Abstract
Enterotoxigenic Escherichia coli (ETEC) is the primary aetiologic agent of traveller’s diarrhoea and a significant cause of diarrhoeal disease and death in developing countries. ETEC O169:H41 strains are known to cause both traveller’s diarrhoea and foodborne outbreaks in developed countries and are cause for concern. Here, whole-genome sequencing (WGS) was used to assemble 46 O169:H41 (ST182) E. coli draft genomes derived from two airplane waste samples sourced from a German international airport. The ST182 genomes were compared with all 84 publicly available, geographically diverse ST182 genomes to construct a core genome-based phylogenetic tree. ST182 isolates were all phylogroup E, the majority serotype O169:H41 (n = 121, 93%) and formed five major clades. The airplane waste isolates differed by an average of 15 core SNPs (range 0–45) but their accessory genome content was diverse. While uncommon in other ST182 genomes, all airplane-derived ST182 isolates carried: (i) extended-spectrum β-lactamase gene blaCTX–M–15 notably lacking the typical adjacent ISEcp1; (ii) qnrS1 and the S83L mutation in gyrA, both conferring resistance to fluoroquinolones; and (iii) a class 1 integron structure (IS26-intI1Δ648-dfrA17-aadA5-qacEΔ1-sul1-ORF-srpC-padR-IS6100-mphR-mrx-mphA-IS26) identified previously in major extraintestinal pathogenic E. coli STs but not in ETEC. ST182 isolates carried ETEC-specific virulence factors STp + CS6. Adhesin/invasin tia was identified in 89% of aircraft ST182 isolates (vs 23%) and was located on a putative genomic island within a hotspot region for various insertions including PAI I536 and plasmid-associated transposons. The most common plasmid replicons in this collection were IncFII (100%; F2:A-:B-) and IncB/O/K/Z (89%). Our data suggest that potentially through travel, E. coli ST182 are evolving a multidrug-resistant profile through the acquisition of class 1 integrons and different plasmids.
Keywords: Enterotoxigenic Escherichia coli (ETEC), ST182, O169:H41, airplane waste, antimicrobial resistance (AMR), blaCTX–M, traveller’s diarrhoea
Introduction
Throughout history, travel has played a key role in disseminating infectious diseases and with air travel now servicing over four billion passengers annually (ICAO, 2018), travellers in unprecedented numbers and speeds continue to shape the emergence and spread of disease to ever broader geographic locations (Institute of Medicine Forum on Microbial Threats, 2010). More recently, international travel has also been shown to contribute to the rise of antimicrobial resistance (AMR), particularly through the acquisition of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (Hassing et al., 2015; Woerther et al., 2017). Indeed, the rates of ESBL-producing Enterobacteriaceae acquisition during travel are alarming, with a recent study reporting up to 75 and 44% acquisition in travellers returning from Asia and Africa respectively (Arcilla et al., 2017). The same study found a strong correlation between ESBL-producing Enterobacteriaceae acquisition, diarrhoea during travel, and antimicrobial use, particularly fluoroquinolones. Similarly, another study found that 80% of travellers returning from southern Asia that used antimicrobials to treat diarrhoea whilst abroad had subsequently acquired ESBL-producing Enterobacteriaceae, as opposed to 47% of travellers that experienced diarrhoea without antimicrobial treatment, and 23% of travellers that reported neither diarrhoea nor antimicrobial use (Kantele et al., 2015). Another recent study involving real-time colonisation dynamics indicated that 70% of participant travellers returning from southeast Asia were colonised with ESBL-producing Enterobacteriaceae but that all had acquired ESBL-producing Enterobacteriaceae at one point in their travels (Kantele et al., 2021). Perhaps unsurprisingly, airplane sewage has been found to be an extraordinary source of antimicrobial resistance genes (ARGs), including ESBL genes, such as blaCTX–M (Heß et al., 2019).
Traveller’s diarrhoea is the most common health problem afflicting visitors to lower socioeconomic regions, affecting up to 40% of travellers (Leung et al., 2019). While most cases are mild and self-limiting, typically resolving within two days (Tribble, 2017), severe cases can present with cholera-like watery stools, result in the loss of 10 L of fluids daily, and can lead to hospitalisation and even death (Margulieux et al., 2018; Mirhoseini et al., 2018; Hosangadi et al., 2019). Antimicrobial treatment for traveller’s diarrhoea reduces both symptoms and duration of the illness (Diemert, 2006). Thus, fluoroquinolones, macrolides and bacterial RNA polymerase-binding rifaximin are antimicrobials commonly prescribed prior to departure for travellers to carry and use in the event of moderate to severe illness (Taylor et al., 2017; Schweitzer et al., 2019). While the provision of an empiric self-treatment option can be appealing to travellers hoping to avoid travel plan disruptions and hospitalisations in developing countries, the practice can lead to inappropriate usage in mild cases (Vilkman et al., 2019) and as previously noted, antimicrobial treatment of traveller’s diarrhoea increases the risk of acquiring ESBL-producing Enterobacteriaceae.
The leading aetiological agent of traveller’s diarrhoea is enterotoxigenic Escherichia coli (ETEC), accounting for approximately 60% of all cases (Mirhoseini et al., 2018). However, ETEC does not only afflict travellers, but is a leading cause of diarrhoeal disease in developing countries and causes significantly more deaths than cholera and typhoid fever (Lamberti et al., 2014; Hosangadi et al., 2019). In humans, ETEC are distinguished from other E. coli pathotypes by the production of heat-labile enterotoxins (LT; LTIh, LTIIa, LTIIb, LTIIc variants) and/or heat-stable enterotoxins (STa; STp, STh variants), and colonisation factors (CFs), which are also known as coli surface (CS) antigens (Wang et al., 2019). There are currently over 29 different CFs identified in ETEC strains with more believed yet to be characterised (Nada et al., 2011; Ban et al., 2015; Zeinalzadeh et al., 2017), however, CFA/I and CS1-CS6 are the most prevalent (Cho et al., 2014). Apart from some LT variants, which are carried on prophages, all CFs and other enterotoxins are carried on plasmids (F-:A-:B-) in various combinations (Regua-Mangia et al., 2004; Wang et al., 2019).
While ETEC infections are predominately associated with visitors to, and residents of, developing countries, some ETEC serotypes have been linked to food-borne outbreaks in developed countries. One such serotype is O169:H41 (sequence type [ST] 182), which was first described causing an outbreak in 1991 in Japan and 15 subsequent outbreaks in Japan between 1991 and 1994 (Nishikawa, 1995; Nishikawa et al., 1998). In the United States, of 16 documented ETEC outbreaks between 1996 and 2003, O169:H41 was identified in ten, and was solely responsible for six (Beatty et al., 2004). In 2006, another O169:H41 outbreak occurred in the United States affecting 36 people and was linked to contaminated coleslaw at a catered event (Devasia et al., 2006). A more recent O169:H41 outbreak in Japan affected 102 people during a 2 day festival in 2012 (Harada et al., 2013) and in the same year, this serotype caused an outbreak linked to contaminated kimchi provided at seven schools in Korea, affecting 1642 people (Cho et al., 2014).
Despite the strong association with gastroenteritis outbreaks, to date no genomic comparisons of O169:H41/ST182 strains, nor reports on potential AMR gene carriage have been published. Here we used whole genome sequencing (WGS) to characterise 46 ST182 isolates collected from the sewage of aircraft landing at a large German international airport in 2016. We performed phylogenetic and pangenomic comparative analyses with other publicly available ST182 genomes and screened for the presence of virulence-associated genes (VAGs), antimicrobial resistance genes (ARGs), chromosomal mutations conferring AMR, and plasmid replicons. Furthermore, AMR regions and an invasin-rich genomic island present in airplane-derived isolates were characterised.
Materials and Methods
Sample Collection and DNA Extraction
Airplane-borne sewage was sampled at two locations on a single day in 2016: one sample was gathered from a vacuum truck which collected waste from aircraft arriving from Mombasa, Canada, and Singapore. The second sample was taken from sewerage where the contents of aircraft tanks was piped to a sewage treatment plant. Thus, the second sample comprised waste from multiple airplanes landing at an international airport in Germany. The two samples were stored in 1 L sterile glass bottles at 4°C and processed within 24 h. To isolate E. coli, suitable dilutions were plated on mFC agar (Carl Roth, Karlsruhe, Germany). After 18 ± 2 h of incubation at 44°C, blue colonies were streaked on Brilliance agar (Oxoid, Wesel, Germany) and grown overnight at 37°C to obtain pure cultures. To identify the isolates as E. coli, colony PCR was performed as described previously to amplify a species-specific fragment of the yccT gene (Clifford et al., 2012; Heß et al., 2018). The isolates were subsequently grown overnight in LB-broth (Carl Roth, Karlsruhe, Germany) and the DNA was isolated using the DNeasy 96 Blood and Tissue Kit (Qiagen, Hilden, Germany) following manufacturer instructions.
Phenotypic Resistance Testing
The susceptibility of the E. coli isolates was tested against 24 antibiotics commonly used to treat E. coli infections (namely, ampicillin, amoxicillin-clavulanic acid, piperacillin, ticarcillin, cefepime, cefuroxime, cefoxitin, cefpodoxime, ceftazidime, cefuroxime, doripenem, ertapenem, imipenem, meropenem, ciprofloxacin, levofloxacin, norfloxacin, amikacin, gentamicin, netilmicin, tobramycin, tigecycline, chloramphenicol, sulfamethoxazole-trimethoprim). The agar diffusion tests followed the EUCAST v6.0 guidelines (eucast.org/) using E. coli ATCC 25922 as a quality control. Applying the clinical breakpoints defined by EUCAST, the isolates were classified as resistant, intermediate, or susceptible.
Whole-Genome Sequencing
Library preparation was done by the iThree Core Sequencing facility, University of Technology Sydney, following the adapted Nextera Flex library preparation kit process, Hackflex (Gaio et al., 2019). Briefly, genomic DNA was quantitatively assessed using Quant-iT picogreen dsDNA assay kit (Invitrogen, United States). The sample was normalised to the concentration of 1 ng/μl. 10 ng of DNA was used for library preparation. After tagmentation, the tagmented DNA was amplified using the facility’s custom designed i7 and i5 barcodes, with 12 cycles of PCR.
