Abstract
In unstimulated cells, NF-κB transcription factors are retained in the cytoplasm by inhibitory IκB proteins. Upon stimulation by multiple inducers including cytokines or viruses, IκBα is rapidly phosphorylated and degraded, resulting in the release of NF-κB and the subsequent increase in NF-κB-regulated gene expression. IκBα gene expression is also regulated by an NF-κB autoregulatory mechanism, via NF-κB binding sites in the IκBα promoter. In previous studies, tetracycline-inducible expression of transdominant repressors of IκBα (TD-IκBα) progressively decreased endogenous IκBα protein levels. In the present study, we demonstrate that expression of TD-IκBα blocked phorbol myristate acetate-phytohemagglutinin or tumor necrosis factor alpha-induced IκBα gene transcription and abolished NF-κB DNA binding activity, due to the continued cytoplasmic sequestration of RelA(p65) by TD-IκBα. In vivo genomic footprinting revealed stimulus-responsive protein-DNA binding not only to the −63 to −53 κB1 site but also to the adjacent −44 to −36 Sp1 site of the IκBα promoter. In vivo protection of both sites was inhibited by tetracycline-inducible TD-IκBα expression. Prolonged NF-κB binding and a temporal switch in the composition of NF-κB complexes bound to the −63 to −53 κB1 site of the IκBα promoter were also observed; with time after induction, decreased levels of transcriptionally active p50-p65 and increased p50–c-Rel heterodimers were detected at the κB1 site. Mutation of either the κB1 site or the Sp1 site abolished transcription factor binding to the respective sites and the inducibility of the IκBα promoter in transient transfection studies. These observations provide the first in vivo characterization of a promoter proximal transcriptional switch involving NF-κB and Sp1 that is essential for autoregulation of the IκBα promoter.
The NF-κB/Rel family of transcription factors participates in the regulation of the immunomodulatory genes and activates numerous cellular genes as well as viral genes including the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) (6, 7, 52, 60). The NF-κB/Rel family members can be subdivided into two subgroups according to their structure and function: the DNA binding proteins NF-κB1(p50), NF-κB2(p52), RelA(p65), c-Rel, and RelB and the NF-κB1(p105) and NF-κB2(p100) precursors which are proteolytically cleaved to generate DNA binding proteins (p50 and p52, respectively). All members of the family share an N-terminal 300-amino-acid domain known as the NF-κB/Rel/dorsal homology region which is responsible for binding to DNA (consensus sequence GGGRNNYYCC [6, 7, 52, 60]), dimerization, and nuclear translocation of NF-κB (5). The dimer composition of different NF-κB subunits and the sequence context of NF-κB sites in different promoters contribute to the differential specificity of gene activation (22, 34, 38, 46).
The members of the IκB family include IκBα (26), IκBβ (58), IκBɛ (61), IκBγ (24), and Bcl-3 (27), as well as the NF-κB proteins p105 (39) and p100 (41), which contain ankyrin repeats in the C-terminal portion of the molecule and bind to NF-κB, masking the nuclear localization sequence (10, 11). The most extensively characterized of the IκB proteins is IκBα. Upon stimulation by many activating agents, including tumor necrosis factor (TNF) and phorbol 12-myristate 13-acetate (PMA), IκBα is rapidly phosphorylated by the recently identified 700- to 900-kDa complex containing IκB kinase (IKK) (19, 48, 62). Phosphorylation targets IκBα for ubiquitination and degradation by the 26S proteasome, resulting in the release of NF-κB (16). The substitution of alanine for Ser-32 and Ser-36 within the N-terminal signal response domain abolished the signal-induced IκBα phosphorylation and degradation, resulting in a blockage of NF-κB activation (13, 14, 59). These mutations also abrogated in vitro ubiquitination of the IκBα protein (16, 50, 55). The amino terminus of IκBα is necessary for signal-induced degradation, but degradation of IκBα also requires the C-terminal domain of the protein (9, 14, 21, 30, 37, 38, 49) which is constitutively phosphorylated by casein kinase II (8, 37, 40). Once released, NF-κB is able to activate target genes until new IκBα is synthesized.
Since the IκBα gene contains NF-κB binding sites in its promoter, NF-κB is able to autoregulate the transcription of its own inhibitor (15, 17, 29, 35, 57). This autoregulatory control of IκBα expression is in part responsible for the transient nature of the NF-κB activation of gene expression; newly synthesized IκB can localize to the nucleus and directly interfere with gene expression by dissociating protein-DNA complexes (3, 53). The IκBα gene has also been shown to be regulated by RelA(p65) at both the mRNA and protein levels: RelA(p65)-IκBα protein interactions increased the half-life of the inhibitory protein, and IκBα mRNA was induced by RelA(p65) as a consequence of increased IκBα gene transcription (12, 56). Stimulation of Jurkat T cells by TNF-α or PMA induced degradation of IκBα protein concomitant with NF-κB release and activation (36, 42). Activation was followed by de novo IκBα synthesis in an NF-κB-dependent manner, and cycloheximide treatment prior to induction resulted in the inhibition of IκBα resynthesis, as well as prolonged NF-κB DNA binding (57).
Previous analysis of the human IκBα promoter (29, 35) identified three NF-κB sites (−63 to −53, −225 to −216, and −319 to −310) and two NF-κB-like sites (−159 to −150 and −34 to −24) (see Fig. 2C). By deletion and point mutagenesis only the κB1 site from −63 to −53 was shown to be functionally important for inducibility, since disruption of the κB1 site completely abolished IκBα promoter activity, whereas deletion of the κB2 site from −319 to −310 and κB3 site from −225 to −216 had no effect. Another study demonstrated that in addition to the κB1 site, the κB-like site located between −34 to −24 is essential for IκBα gene expression since activation of the IκBα promoter by TNF-α was abolished when κB1 and κB-like sites were mutated (29). Both sites bound p50 homodimers in resting HeLa cells and p50, p65, and c-Rel complexes in TNF-α-induced cells.
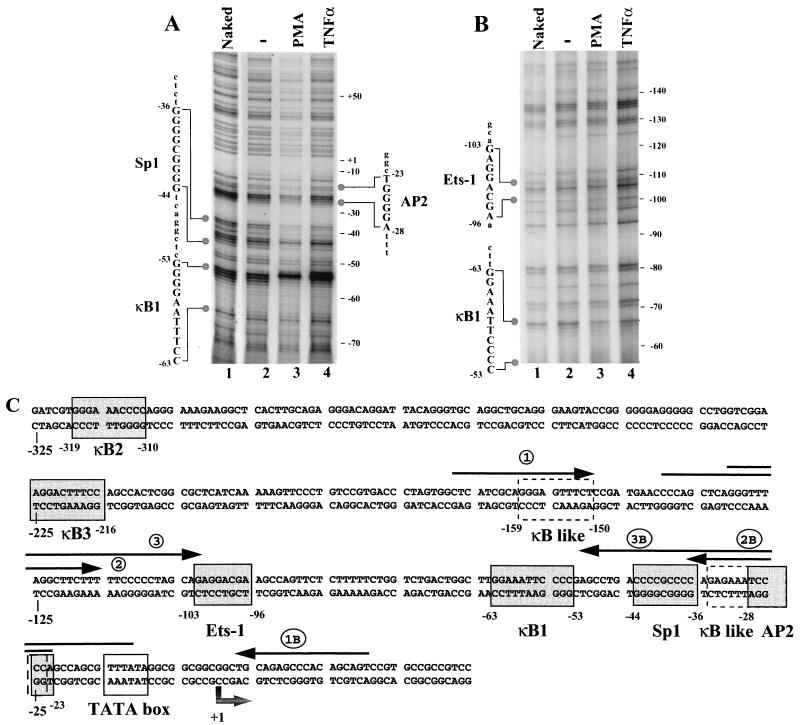
FIG. 2.
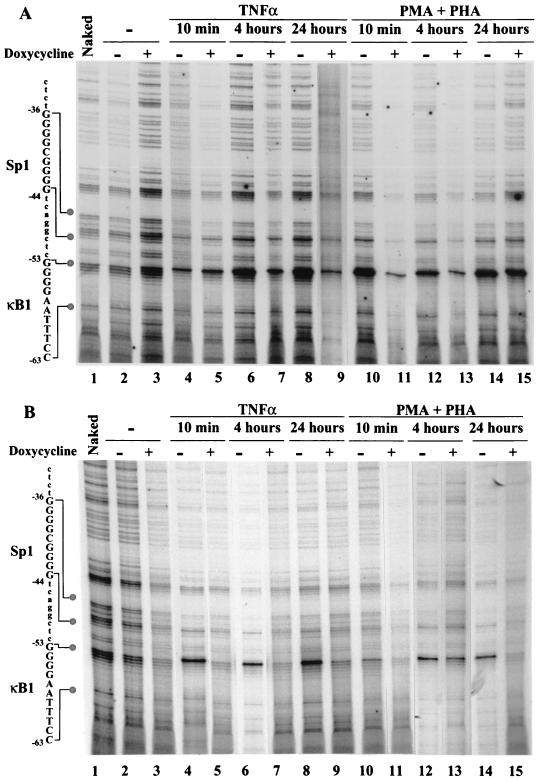
In vivo footprinting of the proximal IκBα gene promoter in Jurkat T cells. (A) Noncoding strand analysis; (B) coding strand analysis. Naked DNA was treated in vitro with DMS (lane 1). Cells were either nonstimulated (lane 2) or treated with PMA-PHA (lane 3) or by TNF-α (lane 4) for 40 min and then were treated with DMS. Genomic DNA was extracted and treated with piperidine. All DNA samples were amplified by LM-PCR and visualized on a Long-Ranger sequencing gel. (C) Sequence of the IκBα promoter. Major consensus sites for protein binding such as the NF-κB sites and sites of Ets-1, Sp1, and AP2 are enclosed in boxes. The mRNA start site and TATA box are also shown. Arrows correspond to primers used in genomic footprinting. Primers 1, 2, and 3 were designed to characterize the noncoding strand; primers 1B, 2B, and 3B were used for the coding strand.
