enhance cocaine reward behavior. We conclude that H3K36me3 functions directly in alternative splicing of Srsf11, and that this mechanism contributes to the underlying reward neurobiology.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Elizabeth A Heller (eheller@pennmedicine.upenn.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and Code Availability
The transcriptomic analyses will likely be utilized by other investigators. All gene lists and sequencing data obtained as part of this manuscript have been deposited to GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/). Accession numbers are available in the accompanying Key Resources Table. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H3K36me3 antibody | Abcam | Cat# ab9050, RRID:AB_306966 |
| Mouse monoclonal anti-H4 antibody | Abcam | Cat# ab31830, RRID:AB_1209246 |
| Rabbit polyclonal anti-SRSF11 antibody | Thermo Fisher Scientific | Cat# PA5-52606, RRID:AB_2647939 |
| Mouse anti-GAPDH | Abcam | Cat# ab9484, RRID:AB_307274 |
| Anti-mouse IgG | Cell Signaling Technology | Cat# 7076S, RRID:AB_330924 |
| Anti-rabbit IgG | Cell Signaling Technology | Cat# 7074S, RRID:AB_2099233 |
| Bacterial and Virus Strains | ||
| HSV-Set2 | This paper | N/A |
| HSV-Set2(R195G) | This paper | N/A |
| hSyn-GW-dCas9-Set2-IRES2-mCherry | This paper | N/A |
| hSyn-GW-dCas9-Set2(R195G)-IRES2-mCherry | This paper | N/A |
| P1005-GFP-Srsf11-sgRNA | This paper | N/A |
| P1005-GFP-NT-sgRNA | This paper | N/A |
| Deposited Data | ||
| RNA-sequencing of mouse NAc following 21 days of cocaine self-administration | This paper | GSE141520 |
| RNA-sequencing of mouse NAc following dCas9-Set2 + Srsf11-sgRNA | This paper | GSE180988 |
| RNA-sequencing of mouse NAc following dCas9-Set2 + Srsf11-sgRNA | This paper | GSE180988 |
| RNA-sequencing of mouse NAc following 21 days of saline self-administration | This paper | GSE141520 |
| Cut&Run sequencing of H3K36me3 of dCas9-SET2 transfected N2a cells | This paper | GSE180988 |
| ChIP-sequencing of H3K36me3 of mouse NAc following cocaine SA | This paper | GSE180988 |
| ChIP-sequencing of H3K36me3 of mouse NAc following HSV-Set2 transfection | This paper | GSE180988 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664) |
| Oligonucleotides | ||
| PCR Primers, see Table S1 | This paper | N/A |
| sgRNA, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Prism v8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| R v4.0.2 | The R Project for Statistical Computing | https://www.r-project.org/ |
| Factor Analysis - Cocaine Self Administration | Walker et al., 2018 | N/A |
| CUT&RUN Analysis | Skene and Henikoff, 2017 | N/A |
| Splicing analysis rMATS | Shen et al., 2014 | http://rnaseq-mats.sourceforge.net/ |
| Splicing analysis MAJIQ | Norton et al., 2017 | https://majiq.biociphers.org/ |
| DiffBind | Ross-Innes et al., 2012 | Bioconductor https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Deseq2 | Love et al., 2014 | Bioconductor https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| SICER | Xu et al., 2014b | https://zanglab.github.io/SICER2/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/motif/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Adult male and female, 8-week old C57BL/6J mice (Jackson) were used in this study. There was no influence/association of sex on the findings. Mice were housed five per cage on a 12-h light-dark cycle at constant temperature (23°C) with access to food and water ad libitum. Animals were habituated to experimenter handling for at least 1 week before experimentation. All animal protocols were approved by the Institutional Animal Care and Use Committee of University of Pennsylvania.
METHOD DETAILS
All experiments were carried out one to three times, and data replication was observed in instances of repeated experiments, or otherwise reported.
Cocaine conditioned place preference
Stereotactic surgery was performed as detailed below. After surgeries, mice were given 3 days to recover before being placed in the chamber and allowed to explore the conditioning apparatus. The paradigm consists of 2 distinct environment chambers (white/black stripe side, and gray side). The initial preference was recorded (Pre-test). Mice with pre-conditioning preference (more than 30% of total time spent in either of the 2 chambers) were excluded from the study. During training, mice received two parings per day: cocaine (15 mg/kg; i.p.) in the morning while confined to the less preferred chamber (pre-test) and saline (0.9%; 1 ml/kg; i.p.) in the afternoon while confined on the opposite chamber of the place preference chamber. On post-test day, mice were placed again in the CPP box with free access to both chambers and the time spent in each side was quantified (Post-test). Data are expressed as time spent on the cocaine-paired side in post-test as well as time spent on the same side in the pre-test (displaying CPP percentage time change). The amount of time spent on the cocaine paired side in the pre- and post-test was compared between animals using a using a repeated measures, two-way ANOVA. For wild-type, HSV-Set2 and HSV-Set2(R195G) injected animals that underwent CPP (see supplement), data was analyzed as a CPP score, subtracting the time spent on the saline-paired side from the cocaine-paired side. For this analysis, CPP score from the pre- and post-test was compared between animals using a repeated measures, two-way ANOVA. S.I.L performed the behavior wile S.J.X. performed all analysis.
Food self-administration
Operant conditioning used in cocaine self-administration was facilitated in naïve mice by first successfully undergoing nose poke training with food (food SA). All mice were habituated to the researcher by daily handling sessions for 7 days prior to the start of training. Mice were introduced to the grain-based food pellets (20 mg, BioServ, Product #F0071) and were food restricted 3 days prior to the start of the operant training. Food SA was conducted in 1-hour sessions for 10 days using Med Associates custom made drug-self administration boxes with nose-poke holes. Animals were run on a fixed ratio 1 (FR1) schedule for the entirety of training. An ‘active poke’ was defined by the triggering of the active hole sensor via animal nose-poke and the subsequent the presentation of a single grain-pellet reward with light (5 second) and tone cues. An ‘inactive poke’ was defined by triggering of the inactive hole sensor with no scheduled consequences. A time out interval of 20 seconds occurred after the engagement of the active nose poke. Any response during the timeout interval had no consequences but was recorded. Following the completion of food SA, mice were then assigned to self-administration groups (cocaine or saline) and were fed ad libitum for the remainder of the experiment.