Due to the number of samples, the quality control for the samples was done by sequencing a pool of samples using MiSeq V2 nano kit – 300 cycles. Briefly, after library amplification, 3 μl of each library was pooled into a library pool. The pool was then cleaned up using SPRIselect beads (Beckman Coulter, United States). The pool was sequenced using MiSeq V2 nano kit (Illumina, United States). Based on the sequencing data generated, the read count for each sample was used to identify any failed libraries (<100 reads) and normalised sample amounts to ensure equal representation in the final pool. The final pool was sequenced on one lane of Illumina Novaseq S4 flow cell, 2 × 150 bp at the Ramaciotti Centre for Genomics (University of New South Wales, Australia).
The resulting E. coli draft genomes were assembled using Shovill v1.0.4 (github.com/tseemann/shovill) with default settings and trimming option and underwent quality control (QC) using assembly-stats (github.com/sanger-pathogens/assembly-stats).
Genome Annotation
Automated annotations were generated using Prokka v1.14.6 (Seemann, 2014) and managed via SnapGene v4.1.9 (snapgene.com). Genomic islands (GIs) were identified using IslandViewer 4 (Bertelli et al., 2017) using E. coli ST182 strain 2014EL-1345-2 as the reference genome, and phage elements were identified using the Phage Search Tool (PHAST) (Zhou et al., 2011).
Phylogenetic Analysis
Additional publicly deposited E. coli ST182 genomes were sourced from the Enterobase database (Zhou et al., 2019) (n = 87; extracted 02/05/20) and Sequence Read Archive (SRA) FASTQ files were downloaded using parallel-fastq-dump (github.com/rvalieris/parallel-fastq-dump). Reads were then assembled using Shovill. E. coli ST182 derived from Enterobase are referred to throughout the manuscript as “strains,” while from this collection they are referred to as “isolates” of a novel ST182 lineage.
The E. coli ST182 pangenome was calculated using Panaroo (Tonkin-Hill et al., 2020) in strict mode after a pre-processing QC step using a Panaroo packaged Mash wrapper script. Any genome outliers with comparatively unusual number of contigs or genes (i.e., number of genes >5100 or <4400; and number of contigs >300) were removed from all downstream analyses. A core gene alignment (4,123,297 bp in length) was also generated using Panaroo (default settings) and used to build a maximum-likelihood phylogenetic tree of all E. coli ST182 sequences via IQ-TREE 2 (Minh et al., 2020) in which ModelFinderPlus (-m MFP) was used to determine the best-fit model and single branch test performed using ultrafast bootstrap (1000 iterations). The pangenome was visualised using Phandango v1.3.0 (Hadfield et al., 2018) and the gene presence/absence matrix generated by Panaroo was used in Scoary (Brynildsrud et al., 2016) for pangenome-wide association studies.
Single nucleotide polymorphism (SNP)-based phylogenetic analyses were performed using Parnsp v1.1.2 (Treangen et al., 2014) with recombination filtering (-x). E. coli ST182 SS_2_A12 (GCA_015200705.1) was used as the reference to produce an alignment (85% of total length; 4,325,083 bp) to airplane-only isolates and the completed genome of ST182 strain F6326-C1 (GCA_002741495.1) was the reference used to produce an alignment to all ST182 sequences. Snp-dists v0.7.0 was then used on both alignments to calculate pairwise SNP distances. All trees were visualised using the Interactive Tree of Life (iTOL v4) (Letunic and Bork, 2019).
Genotyping, Serotyping and Phylogenetic Classification
STs, serogroups and phylogroups for all genomes analysed in this study were determined in silico using Achtman 7 multilocus sequencing typing (MLST) v2.0 (Larsen et al., 2012), SerotypeFinder v2.0 (Joensen et al., 2015), and Clermont Typing (Clermont et al., 2013), respectively. Typing of fimH alleles was performed using FimTyper v1.0 (Roer et al., 2017). ABRicate (github.com/tseemann/abricate) was used to screen for VAGs, ARGs, chromosomal point mutations conferring AMR, and plasmid replicons using the following reference databases: Virulence Factor DataBase (VFDB) (Liu et al., 2019), ResFinder v4.1 (Zankari et al., 2012), the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2020), PointFinder v4.1 (Zankari et al., 2017) and PlasmidFinder v2.1 (Carattoli and Hasman, 2020). An additional custom database with AMR-associated insertion sequences (IS), class 1, 2 and 3 integrases, and additional extraintestinal pathogenic E. coli (ExPEC)-associated VAGs was also utilised and can be accessed at github.com/CJREID/custom_DBs. ABRicate was executed using default parameters (minimal coverage ≥ 80%; minimal identity ≥ 80%), except when screening for truncated intI1 genes where minimal coverage was lowered to > 60%.
The tia-containing region was screened for in E. coli ST182 genomes using the nucleic sequence of this region derived from isolate SS_1_H2 contig 15 (start: 72, 824, end: 133, 647). This sequence was also used to search the NCBI database using BLASTn. A comparison of identified tia-containing regions was made using EasyFig (Sullivan et al., 2011). BLASTn was also used to determine whether AMR-regions characterised in this collection were present in other genomes deposited into NCBI. Associated metadata for 100% identity hits was pulled from Genbank (Leray et al., 2019) and PLSDB v2020_03_04 (Galata et al., 2019).
To infer E. coli ST182 strain 2014EL-1345-2 plasmid carriage in this collection, short-reads from each isolate were mapped to plasmid unnamed 1 (NZ_CP024224.1), unnamed 2 (NZ_CP024225.1), unnamed 3 (NZ_CP024226.1) and unnamed 4 (NZ_CP024227.1) using Burrows-Wheeler (BWA) v0.7.17 (Li and Durbin, 2009) and converted to BAM files using SAMtools v01.1.18 (Li et al., 2009). A bespoke script, available at github.com/maxlcummins/APEC-MGEN-2018, was used to produce a histogram of read-depth as a function of reference coordinate and clustered based on their Euclidean distances, and then used to generate a heatmap.
Statistical Analysis
A pairwise genome distance matrix was generated using Mash (Ondov et al., 2016) and used to create a classical (metric) multidimensional scaling (MDS) plot using R Studio v4.0.2 and the gglot2 v3.3.0 package (ggplot2.tidyverse.org/). MDS plots for VAGs and ARGs were also created in R Studio, using standard R package functions dist and cmdscale in conjunction with gene presence/absence matrices (1 = present; 0 = absent).
Results
Genome Selection, Assembly, and QC
A total of 71 isolates (13 ST lineages) from the two airplane waste samples were sequenced. ST182 made up 65% (n = 46) of all the isolates and are the subject of this study. The 46 ST182 genomes ranged in size from 4,988,334 bp to 5,327,796 bp, with a mean size of 5,208,154 bp. The number of contigs per genome ranged from 145 to 254, with a mean of 183. Read depth ranged from 58× to 124×, with a mean of 85×. Full assembly statistics can be viewed in Supplementary Data 1.
Escherichia coli ST182 Phylogeny
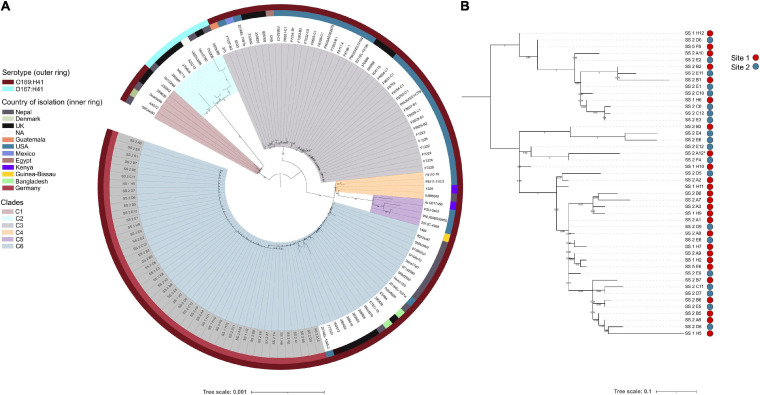
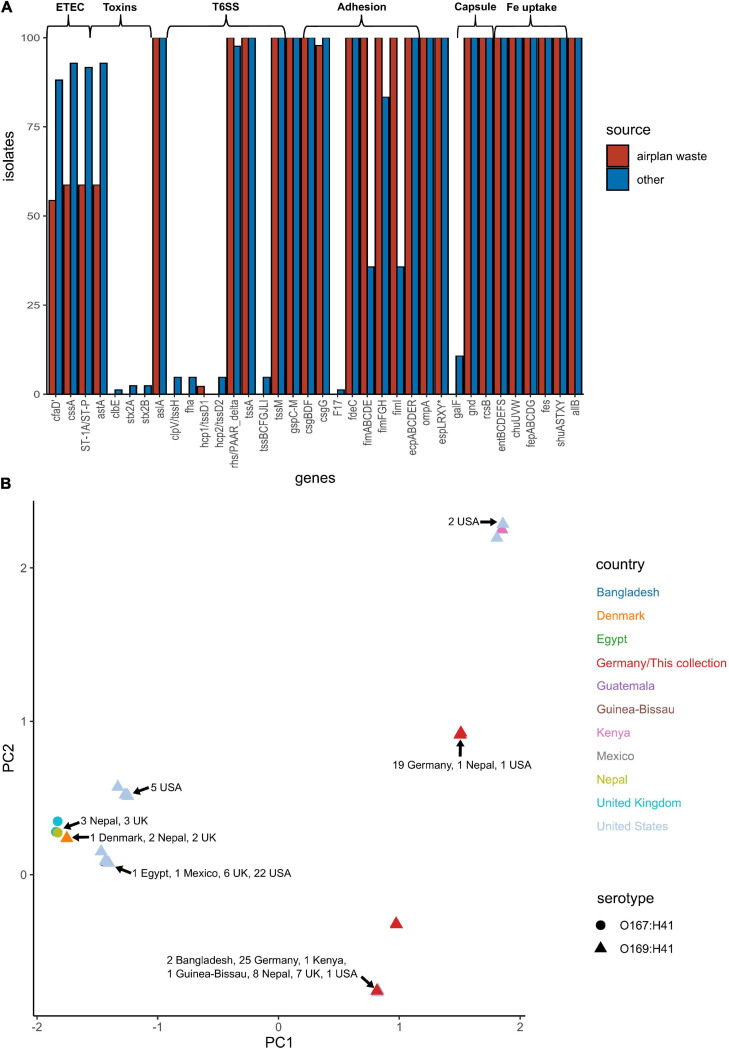
To ascertain the genetic relatedness of the airplane waste isolates to other E. coli ST182 strains all publicly available ST182 genomes (n = 84; metadata available in Supplementary Data 2) were used to construct a core genome-based phylogenetic tree using a core gene alignment 4,123,297 bp in length (Figure 1A). All airplane waste isolates were serotype O169:H41, as were the majority of ST182 strains (n = 121, 93%). These formed five major clades (Figure 1A) – however, a branch comprising of nine O167:H41 strains, formed its own clade (clade 2/C2). Airplane waste isolates were all situated in clade 6, with the closest relative outside the collection was 2014EL-1345-2 a human-sourced ETEC from the United States collected in 2014. All E. coli ST182 were typed as phylogroup E. Regarding fimH alleles, 58% (n = 75) carried fimH30, 31% (n = 40) carried fimH54, and 12% (n = 15) had no identified fimH gene. All airplane waste ST182 isolates carried fimH30. From available metadata, E. coli ST182 were most frequently isolated in Germany (n = 46, 35%; this collection), the United States (n = 38, 29%), the United Kingdom (n = 20, 15%) and Nepal (n = 16, 12%), and most originated from human samples (69%), however, one strain came from lettuce (strain PSU-0403; United States) and one from poultry (strain ALQ017456; Kenya), both of which reside in clade 5.