In Jurkat T cells that express transdominant repressors of IκBα (TD-IκBα) under the control of a tetracycline (Tet)-inducible system, endogenous IκBα protein expression was blocked by TD-IκBα induction (31). We now demonstrate that inducer-dependent induction of IκBα gene transcription was blocked by the transdominant repressor expression at the transcriptional level. To further analyze the autoregulatory control of IκBα expression, dimethyl sulfate (DMS) genomic footprinting was used to determine the pattern of protein-DNA interactions at the IκBα locus in stimulated Jurkat T cells and in TD-IκBα-expressing cells. These studies permit the first in vivo characterization of IκBα transcriptional autoregulation by NF-κB and identify the promoter-proximal NF-κB/Sp1 transcriptional switch as an essential component in the regulation of the IκBα promoter.
MATERIALS AND METHODS
Cell lines and reagents.
Jurkat cells and Jurkat cells stably expressing TD-IκBα were described previously (31). IκBα(2NΔ4) contains alanine substitutions at the Ser-32 and Ser-36 inducible phosphorylation sites as well as a 22-amino-acid deletion of the C-terminal domain of IκBα, a region of the PEST domain that is dispensable with regard to binding of NF-κB subunits but is important for IκBα degradation (9, 37). All cells were grown in RPMI 1640 containing 10% fetal bovine serum (FBS), 2 mM glutamine, and 10 μg of gentamicin per ml. Cells were stimulated by PMA (50 ng/ml; ICN, Costa Mesa, Calif.) plus phytohemagglutinin (PHA; 1 μg/ml; Sigma, St. Louis, Mo.) or TNF-α (10 μg/ml; R&D System, Minneapolis, Minn.).
Immunoblot analysis.
To characterize kinetics of expression, Jurkat T cells transfected with rtTA-Neo (rtTA-Neo Jurkat cells) and rtTA-IκBα(2NΔ4) Jurkat cells were cultured in the presence of doxycycline (Dox; 1 μg/ml; Sigma) for various times. Cells were then washed with phosphate-buffered saline (PBS) and lysed in a mixture containing 10 mM Tris-HCl (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5% Nonidet P-40 (NP-40), 0.5 mM phenylmethysulfonyl fluoride, leupeptin (10 μg/ml), pepstatin (10 μg/ml), and aprotinin (10 μg/ml). Equivalent amounts of whole-cell extract (20 μg) were subject to sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 10% polyacrylamide gel. After transfer, the Hybond membrane (Amersham, Cleveland, Ohio) was incubated overnight with N-terminal IκBα monoclonal antibody MAD10B (30) at 4°C. After four 10-min washes with PBS, membranes were reacted with a peroxidase-conjugated secondary goat anti-mouse antibody (Kierkegaard & Perry Laboratories, Gaithersburg, Md.) at a dilution of 1:1,000. The reaction was then visualized with an enhanced chemiluminescence detection system as recommended by the manufacturer (NEN Life Science, Boston, Mass.).
RNase protection assay.
A 221-bp XbaI-PstI fragment was obtained by PCR amplification with an IκBα cDNA clone (cloned into pSVK3) by using specific primers containing restriction enzyme sites corresponding to positions 824 to 839 (5′ATCATCTAGAAACAGAGTTACCTACC3′) and 1030 to 1045 (5′ATCACTGCAGTAACGTCAGACGCTGG3′); the XbaI-PstI fragment of the PCR product was cloned into the XbaI-PstI site of the pDP18-T7/T3 transcription vector (Ambion, Inc., Austin, Tex.) to generate pDP18CU-/CTERM. 32P-labeled antisense RNA probe was transcribed by using an in vitro transcription kit (Pharmingen, San Diego, Calif.), and RNase protection was carried out with an RNase protection kit (Pharmingen). A β-actin antisense probe (pTRI-β-actin; Ambion) was synthesized by the same protocol and used in the same reaction with an IκBα probe; 5 to 10 μg of total RNA extracted by using an RNeasy mini kit (Qiagen, Valencia, Calif.) from unstimulated or stimulated rtTA-Neo or rtTA-IκBα(2NΔ4) Jurkat cells was used. The resulting protected RNAs were resolved on a 5% denaturing gel and exposed to X-ray film.
EMSA.
Following the addition of 1 μg of Dox per ml to the culture medium for 24 h, nuclear extracts were prepared from rtTA-Neo or rtTA-IκBα(2NΔ4) Jurkat T cells or Jurkat T cells after induction with TNF-α or PMA-PHA for times ranging from 10 min to 24 h. Nuclear extracts were prepared as previously described (31) and subjected to electrophoretic mobility shift assay (EMSA) by using 32P-labeled probes corresponding to the IκBα promoter either in NF-κB DNA binding buffer (20 mM HEPES [pH 7.9], 5% glycerol, 0.1 M KCl, 0.2 mM EDTA [pH 8.0], 0.2 mM EGTA [pH 8.0]) or in NF-κB/Sp1 DNA binding buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 20% glycerol, 0.1 mM EDTA, 0.25 mM ZnSO4, 0.05% NP-40), together with 5 or 0.5 μg of poly(dI-dC), respectively. Oligonucleotides used are as follows: κB1, 5′-GATCTTGGAAATTCCCCGA-3′; Sp1, 5′-TCGAGACCCCGCCCCAG-3′; consensus Sp1, 5′-ATTCGATCGGGGCGGGGCGAGC-3′; mutated Sp1 probe, 5′-ATTCGATCGGTTCGGGGCGAGC-3′; κB1/Sp1, 5′-TCGATTGGAAATTCCCCGAGCCTGACCCCGCCCCAG-3′; mutκB1/Sp1, 5′-TCGATTGTCAATTCCCCGAGCCTGACCCCGCCCCAG-3′; κB1/mutSp1, 5′-TCGATTGGAAATTCCCCGAGCCTGACCAAGCCCCAG-3′; and +5 κB1/Sp1, 5′-TCGATTGGAAATTCCCCGAGCTGCAGCTGACCCCGCCCCAG-3′. Underlining delineates the Sp1 site, and boldface letters indicate mutations in either κB1 or Sp1 sites. Recombinant proteins (glutathione S-transferase [GST]–NF-κB fusion proteins [38, 47] and Sp1 [Promega Inc.]) were also incubated with the probes in a different DNA binding buffer [10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 0.1 mg bovine serum albumin per ml, 50 μM MgCl2, 1 mM ATP, 5 μg of poly(dI-dC) per ml]. The resulting protein-DNA complexes were resolved on 5 to 6% polyacrylamide–1× Tris-borate-EDTA gels and exposed to X-ray film. To demonstrate the specificity of protein-DNA complex formation, 125-fold molar excess of unlabeled oligonucleotide was added to the nuclear extract before addition of labeled probe. Supershift analysis was performed with anti-p65, anti-p50, anti-c-Rel, and anti-Sp1 antibodies (Santa Cruz Biotechnology Inc.).
In vivo genomic footprinting.
Jurkat cells (108) were harvested and resuspended in 1 ml of RPMI 1640–10% FBS containing 20 mM HEPES (pH 7.3). The methylation reaction was performed in presence of 10 μl of concentrated DMS (Aldrich Chemical Company, Milwaukee, Wis.) for 1 min. The reaction was then quenched by two washes in cold PBS containing 2% β-mercaptoethanol. Genomic DNA extraction was performed as previously described (1). Briefly, cells were lysed in 2 ml of Tris buffer (pH 7.5) containing 10 mM NaCl and 10 mM EDTA and supplemented with 100 μl of proteinase K (20 mg/ml), 100 μl sodium dodecyl sulfate (20%), and 100 μl of NP-40 (10%) and then incubated at 50°C overnight. Proteins were precipitated by adding 1.2 ml of 5 M NaCl and centrifuged for 40 min at 7,500 × g (SS34 rotor, Sorval RS5 Superspeed). Cleared supernatant was ethanol precipitated to obtain genomic DNA; the pellet was resuspended in 200 μl H2O with 20 μl of piperidine (Aldrich) and incubated 30 min at 90°C to provide cleavage of methylated G (or A) residues. The DNA control (naked DNA) was first extracted from cells and then submitted to DMS treatment and subsequently to piperidine cleavage to allow methylation and cleavage to all G residues of the sequence. For each sample, 2 μg of DNA were submitted to ligation-mediated PCR (LM-PCR) using Vent DNA polymerase (New England Biolabs, Mississauga, Ontario, Canada) as described elsewhere (23, 43). PCR amplification was for 2 min for the first cycle and was progressively increased to 10 min in the last cycle. A total of 18 cycles were performed for DNA amplification. The third primer was radiolabeled by end labeling using T4 polynucleotide kinase (Pharmacia Biotech) and [γ-32P]ATP (ICN). Two more PCR cycles were performed to radiolabeled elongated DNA. The final labeled PCR product was analyzed on a 5% Hydrolink Long Ranger sequencing gel (Baker, Phillipsburg, N.J.) in 1.0× Tris-borate-EDTA at 65 W and exposed for 12 to 36 h with BioMax sensitive film (Eastman Kodak, Rochester, N.Y.). For the LM-PCR, two sets of oligonucleotides were used: for the noncoding strand, primers 1 (5′-CTCATCGCAGGGAGTTTCT-3′; melting temperature [Tm], 55°C; 2 (5′-CCCAGCTCAGGGTTTAGGCTTCTTT-3′; Tm, 63°C); and 3 (5′-GGGTTTAGGCTTCTTTTTCCCCCTAGCAG-3′; Tm, 66°C); for the coding strand, primers 1B (5′-ACTGCTGTGGGCTCTGCA-3′; Tm, 63°C); 2B (5′-TAAACGCTGGCTGGGGATTTCTCTG-3′; Tm, 63°C); and 3B (5′-TGGGGATTTCTCTGGGGCGGGGTCAGGCT-3′; Tm, 71°C) (see Fig. 2C).