Catheter implantation and bilateral cannula surgery
Following operant food SA training, mice were subjected to an IV catheter implantation and bilateral cannula surgery. Animals were anesthetized with 100mg/kg Ketamine (i.p) and 10mg/kg Xylazine (i.p) and received 5mg/mL Meloxicam (s.c) prior to the surgery. The indwelling catheter (Instech, Product #VABM1B/25) was implanted into the right external jugular vein along with an intercranial bilateral cannula (Plastics One, custom made targeting the Nucleus Accumbens (NAc) (Anterior/Posterior:+1.6, Medial/Lateral:+1.5, Dorsal/Ventral:−4.4). Following surgery, catheters were flushed daily with Herapin (0.05mL/day) and Timentin (3.33mg/day) in saline to prevent clotting and reduce the risk of infection. Mice were allowed 4-6 days to recover, during which time they were monitored and handled daily.
dCas9-Set2 + sgRNA with cocaine self-administration
Prior to the start of cocaine and saline self-administration, catheter patency was checked using ketamine (100 mg/ml, 0.05 ml i.v.). The same active/inactive definitions and paired cues as in food SA were used with the replacement of the grain-pellet reward with an infusion of cocaine or saline. Mice intravenously self-administered either sterile saline or 0.5 mg/kg/infusion of cocaine (Saline IVSA, Cocaine IVSA, respectively) over 5 seconds during daily FR1 scheduled 2-hour sessions for 10 consecutive days in the same Med Associates boxes used during food SA.
After the fourth IVSA session, animals were randomly assigned into a targeting (SRSF11) or non-targeting (NT) group, resulting in four treatment groups: SRSF11-Saline, SRSF11-Cocaine, NT-Saline, NT-Cocaine. Cocaine and saline groups were tested via a 2-way ANOVA to ensure no biases in Discrimination Index (DI), rewards, active pokes or inactive pokes were present prior to plasmid infusion. SRSF11 or NT plasmid was infused using a syringe pump (Chemyx, Product #F101) with a rate of 0.3 uL plasmid/minute resulting in 1.5 uL of plasmid being infused over five minutes. Animals were continued on the IVSA paradigm for six days after infusion of plasmid. The six-day time point was determined by a separate time-course experiment. During day 4-7, we observed high levels of plasmid expression. After final session IVSA, mice were rapidly euthanized by cervical dislocation and decapitated, brains were removed and NAc, VTA, PFC was dissected using aluminum Harris micro-punch (Sigma-Aldrich). Tissue was frozen on dry ice and stored at −80 until downstream analysis. Data were excluded from mice (cocaine n = 4, saline n = 2) that did not complete the total training duration of 10 days due to catheter malfunction or postoperative complications.
HSV-Set2 cocaine self-administration and tissue collection
Mice were habituated, handled, food trained and surgerized as described above. Cocaine self-administration was also performed as described above with the exception that rather than nose holes, wheels were used for active vs inactive response triggers. The wheels were defined as paired if a 90° rotation of the wheel resulted in the presentation of a reward, with light and tone cues, and unpaired if spinning had no scheduled consequences. Mice were then assigned to self-administer either saline or cocaine on a fixed ratio (FR) schedule during daily 2-hour sessions for 21 consecutive days. Acquisition of cocaine SA was defined as three consecutive sessions during which ≥ 10 infusions occurred, infusions did not vary by more than 20%, and at least 80% of spins were on the paired wheel. For self-administration studies used for molecular biochemistry and sequencing, mice were run on a FR1 schedule for two-hour sessions over 21 consecutive days. HSV-Set2 or -Set2(R195G) were given at various time points in the two cohort. Animals used in Set2-virus paired self-administration, an initial FR1 schedule was used. Once animals met learning criteria, they were moved to fixed-ratio 5 (FR5) and subsequently to progressive ratio (PR) schedule. 24 hours after the last cocaine session, mice were rapidly euthanized by cervical dislocation and decapitated, brains were removed and NAc was dissected using aluminum Harris micro-punch (Sigma-Aldrich). Tissue was frozen on dry ice and stored at −80 until downstream analysis. Data were excluded from mice (cocaine n = 3) that did not complete the total training duration of 10 days due to catheter malfunction or postoperative complications.
Statistics and factor analysis for cocaine self-administration
The appropriate statistical test were determined based on the number of comparisons being done. The Grubbs test was used when appropriate to identify outliers. Sample sizes were selected using G*Power to match effects in preliminary data using a Type I error of 5% (Bonferroni corrected for multiple outcomes) and yielding at least 80% power. Power-calculations are conducted either in PASS (Power and Sample Size, NCSS, Kaysville, UT) modeling z-scores for one-sample tests, or by data simulation in Stata (Stata Corporation, College Station, TX). Data are displayed as mean +/− standard error of mean (SEM) for each group (SRSF11-Saline, SRSF11-Cocaine, NT-Saline, NT-Cocaine). Statistical analyses were performed using Prism GraphPad software (version 8.4.3). Data were analyzed using two-, or three-way analysis of variance (ANOVA). Where appropriate, within-subjects repeated-measures (RM) ANOVA factors was utilized. For all ANOVAs, statistical significance (alpha) was defined as p < 0.05, indicated by * in graphs. The number of asterisks denotes significance level and is also reported within all figure legends.
For confirming plasmid groups (Supplement figure 4C-D), we first confirmed that the active and inactive poke data from IVSA sessions one-four were normally distributed using the Shapiro-Wilks test. We then averaged those data and used a two-way ANOVA with factors of potential group (SRSF11 vs. NT), and engagement of poke type (active poke vs. inactive poke). The cocaine and saline-self administration groups were analyzed separately.
To determine if Srsf11 impacts cocaine taking (Figure 5C) as well as to determine if SRSF11 impacts taking generally (saline, control; Figure 5C), we first confirmed data was normally distributed using the Shapiro-Wilks test. We then used a three-way repeated measures ANOVA with factors of plasmid group (SRSF11 vs. NT), engagement of poke type (active poke vs. inactive poke), and session (one vs. two vs. three, vs. four vs. five vs. six). Matched factors included poking holes and plasmid. If violations in sphericity were observed during the repeated measures-ANOVAs, the Greenhouse–Geisser estimate of sphericity was used to decrease the degrees of freedom and correct the violation. All epsilon values, which quantify this correction, are reported within the text. When significant interactions and corresponding main effects were observed, significance levels for multiple post hoc comparisons were determined using Tukey’s multiple comparisons test. The cocaine and saline-self administration groups were analyzed separately.
To analyze if there were differences in overall infusions between plasmid groups, we confirmed that the infusion data from IVSA sessions one-six post infusion were normally distributed using the Shapiro-Wilks test. We conducted a repeated measures two-way ANOVA with factors of plamid group (SRSF11 vs. NT), and session (one vs. two vs. three, vs. four vs. five vs. six). The cocaine and saline-self administration groups were analyzed separately. Factor analysis on cocaine SA was carried out based on published method (Walker et al., 2018). Briefly, A standard factor analysis was performed on cocaine SA behavioral variables using the scikit-learn package. This is to reduce the dimensions of behavioral variables in cocaine SA and generate a composite Addiction Index (AI) of 3 factors (Figure 7, Supplement Figure 6).