FIGURE 1.
E. coli ST182 phylogeny. (A) Mid-point root maximum-likelihood tree by core genome alignment showing genetic relatedness between 130 ST182 genomes. Isolates from this collection have tip label shaded in gray. Tree generated using IQTree2. (B) Mid-point rooted SNP-based phylogeny tree showing topology of the 46 airplane waste isolates only. Site 1: vacuum truck with waste from aircraft arriving from Mombasa, Canada, and Singapore. Site 2: Sewage from mixed aircraft tanks. Tree generated using Parsnp. Both trees visualised using iTOL.
To view airplane waste isolate topology, a SNP-based phylogeny was also constructed (Figure 1B) using 85% (4325083 bp) of the reference genome SS_2_A12 to identify SNP sites. Despite being sampled from two different sites, these isolates differed by an average of 15 core SNPs (range 0–45) (Pairwise SNP distance matrix in Supplementary Data 3). A SNP analysis across all ST182 genomes was performed using 84% (4332006 bp) of the oldest completed genome as reference (strain F6326-C1; collected 1998; clade 3). With the exception of clade 5 isolates, SNP distances within clades were generally low, ranging from on average 51 SNPs for clade 6 to 111 SNPs for clade 2 (Table 1), with the exception of clade 5 isolates which differed by an average of 302 SNPs. SNP distances between clades were much higher, the greatest distance being between clade 2 and clade 5 isolates at an average of 6300 SNPs (Table 1).
TABLE 1.
Average SNP distances across E. coli ST182 clades (Range in brackets).
| Clade 1 | Clade 2 | Clade 3 | Clade 4 | Clade 5 | Clade 6 | |
| Clade 1 | 65 (20–95) | |||||
| Clade 2 | 3410 (3325–3469) | 111 (7–199) | ||||
| Clade 3 | 2088 (2028–2200) | 5417 (5331–5535) | 86 (0–235) | |||
| Clade 4 | 2486 (2418–2540) | 5815 (5721–5875) | 1815 (1747–1926) | 77 (8–108) | ||
| Clade 5 | 2980 (2922–3019) | 6300 (6215–6344) | 2309 (2247–2405) | 597 (534–637) | 302 (1–462) | |
| Clade 6 | 2562 (2459–2618) | 5893 (5762–5948) | 1895 (1788–2004) | 169 (59–223) | 680 (575–717) | 51 (0–202) |
Escherichia coli ST182 Pangenome
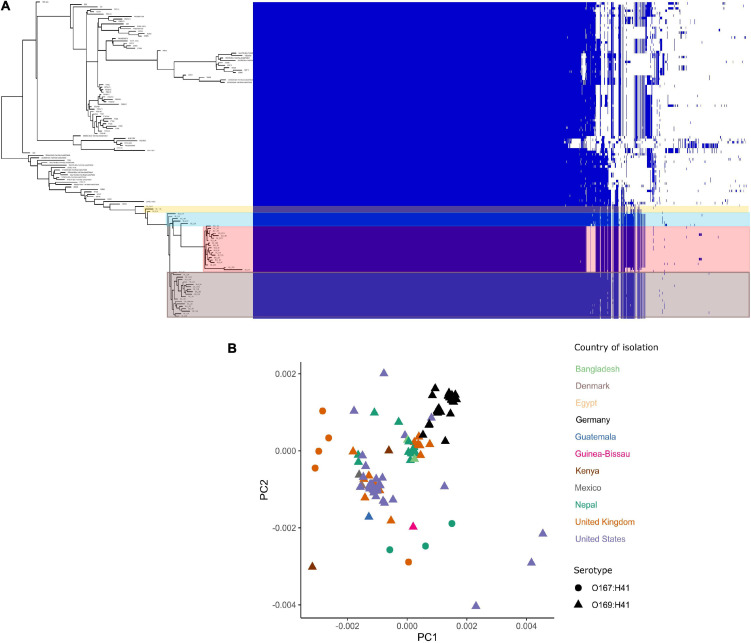
The pangenome for the 130 E. coli ST182 genomes used in the phylogenetic analysis consists of 6339 genes made up of a core genome of 4053 genes (64%) and an accessory genome of 2286 genes. The accessory genome was further broken down into a soft-core genome of 162 genes (present in 95–99% of genomes), a shell genome of 942 genes (15–94%) and a cloud genome of 1182 (<15%). A genome-wide association study identified 1057 genes associated with the airplane waste ST182 lineage, including both genes positively and negatively correlated (Supplementary Data 4). Despite having conserved core genomes, the airplane waste isolates formed 4 clusters of their own based on accessory genome content (Figure 2A). A pairwise genome distance MDS plot (Figure 2B; airplane waste isolates in black) shows that most of the airplane waste isolates formed a clonal group, however, several airplane waste ST182 isolates were in closer proximity to isolates originating from United Kingdom and one isolate from the United States (2014EL-1345-2). Notably, two other clonal groups were evident consisting of isolates originating from diverse geographical regions, including Bangladesh, Nepal and the United Kingdom in one group, and Egypt, United Kingdom and United States in the other.
FIGURE 2.
E. coli ST182 pangenome. (A) Gene presence/absence matrix clustered by accessory genome visualised using Phandango. Airplane waste isolate clusters highlighted. (B) MDS illustrating pairwise genome distances calculated using Mash. Isolates colored by country of isolation. All originating from Germany (black) pertain to this collection. Shapes distinguish serotypes.
Escherichia coli ST182 Antimicrobial Resistance Profile
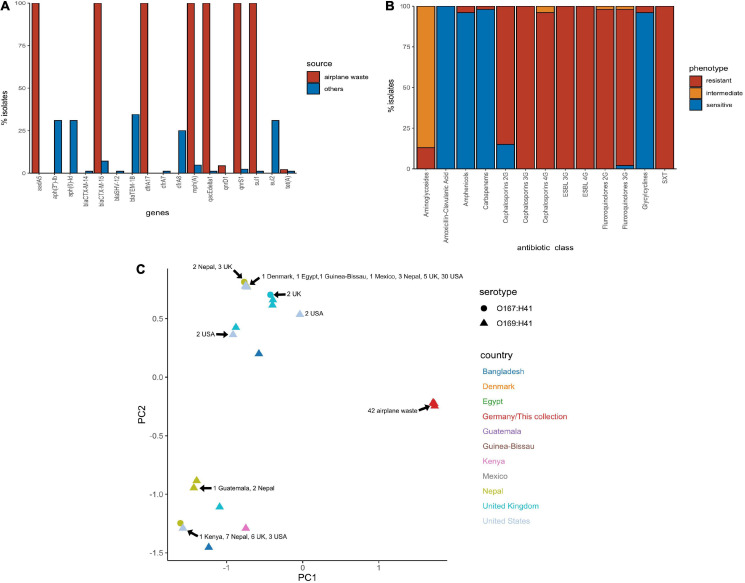
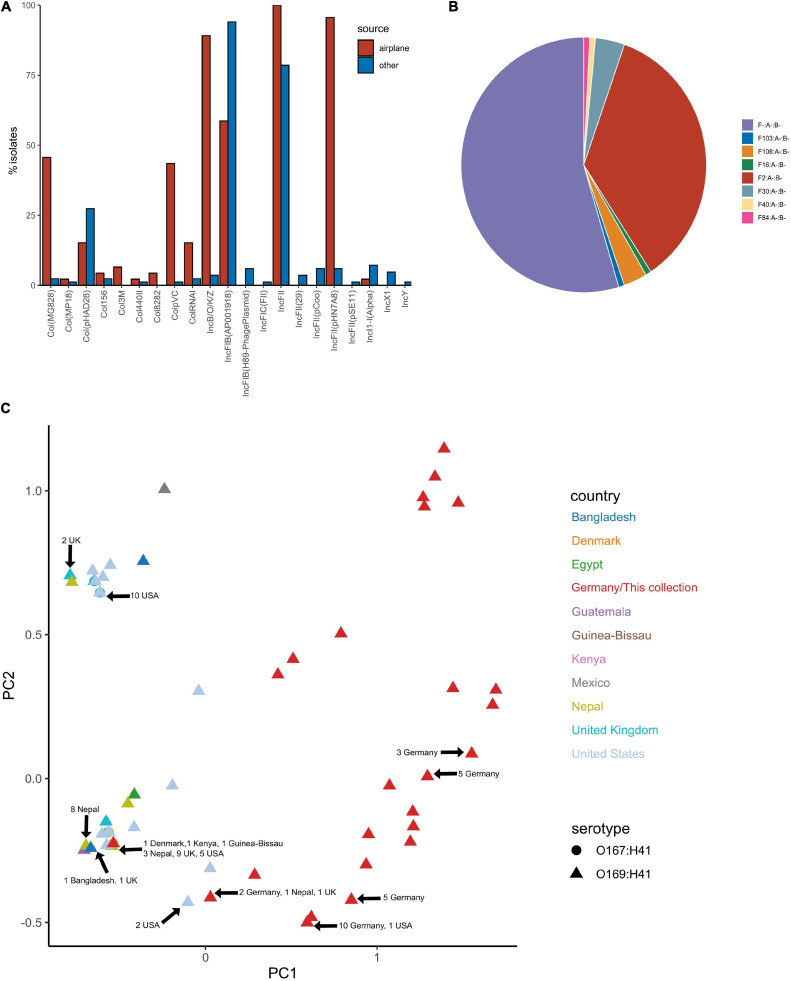
All 130 E. coli ST182 genomes were screened for ARGs (Supplementary Data 5). The presence of ARGs typically associated with horizontal gene transfer (HGT) are presented in Figure 3A. Of these HGT-associated ARGs, the airplane waste derived ST182 isolates carried an average of eight while other ST182 strains carried an average of three, and all airplane-derived isolates carried ARGs conferring resistance to ESBLs (blaCTX–M–15), streptomycin (aadA5), macrolides (mphA), fluoroquinolone (qnrS1), sulphonamide (sul1) and trimethoprim (dfrA17) (Figure 3A). The genotypic data was congruent with phenotypic data, and according to the criteria defined by Magiorakos et al. (2012), all E. coli ST182 airplane waste isolates were classified as multidrug resistant (Figure 3B and Supplementary Data 6). Conversely, these six specific genes were far less common in other E. coli ST182 strains, ranging from 0% for aadA5 and dfrA17 to 6% (n = 5) for both blaCTX–M–15 and mphA. Only one other intact gene conferring resistance to ESBLs was identified in other ST182 strains (i.e., blaCTX–M–14) identified in one strain (069d2850; Nepal), and only one carbapenem resistance gene (i.e., blaKPC–3) also in one strain (2016EL-1001a; United States). The most common ARGs in other ST182 strains were blaTEM–1B (β-lactam, 34% vs 0% in airplane waste isolates), aph(3”)-Ib (aminoglycoside, 33 vs 0%) and sul2 (sulphonamide, 30 vs 4%) (Figure 3B). An MDS analysis of all ARGs identified using the comprehensive antimicrobial resistance database (CARD), which includes transmissible as well as intrinsic ARGs (such as efflux pumps), demonstrated that overall airplane waste isolates had the most distinct and divergent ARG profiles (Figure 3C; red triangles).