Plasmid construction and mutagenesis.
0.4SK-pGL3 Luc was obtained by subcloning the 0.4-kb fragment of the IκBα promoter (a kind gift from A. Israël) into KpnI/SacI-digested pGL3. Plasmids carrying point mutations in the κB1 or Sp1 site or in both sites were obtained by the subcloning of PCR-amplified fragments into KpnI/SacI-digested pGL3. Briefly, these constructs were obtained in two steps by a procedure previously described (33). The first round of amplification used 0.4SK-pGL3 Luc as template, 5′-CACGCGTAAGAGCTCCACCG-3′ (SacI primer) as 3′ primer, and 5′-GGAAATTCaaCGAGCCTGAC-3′ (nucleotides in lowercase indicate point mutations) as 5′ primer for κB1 site mutagenesis or 5′-CCTGACCaaGCCCCAGAGAA-3′ as 5′ primer for Sp1 site mutagenesis. The amplified fragments were used as the 3′ primer in a second PCR using 0.4SK-pGL3 Luc as template and 5′-CTATCGATAGGTACCGGGCC-3′ (KpnI primer) as 5′ primer. In each case, the final products were purified, digested by KpnI and SacI, and inserted between these sites in the pGL3 polylinker. The construct carrying both κB1 and Sp1 mutations was similarly obtained by using Sp1-mutated IκBα promoter as template. The promoter constructs carrying internal deletions or insertions were obtained by the ligation of two separately amplified fragments, one digested by SacI and the other digested by KpnI, to KpnI/SacI-digested pGL3. Thus, plasmid Δ8-IκBα was constructed by ligation of two fragments amplified with 5′-CCCCGCCCCAGAGAAATC-3′/SacI primers and KpnI/5′-GGGGAATTTCCAAGCCAGT-3′ primers, in the presence of 0.4SK-pGL3 Luc as template. The +5- and +9-IκBα plasmids were similarly constructed with 5′-TGCAGCTGACCCCGCCCCAGAGAAA-3′/SacI (inserted nucleotides are underlined) and KpnI/5′-GCTCGGGGAATTTCCAAGCCA-3′ primers and with 5′-GCAGCATCGCTGACCCCGCCCCAGAGAAA-3′/SacI and KpnI/5′-GCTCGGGGAATTTCCAAGCCA-3′ primers, respectively. The correct sequences of the constructs presented were confirmed by DNA sequence analysis. Information concerning PCR is available upon request.
Transfection and luciferase assay.
Jurkat T cells were transiently transfected by the DEAE-dextran method (32). One microgram of 0.4SK-pGL3 (wild-type IκBα promoter) Luc reporter plasmids or mutant 0.4SK-pGL3 (mutκB1, mutSp1, and mutκB1/Sp1) along with pRL-TK (for transfection normalization; Promega) was resuspended in TS solution (8 mg of NaCl per ml, 0.38 mg of KCl per ml, 0.1 mg of Na2HPO4 · 7H2O per ml, 3.0 ml of Tris, 0.1 mg of MgCl2 per ml, 0.1 mg of CaCl2 per ml [pH 7.4]) with 25 μg of DEAE-dextran (Pharmacia). For each transfection, 5 × 105 cells in exponential phase were washed once in 100 μl of TS, resuspended with the DNA solution, and incubated at room temperature for 20 min. Cells were then incubated at 37°C for 30 min in 0.5 ml with medium containing 10% FBS and 0.1 mM chloroquine (Sigma), after which they were centrifuged and resuspended in fresh medium and serum. At 30 h after transfection, cells were induced with TNF-α or PMA-PHA. At 16 h after induction, cells were harvested and lysed by 1× passive lysis buffer, and then luciferase activity was analyzed by the Dual-Luciferase reporter assay system (Promega) as specified by the manufacturer. The background obtained from mock-transfected cells was subtracted from each experimental value. The experiments were performed in triplicate in 24-well plates, and the average fold induction was calculated.
RESULTS
Tet-induced TD-IκBα blocks expression of the IκBα gene.
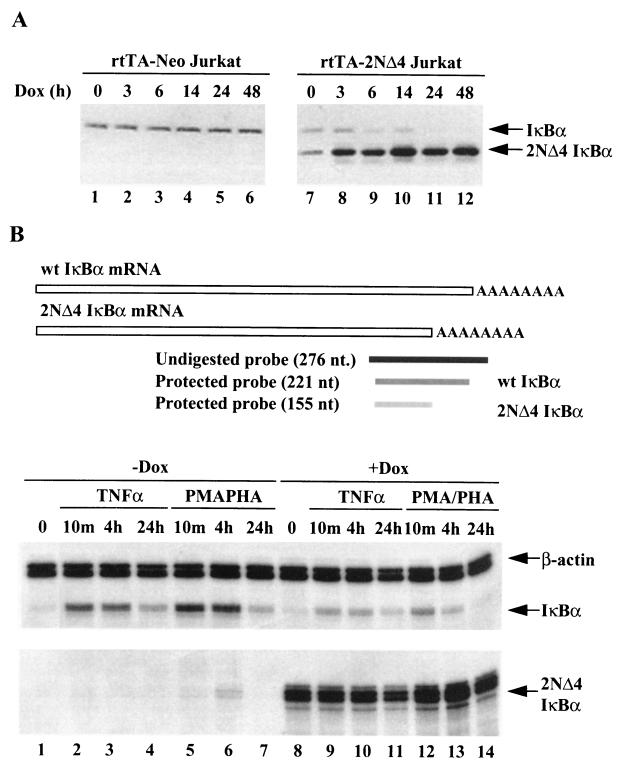
As shown previously (31), endogenous IκBα protein expression was abolished after TD-IκBα induction for 24 h (Fig. 1A, lane 7 to 11). To determine whether IκBα gene transcription was downregulated by TD-IκBα, rtTA-IκBα(2NΔ4) Jurkat cells were treated with TNF-α or PMA in the presence or absence of Dox, and endogenous IκBα and TD-IκBα mRNAs were analyzed by RNase protection analysis with a 27-nucleotide (nt) 3′ cDNA probe that specifically recognized the C terminus of endogenous IκBα mRNA as well as the truncated IκBα(2NΔ4) mRNA (Fig. 1B). After 10 min of TNF-α or PMA-PHA induction, we detected 20- and 40-fold increases in IκBα mRNA, respectively (Fig. 1B, lanes 2 and 5); subsequently the level of IκBα mRNA declined with time in rtTA-IκBα(2NΔ4) Jurkat cells (Fig. 1B, lanes 4 and 7). In TD-IκBα-expressing cells, endogenous IκBα mRNA induction was decreased fivefold by Dox addition, whereas high-level expression of the IκBα(2NΔ4) transgene was easily detected by RNase protection (Fig. 1B, lanes 8 to 14). These results indicate that Dox-induced TD-IκBα expression interfered with the induced but not the basal level of endogenous IκBα mRNA transcription.
FIG. 1.
Tet-induced TD-IκBα inhibits endogenous IκBα expression. (A) rtTA-Neo (lanes 1 to 6) and rtTA-IκBα(2NΔ4) (lanes 7 to 12) Jurkat cells were incubated with Dox (1 μg/ml) for 0, 3, 6, 14, 24, and 48 h. Endogenous IκBα (top arrow) and TD-IκBα (bottom arrow) were detected by immunoblotting with antibody MAD10B. (B) Schematic representation of C-terminal IκBα probe used in RNase protection analysis. rtTA-IκBα(2NΔ4) Jurkat cells were treated with TNF-α (10 ng/ml; lanes 2 to 4 and 9 to 11) or PMA (50 ng/ml) plus PHA (1 μg/ml) (lanes 5 to 7 and 12 to 14) for 0, 10 min, 4 h, or 24 h in the absence (lanes 1 to 7) or presence (lanes 8 to 14) of Dox (1 μg/ml, 24 h). Endogenous (wild-type [wt]) IκBα and TD-IκBα mRNAs were detected with using the 276-nt 3′ cDNA probe by RNase protection assay. Arrows indicate β-actin, IκBα (221-nt band), and IκBα(2NΔ4) (155-nt band). The results shown are representative of at least three independent experiments.
Using the κB1 site of the IκBα promoter as the probe in EMSA, we showed that TNF-α- or PMA-PHA-induced NF-κB binding activity was completely blocked in rtTA-IκBα(2NΔ4) Jurkat cells when TD-IκBα is expressed (reference 31 and data not shown). Coimmunoprecipitation studies performed with anti-p65 and anti-IκBα antibodies further demonstrated that inhibition of NF-κB DNA binding activity and endogenous IκBα transcription in TD-IκBα-inducible cells are due to the tight association between the NF-κB transactivator p65 and TD-IκBα, which is resistant to inducer-mediated degradation (data not shown).
In vivo genomic footprinting of the IκBα promoter.