Intra-NAc virus transfusion and plasmid transfection
Following anesthesia with a mixture of oxygen and isoflurane, mice were stereotactically infused with HSV virus encoding Set2 plasmid or catalytically dead Set2(R195G) control (Strahl et al., 2002). Nucleus accumbens (NAc) was targeted bilaterally using the following stereotaxic coordinates: Anterior/Posterior +1.6, Medial/Lateral +1.5, and Dorsal/Ventral −4.4 at an angle of 10° from the midline (relative to Bregma). A total of 1 μL of virus was delivered on each side over a 5-min period, followed by 5 min of rest. For Set2 with inhibitor Bay598 (SML1603) (10uM) treatment, inhibitor was dissolved in DMSO and mixed with virus. 1 μL of virus and inhibitor mixture was delivered to NAc on each side at a rate of 0.2 ul per minute, followed by 5 min of rest. dCas9-Set2 in-vivo transfection was conducted using the transfection reagent Jet-PEI (Polyplus transfection), prepared according to manufacturer’s instructions. 12.5 μL DNA plasmid (1.0 μg/uL) was diluted in 12.5 μL of sterile 10% glucose and added to diluted Jet-PEI, mixed by pipetting and incubated at room temperature (RT) for 15 min. A total of 1.5 μL of Jet-PEI/Plasmid solution was delivered NAc at a rate of 0.2 ul per minute, followed by 5 min of rest. In all molecular and behavioral experiments, proper NAc targeting of virus infusion was confirmed post hoc by preparing brain slices and visual confirmation of both needle track and GFP expression by microscopy. The mice were subjected to behavioral, immunohistochemical or DNA/RNA analysis after 2 days for virus transfection and 3 days for Jet-PEI transfection.
Protein extraction, western blotting, and analysis
NAc was surgically dissected from dCas9-Set2 plus sgRNAs, cocaine, and/or saline treated animals and flash frozen on dry ice. Frozen NAc tissue was lysed in RIPA buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.1% SDS) plus complete protease (Roche #11697498001) and phosphatase (Sigma-Aldrich #4906837001) inhibitor cocktails. Protein lysates were centrifuged and supernatants were quantified using Pierce Bicinchoninic Acid Kit (ThermoFisher #23225). 25 ug of total protein were loaded onto 4 to 12%, Bis-Tris protein gels (Invitrogen #NP0329BOX) along with control ladder (PageRuler #26616). Gels were run at 50 mV for 30 min before switching to 150 mV for 1.5 hrs. Western blots were transferred for 25 min at 400 constant mA using NuPAGE Transfer Buffer (#NP0006) and PVDF Transfer Membrane (VWR #29301-854). Transfer was verified using Ponceau (Amresco #A611-K793) and Coomassie Blue staining. Membranes were blocked for 1 hr at RT in 5% nonfat milk (VWR #M-0841) or 1x Rapid Block (VWR #97064-124) in Tris Buffer Saline with 1% Tween (TBST). Primary antibodies were prepared in 5% nonfat milk in 1x-TBST and incubated o/n at 4°C shaking. Primary antibodies were as follows: rabbit polyclonal anti-H3K36me3 antibody (1:1,000, Abcam Cat# ab9050, RRID:AB_306966), mouse monoclonal anti-H4 antibody (1:2,000, Abcam Cat# ab31830, RRID:AB_1209246), rabbit polyclonal anti-SRSF11 antibody (1:500, Thermo Fisher Scientific Cat# PA5-52606, RRID:AB_2647939), and mouse anti-GAPDH (1:2,000, Abcam Cat# ab9484, RRID:AB_307274). Secondaries were prepared in 5% nonfat milk in 1x-TBST and incubated for 3 hrs at RT shaking. Secondary antibodies were as follows: HRP-linked, Anti-mouse IgG (1:5,000, Cell Signaling Technology Cat# 7076S, RRID:AB_330924), HRP-linked, Anti-rabbit IgG (1:1,000, Cell Signaling Technology Cat# 7074S, RRID:AB_2099233). The blots were washed 6 times for 5 min in 1x-TBST after each antibody incubation. The blots were developed using an enhanced chemiluminescence kit (Thermo Scientific #34075). Levels of proteins of interest were quantified using ImageJ relative to within sample loading control gene, GAPDH, and compared using Student’s t-test. Each western blot was reproduced at least two times. To accomplish blinded analysis, protein isolation, quantification, western blot, and transfer was performed by S.I.L, and analysis was performed by D.F.K.
Chromatin immunoprecipitation (ChIP) and ChIP-sequencing analysis
ChIP was performed on frozen bilateral NAc 2 mm punches pooled from 2-3 mice, dissected as described above. Chromatin was prepared as described previously (Heller et al., 2014) and sheared using a diogenode bioruptor XL at high sonication intensity for 30 minutes (30 s on/30 s off). Fragment size was verified at 150–300 bp with an Agilent 2100 bioanalyzer. Sheared chromatin was incubated overnight with the H3K36me3 (Abcam ab9050) antibody previously bound to magnetic beads (Dynabeads M-280, Life Technologies. After reverse cross-linking and DNA purification (Qiagen Spin Column), ChIP-sequencing libraries were prepared using the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (E6240L) using an adaptor oligo dilution of 1:20 for all samples. Prepared samples were pooled and sequenced on Illumina Hiseq 4000 platform to achieve ~30 million reads per sample. Raw sequencing data was processed to generate fastq files of 50 bp single-end reads for further processing. Sequences were aligned using Bowtie v2.1.0, allowing for ≤2 mismatches to the reference in 50 bp.
High quality and uniquely aligned reads were confirmed for H3K36me3 ChIP-sequencing. Uniquely mapped reads were used for subsequent analysis. Peak calling was performed using SICER (Xu et al., 2014b), whereas DiffBind (Ross-Innes et al., 2012) was used to identify the differential histone modification sites between treatment and control groups.
Integrated profiles were created by averaging the observed signal for each bin for the selected set of relevant genes using deepTools computeMatrix; values displayed indicate fragments per 50-bp bin per million mapped reads (Figures 2A-B, 4C-E, 6D-E, Supplement figure 4F, D-E). Integrated profile of H3K36me3 enrichment at splice junctions (Figure 3C, 6E, Supplement figure 3) was created similarly using deepTools computeMatrix. Splice junctions were defined using 150bp up- and down-stream of spliced exon start/end site. Permuted splicing junctions were generated by randomly permuting the genomic locations of the splice junctions using Bedtools shuffle. H3K36me3 Enrichment of islands over genome was assessed using HOMER software with a false discovery rate (FDR) of 5% as a cutoff and a significance score (fold-change score over input) of 1.5. By default, HOMER used DESeq2's rlog transform to create normalized log2 read counts, thereafter generate log2 transformed enrichment relative to a specific genomic category (Heinz et al., 2010). Gene Ontology was analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource specifying for biological processes. UCSC genome browser was used to visualize the track coverage.