FIGURE 3.
Antimicrobial resistance (AMR) profile of E. coli ST182 strains. (A) Bar graph comparing distribution of the most common ARGs identified in both ST182 airplane waste isolates (red) and other ST182 strains (blue). (B) Phenotypic AMR profile of ST182 airplane waste isolates. Red = resistant isolates (%), orange = intermediate (%), blue = sensitive (%). (C) MDS analysis of ARGs detected in ST182 isolates. Where identical profiles cause shapes to overlap arrows have been added detailing the country of isolation for each clustered isolate.
Chromosomal point mutations conferring AMR were also screened for in airplane-derived isolates and all were observed having the S83L mutation (TCG→TTG) in DNA gyrase subunit A gene gyrA conveying resistance to nalidixic acid and ciprofloxacin (fluoroquinolone). The same mutation was also found in 25 (30%) other ST182 isolates. No other AMR-associated mutations were identified (Supplementary Data 5). Interestingly, while airplane waste ST182 isolates were resistant to ciprofloxacin, most were less resistant to norfloxacin, including seven isolates that were sensitive (Supplementary Data 6).
Class 1 integrase (intI1), as well as transposons and insertion sequences (ISs) associated with mobilising AMR regions were also screened for Supplementary Data 5. Notably, all airplane-derived isolates had at least partial IS26 and/or the 3 SNP variant IS15DI (32 intact [70%], 13 incomplete [30%]), while less than half (45%) of other ST182 strains possessed IS26, and none had IS15DI. Similarly, while all airplane-derived isolates carried at least a partial intI1 gene, this gene was only present in three other ST182 strains (3%, 2 intact, 1 partial).
Escherichia coli ST182 AMR Regions
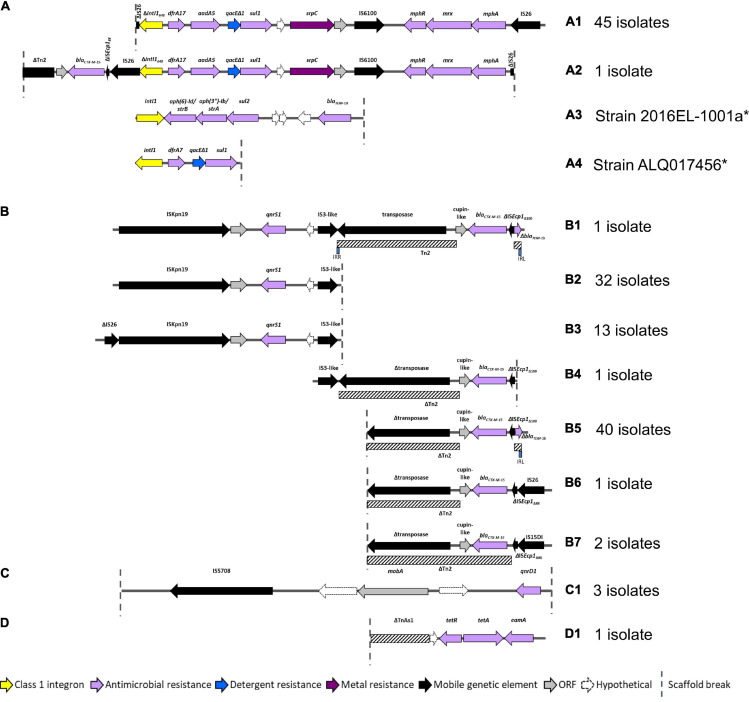
As AMR regions are often abundant in repetitive sequences, assembling complete structures using short-read sequencing data can be challenging. Despite this limitation we resolved four main structures with several variants (Figure 4). BLASTn and NCBI were used to determine whether these AMR regions had been previously deposited into public databases (Table 2).
FIGURE 4.
AMR regions in ST182 isolates. (A) Structures containing class 1 integrons. *Two structures not from this collection but present in two other ST182 strains analysed in this study. (B) Structures containing blaCTX–M–15 and/or qnrS1. (C) Structure carrying qnrD1. (D) Structure containing tetA.
TABLE 2.
Summary of BLAST hits (100%) to AMR regions found in this collection.
| IS26 flanked class 1 integron, chromate resistance and macrolide inactivation structure (Figure 4A1) | ||||||||
| Species | Strain | ST | Plasmid name | Accession | pMLST | Host | Location | Date |
| E. coli | 118UI | 69 | pEco118UIb | NZ_CP032516.1 | IncF NT | Human | Mexico | 2009 |
| E. coli | CA08 | 131 | pCA08 | CP009233.1 | F2:A-:B20 | Human | United States | NA |
| E. coli | Ecol_867 | 131 | pEC_867_1 | CP018982.1 | F-:A-:B20 | Human | Canada | 2013 |
| E. coli | AR_0104 | 131 | unitig_5 | NZ_CP020117.1 | F1:A-:B- | NA | NA | NA |
| E. coli | DA33135 | 131 | pDA33135-139 | NZ_CP029577.1 | F29:A-:B10 | Human | Sweden | NA |
| E. coli | E41-1 | 131 | p1 | NZ_CP028484.1 | F1:A-:B20 | Human | China | 2017 |
| E. coli | TO60 | 131 | p2 | NZ_LS992169.1 | F1:A-:B20 | NA | NA | NA |
| E. coli | U15A | 131 | pU15A_A | NZ_CP035721.1 | IncI1-Iγ | Human | United States | 2015 |
| E. coli | MO | 131 | pMO | MG886288.1 | F1:A-:B20 | Water | NA | NA |
| E. coli | AR_0086 | 131 | pUnnamed1 | NZ_CP032202.1 | F2:A-:B20 | NA | NA | NA |
| E. coli | 131 | 131 | p146-1 | NZ_CP041573.1 | IncF NT | Human | United States | NA |
| E. coli | E303 | 131 | pE303_IMP6 | AP022363.1 | IncF NT | Human | Japan | 2015 |
| E. coli | 425 | 131 | p425 | MK295830.1 | F1:A6:B20 | Human | Israel | 2007 |
| E. coli | HOS15 | 131 | - | SRR11495735 | - | Human | Australia | 2006 |
| E. coli | HOS58 | 131 | - | SRR11495760 | - | Human | Australia | 2007 |
| E. coli | A1_181 | 410 | p_unnamed3 | NZ_CP040070.1 | F1:A-:B- | Seagull | United States | 2016 |
| E. coli | ECONIH1 | 648 | pECO-824 | NZ_CP009860.1 | F1:A-:B1 | Human | United States | 2013 |
| E. coli | MS14385 | 648 | p2 | NZ_LR130556.1 | F1:A-:B- | Human | Australia | NA |
| K. michiganensis | K518 | NA | Chromosome | NZ_CP023185.1 | Chromosome | Human | China | 2017 |
| K. pneumoniae | F93-2 | 17 | pF93-2_1 | NZ_CP026158.1 | F2:A-:B- | Human | China | 2014 |
| K. pneumoniae | FDAARGOS_439 | 147 | pUnnamed | NZ_CP023918.1 | F2:A22:B- | Human | Canada | 2014 |
| blaCTX–M–15 and qnrS1-containing structure (Figure 4B1) | ||||||||
| Species | Strain | ST | Plasmid name | Accession | pMLST | Host | Location | Date |
| E. coli | MRSN346647 | 38 | pMRSN346647_113.1 | NZ_CP018207.1 | F-:A-:B53 | Human | United States | 2016 |
| E. coli | 2014EL-1345-2 | 182 | pUnnamed 3 | CP024226.1 | IncB/O/K/Z | Human | United States | 2014 |
| tetA-containing structure (Figure 4D1) | ||||||||
| Species | Strain | ST | Plasmid name | Accession | pMLST | Host | Location | Date |
| E. coli | ECCWS199 | 48 | pTB222 | NZ_CP032239.1 | IncN | Chicken | China | 2017 |
| K. pneumoniae | TH164 | 873 | pTH164-1 | NZ_CP035212.1 | IncR | Human | Thailand | 2012 |
| S. enterica | SCSM4.1 | 34 | plas4.1.1 | NZ_CP047116.1 | IncHI2 | Chicken | China | 2019 |
| S. enterica | sg_wt7 | 36 | Chromosome | NZ_CP036168.1 | Chromosome | Food | Singapore | 2016 |
All 46 isolates contained an IS26 flanked AMR region comprised of a class 1 integron cassette, chromate resistance gene srpC (chrA homologue), and macrolide inactivation gene cluster mphA-mrx-mphR (Figures 4A1,A2). The class 1 integron structure consisted of an IS26-mediated intI1 truncation (ΔintI1648), dfrA17, aadA5, qacEΔ1 (quaternary ammonium compounds resistance), and sul1. The class 1 integron structure present in airplane isolates differed from those identified in other E. coli ST182 strains (n = 2, one strain each) (Figures 4A3,A4). One isolate (SS_2_D8) also carried blaCTX–M–15 upstream to the class 1 integron (Figure 4A2). While the ARG configuration described in Figure 4A2 appears novel (no 100% BLASTn hits), the Figure 4A1 structure found in all airplane isolates also features in three Klebsiella spp. strains, and 18 E. coli strains (Table 1). Of the E. coli strains, all were from pandemic ExPEC lineages (ST69, ST131, ST410, ST648) isolated mainly from human hosts from various geographical locations. The structure was most frequently located on IncF plasmids of varying pMLSTs, but also chromosomally in one Klebsiella michiganensis strain (Table 2).