To analyze the in vivo occupancy of the IκBα promoter, genomic footprinting analysis was performed in Jurkat and IκBα(2NΔ4)-expressing Jurkat cells. Following stimulation by either TNF-α or PMA-PHA, living cells were submitted to DMS treatment, which methylates G residues and to a lesser extent A residues; genomic DNA was then extracted and submitted to piperidine treatment to cleave methylated residues. Then, piperidine-cleaved DNA was amplified by LM-PCR using specific primers for IκBα promoter as detailed in Fig. 2C. A G-specific sequence ladder was also generated as reference and analyzed by sequencing.
Footprinting primers were initially designed to analyze in vivo protein-DNA interactions occurring in the proximal −10 to −170 region of the IκBα promoter (primers 1, 2, and 3 for the noncoding strand; primers 1B, 2B, and 3B for the coding strand [Fig. 2C]). In resting Jurkat T cells, the proximal G residues of the κB1 site at bp −53 to −56 were cleaved and easily detected by comparison with the G ladder, revealing no protein-DNA interaction in the absence of stimulation (Fig. 2A, lanes 1 and 2). In the absence of stimulation, a weak interaction was detected at the Sp1 site located between bp −44 and −36, adjacent to the κB1 site (Fig. 2A, lanes 1 and 2). In response to PMA-PHA or TNF-α induction for 40 min, the κB1 site was strongly protected by protein-DNA interactions, resulting in very limited cleavage of the −54G, −55G, and −56G residues and a hypersensitive cleavage at −53G (Fig. 2A, lanes 3 and 4). Similarly, modifications of the pattern of the Sp1 site were observed; with naked DNA or DNA from unstimulated cells, we detected cleavage of −44G, −43G, −42G, and −41G as well as −39G, −37G, −36G (Fig. 2A, lanes 1 and 2), while following induction, the G residues of the Sp1 site were protected with the exception of the −42G residue, which was hypersensitive to cleavage (Fig. 2A, lanes 3 and 4).
Changes in the coding strand methylation pattern of the κB1 site were also detected with specific primers, although PCR amplification was difficult to obtain because of the GC-rich nature of the region, allowing detection of κB1 and Ets but not Sp1 sites (Fig. 2C). The two G residues of the κB1 site located at −63 and −62 were methylated on naked DNA (Fig. 2B, lane 1 and 2). Following stimulation of Jurkat T cells with PMA-PHA or TNF-α, the −63G residue was methylated and cleaved whereas the −62G residue was protected from methylation (Fig. 2B, lanes 3 and 4), thus demonstrating protection of the κB1 site on both coding and noncoding strands. Two other potential binding sites in the −10 to −110 region, AP2 and Ets-1, were identified in vivo (Fig. 2A and B); hypermethylation of the −24G and −25G residues was observed at the AP2 site in both unstimulated and stimulated cells (Fig. 2A; compare lane 1 to lanes 2 to 4). Similarly, methylation of the −99A residue at the Ets-1 site appeared slightly enhanced in resting and stimulated cells (data not shown); for both sites, induction-specific changes in promoter occupancy were not detected.
Scanning analysis of the methylation pattern of the −20 to −70 IκBα promoter region.
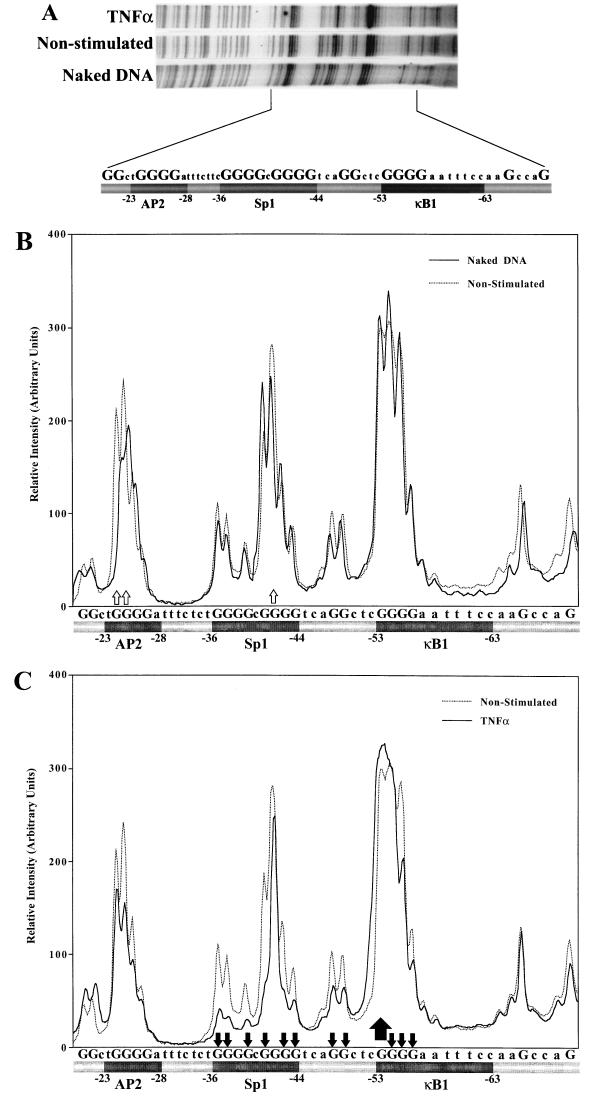
Since the modifications at the Sp1 site were more discrete than those detected at the κB1 site, changes in the methylation pattern of the −20 to −70 region of IκBα promoter which includes the AP2, Sp1, and κB1 sites were analyzed by densitometry scanning (Fig. 3). A representative autoradiograph is presented in Fig. 3A. Comparison of nonstimulated and naked DNA patterns revealed increased methylation of the −42G residue and a slight decrease in −41G methylation at the Sp1 site, whereas no significant differences in methylation were observed for the bordering −20G and −66G residues, indicating an even methylation pattern in the scanned region (Fig. 3B). These data indicate that the Sp1 site was constitutively occupied in resting Jurkat T cells. Similarly, hypermethylation of −24 and −25G residues in nonstimulated Jurkat cells suggests a constitutive binding at the AP2 site. In contrast, no binding was detected on the κB1 site in unstimulated conditions. Following stimulation by either TNF-α, as shown in Fig. 3C, or PMA-PHA (data not shown), protection of the Sp1 site was modified, as observed by a strong decrease in methylation at residues −36G, −37G, −39G, −41G, −43G, and −44G; only −42G remained methylated. As clearly seen in Fig. 2A, inducible binding at the κB1 site is characterized by decreased methylation of −54G, −55G, and −56G as well as a very strong increase in −53G methylation, detected as a broad peak by densitometric scanning (Fig. 3C). Interestingly, methylation of −48G and −49G, which are located between Sp1 and κB1 sites, was also significantly decreased after stimulation, indicating that the inducible changes at the κB1 and Sp1 sites affect the whole region delimited by these two sites (Fig. 3C). In contrast, no significant modifications of −66G and −21G were observed after stimulation, thus restricting the inducible region to the Sp1 and κB1 sites.
FIG. 3.
Scanning analyses of In vivo footprinting of the proximal IκBα gene promoter in Jurkat T cells. Methylation patterns observed on the −20 to −70 region of noncoding strand of IκBα promoter with naked DNA or from nonstimulated or TNFα stimulated cells for 4 h (A) were analyzed by densitometry scanning using a Hewlett-Packard Scan Jet 4c scanner and NIH Image 1.60 software. Comparison of profiles obtained with naked DNA versus DNA from resting Jurkat T cells (B) or with DNA from resting cells versus TNF-α-stimulated cells (C) corresponds to profiles obtained in at least three independent experiments. Open arrows represent constitutive modifications; filled arrows correspond to inducible changes. Arrows pointing up or down represent increased or decreased methylation on G residues. The sequence of the scanned region, where the methylated G residues are in capital letters, is indicated below each graph.
Together, these results demonstrate the in vivo occupancy of the −63 to −53 κB1 site of the IκBα promoter after stimulation with either PMA-PHA or TNF-α and also indicate that the Sp1 site located 10 bp downstream from the κB1 site may play a role in the inducible transcription of the IκBα promoter.
Footprinting analysis of the IκBα upstream promoter.
The potential role of other upstream NF-κB sites that may play a role in IκBα inducibility (29, 35), including κB2 and κB3 sites was also evaluated. No significant modification of the patterns was observed in the region corresponding to the κB2 site at −319 to −310 or in the putative κB site at −34 to −24, located downstream of κB1. However, the A residue at position −222 in the κB3 site appeared methylated in both unstimulated and TNF-α-stimulated cells, suggesting constitutive protein binding to this site (data not shown). These data indicate that although other κB sites in the IκBα promoter are recognized in vitro by NF-κB complexes (29, 35) and in vivo by constitutive binding complexes, they are not modified in vivo after stimulation of Jurkat T cells. Only the κB1 site appears to be targeted by inducible NF-κB binding proteins as detected by in vivo genomic footprinting (Fig. 2).
NF-κB protection of the κB1 site is blocked in TD-IκBα-expressing cells.