RNA extraction, splicing PCR and qPCR
Total RNA was extracted from tissue and cells using the RNeasy Mini Kit (Qiagen 74134) as recommended by the manufacturer. Splicing PCR was used to biochemically validate bioinformatic splicing results based on previously published methods (Gonatopoulos-Pournatzis et al., 2018; Lee et al., 2016; Parikshak et al., 2016; Zhang et al., 2016). Total RNA (600 ng) was reverse-transcribed using Invitrogen Superscript IV reverse transcriptase. cDNA (50 ng) was amplified using OneStep RT-PCR kit (Qiagen 210210) according to the manufacturer’s recommendations with 20-25 cycles. Forward and reverse primers were designed to anneal to the constitutively included exons upstream and downstream of the alternative exon, respectively. PCR products were resolved on 1% agarose gels (Lonza) and counterstained with SYBR Gold for visualization. Samples were randomized for the PCR run. Gels were quantified using ImageJ (NIH). First the exon-included and exon-excluded band intensities were corrected by subtracting background. Then, intensity of the exon-included band was divided by the sum of the exon- included and exon-excluded bands. The result was multiplied by 100% to obtain the PSI value. Differential gene expression was measured by qPCR (Heller et al., 2016), and data analysis was performed by comparing Ct values of the experimental group to control using the ΔΔCt method (Livak and Schmittgen, 2001).
RNA-sequencing and analysis
RNA quality control assays and library preparation were performed by the University of Pennsylvania Next Generation Sequencing core. Total RNA was quantified using fluorescent chemistry contained in the Qubit RNA HS Assay Kit (Thermo Fisher Scientific Q10211). 1 ul was used to assess RNA Integrity Number (RIN) using the Agilent Bioanalyzer RNA nano kit (Agilent 5067-1513). The RIN range was between 8 and 9.8. RNA-seq was carried out on an Illumina HiSeq 4000. Sequence alignment was performed using STAR (Dobin et al., 2013). The FASTQ sequence was aligned to the mouse genome mm9 or mm10 and unique read alignments were used to quantify expression and aggregated on a per-gene basis using the Ensembl (GRCm38.67) annotation. We used DESeq2 (Love et al., 2014) to assess differential expression between sample groups, and both rMATS (Shen et al., 2014) and MAJIQ (Green et al., 2017; Norton et al., 2018) to assess alternative splicing. Alternative splicing was quantified using the per cent spliced in (PSI) metric using Multivariate Analysis of Transcript Splicing (MATS, v3.2.4). For each event, rMATS reports PSI supporting the inclusion (I) or splicing (S) of an event. For these events, the PSI is calculated as PSI = I/(I + S)). To account for estimation uncertainty in individual replicates and variability among replicates, PSI values was first filtered by 99.9% confidence interval. In Figure 1, ΔPSI quantified the difference in relative isoform abundance between cocaine and saline SA. In Figure 4I-K, differential alternative splicing was performed using unpaired t-test, given that each sample was collected from the two hemispheres of the same animal; significant results are reported at Benjamini–Hochberg FDR < 0.05. Cocaine SA splicing data (Figure 4A-B) was defined as MAJIQ identified alternative spliced transcripts common to cocaine SA plus 1-day and 28-day abstinence.
De novo motifs (recurring, fixed-length patterns) in the top 1000 (rank by “inter’ treatment difference) alternative splicing junctions (+−150bps of splice site) were identified using HOMER software with scrambled sequence as control. Similar motif analysis was done using MEME. Searched motifs were then submitted as input to the MEME-Tomtom program to rank the motifs in the database and produce an alignment for each significant match for splice factors using Ray2013 RBP databases (Ray et al., 2013).
Extraction of histones from NAc tissue
Tissue was lysed in nuclear isolation buffer (15 mM Tris pH 7.5, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose, 10 mM sodium butyrate, 1 mM DTT, 500 μM AEBSF, 5 nM microcystin) containing 0.3% NP-40 alternative on ice for 5 min. Nuclei were pelleted and resuspended in 0.4 N H2SO4 followed by 1.5 hr rotation at 4°C. After centrifugation, supernatants were collected, proteins were precipitated in 33% TCA overnight on ice, washed with acetone, and resuspended in deionized water.
Acid-extracted histones (5-10 μg) were resuspended in 100 mM ammonium bicarbonate (pH 8), derivatized using propionic anhydride and digested with trypsin as previously described (Sidoli et al., 2016). After a second round of propionylation the resulting histone peptides were desalted using C18 Stage Tips, dried using a centrifugal evaporator, and reconstituted using 0.1% formic acid in preparation for LC-MS analysis.
Histone mass spectrometry
Nanoflow liquid chromatography was performed using a Thermo ScientificTM Easy nLCTM 1000 equipped with a 75 μm x 20 cm column packed in-house using Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH, Germany). Buffer A was 0.1% formic acid and Buffer B was 0.1% formic acid in 80% acetonitrile. Peptides were resolved using a two-step linear gradient from 5% to 33% B over 45 min, then from 33% B to 90% B over 10 min at a flow rate of 300 nL/min. The HPLC was coupled online to an Orbitrap Elite mass spectrometer operating in the positive mode using a Nanospray Flex™ Ion Source (Thermo Scientific) at 2.3 kV.
For MS, a full scan (m/z 300-1100) was acquired in the Orbitrap mass analyzer with a resolution of 120,000 (at 200 m/z) followed by 8 MS2 scans using precursor isolation windows of 50 m/z each (e.g. 300-350, 350-400…650-700). A second full scan was then performed followed by the remaining 8 MS2 scans (800-850, 900-950…). MS/MS spectra were acquired in the ion trap operating in normal mode. Fragmentation was performed using collision-induced dissociation (CID) in the ion trap mass analyzer with a normalized collision energy of 35. AGC target and maximum injection time were 5e5 and 50 ms for the full MS scan, and 3e4 and 50 ms for the MS2 scans, respectively. Raw files were analyzed using EpiProfile 2.0 (Yuan et al., 2018). The area under the curve (AUC) for each modified state of a peptide was normalized against the total of all peptide AUCs to give the relative abundance of the histone modification.
Plasmid construction and sgRNA design
The Set2 and Set2 mutant R195G plasmid used for this study were obtained from Dr. Brian Strahl (UNC). Plasmids were then packaged into HSV-GFP virus. dCas9-Set2 construct was optimized, synthesized and cloned into pENTR-dTOPO by GENEWIZ. Then the entry plasmid pENTR-dTOPO-dCas9-Set2 was packaged into destination vector hSyn-GW-IRES2-mCherry using Gateway LR Clonase II enzyme mix (ThermoFisher #11791020). The hSyn-GW-dCas9-Set2-IRES2-mCherry vector, encoding mammalian codon-optimized Streptococcus pyogenes dCas9 fused to a human SET2 domain were used for neuronal cell line and neuronal tissue transfections.
Guide RNAs were designed and screened using the web tool http://crispr.mit.edu/. The PAM sequence of these guide RNAs was used to determine the minimal off-target effect. The three sgRNAs with the highest scores (suggest least off-target effect) were designed based on H3K36me3 enriched region and strand specificity. Then the 19bp target sequence was synthesized into gBlock that contains U6 promoter, guide RNA scaffold and termination signal. Then gBlock was cloned into an empty backbone vector pENTR/d-TOPO (ThermoFisher K240020) following manufacture instructions. Additionally, the entry plasmid was packaged into destination vector P1005-GFP using Gateway LR Clonase II enzyme mix (ThermoFisher #11791020).