Unlike SS_2_D8, most isolates (n = 44) carried blaCTX–M–15 on contigs without a downstream integron, and a scaffold break truncating the Tn2 transposon harbouring this gene (Figures 4B4–7). The Tn2 transposon typically carries blaTEM–1B and a 169 bp fragment of blaTEM–1B was identified (Figures 4B1,B5), but the fragment is removed by IS26/IS15DI-mediated Tn2 truncations seen in Figures 4B6,B7. The blaTEM–1B fragment is immediately adjacent to a ISEcp1 fragment, which is either ISEcp1Δ88 in structures 4B6 and B7 (with IS26/IS15D1) or ISEcp1Δ100 in the absence of adjacent IS elements. In one isolate, SS_2_C11, a continuation of the contig upstream revealed an additional AMR region containing fluoroquinolone resistance gene qnrS1 (Figure 4B1). Given that this region, spanning 5684 bp from ISKpn to an IS3-like element, is identical to the qnrS1-containing contigs in all other isolates (Figures 4B2,B3), suggests that the structure presented in Figure 4B1 may be common to all isolates, though future long-read sequencing is needed for confirmation. This SS_2_C11 blaCTX–M–15 and qnrS1- containing structure (Figure 4B1) has only been observed in two publicly deposited genomes; one E. coli ST38 strain isolated from a human in the United States (NZ_CP018207.1) and carried on an IncF plasmid, and the other in an IncB/O/K/Z plasmid from ST182 strain 2014EL-1345-2 (closest relative to this collection, Figure 1A).
In addition to qnrS1, three isolates, SS_2_B2, SS_2_E8 and SS_2_D8, also carried another fluoroquinolone resistance gene – qnrD1. In each instance, this gene was located on the structure described in Figure 4C1 and appears unique to this collection. Only one isolate, SS_2_E8, carried an intact tetracycline gene tetA, the genetic context of which is described in Figure 4D1.
Escherichia coli ST182 Virulence Profile
E. coli ST182 genomes were screened for VAGs using ABRicate in conjunction with the VFDB database (Supplementary Data 5). ETEC are defined by the ability to produce heat-liable toxin (LT) and/or heat-stable toxin (ST, subtypes STh and STp), and are characterised by antigenically distinct colonisation factors (CF) and coli surface antigens (CS). The only defining ETEC virulence combination identified in any E. coli ST182 was CS6 (cssA) + STp, present in 59 and 93% of airplane waste isolates and other ST182 strains, respectively. Similarly, the EAST1 toxin, (astA), was also less prevalent in the airplane waste collection (59%) compared to other ST182 strains (93%). Notably, while the type 1 pili operon was present in all airplane waste isolates, fimABCDEI were identified in only 36% of other ST182 stains, though fimFGH counts were higher at 83% (Figure 5A). An MDS analysis of VAGs showed that 25 airplane waste isolates shared identical virulence profiles to 20 ST182 strains isolated in Bangladesh, Kenya, Guinea-Bissau, Nepal, the United Kingdom and the United States, and that 19 isolates shared identical virulence profiles to two other ST182 strains, one isolated in Nepal, the other in the United States. The remaining 2 airplane waste isolates had unique virulence profiles (Figure 5B).
FIGURE 5.
Virulence-associated gene (VAG) profile of E. coli ST182 strains. (A) Bar graph comparing distribution of VAGs identified in both airplane waste ST182 isolates (red) and other ST182 strains (blue). (B) MDS analysis of VAGs detected in ST182 isolates. Where identical profiles cause shapes to overlap, arrows have been added detailing the country of isolation for each clustered isolate.
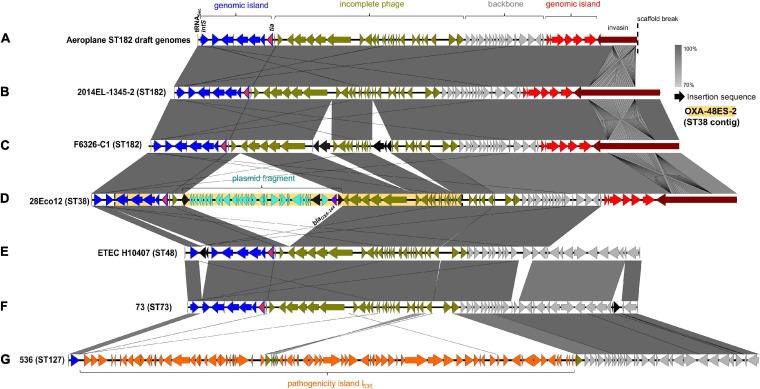
In addition to the fim operon, airplane waste isolates also had a greater occurrence of adhesin/invasin tia (89 vs 23%). This virulence factor was found situated on a predicted GI of 10,010 bp, attached at tRNASec. The GI contained a prophage integrase (intS) with 70% amino acid sequence identity to the Enterobacteria phage phiR73 integrase, phosphoethanolamine transferase (psiE), GTPase era, and four hypothetical protein encoding genes. The GI was adjacent to a 26,477 phage-like region (predicted to be an incomplete phage by PHAST) and 11,351 bp downstream from the phage-like region was another predicted GI of 6,962 bp. This second GI contained VAGs including invasin ipaB and associated chaperone sicA, cell invasion protein gene sipD and virulence transcriptional regulator hilA and was boarded by a gene encoding a protein with invasin, intimin and inverse autotransporter conserved domains. This entire 60,824 bp region (Figure 6A) was present at 100% identity and 99.89–100% coverage in all airplane isolates that carried the tia gene, and in other tia-carrying ST182 strains at 99.69–100% identity and 80.5–100% coverage (mean coverage of 95.62%) (BLAST results Supplementary Data 7). The region in airline waste isolates shared high nucleotide sequence homology to the same region situated in the complete chromosomes of E. coli ST182 strains 2014EL-1345-2 (Figure 6B) and F6326-C1 (Figure 6C) with the main variations occurring due to a scaffold break in our isolates resulting in a truncated boarder invasion/intimin/autotransporter gene (∼5,300 bp vs ∼12,000 bp), and ISSo4-like (IS21 family) and IS679-like (IS66 family) insertions within the phage-like region in E. coli strain F6326-C1 (Figure 6C). The region was also located in E. coli strains ETEC H10407 (ST48) and 73 (ST73), however, these strains lacked the second GI containing ipaB and sipD, and the large invasion/intimin/autotransporter gene (Figures 6E,F).
FIGURE 6.
Invasin-enriched region containing tia. (A) Region represented in isolates from this collection. (B) Region located E. coli ST182 strain 2014EL-1345-2 (CP024223.1, location: 2157800 – 2225307). (C) Region located in E. coli ST182 strain F6326-C1 (CP024263.1, location: 3353753 – 3426635). (D) Region in E. coli ST38 strain 28Eco12 shown in reversed orientation (CP038505.1, location: 48178 – 136524). Shaded yellow region represents genes present a sequenced E. coli ST38 contig (KT444705.1). The full contig is represented, which lacks four genes present in 28Eco12, including tia, and has no additional genes. (E) Region in E. coli ST48 (CC10) strain ETEC H10407 (FN69414.1, location: 4215539 – 4277567). (F) Region in E. coli ST73 strain 73 shown in reverse (CP041538.1, location: 1778685 – 1839792). (G) Region in E. coli ST127 strain 536 (CP000247.1, location 3947789 – 4041729). Image created using EasyFig.
Interestingly, this chromosomal region appears to be a hotspot for various insertions, including a typically IncL/M plasmid-associated 20,327 bp Tn6237 flanked by two IS1R elements (Beyrouthy et al., 2014), containing carbapenem-resistance gene blaOXA–244 (1 SNP variant of blaOXA–48), found in two E. coli ST38 strains (Figure 6D) and the uropathogenic E. coli (UPEC)-associated pathogenicity island (PAI) PAI I536 (Figure 6G).
Plasmid Replicon Profile
Plasmid replicons were screened for using ABRicate in conjunction with the PlasmidFinder database. The plasmid replicon profile for isolates in this collection varied considerably in terms of incompatibility (Inc) types, prevalence and combinations compared to other ST182 strains analysed, as well as within the collection (Figures 7A,C). The number of replicons per airplane waste isolate ranged from 2 to 10, with an average of 5, while in other E. coli ST182 strains the number ranged from 0 to 6, with an average of 3 replicons. The most common replicon in this collection was IncFII (AYA58016) (n = 46, 100%; vs n = 66, 79% of other ST182 strains), followed by IncFII_pHN7A8 (JN232517) (n = 44, 96%; vs n = 5, 6%) and IncB/O/K/Z (FN868832) (n = 41, 89%; vs n = 2, 2%). Conversely, the most common replicon in other ST182 strains was IncFIB (AP001918) (n = 79, 94%; vs n = 27, 59% in airplane isolates), followed by the previously mentioned IncFII (AYA58016, present in all airplane isolates) and Col_pHAD28 (n = 23, 27%; vs n = 7, 15%). For F plasmids, pMLST identified that all airplane waste isolates carried F2:A-:B- plasmids, and that this pMLST was unique to this group, while F-:A-:B- was the most common pMLST in other ST182 strains (n = 71, 85%) (Figure 7B).