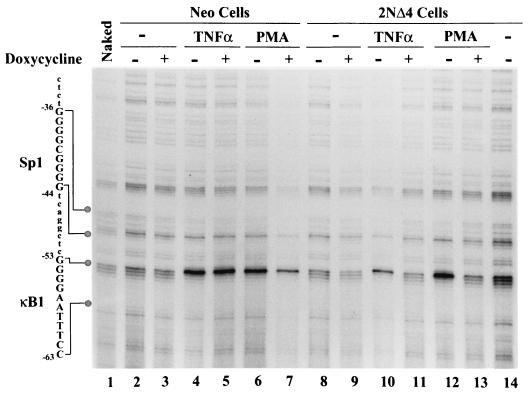
Next, control (Neo) Jurkat and TD-IκBα(2NΔ4)-expressing cells were treated with TNF-α or PMA-PHA for 40 min following 24 h of Dox induction (Fig. 4) and then subjected to genomic footprint analysis using noncoding specific primers 1, 2, and 3. In control cells, the 4G ladder was easily identified (Fig. 4, lanes 1 to 3), and following TNF-α or PMA-PHA addition, the characteristic hypersensitive cleavage of −53G was detected (lanes 4 to 7). In IκBα(2NΔ4)-expressing cells, TD-IκBα induction resulted in complete inhibition of inducible binding complexes to the κB1 site in a Dox-dependent manner; TNF-α or PMA-PHA stimulation in the absence of Dox-induced TD-IκBα resulted in a footprint at the κB1 site that was indistinguishable from that of stimulated Jurkat cells (lanes 4 to 7, 10, 12, and 14), whereas in the presence of Dox-induced TD-IκBα, the observed footprint pattern resembled that for unstimulated control Jurkat cells (lanes 2, 3, 8, 9, 11, and 13). Although less clearly resolved, the pattern of methylation and cleavage of the adjacent Sp1 site was also sensitive to Dox induction. For example, in TD-IκBα-inducible cells, PMA-PHA treatment resulted in protection of the Sp1 site with the exception of −42G (Fig. 4, lane 12), whereas Dox induction resulted in a pattern of protection at the Sp1 site that was characteristic of unstimulated control Jurkat cells (Fig. 4, lane 13; compare with lanes 2, 3, and 8). These results further indicate that binding of complexes to the Sp1 site may be coordinately regulated by the adjacent κB1 site in a TD-IκBα-inducible manner.
FIG. 4.
In vivo footprint of the IκBα gene promoter in rtTA-Neo and Tet-inducible TD-IκBα-expressing cells. Noncoding strand analysis was performed with rtTA-Neo (lanes 2 to 7) or rtTA-IκBα(2NΔ4) (lanes 8 to 14) Jurkat cells, either unstimulated (lanes 2, 3, 8, 9, and 14) or stimulated by TNF-α (10 ng/ml; lanes 4, 5, 9, and 10) or PMA-PHA (50 ng/ml and 1 μg/ml, respectively; lanes 6, 7, 12, and 13) for 40 min. Naked DNA was methylated in vitro (lanes 1). Where indicated (+), cells were pretreated with Dox (1 μg/ml, 24 h).
Prolonged NF-κB binding and temporal switch in the composition of NF-κB complexes at the κB1 site.
To determine the kinetics of the in vivo occupancy of κB1 and Sp1 sites of the IκBα promoter, control Neo (Fig. 5A) and TD-IκBα-expressing (Fig. 5B) cells were analyzed at different times after stimulation by TNF-α or PMA-PHA. Surprisingly, the same protection of the κB1 site was observed from 10 min to 24 h following TNF-α or PMA-PHA treatment in both cell types (Fig. 5A, lanes 4 to 15; Fig. 5B, lanes 4, 6, 8, 10, 12, and 14). In TD-IκBα-expressing cells pretreated with Dox, the pattern of methylation and cleavage of the κB1 site remained characteristic of unstimulated cells (Fig. 5B, lanes 5, 7, 9, 11, 13, and 15), regardless of the time of TNF-α or PMA-PHA stimulation. Prolonged protection of the adjacent Sp1 site was also observed from 10 min to 24 h in stimulated control or TD-IκBα-expressing cells (Fig. 5A, lanes 4, 5, 7, 10, 12, 14, and 15; Fig. 5B, lanes 4, 8, and 12). Again, Dox induction of TD-IκBα expression resulted in a methylation pattern at the Sp1 site that was characteristic of unstimulated cells (Fig. 5B, lanes 5, 7, 9, and 13).
FIG. 5.
Kinetics of protein-DNA interactions on the proximal IκBα promoter. For noncoding strand analysis of rtTA-Neo (A) and rtTA-IκBα(2NΔ4) (B) Jurkat T cells, naked DNA was methylated in vitro (lane 1). Cells were either unstimulated (lanes 2 and 3) or stimulated with TNF-α (10 ng/ml; lanes 4 to 9) or PMA-PHA (50 ng/ml and 1 μg/ml, respectively; lanes 10 to 15). The time of cell harvesting following TNF-α or PMA-PHA stimulation is indicated above the lanes. Where indicated (+), cells were pretreated with Dox (1 μg/ml, 24 h).
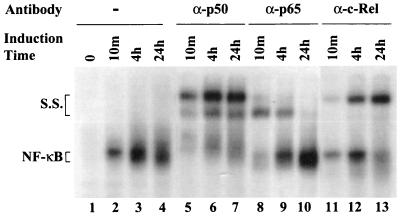
To identify the subunit composition of the NF-κB complexes during IκBα induction, EMSA supershift analysis of extracts from rtTA-Neo Jurkat cells treated with PMA-PHA for 10 min, 4 h, and 24 h was performed with the −66 to −51 κB1 probe and anti-p65, -p50, and -c-Rel antibodies. All PMA-PHA-induced complexes were shifted with anti-p50 antibodies regardless of the time of stimulation (Fig. 6, lanes 5 to 7). Interestingly, at early times after induction (10 min and 4 h), the NF-κB complex contained the p65 subunit (lanes 8 and 9), whereas at 24 h, the NF-κB complex was not shifted by anti-p65 antibodies (lane 10), indicating that p65 was no longer a component of the NF-κB complex. Furthermore, shift analysis demonstrated that at 4 and 24 h after induction, c-Rel was a component of the NF-κB complex; in fact, by 24 h after induction, the c-Rel–p50 heterodimer was the main component of the NF-κB complex (lanes 11 to 13). This temporal switch in NF-κB composition is likely to be responsible for the prolonged protection of the κB1 site observed by in vivo genomic footprinting.
FIG. 6.
Switch in the composition of NF-κB complexes bound to the κB1 site. Mobility shift and supershift analyses were performed with nuclear extracts from rtTA-Neo Jurkat cells treated with PMA-PHA for the times indicated (lanes 1 to 4), or analyzed by using anti-p50 (lanes 5 to 7), anti-p65 (lanes 8 to 10), and anti-c-Rel (lanes 11 to 13) antibodies with a [γ-32P]ATP labeled κB1 probe. The NF-κB and supershifted (S.S.) complexes are indicated.
p65 and Sp1 bind together to the κB1/Sp1 site of IκBα promoter.
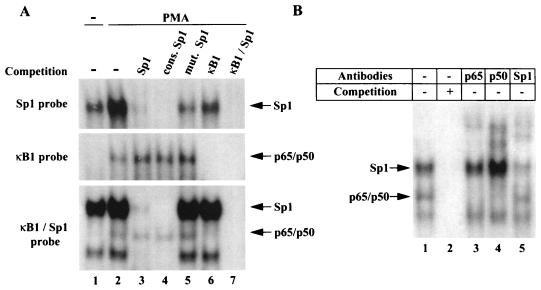
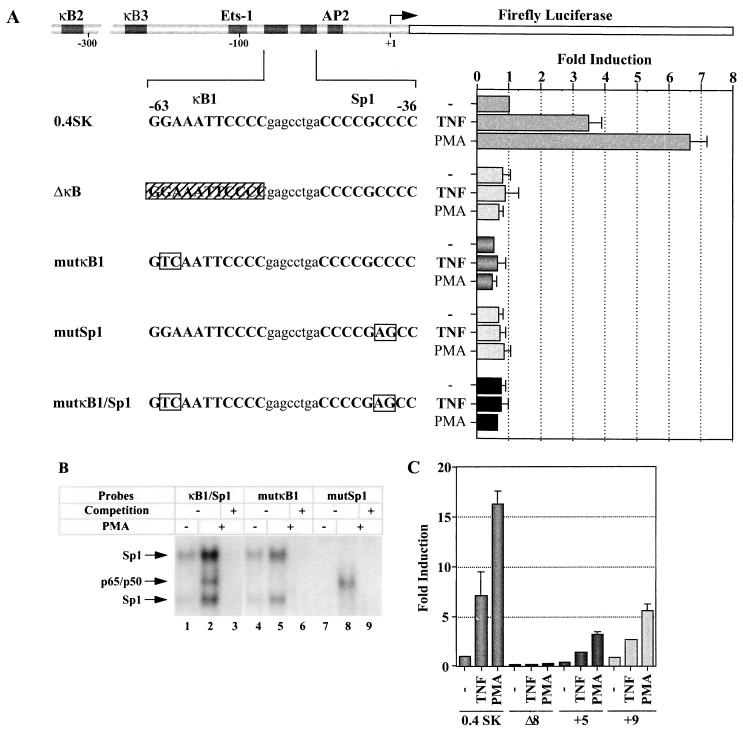
To assess Sp1 binding to the NF-κB/Sp1 region, EMSA analysis was performed with probes encompassing the −44 to −36 Sp1 site, the −63 to −36 κB1 site, or both κB1 and Sp1 sites (κB1/Sp1 probe from the −65 to −34 region of the IκBα promoter). PMA treatment of Jurkat cells resulted in a 10-fold increase in the intensity of the Sp1 binding complex (Fig. 7A, top panel, lanes 1 and 2); complex formation was blocked in competition reactions by both Sp1 and κB1/Sp1 binding sites (Fig. 7A, top panel, lanes 3 and 7) but not by the κB1 site alone (Fig. 7A, lane 6). This complex was further identified as an Sp1 binding activity by its competition in the presence of the consensus Sp1 sequence, whereas only partial inhibition was observed when a mutated Sp1 sequence was used as competitor (Fig. 7A, top panel, lanes 4 and 5). As a control, no inhibition of NF-κB binding to κB1 was detected in the presence of the Sp1 site of the IκBα promoter, consensus Sp1 or mutated Sp1 sequences (Fig. 7A, middle panel, lanes 2 to 5). Identification of the complexes detected by κB1/Sp1 probe was further determined by supershift analysis (Fig. 7B). As expected, anti-p50 and anti-p65 antibodies abolished p65/p50 binding to κB1/Sp1 probe, whereas anti-Sp1 antibodies reacted against the Sp1-containing complex (Fig. 7B, lanes 3 to 5). The faster-migrating band observed with the κB1/Sp1 probe (Fig. 7A, bottom panel, lanes 1, 2, 5, and 6) is likely to be a degradation product of Sp1-containing complex generated during nuclear protein extraction; this band is competed by Sp1-related oligonucleotides (Fig. 7A, lanes 3, 4, and 7) but is not affected by anti-Sp1 antibodies (Fig. 7B, lane 5). This EMSA analysis failed to reveal a complex formed by both endogenous Sp1 and NF-κB bound to the κB1/Sp1 probe.