Cell culture and transfection
Neuro2a cells (ATCC CCL-131) were cultured in EMEM medium with 10% FBS in 6-well plates. The cells were transfected with 400 ng of plasmid DNA (dCas9-Set2/R195G with gRNA) using Effectene reagent (Qiagen 301425), and RNA was isolated using the RNeasy Mini Kit (Qiagen 74134) according to the manufacturer instructions. The cells were maintained at 37 °C and 5% CO2, and harvested in 24 hours. dCas9-Set2 with sgRNA were transfected in six-well plates with 1 mg of each respective dCas9 expression vector, 0.33 mg of individual gRNA expression vectors using Effectene reagent as per the manufacturer’s instruction. The cells were maintained at 37 °C and 5% CO2, and harvested 48 hours after transfection.
CUT&RUN
CUT&RUN was performed based on published method (Skene and Henikoff, 2017; Skene et al., 2018). Transfected N2a cells were first lysed to isolate the nuclei. The nuclei were then centrifuged, washed and incubated with lectin-coated magnetic beads. The lectin-nuclei complexes were then resuspended with anti-H3K36me3 antibody (Abcam Cat# ab9050, RRID:AB_306966) and incubated overnight at 4°C. The nuclei were washed next day to remove unbound antibodies and incubated with Protein-A-MNase for 1 hour, then washed again to remove unbound protein-A-MNase. CaCl2 was added to initiate the calcium dependent nuclease activity of MNase to cleave the DNA around the DNA-binding protein. The protein-A-MNase reaction was quenched by adding chelating agents (EDTA and EGTA). The cleaved DNA fragments were then liberated into the supernatant and incubated with proteinase K for 15 minutes at 70°C. DNA fragments were purified using Qiagen mini-elute and used to construct a sequencing library using NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina® (#E6240L) using an adaptor oligo dilution of 1:20 for all samples. Samples were pooled and sequenced on Illumina Hiseq 4000 platform to achieve ~20 million reads per sample. Raw sequencing data was processed to generate fastq files of 50 bp pair-end reads for further processing. The data was then collected and analyzed using Bowtie2 software with specifics (--local --very-sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700) that aligns sample sequences to mm9 to identify the H3K36me3 targeted DNA fragments.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were carried out one to three times, and data replication was observed in instances of repeated experiments, or otherwise reported. Software packages, Graphpad Prism and R, were used for the statistical analyses in this section.
Cocaine self-administration:
Self-administration data was collected and saved automatically via Med-PC software (Med Associates). Multiple cohorts of animals were run at different times of the year for cocaine SA and cocaine SA with HSV-Set2 (n=12-24 per group). Only one cohort of cocaine SA with dCas9-SET2+sgRNA was run (n=12 per group).
Factor analysis:
Factor analysis was used to reduce the dimensions of the interdependent behavioral variables and help account for variability in the data due to differences in training, cohorts, and different virus administration time points. All animals were included in the analysis. All behavioral measurements were first shifted to convert all data to non-negative values followed by log2(x+1) transformation. For “total intake”, an additional variable referred to as “intake or not” was included to indicate whether total intake > 0. This accounted for the lack of cocaine intake in the saline groups. Following previous publication, the factor number was set to 8 when factor analysis was then applied to the whole dataset. The transformed data from the analysis was then used as a continuous variable for each factor.
Western blot:
All westerns were quantified using ImageJ in a blinded manner. The region of interest was defined using rectangle tool and maintained for all lanes. Area under the curve was calculated and intensity of the gene of interest was normalized to loading control.
PCR:
PCR validation of spliced genes was performed in biological replicate tissue by calculating percent spliced in index (PSI) by comparing the intensity of bands corresponding to respective spliced isoforms in ImageJ. (At least 3 per treatment per gene)
qPCR:
Gene expression changes measured by qPCR were run using standard practices for relative quantification using the 2(-Delta Delta C(T)) method. (Livak and Schmittgen)
Statistics by figure number:
The appropriate statistical tests were determined based on the number of comparisons being done. All statistical analyses were done using R or Prism software. When appropriate, a post-hoc test followed to determine differences across multiple comparisons. Data are expressed as mean ± SEM. The total number (n) of samples or observations in each group and the statistical tests used for comparisons between groups or within groups are indicated on the figures directly or in the figure legends. All experiments were carried out one to three times, and data replication was observed in instances of repeated experiments, or otherwise reported. For comparisons of two groups, means were compared using Student’s t-test or two-way ANOVAs when parameters followed a normal distribution. Repeated measure two-way ANOVA was used for comparison of two groups on different observations, in the analysis of cocaine self-administration and conditioned place preference. One-way ANOVA was used for analysis of three or more experimental groups. Main and interaction effects were considered significant at P < 0.05. P values greater than 0.05 and below 0.1 were considered trends.
Figure 1: Average cocaine and saline infusions per session were compared using a repeated measures two-way ANOVA (Fig 1 B). Alternative splicing statistics were generated by rMATS. The statistical model of rMATS calculates the P-value and false discovery rate of the difference in the isoform ratio of a gene between two conditions (Fig 1C). Hypergeometric tests were used to test the significance of overlap between brain regions (Fig 1 D-E).
Figure 2: Distributions of H3K36me3 enrichment at splice junctions of skipped exon splicing type were compared between cocaine and saline groups and tested by a one-way ANOVA (Fig 2C, E and G). Differences of percent spliced in (PSI) by PCR on selected genes were tested by Studen’s t-test to validate cocaine regulated alternative splicing events identified by rMATS (Fig 2I).
Figure 3: Averaged z-score of Srsf11 binding frequency by Clip-seq on splicing motifs identified in cocaine SA splice junctions was compared to saline SA control and −log(p-value) plotted (Fig 3B). Transcript expression (RNA-seq) and H3k36me3 enrichment (ChIP-seq) at the Srsf11 alternative splice junction was quantified and compared after cocaine and saline SA using a Studen’s t-test (Fig 3D-F). Srsf11 protein expression in NAc after cocaine and saline SA was normalized to within sample loading control and compared using Studen’s t-test (Fig 3G). Pearson correlation coefficients and corresponding P-values were graphed using density plots (Fig 3H). Srsf11 exon skipping was validated in biological replicates of NAc from cocaine and saline SA, quantified in ImageJ and then compared using Studen’s t test (Fig 3I). Pearson correlation was used to compare correlation between Srsf11 inclusion levels (PSI) and cocaine intake within each animal from cocaine SA (Fig 3J).
Figure 4: Srsf11 isoform and protein expression was compared in N2a cells (Fig 4G) and NAc (Fig 4H) transfected with dCas9-Set2 and sgRNAs using Studen’s t-test. Overlap of alternatively spliced genes by cocaine SA and dCas9-SET2-Srsf11 was measured by Fisher’s exact test (Fig 4J).
Figure 5: Cocaine preference measured by CPP was compared between animals treated with control- or Srsf11-sgRNAs using a repeated measures, two-way ANOVA comparing session, treatment, and interaction effect (Fig 5A). Cocaine preference in SA was measured on several different metrics including cocaine infusions and active versus inactive responses using a repeated measures, two-way ANOVA or three-way ANOVA, respectively (Fig 5C and D).