FIGURE 7.
Plasmid replicon profile of E. coli ST182 isolates. (A) Bar graph comparing the distribution of plasmid replicons identified in airplane waste isolates (red) and other ST182 strains (blue). (B) Pie chart illustrating the distribution of F plasmid pMLST. F2:A-:B- (red) is unique to airplane waste isolates. (C) MDS analysis of plasmid replicons. Isolates from this collection depicted as red triangles. Where identical profiles caused overlaps, arrows have been added detailing country of origin for each clustered isolate.
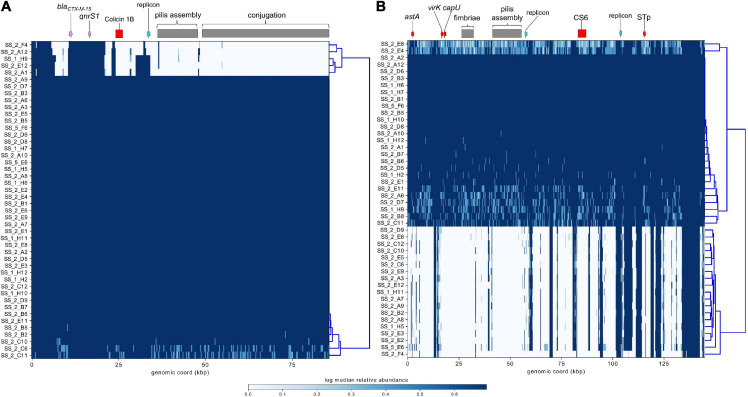
ST182 strain 2014EL-1345-2 is currently the closest relative to the airplane waste isolates and likewise carries blaCTX–M–15. This gene is harboured on an IncB/O/K plasmid (NZ_CP024226.1, 85,864 bp), and was used here as a reference sequence for short read mapping to indicate the presence of a similar plasmid in the airplane waste isolates (Figure 8A). While the AMR region, consisting of blaCTX–M–15 and qnrS1, was present in all airplane waste isolates, five isolates lacked most of the plasmid contents, including the IncB/O/K replicon in two isolates suggesting that blaCTX–M–15 and qnrS1 are carried on a different Inc type plasmid. Plasmid read mapping was also performed on 2014EL-1345-2 plasmid CP024227.1 (IncF plasmid), which harbours the ETEC-specific VAGs STp and CS6 and shares 99.88% nucleic acid sequence identity with virulence plasmid pEntYN10 (AP014654.2) isolated from an O169:H41 strain that caused an outbreak in Japan (Ban et al., 2015) (plasmid comparisons in Supplementary Data 8). Consistent with VAG screening, 24 (52%) of airplane waste isolates appear to carry a similar plasmid, though lower read-depths in some isolates suggests allelic variations (Figure 8B). Furthermore, the pMLST for the 2014EL-1345-2 plasmid is F-:A-:B-, not F2:A-:B- as carried by airplane waste isolates. Future long-read sequencing studies are needed to provide both greater insight into these initial observations, as well as to investigate plasmids harbouring replicons absent in strain 2014EL-1345-2.
FIGURE 8.
Mapping of short-reads indicating the presence of IncB/O/K/Z and IncF plasmids. (A) Schematic and heatmap for IncB/O/K/Z plasmid from E. coli ST182 strain 2014EL-1345-2. (B) Schematic and heatmap for IncF plasmid from E. coli strain 2014EL-1345-2. Clustering of rows is based on the similarity between plasmid coverage profiles. Solid blue color indicates a read depth of over 20, and a light blue color indicates a read depth between 10 and 20.
Discussion
ETEC strains are the leading cause of diarrhoeal disease in humans visiting or residing in developing countries and cause foodborne outbreaks in developed countries. ETEC O169:H41 strains have caused at least 29 outbreaks of gastroenteritis in developed countries, yet to date, no comparative genomic analyses nor reports on AMR pertaining to this ETEC serotype have been published. Here we present 46 draft genomes of O169:H41 (ST182) E. coli isolates originating from sewage taken from aircraft landing at an international airport in Germany on a single day in 2016. By comparing these isolates to 84 publicly available ST182 genomes, several key observations were made including: (i) ST182 ETEC carry heat-stable toxin STp and colonisation factor CS6; (ii) the airplane ST182 isolates have conserved core genomes but carry diverse sets of plasmid replicons; (iii) the airplane ST182 lineage is ESBL-producing and fluoroquinolone resistant; and (iv) the airplane waste isolates carry a class 1 integron-containing structure seen in major ExPEC STs such as ST131.
Escherichia coli ST182 Carry Heat-Stable Toxin STp, Colonisation Factor CS6, and a Putative Tia-Associated Genomic Island
Defining features of human ETEC include the presence of plasmid-associated heat-labile enterotoxins (LT) and/or heat-stable enterotoxins (STa) and CFs. These virulence factors are typically carried on IncFIB and IncFII plasmids and occur in various combinations (Johnson and Nolan, 2009). ETEC expressing STa have been described as more virulent than ETEC expressing only LT (Qadri et al., 2005). STa includes variants STh and STp, the latter being first described in porcine ETEC but has since been identified in human and bovine ETEC (Qadri et al., 2005; Dubreuil et al., 2016; Wang et al., 2019). Here we found that the only combination of classical ETEC virulence factors in E. coli ST182 was STp + CS6. This virulence combination has been reported as most prevalent in patients with diarrhoea in Thailand (Puiprom et al., 2010), and associated with moderate-severe diarrhoea in traveller’s to Mexico and Guatemala (Nicklasson et al., 2010).
A complete genome of ST182 strain 2014EL-1345-2, isolated in 2014 in the United States (Smith et al., 2018), carrying an IncF plasmid containing both STp and CS6, enabled short-read plasmid mapping to be performed and suggested that approximately half of the airplane-derived ST182 isolates carried similar plasmid cargo. This was congruent with the observation that only 52% of airplane-derived ST182 isolates carried STp + CS6. However, it is important to note that ETEC virulence plasmids have demonstrated instability under laboratory conditions (Crossman et al., 2010; Hazen et al., 2019). Indeed, the pEntYN10 plasmid from a Japanese O169:H41 outbreak strain, sharing high sequence homology to the mapped 2014EL-1345-2 virulence plasmid (99.88%), is unstable in vitro (Ban et al., 2015) and other STp + CS6 carrying plasmids have been lost after one overnight passage (Tobias et al., 2016). Therefore, it is possible that the virulence plasmid in some airplane-derived ST182 isolates was lost prior to sequencing.
In addition to enterotoxins and CFs, ETEC are known to carry a number of non-classical virulence factors such as serine protease autotransporter EatA (Patel et al., 2004), adhesins EtpA (Roy et al., 2008), TibA (Lindenthal and Elsinghorst, 2001) and Tia (Mammarappallil and Elsinghorst, 2000), dynamin LeoA (Michie et al., 2014), and enteroaggregative E. coli heat-stable enterotoxin EAST1 (Yamamoto and Echeverria, 1996). In agreement with other studies screening non-classical VAGs in ETEC, EAST1 (encoded by astA) was the most common in E. coli ST182 outside the airplane waste collection (92%), though less common within the collection (59%) (Rivera et al., 2013). However, only one non-airplane waste isolate carried eatA, and no E. coli ST182 strain or isolate carried genes encoding TibA, EtpA or LeoA. This is consistent with a previous study reporting that TibA and EtpA are rare in STa + CS6 ETEC (Hazen et al., 2019). Nevertheless, we found that tia was prevalent in airplane ST182 isolates (89 vs. 23% in other ST182 strains). In addition to a role in adhesion, Tia has been demonstrated to facilitate invasion of intestinal cells (Fleckenstein et al., 1996). The tia gene is often situated on a subtilase-encoding pathogenicity island (SE-PAI) (Bondì et al., 2017; Wyrsch et al., 2020), however, in all tia-positive ST182 strains and isolates we found the gene on a putative genomic island (GI) attached to tRNASEC. Our comparative analysis of this GI and the surrounding region confirmed a previous report that the tRNASEC appears to be a hotspot for various insertions, including AMR plasmid fragments and the entire ExPEC-associated PAI I536 (Abril et al., 2019). This locus is significant in the evolution of ETEC.
Escherichia coli ST182 Diversity
Of the 130 E. coli ST182 genomes analysed in this study, the majority (93%) were of serotype O169:H41, though a small portion were O167:H41 (7%). Interestingly, the O167 serogroup is associated with both ETEC and enteroinvasive E. coli (EIEC) and related to a Shigella boydii O-antigen (Gross et al., 1983). All E. coli ST182 strains were typed as phylogroup E, which is uncommon, as most ETEC are of phylogroup A and B1 (von Mentzer et al., 2014; Mosquito et al., 2015; Sahl et al., 2017). Indeed, a phylogenetic analysis of 362 ETEC isolates found that isolates of serogroup O169 containing STp + CS6 formed their own clade, L7, and were the only ETEC lineage associated with phylogroup E (von Mentzer et al., 2014).
Our pangenome analysis demonstrated that ST182 genomes share a core genome of 4053 genes and have an accessory genome of 2286 genes. Despite being clonal in nature, the airplane waste isolates could be discerned by their accessory genomes and carried diverse sets of plasmid replicons. ETEC strains have been reported carrying up to six plasmids (Rasko et al., 2008; Crossman et al., 2010) and up to eight plasmid replicons (Hazen et al., 2019). Here we found that airplane ST182 isolates carried two to 10 plasmid replicons with an average of five per isolate. This was significantly higher than other ST182 strains which carried zero to six replicons with an average of three. The types of replicons also differed, with the most prevalent in airplane isolates being IncFII and IncB/O/K/Z versus IncFIB in other ST182, and indeed other ETEC (Hazen et al., 2019). pMLST indicated that all airplane waste isolates carried a F2:A-:B- plasmid, which was not present in any other ST182 strain. F2:A-:B- plasmids have been reported carrying various blaCTX–M genes and other ARGs in both humans and animals (Deng et al., 2011; Dahmen et al., 2013; Grami et al., 2014; Díaz-Jiménez et al., 2020).