FIG. 7.
NF-κB and Sp1 bind to the −63 to −36 region of the IκBα promoter. (A) EMSA analysis was performed with radiolabeled oligonucleotide probes specific to Sp1 (top panel), κB1 (middle panel), and κB1/Sp1 (bottom panel). Nuclear extracts prepared from Jurkat cells were either unstimulated (lane 1) or treated with PMA (50 ng/ml) for 2 h (lanes 2 to 7). Competition was performed in the presence of a 125-fold excess of unlabeled oligonucleotide: Sp1 site of IκBα promoter (lane 3), Sp1 consensus (cons. Sp1; lane 4), mutant Sp1 (mut. Sp1; lane 5), κB1 (lane 6), or κB1/Sp1 (lane 7). To facilitate detection of simultaneous binding of NF-κB and Sp1, EMSA buffer conditions were modified as described in Materials and Methods and the amount of extract used in the binding reactions was varied between 150 ng and 3 μg; for the binding reactions shown, 150 ng of Jurkat nuclear extract and 500 ng of poly(dI-dC) were used. (B) Complex composition was analyzed by supershift analysis. PMA-induced nuclear extracts were incubated with anti-p65 (lane 3), anti-p50 (lane 4), and anti-Sp1 (lane 5) antibodies.
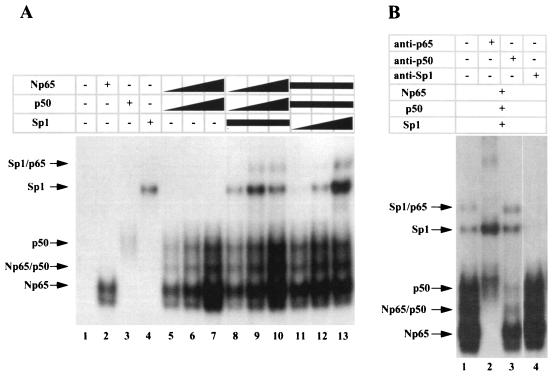
To determine whether Sp1 and p65-p50 bind to their recognition sites (κB1/Sp1) cooperatively in the IκBα promoter, an EMSA was performed with different amounts of recombinant p65-p50 or Sp1, using the κB1/Sp1 probe from the −65 to −34 region of the IκBα promoter. In the absence of Sp1, p65 and p50 bound to the κB1/Sp1 probe in a dose-dependent manner (Fig. 8A, lanes 2, 3, 5 to 7). In the presence of the same amount of Sp1, no cooperative binding was observed between Sp1 and p65-p50 with increasing amounts of p65 and p50 (Fig. 8A, lanes 8 to 10). However, at high concentration of p65 and p50, we observed an additional complex of slower mobility that also appeared with increasing concentrations of Sp1 when a fixed amount of p65 and p50 was used (Fig. 8A, 11 to 13). This complex was composed of p65 and Sp1 since incubation with specific antibodies eliminated complex formation (Fig. 8B). Similarly, anti-p50 antibody removed the p50-containing complexes (Fig. 8B, lane 3) whereas anti-p65 antibody shifted the complexes containing p65 (Fig. 8B, lane 2). Anti-Sp1 antibody also disrupted complex formation by removing either Sp1 binding alone or Sp1-p65 heterodimers (Fig. 8B, lane 4). Therefore, while Sp1 and p65-p50 do not bind cooperatively to the κB1/Sp1 site, both Sp1 and p65 bind together to the κB1/Sp1 sites of the IκBα promoter. The discrepancy between EMSA performed with Jurkat nuclear extracts and recombinant proteins regarding the formation of NF-κB–Sp1 complex could be due to limiting amount of one of the component in nuclear extract, thus excluding detection of this complex. This hypothesis is consistent with the weakness of the Sp1-p65 complex detected in the presence of high amount of recombinant proteins compared to the p50- and/or p65-containing complexes (Fig. 8A, lane 13).
FIG. 8.
NF-κB and Sp1 can co-occupy the κB1/Sp1 site of the IκBα promoter. (A) EMSA was performed with recombinant p65 (GST-Np65), p50 (GST-p50), and Sp1 proteins, using radiolabeled κB1/Sp1 site [γ-32P]ATP labeled. Each recombinant p65 (2 ng), p50 (2.73 ng), and Sp1 (3 ng) was used alone in lanes 2 to 4. Increasing amounts of p65 (0.5, 1, and 2 ng) combined with increasing amounts of p50 (0.65, 1.3, and 2.73 ng) were incubated in the absence (lanes 5 to 7) or presence (lanes 8 to 10) of Sp1 (3 ng). Increasing amounts of Sp1 (1, 3, and 5 ng) were also tested with the same amount of p65 (1 ng) and p50 (1.3 ng) in lanes 11 to 13. Shifted complexes are indicated by arrows. (B) Combinations of recombinant p65 (2 ng), p50 (2.73 ng), and Sp1 (3 ng) proteins were incubated with radiolabeled κB1/Sp1 oligonucleotides in the presence or absence of specific antibodies for p65, p50, and Sp1 proteins. The composition of the complexes was analyzed by supershift analysis using anti-p65, anti-p50, or anti-Sp1 antibodies (lanes 2 to 4).
IκBα gene expression is dependent on both NF-κB and Sp1 binding.
We next examined the functional role of the NF-κB and Sp1 sites in IκBα gene transcription in Jurkat cells by transient cotransfection with luciferase reporter constructs driven by the wild-type IκBα promoter (0.4SK) (35) or by mutated versions of the IκBα promoter (Fig. 9). Treatment of transfected Jurkat cells with TNF or PMA-PHA resulted in 4- and 7-fold stimulation of gene activity, respectively. Deletion or point mutation of the κB1 site of the IκBα promoter (ΔκB and mutκB1) abrogated TNF- and PMA-PHA-induced gene activation relative to the wild-type promoter (Fig. 9A). Strikingly, point mutation of the Sp1 site (mutSp1) also dramatically decreased induction of IκBα gene expression and also slightly decreased basal-level promoter activity. As expected, mutation of both κB1 and Sp1 sites also completely inhibited gene activity. As shown in Fig. 9B, EMSA analysis demonstrated that impairment of transactivation was due to lack of NF-κB binding (Fig. 9B, lanes 4 to 6) or Sp1 binding (Fig. 9B, lanes 7 to 9) to κB1/Sp1 sites. From these results, we conclude that both κB1 and Sp1 sites are required for full induction of IκBα promoter. To further analyze whether activation requires direct contact between NF-κB and Sp1, mutant luciferase reporter plasmids containing deletions or insertions between κB1 and Sp1 sites were tested (Fig. 9C). Interestingly, the introduction of 5 or 9 nt between the κB1 and Sp1 sites which alters the helical relationship between the two sites decreased but did not eliminate IκBα inducibility (Fig. 9C). When 8 nt (Δ8) between κB1 and Sp1 sites were deleted, transcriptional inducibility was completely abolished. Together, these results indicate that both κB1 and Sp1 sites are required for full induction of IκBα promoter.
FIG. 9.
Both κB and Sp1 are required for full TNF-α- or PMA-induced IκBα promoter activity. (A) Jurkat cells were transfected with 1 μg of luciferase reporter plasmid containing wild-type (0.4SK), mutant κB1 (ΔκB or mutκB1), mutant Sp1 (mutSp1), or mutant κB1/Sp1 (mutκB1/Sp1) IκBα promoter. Twenty four hours after transfection, cells were treated with TNF-α (10 ng/ml) or PMA (50 ng/ml) or left untreated for an additional 16 h. Transfection efficiency was normalized to that of Renilla luciferase (see Materials and Methods). The experiments were performed in triplicate, and the average fold induction was calculated. (B) EMSA was performed with different radiolabeled oligonucleotide probes (κB1/Sp1, mutated κB1/Sp1 [mutκB1], and κB1/mutated Sp1 [mutSp1]), using uninduced or PMA (50 ng/ml for 2 h)-induced Jurkat nuclear extract. (C) Luciferase assays were performed as described for panel A, in using 1 μg of reporter plasmids containing an 8-nt deletion (Δ8), 5-nt addition (+5), and 9-nt addition (+9) between κB1 and Sp1 sites of the IκBα promoter. Transfection efficiency was normalized to that of Renilla luciferase (see Materials and Methods). The experiments were performed in triplicate, and the average fold induction was calculated.