Figure 6: Srsf11 isoform expression, protein expression and PCR validation of target genes were performed in biological replicates using within-animal control via bilateral injections of HSV-Set2 and -Set2(R195G) and compared using Studen’s t-test (Fig 6B, G, H, I, J).
Figure 7: Cocaine preference measured by CPP was compared between animals treated with HSV-Set2 and -Set2(R195G) using a repeated measures, two-way ANOVA comparing session, treatment, and interaction effect (Fig 7A). Similarly, the effect of inhibiting Set2 methyltransferase activity on cocaine CPP was measured using a repeated measures, two-way ANOVA comparing session, treatment, and interaction effect (Fig 7B). SA was used to measure cocaine preference and addiction-like phenotype in HSV-Set2 and -Set2(R195G) treated animals comparing factors individually (Studen’s t-test, Fig 7 D-G, F) or collectively. Factor loading showed Factor 2 was positively associated with SA behavior cocaine intake (Fig 7E).
S1: Discrimination index, paired responses and unpaired responses were compared between cocaine and saline exposed animals in SA using a repeated measures, two-way ANOVA comparing drug, session and interaction effects (Supp Fig 1 A-C). Comparisons of alternatively spliced and total spliced transcripts between cocaine and saline treated biological replicates was performed by rMATs in three different tissue types and splicing types using hypergeometric tests of significance for each overlap (Supp Fig 1 D-G).
S4: Quantitative mass spectrometry of H3.1 and H3.3K36me3 in HSV-Set2, Set2(R195G) and -GFP injected NAc was compared using a two-way ANOVA comparing histone variant, virus, and interaction effects (Supp Fig 4B). H3K36me3 enrichment was compared between WT, HSV-Set2 and -Set2(R195G) with and without inhibitor using a one-way ANOVA with Tukey’s multiple comparisons.
S5: Distance traveled in CPP was measured using a Student’s t-test (Supp Fig 5A). Average infusions, active, inactive and total responses were compared for cocaine and saline SA animals treated with dCas9-Set2 and either Srsf11-sgRNA or ctrl-sgRNA using repeated measures two-way ANOVA (Supp Fig 5A-E). Animal weights were measured SA and compared using a one-way ANOVA (Supp Fig 5F). Average active and inactive responses per session after viral infusion for saline SA was measured using a three-way ANOVA (Supp Fig 5H).
S6: Srsf11 expression levels were compared between HSV-Set2 and -Set2(R195G) treated biological replicates using a Student’s t-test. (Supp Fig 6B, J, K) Overlaps were assessed by Fisher exact test (Supp Fig 6E, H, L).
S7: CPP score was compared between animals treated with HSV-Set2 and -Set2(R195G) using a repeated measures, two-way ANOVA measuring virus, drug and interaction effect (Supp Fig 7A). Percent change of CPP was compared between virus groups treated with inhibitor using a Student’s t-test (Supp Fig 7B). PSI of Set2 target genes was compared between HSV-Set2 and -Set2(R195G) using a student’s t-test (Supp Fig 7 D-F, N, O). Srsf11 protein levels between saline and cocaine or control- and Srsf11-sgRNAs were compared using a Student’s t-test (Supp Fig 3, Data S1). Factor loading for behavioral endpoints used in factor analysis. The association of each factor with each behavioral endpoint included in the analysis is displayed with yellow being positively associated, blue being negatively associated and black being not associated with each endpoint (Supp Fig 7G).
Supplementary Material
Highlights.
Cocaine self-administration regulates alternative splicing and H3K36me3 enrichment
Alternative splicing of splice factor, Srsf11, is accompanied by enrichment of H3K36me3
CRISPR epigenetic editing of H3K36me3 drives alternative splicing of Srsf11 in brain
Srsf11 regulates downstream splice events and augments cocaine reward-behavior
ACKNOWLEDGEMENTS
The authors wish to thank Dr. Brian D. Strahl (University of North Carolina) for generously donating the Set2 and Set2(R195G) plasmids used in this study. CUT&RUN protocols and pA-MNase was kindly provided by Dr. Kavitha Sarma and Dr. Qingqing Yan (The Wistar Institute). The authors thank Dr. Erica Korb (University of Pennsylvania) for critical reading of the manuscript. Financial support is kindly acknowledged from the Charles E. Kaufman Foundation Young Investigator Award (to E.A.H.), a Whitehall Foundation grant (to E.A.H.), NIH-NIDA Avenir Director’s Pioneer Award DP1 DA044250 (to E.A.H.), Pharmacology T32 Predoctoral Training Grant T32GM008076 (to S.I.L.), Research Supplements to Promote Diversity in Health-Related Research DP1 DA044250-01 (to E.A.H. and M.D.C.), T32 Predoctoral Training Grant in Pharmacology T32GM008076 (to M.D.C.), T32 Predoctoral Training Grant in Addiction T32DA028874 (to D.K.F.), NIDA Research Project Grants NIDA R01 DA33641 and NIDA R01 DA15214 (to R.C.P.), and NIDA K01 Mentored Research Scientist Career Development Award DA039308 (to M.E.W.). The cocaine used in this study was kindly provided by the NIDA drug supply program. dCas9 expression assessed by immunochemistry imaging was adapted from a previous collaboration with Dr. Alison Bond and Dr. Hongjun Song (Perelman School of Medicine at the University of Pennsylvania).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- De Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. (2011). Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol 18, 977–983. [DOI] [PubMed] [Google Scholar]
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, et al. (2013). ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. U. S. A 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzalluz-Luqueángeles, and Conesa A (2018). Single-cell RNAseq for the study of isoforms-how is that possible? Genome Biol. 19, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Levy MJ, Zhang N, Li H, Florens L, Washburn MP, and Workman JL (2021). The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain. Nat. Commun 12, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee A, Djekidel MN, Chen R, Chen W, Tuesta LM, and Zhang Y (2019). Cell type-specific transcriptional programs in mouse prefrontal cortex during adolescence and addiction. Nat. Commun 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchta WC, Moutal A, Hines B, Garcia-Keller C, Smith ACW, Kalivas P, Khanna R, and Riegel AC (2020). Dynamic CRMP2 Regulation of CaV2.2 in the Prefrontal Cortex Contributes to the Reinstatement of Cocaine Seeking. Mol. Neurobiol 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, and Gozani O (2017). RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J. Biol. Chem 292, 13381–13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MD, Hu Q, Bond AM, Lombroso SI, Czarnecki KS, Lim CJ, Song H, Wimmer ME, Pierce RC, and Heller EA (2020). Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat. Commun 11, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cai Z, Bai M, Yu X, Zhang C, Cao C, Hu X, Wang L, Su R, Wang D, et al. (2018). The RNA-binding protein ROD1/PTBP3 cotranscriptionally defines AID-loading sites to mediate antibody class switch in mammalian genomes. Cell Res. 28, 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Aoto J, and Südhof TC (2019). Alternative Splicing of Presynaptic Neurexins Differentially Controls Postsynaptic NMDA and AMPA Receptor Responses. Neuron 102, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, and Kellis M (2017). Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc 12, 2478–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, et al. (2014). Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Wu M, Braunschweig U, Roth J, Han H, Best AJ, Raj B, Aregger M, O’Hanlon D, Ellis JD, et al. (2018). Genome-wide CRISPR-Cas9 Interrogation of Splicing Networks Reveals a Mechanism for Recognition of Autism-Misregulated Neuronal Microexons. Mol. Cell 72, 510–524.e12. [DOI] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Niibori R, Salter EW, Weatheritt RJ, Tsang B, Farhangmehr S, Liang X, Braunschweig U, Roth J, Zhang S, et al. (2020). Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions. Mol. Cell 1176–1192. [DOI] [PubMed] [Google Scholar]