Escherichia coli ST182 Found in Airplane Waste Is ESBL-Producing
A standout feature of this E. coli ST182 lineage was its AMR profile. All airplane-derived isolates were resistant to 3rd and 4th generation extended-spectrum β-lactams, and all carried ESBL gene blaCTX–M–15. Conversely, blaCTX–M–15 was only identified in six other ST182 strains (7%) and blaCTX–M–14 in one other ST182 strain. The prevalence of ESBL-producing ETEC is growing, particularly in association with traveller’s diarrhoea. A study investigating antimicrobial susceptibilities in traveller’s diarrhoea isolates from 2006 to 2008 found ESBL-producing ETEC in 6% of isolates originating from India (Ouyang-Latimer et al., 2011), while in 2011–2017, another study found 43% of traveller’s diarrhoea isolates from visitors to India and Southeast Asia were ESBL-producing ETEC (Guiral et al., 2019). In the latter study, ESBL-production was attributed to blaCTX–M–15 and blaCTX–M–27 carriage. Similarly, a study surveying clinical ETEC in Nepal from 2001 to 2016 found only one ESBL-producing ETEC isolate between 2001 and 2009 while over 30% of ETEC isolates collected post 2013 were ESBL-producing, 80% of which carried blaCTX–M–15 (Margulieux et al., 2018). These significant increases in ESBL-producing ETEC from the early-mid 2000s to the following decade coincides with the period of rapid global expansion of pandemic ESBL-producing ExPEC lineages, most notably ST131 H30Rx (Zilberberg and Shorr, 2013; Poolman and Wacker, 2016). Additionally, ExPEC have recently been identified as the most common pathotype associated with travel-acquired ESBL-producing E. coli (Kantele et al., 2020). Given that ExPEC can colonise the gastrointestinal tract (Manges and Johnson, 2015) and that blaCTX–M–15 and blaCTX–M–14 (both strongly associated with ExPEC) are most frequently carried on IncF plasmids (Bevan et al., 2017), it is possible that ExPEC have contributed to the evolution of ESBL-production in ETEC (or vice versa) via horizontal gene transfer within a shared environment. However, in ExPEC the majority of blaCTX–M genes are associated with ISEcp1 (also known as ISEc9), which can capture and mobilise blaCTX–M genes and other ARGs (Carattoli, 2013, 2009; Li et al., 2015), but in the airplane-derived ST182 lineage we found that only a 88 bp or 100 bp fragment of ISEcp1 adjacent to the blaCTX–M–15 genes. In each isolate, blaCTX–M–15 was found situated within a Tn2 transposon and adjacent to a cupin-like gene. Downstream to blaCTX–M–15 we identified a 169 bp fragment of blaTEM–1B, the passenger gene of Tn2 (Siguier et al., 2006; Bailey et al., 2011). This blaCTX–M–15 context, including the blaTEM–1B fragment, has only been described once in the literature and was present in ETEC O159:H20 strains responsible for the first ESBL-associated foodborne outbreak in South Korea in 2016 (Kim et al., 2017).
Escherichia coli ST182 Found in Airplane Waste Is Fluoroquinolone Resistant
Like ESBL-production, fluoroquinolone resistance in ETEC strains has also been increasing, particularly in isolates originating from traveller’s visiting India, Southeast Asia and Africa (Mendez Arancibia et al., 2009; Guiral et al., 2019). Between 2001 to 2007, it was reported that 8% of ETEC strains from India and Southeast Asia were resistant to fluoroquinolones (Mendez Arancibia et al., 2009), increasing to 43% in 2011–2017 (Guiral et al., 2019). In this study, all airplane-derived ST182 isolates were phenotypically resistant to fluoroquinolones and all carried the plasmid-associated fluoroquinolone resistance gene qnrS1, with three isolates also carrying qnrD1. The plasmid-associated qnrD1 gene is relatively uncommon and generally only reported in Proteeae (Proteus spp., Morganella morganii and Providencia stuartii); however, a recent study on 183 Enterobacteriaceae sourced from health care facilities in India found 23.5% harboured qnrD1 indicating dissemination to other species including E. coli and Klebsiella spp. (Dasgupta et al., 2017). Only two ST182 strains outside this collection (2%) carried qnrS1, and no other fluoroquinolone resistant genes were identified. One qnrS1-carrying ST182 strain was again 2014EL-1345-2, the closest relative to this collection. The qnrS1 gene in 2014EL-1345-2 is situated on a resolved IncB/O/K/Z plasmid, along with blaCTX–M–15 upstream to qnrS1. Short-read plasmid-mapping exercises suggested that most airplane-derived isolates carried a similar plasmid. However, future long-read sequencing is required to confirm this observation as mapped reads are taken from the entire genome and do not confirm genetic context.
In addition to plasmid-associated fluoroquinolone genes, we identified a fluoroquinolone-conferring S83L mutation in gyrA (Ruiz, 2003) in all airplane isolates and 30% of other ST182 isolates. Mutations conferring resistance to fluoroquinolone in gyrA and in DNA topoisomerase 4 subunit A parC have been described in ETEC previously (Mendez Arancibia et al., 2009), and specifically the S83L mutation was identified in ETEC outbreak strains of various serotypes in India (Chakraborty et al., 2001; Pazhani et al., 2011).
Airplane Waste Escherichia coli ST182 Carry a Class 1 Integron Found in ExPEC
Class 1 integrons are considered a reliable proxy for a multidrug resistant genotype and have played an important role in the spread of AMR in Enterobacteriaceae (Gillings, 2014). Class 1 integrons are often associated with mobile genetic elements, such as plasmids and IS elements, particularly IS26 (Partridge et al., 2018), which enable lateral dissemination of ARG cargo. Here we found that, while class 1 integrons were rare in other ST182 (3%) strains, all airplane waste ST182 isolates carried a class 1 integron upstream of a macrolide inactivation gene cluster (mphR-mrx-mphA). This complex resistance region (CRR) was flanked by IS26 elements and was comprised of IS26-ΔintI1648-dfrA17-aadA5-qacEΔ1-sul1-ORF-srpC-padR-IS6100-mphR-mrx-mphA-IS26. The dfrA17-aadA5 integron (In54) cassette was first characterised in 2000 after its discovery in an E. coli isolated from a urinary tract infection (UTI) patient in Australia (White et al., 2000). Since then, dfrA17-aadA5 has become one of the most prevalent cassette arrangements worldwide in E. coli (Yu et al., 2004; Xia et al., 2016; Kaushik et al., 2019) and in other Enterobacteriaceae including Salmonella species (Meng et al., 2017), and is thought to have driven trimethoprim resistance in ExPEC ST69 strains (also known as clonal group A) (Solberg et al., 2006). While the dfrA17-aadA5 cassette has been previously identified in integrons present in clinical ETEC strains (Kartsev et al., 2015), a BLASTn search demonstrated that the entire IS26 flanked CRR containing the integron found in ST182 airplane isolates has not been observed in ETEC previously, but in Klebsiella, Salmonella and ExPEC-associated STs, most commonly in ST131. In ST131 strains, this CRR is distributed globally, is carried on various F plasmids, and is harboured by isolates of various origins including humans (Li et al., 2020) and oysters (Fernandes et al., 2020).
In conclusion, within the context of growing reports of rising antimicrobial resistance in travel acquired Enterobacteriaceae, we provide the first WGS analysis of antimicrobial and virulence gene carriage in ETEC ST182 isolates originating from airline passengers arriving in Germany from abroad. Our data indicates that these isolates constitute a new lineage of ST182 that is resistant to multiple, clinically important antibiotics. Overall, this study contributes to a better understanding of the spread of ETEC as well as AMR worldwide.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/639663, SAMN15244685 – SAMN15244730.
Author Contributions
VJ: conceptualisation, formal analysis, investigation, visualisation, and writing – original draft. SH: data curation and investigation. KA: investigation. TB: funding acquisition and project administration. SD: conceptualisation, funding acquisition, project administration, and supervision. All authors contributed to writing – review and editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
DNA isolation was performed by our colleagues U. Mogck and A.S. Reichelt from the Institute of Hydrobiology, Technische Universität Dresden, Germany.