DISCUSSION
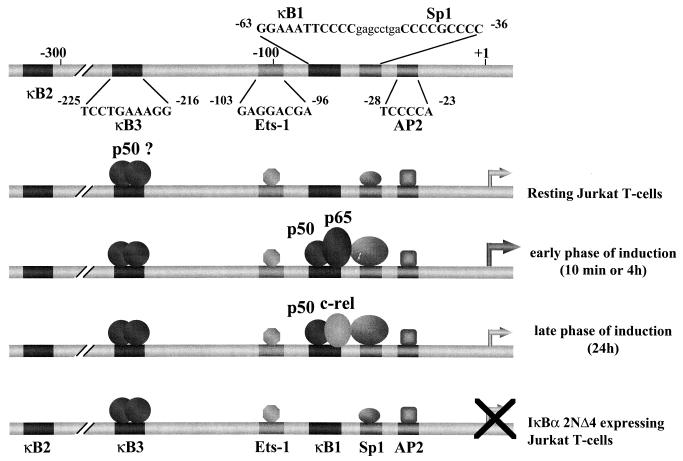
This report presents, for the first time, an in vivo genomic footprinting analysis of the IκBα promoter and characterizes the autoregulatory control of IκBα transcription by an NF-κB/Sp1 transcriptional switch. In previous studies, TD-IκBα expression was shown to inhibit endogenous IκBα at the protein level as well as to interfere with NF-κB binding and HIV-1 multiplication in Jurkat cells (31). We now demonstrate that induction of endogenous IκBα after TNF-α or PMA-PHA treatment is suppressed by TD-IκBα at the transcriptional level. Tet-induced TD-IκBα expression blocked NF-κB binding activity at the proximal −63 to −53 κB1 site of the IκBα promoter. In vivo genomic footprinting revealed multiple protein-DNA interactions in the region of the IκBα promoter between −250 to +100 bp in Jurkat T cells; protection of Sp1, AP2, Ets-1, and κB3 sites in unstimulated cells indicates that these sites participate in basal-level IκBα transcription. In response to stimulation of Jurkat T cells by PMA-PHA or TNF-α, changes in methylation of the κB1 site and the adjacent Sp1 site were observed, whereas no inducible changes were detected at κB3 or other sites (data not shown). The protection observed at κB1 and Sp1 sites was sustained from 10 min to 24 h and together with the EMSA results demonstrated that binding of p50-p65 heterodimer correlated with early transcriptional induction of the IκBα gene; at later times, the switch in composition of the NF-κB complexes to predominantly p50–c-Rel heterodimers correlated with transcriptional downregulation. Deletion and point mutagenesis demonstrated that both κB1 and Sp1 sites were absolutely required for IκBα promoter induction, whereas only Sp1 was involved in basal transcription of this promoter; a strict spacing requirement between κB1 and Sp1 sites was also essential for full activation of the IκBα promoter. Together, these studies suggest a model for IκBα transcriptional regulation in Jurkat T cells, as summarized in Fig. 10. Early activation of IκBα promoter activity is accompanied by p65-p50 binding to the κB1 site of the promoter, as well as by modulation of Sp1 binding at the adjacent Sp1 site. Downregulation of IκBα transcription, occurring at later times after induction, is associated with a switch in the composition of NF-κB complexes, from p50-p65 to p50–c-Rel heterodimers. This mechanism is in agreement with inhibition of p65-mediated transcription of HIV-1 LTR and interleukin-2 receptor alpha-chain promoters by c-Rel (20). Furthermore, c-Rel is induced with delayed kinetics compared to p65 (20) and may inhibit IκBα transcription by competition with p50 and p65 for occupancy of the κB1 binding site (Fig. 10).
FIG. 10.
Schematic representation of the protein-DNA interactions regulating the IκBα promoter. IκBα promoter organization, including κB1 to κB3 as well as Sp1, Ets-1, and AP2 binding sites, is shown at the top. In resting Jurkat T cells, protection is observed at the κB3, Ets-1, Sp1, and AP2 sites, which likely contribute to basal transcription. Early after induction by PMA-PHA or TNF-α, the κB1 site (−63 to −53) is occupied by p65-p50 heterodimers. At later times, a switch in complex composition to p50–c-Rel heterodimers correlates with downregulation of IκBα transcription. Protection is also observed at the adjacent Sp1 site (−44 to −36) and is also modulated by inducer-mediated stimulation or by activation of Tet-induced TD-IκBα expression which sequesters p65 in the cytoplasm. Binding to the Sp1 site may be related to the occupancy of the κB1 site by inducible NF-κB complexes. No changes were observed on κB3, Ets-1, and AP2 sites during cell activation.
The EMSA and genomic footprinting data are consistent with inhibition of IκBα promoter activity identified by deletion of the κB1 site (35); the κB2 and κB3 sites appear to play no role in the inducibility of the IκBα promoter at least in Jurkat T cells stimulated by PMA-PHA or TNF-α. Our results are also in agreement with the mutagenesis analysis performed by Ito et al. (29), showing a predominant role for the κB1 site. This analysis had also suggested that full activation of the IκBα promoter also required another κB-like site located downstream of κB1 between nt −34 and −24, as well as the upstream κB2 site. In the present study, no inducible in vivo protein-DNA interactions were observed at either of these sites. Although a role for κB2 and κB3 sites cannot be excluded, our in vivo data clearly demonstrate that κB1 and Sp1 sites play the major role in the inducibility of the IκBα promoter in Jurkat T cells. Furthermore, in vivo genomic footprinting experiments performed with the U937 promonocytic cell line revealed the same in vivo protection pattern as observed with Jurkat T cells: only the Sp1 site was protected before stimulation and both Sp1 and κB1 sites were targeted by inducible complexes after induction (data not shown). Thus, a common mechanism of IκBα regulation involving the NF-κB/Sp1 transcriptional switch is likely active in multiple cell types, including T cells and monocytes/macrophages.
Many genes regulated by NF-κB also contain adjacent Sp1 sites, and direct interaction between NF-κB proteins and Sp1 has been demonstrated (45); a recent study also identified in vitro binding of Sp1 to the κB sites located on promoters such as the interleukin-6 and P-selectin (28). We have demonstrated binding of p50–c-Rel and p50-p65 heterodimers to the κB1 site in response to cell induction, as well as binding of Sp1 to its own site in IκBα promoter. Furthermore, the Sp1 protection observed was reversed with TD-IκBα activation (Fig. 10). This coordinate change suggests that the binding of NF-κB inducible complexes to the κB1 site may also facilitate an increased Sp1 binding affinity to the adjacent Sp1 site. Interestingly, the residues at positions −48 and −49, located between the NF-κB and Sp1 sites, became more sensitive to methylation and cleavage, suggesting that the protein-DNA conformation of the entire NF-κB/Sp1 region is modified after stimulation (Fig. 3).
Sp1 binding to its consensus site prior to stimulation indicates that it may contribute to basal transcription of the IκBα gene. Interestingly, a different methylation and cleavage pattern was observed at the Sp1 site after stimulation. Direct Sp1 conformational changes and/or alterations in Sp1 binding affinity may be induced after stimulation via Sp1 posttranslational modification. Sp1 has been described as a zinc finger phosphoprotein which upon cell activation undergoes specific phosphorylations and dephosphorylations that regulate its DNA binding activity and its interactions with other proteins (4, 51, 54). Following cell stimulation, Sp1 can be phosphorylated by casein kinase II, by protein kinase A, and by a recently described 60-kDa kinase, activated in response to Neu differentiation factors (2). The inducible change at the Sp1 binding site in response to Jurkat T-cell stimulation may reflect such Sp1 modifications leading to increased binding on DNA.
The critical role of κB1 site and the adjacent Sp1 site in the inducibility of the IκBα promoter is further supported by the conservation of these two sites in the murine and porcine homologs of the IκBα promoter (17, 18). Moreover, not only are the exact sequences conserved, but also the 10-bp spacing between both sites is maintained between species. This distance, corresponding to one helical turn of DNA, may permit a physical interaction between proteins bound to the Sp1 site and the p65-p50 complex on the same face of chromatin in vivo, as shown for the HIV-1 LTR promoter (45). Although no cooperativity in the binding of NF-κB and Sp1 was observed by EMSA, transfection studies using hybrid IκBα promoters in which nucleotides between κB1 and Sp1 sites were inserted or deleted revealed a strict spacing requirement for maximal inducibility of IκBα promoter. Deletion of the 8 nt between both sites or insertion of 5 or 9 nt significantly lowered IκBα promoter inducibility. The fact that addition of an half helical turn or a complete helical turn led to a similar decrease in IκBα gene inducibility argues against a direct physical interaction between NF-κB and Sp1 and suggests instead a requirement for the interaction of NF-κB and/or Sp1 with basal transcription factors such as TATA binding protein (TBP)-associated factors or with the transcription machinery for maximal induction of IκBα promoter. Sp1 has been found to associate with CBP/p300 in a multiprotein complex (44), and the NF-κB p65 subunit is able to interact with the amino-terminal region of the coactivator p300, resulting in gene activation of E-selectin and VCAM-1 (25). Further studies are required to characterize the association of NF-κB and Sp1 with TBP-associated factors, TBP, or CBP/p300 and their role in IκBα regulation.
ACKNOWLEDGMENTS
M.A., H.J.K., and P.G. contributed equally to this work.
We thank Alain Israël and Ron Hay for reagents used in this study. We also thank members of the Molecular Oncology Group, Lady Davis Institute for helpful discussions.