- Green CJ, Gazzara MR, and Barash Y (2017). MAJIQ-SPEL: Web-Tool to interrogate classical and complex splicing variations from RNA-Seq data. Bioinformatics doi: 10.1093/bioinformatics/btx565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, and Noble WS (2007). Quantifying similarity between motifs. Genome Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Li J, Pelkey KA, Pandey S, Chen X, Wang YX, Wu K, Ge L, Li T, Castellano D, et al. (2019). Shisa7 is a GABAAreceptor auxiliary subunit controlling benzodiazepine actions. Science (80- . ) 366, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Epstein DH, Nader MA, and Shaham Y (2016). Time to connect: Bringing social context into addiction neuroscience. Nat. Rev. Neurosci 17, 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Pena CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robinson MS, et al. (2014). Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci 17, 1720–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Hamilton PJ, Burek DD, Lombroso SI, Peña CJ, Neve RL, and Nestler EJ (2016). Targeted epigenetic remodeling of the cdk5 gene in nucleus accumbens regulates cocaine- and stress-evoked behavior. J. Neurosci 36, 4690–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishimoto A, Pletnikova O, Lang DL, Troncoso JC, Egan JM, and Liu QR (2019). Neurexin 3 transmembrane and soluble isoform expression and splicing haplotype are associated with neuron inflammasome and Alzheimer’s disease. Alzheimer’s Res. Ther 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicová J, Hozeifi S, Dušková E, Icha J, Tománková T, and Staněk D (2011). Histone Deacetylase Activity Modulates Alternative Splicing. PLoS One 6, e16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicová J, Hozeifi S, Stejskalová E, Dušková E, Poser I, Humpolíčková J, Hof M, and Staněk D (2013). The C-terminal domain of Brd2 is important for chromatin interaction and regulation of transcription and alternative splicing. Mol. Biol. Cell 24, 3557–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kim EJ, Feng J, Grant GR, and Heller EA (2017). Histone posttranslational modifications predict specific alternative exon subtypes in mammalian brain. PLOS Comput. Biol 13, e1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Greene CS, and Heller EA (2020). Specific histone modifications associate with alternative exon selection during mammalian development. Nucleic Acids Res. 48, 4709–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Taguchi R, Deguchi M, Ogasawara H, and Inoue E (2020). Extensive splicing changes in an ALS/FTD transgenic mouse model overexpressing cytoplasmic fused in sarcoma. Sci. Rep 10, 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier XE, Soto L, Gandal MJ, Thomas Gonatopoulos-Pournatzis X, Heller EA, Luo XD, and Zheng S (2019). Mechanisms of Neuronal Alternative Splicing and Strategies for Therapeutic Interventions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, and Ahringer J (2009). Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet 41, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrigand K, Magnone V, Barbry P, and Waldmann R (2020). High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, and Martin KC (2016). Cytoplasmic Rbfox1 Regulates the Expression of Synaptic and Autism-Related Genes. Neuron 89, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CS, Douglass SM, Morselli M, Obusan MB, Pavlyukov MS, Pellegrini M, and Johnson TL (2019). H3K36 Methylation and the Chromodomain Protein Eaf3 Are Required for Proper Cotranscriptional Spliceosome Assembly. Cell Rep. 27, 3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, and Wong PC (2015). TDP-43 repression ofnonconserved cryptic exons is compromised in ALS-FTD. Science (80-. ) 349, 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D (2005). Neuronal proteins custom designed by alternative splicing Introduction: custom tuned proteins for fine control. Curr. Opin. Neurobiol 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Lipscombe D, and Lopez Soto EJ (2019). Alternative splicing of neuronal genes: new mechanisms and new therapies. Curr. Opin. Neurobiol 57, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorsch ZS, Hamilton PJ, Ramakrishnan A, Parise EM, Salery M, Wright WJ, Lepack AE, Mews P, Issler O, McKenzie A, et al. (2019). Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci 22, 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RRF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, and Misteli T (2010). Regulation of alternative splicing by histone modifications. Science (80-. ) 327, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA (2007). Circadian rhythms, the mesolimbic dopaminergic circuit, and drug addiction. ScientificWorldJournal. 7, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SL, and Strahl BD (2017). Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci 74, 3317–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Henriques T, Lavender CA, McKay DJ, Strahl BD, Duronio RJ, Adelman K, and Matera AG (2017). Histone gene replacement reveals a posttranscriptional role for H3K36 in maintaining metazoan transcriptome fidelity. Elife 6, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton SS, Vaquero-Garcia J, Lahens NF, Grant GR, and Barash Y (2018). Outlier detection for improved differential splicing quantification from RNA-Seq experiments with replicates. Bioinformatics 34, 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Severing E, Angenent GC, and Immink RGH (2017). Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 18, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta L, de la T, Huang J, et al. (2016). Genome-wide changes in lncRNA, alternative splicing, and cortical patterning in autism. Nature 540, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013). Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. (2013). A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodor J, FitzPatrick DR, Eyras E, and Cáceres JF (2017). The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 14, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Vakili G, Kumaresan V, Schmidt HD, Famous KR, Chawla P, Vassoler FM, Overland RP, Xia E, Bass CE, Terwilliger EF, et al. (2010). Cocaine-Induced Chromatin Remodeling Increases Brain-Derived Neurotrophic Factor Transcription in the Rat Medial Prefrontal Cortex, Which Alters the Reinforcing Efficacy of Cocaine. J. Neurosci 30, 11735–11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yuan Y, Zucker-Scharff I, Fak JJ, Jereb S, Tajima Y, Licatalosi DD, and Darnell RB (2019). Differential NOVA2-Mediated Splicing in Excitatory and Inhibitory Neurons Regulates Cortical Development and Cerebellar Function. Neuron 101, 707–720.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, and Pierce RC (2010). Cocaine-induced neuroadaptations in glutamate transmission: Potential therapeutic targets for craving and addiction. Ann. N. Y. Acad. Sci 1187, 35–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Fagnocchi L, Mastrototaro G, Massimino L, Zaghi M, Indrigo M, Cattaneo S, Martini D, Gabellini C, Pucci C, et al. (2019). SETD5 Regulates Chromatin Methylation State and Preserves Global Transcriptional Fidelity during Brain Development and Neuronal Wiring. Neuron 104, 1–19. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, and Xing Y (2014). rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci 111, E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Bhanu NV, Karch KR, Wang X, and Garcia BA (2016). Complete Workflow for Analysis of Histone Post-translational Modifications Using Bottom-up Mass Spectrometry: From Histone Extraction to Data Analysis. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, and Henikoff S (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc 13, 1006–1019. [DOI] [PubMed] [Google Scholar]