This research was supported by grants from the Medical Research Council of Canada and CANFAR. M.A. and P.G. were supported by FRSQ postdoctoral fellowships, H.K. was supported by an FCAR studentship and J.H. was supported by an MRC Senior Scientist award.
REFERENCES
- 1.Algarté M, Lécine P, Costello R, Plet A, Olive D, Imbert J. In vivo regulation of interleukin-2 receptor alpha gene transcription by the coordinated binding of constitutive and inducible factors in human primary T cells. EMBO J. 1995;16:5060–5072. doi: 10.1002/j.1460-2075.1995.tb00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alroy I, Soussan L, Seger R, Yarden Y. Neu differentiation factor stimulates phosphorylation and activation of the Sp1 transcription factor. Mol Cell Biol. 1999;19:1961–1972. doi: 10.1128/mcb.19.3.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Fernandez B, Dominguez I, Jacqué J M, Thomas D, Diaz-Meco M T, Moscat J, Virelizier J L. Phosphatidylcholine hydrolysis activates NF-κB and increases human immunodeficiency virus replication in human monocytes and T lymphocytes. J Virol. 1993;67:6596–6604. doi: 10.1128/jvi.67.11.6596-6604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong S A, Barry D A, Leggett R W, Mueller C R. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J Biol Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 8.Barroga C F, Stevenson J K, Schwarz E M, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauparlant P, Lin R, Hiscott J. The role of the C-terminal domain of IκBα in protein degradation and stability. J Biol Chem. 1996;271:10690–10696. doi: 10.1074/jbc.271.18.10690. [DOI] [PubMed] [Google Scholar]
- 10.Beg A A, Baldwin S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 11.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequence of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 12.Bitar R, Beauparlant P, Lin R, Pitha P, Hiscott J. Retrovirus-mediated transfer of nuclear factor-κB subunit genes modulates IκBα and interferon β expression. Cell Growth Differ. 1995;6:965–976. [PubMed] [Google Scholar]
- 13.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown K, Franzoso G, Baldi L, Carlson L, Mills L, Lin Y-C, Gerstberger S, Siebenlist U. The signal response of IκBα is regulated by transferable N- and C-terminal domains. Mol Cell Biol. 1997;17:3021–3027. doi: 10.1128/mcb.17.6.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown K, Park S, Kanno T, Franzoso G, Siebenlist U. Mutual regulation of the transcriptional activator NF-κB and its inhibitor, IκBα. Proc Natl Acad Sci USA. 1993;90:2532–2536. doi: 10.1073/pnas.90.6.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 17.Chiao P J, Miyamoto S, Verma I M. Autoregulation of IκBα activity. Proc Natl Acad Sci USA. 1994;91:28–32. doi: 10.1073/pnas.91.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Martin R, Vanhove B, Cheng Q, Hofer E, Csizmadia V, Winkler H, Bach F H. Cytokine-inducible expression in endothelial cells of an IκBα-like gene is regulated by NF-κB. EMBO J. 1993;12:2773–2779. doi: 10.1002/j.1460-2075.1993.tb05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 20.Doerre S, Sista P, Sun S-C, Ballard D W, Greene W C. The c-rel protooncogene product represses NF-κB p65-mediated transcriptional activation of the long terminal repeat of type 1 human immunodeficiency virus. Proc Natl Acad Sci USA. 1993;90:1023–1027. doi: 10.1073/pnas.90.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst M K, Dunn L L, Rice N R. The PEST-like sequence of IκBα is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol Cell Biol. 1995;15:872–882. doi: 10.1128/mcb.15.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita T, Nolan G P, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 23.Garrity P A, Wold B. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci USA. 1992;89:1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerondakis S, Morrice N, Richardson I B, Wettenhall R, Fecondo J, Grumont R J. The activity of a 70 kilodalton IκB molecule identical to the carboxyl terminus of the p105 NF-κB precursor is modulated by protein kinase A. Cell Growth Differ. 1993;4:617–627. [PubMed] [Google Scholar]
- 25.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;7:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haskill S, Beg A A, Tompkins S M, Morris J S, Yurochko A D, Sampson-Johannes A, Mondal K, Ralph P, Baldwin A S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 27.Hatada E N, Nieters A, Wulczyn F G, Naumann M, Meyer R, Nucifora G, McKeithan T W, Scheidereit C. The ankyrin repeat domains of the NF-κB precursor p105 and the proto-oncogene bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc Natl Acad Sci USA. 1992;89:2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano F, Tanaka H, Hirano Y, Hiramoto M, Handa H, Makino I, Scheidereit C. Functional interference of Sp1 and NF-κB through the same DNA binding site. Mol Cell Biol. 1998;18:1266–1274. doi: 10.1128/mcb.18.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito C Y, Kazantsev A G, Baldwin A S., Jr Three NF-κB sites in the IκB-α promoter are required for induction of gene expression by TNFα. Nucleic Acids Res. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaffray E, Wood K M, Hay R T. Domain organization of IκBα and the sites of interaction with NF-κB p65. Mol Cell Biol. 1995;15:2166–2172. doi: 10.1128/mcb.15.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg M A, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J Biol Chem. 1998;273:7431–7440. doi: 10.1074/jbc.273.13.7431. [DOI] [PubMed] [Google Scholar]
- 32.Lacoste J, D’Addario M, Roulston A, Wainberg M A, Hiscott J. Cell-specific differences in activation of NF-κB regulatory elements of human immunodeficiency virus and beta interferon promoters by tumor necrosis factor. J Virol. 1990;64:4726–4734. doi: 10.1128/jvi.64.10.4726-4734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landt O, Grunert H P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 34.La Rosa F A, Pierce J W, Sonenshein G E. Differential regulation of the c-myc oncogene promoter by the NF-κB rel family of transcription factors. Mol Cell Biol. 1994;14:1039–1044. doi: 10.1128/mcb.14.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Bail O, Schmidt-Ullrich R, Israël A. Promoter analysis of the gene encoding the IκB-α/MAD 3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C-C, Korner M, Ferris D K, Chen E, Dai R-M, Longo D L. NF-κB/Rel family members are physically associated phosphoprotein. Biochem J. 1994;303:499–506. doi: 10.1042/bj3030499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin R, Gewert D, Hiscott J. Differential transcriptional activation in vitro by NF-κB/Rel proteins. J Biol Chem. 1995;270:3123–3131. doi: 10.1074/jbc.270.7.3123. [DOI] [PubMed] [Google Scholar]
- 39.Liou H-C, Nolan G P, Ghosh S, Fujita T, Baltimore D. The NF-κB p50 precursor, p105, contains an internal IκB-like inhibitor that preferentially inhibits p50. EMBO J. 1992;11:3003–3009. doi: 10.1002/j.1460-2075.1992.tb05370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercurio F, DiDonato J A, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-κB mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 42.Miyamoto S, Maki M, Schmitt M J, Hatanaka M, Verma I M. Tumor necrosis factor α-induced phosphorylation of IκBα is a signal for its degradation but not dissociation from NF-κB. Proc Natl Acad Sci USA. 1994;91:12740–12744. doi: 10.1073/pnas.91.26.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 44.Owen G I, Richer J K, Tung L, Takimoto G, Horwitz K B. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 45.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins N D, Schmid R M, Duckett C S, Leung K, Rice N R, Nabel G J. Distinct combinations of NF-κB subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petropoulos L, Lin R, Hiscott J. Human T cell leukemia virus type 1 Tax protein increases NF-κB dimer formation and antagonizes the inhibitory activity of the IκBα regulatory protein. Virology. 1996;225:52–64. doi: 10.1006/viro.1996.0574. [DOI] [PubMed] [Google Scholar]
- 48.Régnier C, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez M S, Michalopoulos I, Arenzana-Seisdedos F, Hay R T. Inducible degradation of IκBα in vitro and in vivo requires the acidic C-terminal domain of the protein. Mol Cell Biol. 1995;15:2413–2419. doi: 10.1128/mcb.15.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez M S, Wright J, Thompson J, Thomas D, Baleux F, Virelizier J L, Hay R T, Arenzana-Seisdedos F. Identification of lysine residues for signal-induced ubiquitination and degradation of IκBα in vivo. Oncogene. 1996;12:2425–2435. [PubMed] [Google Scholar]
- 51.Rohlff C, Ahmad S, Borellini F, Lei J, Glazer R I. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J Biol Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- 52.Roulston A, Lin R, Beauparlant P, Wainberg M A, Hiscott J. Regulation of human immunodeficiency virus type 1 and cytokine gene expression in myeloid cells by NF-κB/Rel transcription factors. Microbiol Rev. 1995;59:481–505. doi: 10.1128/mr.59.3.481-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanceau J, Kaisho T, Hirano T, Wietzerbin J. Triggering of the human interleukin-6 gene by interferon-gamma and tumor necrosis factor-alpha in monocytic cells involves cooperation between interferon regulatory factor-1, NF- kappa B, and Sp1 transcription factors. J Biol Chem. 1995;17:27920–27931. doi: 10.1074/jbc.270.46.27920. [DOI] [PubMed] [Google Scholar]
- 55.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott M L, Fujita T, Liou H-C, Nolan G P, Baltimore D. The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 57.Sun S-C, Ganchi P A, Ballard D W, Greene W C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 58.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 59.Traenckner E B, Phal H L, Henkel T, Schmidt N K, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verma I M, Stevenson J K, Schwarz E M, Antwerp D V, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 61.Whiteside S T, Epinat J, Rice N R, Israel A. I kappa B epsilon, a novel member of the IκB family, controls RelA and cRel NF-κB activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]