- Sorenson MR, Jha DK, Ucles S. a, Flood DM, Strahl BD, Stevens SW, and Kress TL (2016). Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto EJL, and Lipscombe D (2020). Cell-specific exon methylation and CTCF binding in neurons regulate calcium ion channel splicing and function. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto XJL, Gandal XJ, Gonatopoulos-Pournatzis X, Heller EA, Luo X, and Zheng S (2019). Mechanisms of neuronal alternative splicing and strategies for therapeutic interventions. J. Neurosci 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD, et al. (2002). Set2 Is a Nucleosomal Histone H3-Selective Methyltransferase That Mediates Transcriptional Repression. Mol Cell Biol 22, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Iida K, Tsubota T, Hosokawa M, Denawa M, Brown JB, Ninomiya K, Ito M, Kimura H, Abe T, et al. (2018). Loss of Sfpq Causes Long-Gene Transcriptopathy in the Brain. Cell Rep. 23, 1326–1341. [DOI] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, and Pierce RC (2013). Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci 16, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong JK, Lin CH, Zhang M, Chen L, Black DL, and Zheng S (2016). PTBP1 and PTBP2 Serve Both Specific and Redundant Functions in Neuronal Pre-mRNA Splicing. Cell Rep. 17, 2766–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Loh YHE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, et al. (2018). Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biol. Psychiatry 84, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, and Bähler J (2011). Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 12, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SJ, and Heller EA (2018). Single sample sequencing (S3EQ) of epigenome and transcriptome in nucleus accumbens. J. Neurosci. Methods 308, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lu Z, Xu M, Pan L, Deng Y, Xie X, Liu H, Ding S, Hurd YL, Gavril Pasternak XW, et al. (2014). Cellular/Molecular A Heroin Addiction Severity-Associated Intronic Single Nucleotide Polymorphism Modulates Alternative Pre-mRNA Splicing of the Opioid Receptor Gene OPRM1 via hnRNPH Interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Grullon S, Ge K, and Peng W (2014b). Spatial Clustering for Identification of ChIP-Enriched Regions (SICER) to Map Regions of Histone Methylation Patterns in Embryonic Stem Cells. Methods Mol. Biol 1150, 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Luo J, Zhao W, and Zhou X (2017). Deep learning of the splicing (epi)genetic code reveals a novel candidate mechanism linking histone modifications to ESC fate decision. Nucleic Acids Res. 45, 12100–12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, et al. (2017). Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J. Clin. Invest 127, 3375–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xie J, Long C, Erdjument-Bromage H, Ding X, Zheng Y, Tempst P, Chen S, Zhu B, and Reinberg D (2009). Heterogeneous Nuclear Ribonucleoprotein L Is a Subunit of Human KMT3a/Set2 Complex Required for H3 Lys-36 Trimethylation Activity in Vivo. J. Biol. Chem 284, 15701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z-F, Sidoli S, Marchione DM, Simithy O, Janssen KA, Szurgot MR, and Garcia BA (2018). EpiProfile 2.0: A Computational Platform for Processing Epi- Proteomics Mass Spectrometry Data. J Proteome Res 17, 2533–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kharchenko PV, Sharp PA, Walsh CA, Zhang X, Chen MH, Wu X, Kodani A, et al. (2016). Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 166, 1147–1162.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lu Y, and Tian W (2012). Epigenetic features are significantly associated with alternative splicing. BMC Genomics 13, doi: 10.1186/1471-2164-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhang Y, Li YE, Lucero J, Behrens MM, and Ren B (2021). Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat. Methods 18, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic analyses will likely be utilized by other investigators. All gene lists and sequencing data obtained as part of this manuscript have been deposited to GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/). Accession numbers are available in the accompanying Key Resources Table. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H3K36me3 antibody | Abcam | Cat# ab9050, RRID:AB_306966 |
| Mouse monoclonal anti-H4 antibody | Abcam | Cat# ab31830, RRID:AB_1209246 |
| Rabbit polyclonal anti-SRSF11 antibody | Thermo Fisher Scientific | Cat# PA5-52606, RRID:AB_2647939 |
| Mouse anti-GAPDH | Abcam | Cat# ab9484, RRID:AB_307274 |
| Anti-mouse IgG | Cell Signaling Technology | Cat# 7076S, RRID:AB_330924 |
| Anti-rabbit IgG | Cell Signaling Technology | Cat# 7074S, RRID:AB_2099233 |
| Bacterial and Virus Strains | ||
| HSV-Set2 | This paper | N/A |
| HSV-Set2(R195G) | This paper | N/A |
| hSyn-GW-dCas9-Set2-IRES2-mCherry | This paper | N/A |
| hSyn-GW-dCas9-Set2(R195G)-IRES2-mCherry | This paper | N/A |
| P1005-GFP-Srsf11-sgRNA | This paper | N/A |
| P1005-GFP-NT-sgRNA | This paper | N/A |
| Deposited Data | ||
| RNA-sequencing of mouse NAc following 21 days of cocaine self-administration | This paper | GSE141520 |
| RNA-sequencing of mouse NAc following dCas9-Set2 + Srsf11-sgRNA | This paper | GSE180988 |
| RNA-sequencing of mouse NAc following dCas9-Set2 + Srsf11-sgRNA | This paper | GSE180988 |
| RNA-sequencing of mouse NAc following 21 days of saline self-administration | This paper | GSE141520 |
| Cut&Run sequencing of H3K36me3 of dCas9-SET2 transfected N2a cells | This paper | GSE180988 |
| ChIP-sequencing of H3K36me3 of mouse NAc following cocaine SA | This paper | GSE180988 |
| ChIP-sequencing of H3K36me3 of mouse NAc following HSV-Set2 transfection | This paper | GSE180988 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664) |
| Oligonucleotides | ||
| PCR Primers, see Table S1 | This paper | N/A |
| sgRNA, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Prism v8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| R v4.0.2 | The R Project for Statistical Computing | https://www.r-project.org/ |
| Factor Analysis - Cocaine Self Administration | Walker et al., 2018 | N/A |
| CUT&RUN Analysis | Skene and Henikoff, 2017 | N/A |
| Splicing analysis rMATS | Shen et al., 2014 | http://rnaseq-mats.sourceforge.net/ |
| Splicing analysis MAJIQ | Norton et al., 2017 | https://majiq.biociphers.org/ |
| DiffBind | Ross-Innes et al., 2012 | Bioconductor https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Deseq2 | Love et al., 2014 | Bioconductor https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| SICER | Xu et al., 2014b | https://zanglab.github.io/SICER2/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/motif/ |