Footnotes
Funding. This work received funding from the German Federal Ministry of Education and Research (grant 02WRS1377D, HYREKA), DSWAP (EU Prima). This project was also partly funded by the Australian Centre for Genomic Epidemiological Microbiology (AusGEM), a collaborative partnership between the NSW Department of Primary Industries and the University of Technology Sydney.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.731050/full#supplementary-material
References
- Abril D., Bustos Moya I. G., Marquez-Ortiz R. A., Josa Montero D. F., Corredor Rozo Z. L., Torres Molina I., et al. (2019). First Report and Comparative Genomics Analysis of a blaOXA-244-Harboring Escherichia coli Isolate Recovered in the American Continent. Antibiotics 8:222. 10.3390/antibiotics8040222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48 D517–D525. 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcilla M. S., van Hattem J. M., Haverkate M. R., Bootsma M. C. J., van Genderen P. J. J., Goorhuis A., et al. (2017). Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect. Dis. 17 78–85. 10.1016/S1473-3099(16)30319-X [DOI] [PubMed] [Google Scholar]
- Bailey J. K., Pinyon J. L., Anantham S., Hall R. M. (2011). Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66 745–751. 10.1093/jac/dkq529 [DOI] [PubMed] [Google Scholar]
- Ban E., Yoshida Y., Wakushima M., Wajima T., Hamabata T., Ichikawa N., et al. (2015). Characterization of unstable pEntYN10 from enterotoxigenic escherichia coli (ETEC) O169: H41. Virulence 6 735–744. 10.1080/21505594.2015.1094606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty M. E., Bopp C. A., Wells J. G., Greene K. D., Puhr N. D., Mintz E. D. (2004). Enterotoxin-producing Escherichia coli O169:H41. United States. Emer. Infect. Dis. 10 518–552. 10.3201/eid1003.030268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Laird M. R., Williams K. P., Lau B. Y., Hoad G., Winsor G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45 W30–W35. 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan E. R., Jones A. M., Hawkey P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72 2145–2155. 10.1093/jac/dkx146 [DOI] [PubMed] [Google Scholar]
- Beyrouthy R., Robin F., Delmas J., Gibold L., Dalmasso G., Dabboussi F., et al. (2014). IS1R-Mediated Plasticity of IncL/M Plasmids Leads to the Insertion of blaOXA-48 into the Escherichia coli Chromosome. Antimicrob. Agents Chemother. 58 3785–3790. 10.1128/AAC.02669-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondì R., Chiani P., Michelacci V., Minelli F., Caprioli A., Morabito S. (2017). The Gene tia, Harbored by the Subtilase-Encoding Pathogenicity Island, Is Involved in the Ability of Locus of Enterocyte Effacement-Negative Shiga Toxin-Producing Escherichia coli Strains To Invade Monolayers of Epithelial Cells. Infection and Immunity 85 e613–e617. 10.1128/IAI.00613-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O., Bohlin J., Scheffer L., Eldholm V. (2016). Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 17:238. 10.1186/s13059-016-1108-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53 2227–2238. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303 298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Hasman H. (2020). PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2075 285–294. 10.1007/978-1-4939-9877-7_20 [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Deokule J. S., Garg P., Bhattacharya S. K., Nandy R. K., Nair G. B., et al. (2001). Concomitant Infection of EnterotoxigenicEscherichia coli in an Outbreak of Cholera Caused byVibrio cholerae O1 and O139 in Ahmedabad, India. J. Clin. Microb. 39 3241–3246. 10.1128/JCM.39.9.3241-3246.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Kim J., Oh K.-H., Hu J. K., Seo J., Oh S. S., et al. (2014). Outbreak of enterotoxigenic Escherichia coli O169 enteritis in schoolchildren associated with consumption of kimchi, Republic of Korea, 2012‡. Epidemiol. Infect. 142 616–623. 10.1017/S0950268813001477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5 58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- Clifford R. J., Milillo M., Prestwood J., Quintero R., Zurawski D. V., Kwak Y. I., et al. (2012). Detection of Bacterial 16S rRNA and Identification of Four Clinically Important Bacteria by Real-Time PCR. PLoS One 7:e48558. 10.1371/journal.pone.0048558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman L. C., Chaudhuri R. R., Beatson S. A., Wells T. J., Desvaux M., Cunningham A. F., et al. (2010). A Commensal Gone Bad: Complete Genome Sequence of the Prototypical Enterotoxigenic Escherichia coli Strain H10407. J. Bacteriol. 192 5822–5831. 10.1128/JB.00710-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmen S., Madec J.-Y., Haenni M. (2013). F2:A-:B- plasmid carrying the extended-spectrum β-lactamase bla(CTX–M–55/57) gene in Proteus mirabilis isolated from a primate. Int. J. Antimicrob. Agents 41 594–595. 10.1016/j.ijantimicag.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Dasgupta N., Paul D., Dhar D., Ingti B., Bhattacharjee D., Chakravarty A., et al. (2017). An insight into selection specificity of quinolone resistance determinants within Enterobacteriaceae family. Journal of Global Antimicrobial Resistance 10 40–46. 10.1016/j.jgar.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Deng Y., He L., Chen S., Zheng H., Zeng Z., Liu Y., et al. (2011). F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-blaCTX–M–9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob. Agents Chemother. 55 4926–4929. 10.1128/aac.00133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasia R. A., Jones T. F., Ward J., Stafford L., Hardin H., Bopp C., et al. (2006). Endemically Acquired Foodborne Outbreak of Enterotoxin-producing Escherichia coli Serotype O169:H41. Am. J. Med. 119 .e7–.e168. 10.1016/j.amjmed.2005.07.063 [DOI] [PubMed] [Google Scholar]
- Díaz-Jiménez D., García-Meniño I., Herrera A., García V., López-Beceiro A. M., Alonso M. P., et al. (2020). Genomic Characterization of Escherichia coli Isolates Belonging to a New Hybrid aEPEC/ExPEC Pathotype O153:H10-A-ST10 eae-beta1 Occurred in Meat. Poultry, Wildlife and Human Diarrheagenic Samples. Antibiotics 9:192. 10.3390/antibiotics9040192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemert D. J. (2006). Prevention and self-treatment of traveler’s diarrhea. Clin. Microbiol. Rev. 19 583–594. 10.1128/CMR.00052-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil J. D., Isaacson R. E., Schifferli D. M. (2016). Animal Enterotoxigenic Escherichia coli. EcoSal. Plus 7:2016. 10.1128/ecosalplus.ESP-0006-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Cunha M. P. V., Lopes R., Cerdeira L., Lincopan N. (2020). Emergence of CTX-M-27-producing Escherichia coli of ST131 and clade C1-M27 in an impacted ecosystem with international maritime traffic in South America. J. Antimicrob. Chemother. 75 1647–1649. 10.1093/jac/dkaa069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein J. M., Kopecko D. J., Warren R. L., Elsinghorst E. A. (1996). Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64 2256–2265. 10.1128/iai.64.6.2256-2265.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaio D., To J., Liu M., Monahan L., Anantanawat K., Darling A. E. (2019). Hackflex: low cost Illumina sequencing library construction for high sample counts. bioRxiv 2019:779215. 10.1101/779215 [DOI] [Google Scholar]
- Galata V., Fehlmann T., Backes C., Keller A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47 D195–D202. 10.1093/nar/gky1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M. R. (2014). Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 78 257–277. 10.1128/MMBR.00056-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grami R., Dahmen S., Mansour W., Mehri W., Haenni M., Aouni M., et al. (2014). blaCTX–M–15-Carrying F2:A-:B- Plasmid in Escherichia coli from Cattle Milk in Tunisia. Microbial. Drug Res. 20 344–349. 10.1089/mdr.2013.0160 [DOI] [PubMed] [Google Scholar]
- Gross R. J., Thomas L. V., Cheasty T., Day N. P., Rowe B., Toledo M. R., et al. (1983). Enterotoxigenic and enteroinvasive Escherichia coli strains belonging to a new O group, O167. J. Clin. Microbiol. 17 521–523. 10.1128/jcm.17.3.521-523.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiral E., Quiles M. G., Muñoz L., Moreno-Morales J., Alejo-Cancho I., Salvador P., et al. (2019). Emergence of Resistance to Quinolones and β-Lactam Antibiotics in Enteroaggregative and Enterotoxigenic Escherichia coli Causing Traveler’s Diarrhea. Antimicrobial. Agents Chemother. 63:18. 10.1128/AAC.01745-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Croucher N. J., Goater R. J., Abudahab K., Aanensen D. M., Harris S. R. (2018). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34 292–293. 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T., Itoh K., Yamaguchi Y., Hirai Y., Kanki M., Kawatsu K., et al. (2013). A Foodborne Outbreak of Gastrointestinal Illness Caused by Enterotoxigenic Escherichia coli Serotype O169:H41 in Osaka, Japan. Jap. J. Infect. Dis. 66 530–533. 10.7883/yoken.66.530 [DOI] [PubMed] [Google Scholar]
- Hassing R. J., Alsma J., Arcilla M. S., Genderen P. J., van, Stricker B. H., et al. (2015). International travel and acquisition of multidrug-resistant Enterobacteriaceae: a systematic review. Eurosurveillance 20:30074. 10.2807/1560-7917.ES.2015.20.47.30074 [DOI] [PubMed] [Google Scholar]
- Hazen T. H., Nagaraj S., Sen S., Permala-Booth J., Canto F. D., Vidal R., et al. (2019). Genome and Functional Characterization of Colonization Factor Antigen I- and CS6-Encoding Heat-Stable Enterotoxin-Only Enterotoxigenic Escherichia coli Reveals Lineage and Geographic Variation. mSystems 4:18. 10.1128/mSystems.00329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heß S., Berendonk T. U., Kneis D. (2018). Antibiotic resistant bacteria and resistance genes in the bottom sediment of a small stream and the potential impact of remobilization. FEMS Microbiol. Ecol. 94:128. 10.1093/femsec/fiy128 [DOI] [PubMed] [Google Scholar]
- Heß S., Kneis D., Österlund T., Li B., Kristiansson E., Berendonk T. U. (2019). Sewage from Airplanes Exhibits High Abundance and Diversity of Antibiotic Resistance Genes. Environ. Sci. Technol. 53 13898–13905. 10.1021/acs.est.9b03236 [DOI] [PubMed] [Google Scholar]
- Hosangadi D., Smith P. G., Kaslow D. C., Giersing B. K., Who E. T. E. C.Shigella Vaccine Consultation Expert Group. (2019). WHO consultation on ETEC and Shigella burden of disease, Geneva, 6-7th April 2017: Meeting report. Vaccine 37 7381–7390. 10.1016/j.vaccine.2017.10.011 [DOI] [PubMed] [Google Scholar]
- ICAO (2018). International Civil Aviation Organisation Annual Report 2018. Montreal: ICAO. [Google Scholar]
- Institute of Medicine Forum on Microbial Threats (2010). Infectious Disease Movement in a Borderless World: Workshop Summary, The National Academies Collection: Reports funded by National Institutes of Health. Washington,DC: National Academies Press (US). [PubMed] [Google Scholar]
- Joensen K. G., Tetzschner A. M. M., Iguchi A., Aarestrup F. M., Scheutz F. (2015). Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 53 2410–2426. 10.1128/JCM.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Nolan L. K. (2009). Pathogenomics of the Virulence Plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73 750–774. 10.1128/MMBR.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantele A., Kuenzli E., Dunn S. J., Dance D. A. B., Newton P. N., Davong V., et al. (2021). Dynamics of intestinal multidrug-resistant bacteria colonisation contracted by visitors to a high-endemic setting: a prospective, daily, real-time sampling study. Lancet Microbe 2 e151–e158. 10.1016/S2666-5247(20)30224-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantele A., Lääveri T., Mero S., Häkkinen I. M. K., Kirveskari J., Johnston B. D., et al. (2020). Despite Predominance of Uropathogenic/Extraintestinal Pathotypes Among Travel-acquired Extended-spectrum β-Lactamase-producing Escherichia coli, the Most Commonly Associated Clinical Manifestation Is Travelers’ Diarrhea. Clin. Infect. Dis. 70 210–218. 10.1093/cid/ciz182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantele A., Lääveri T., Mero S., Vilkman K., Pakkanen S. H., Ollgren J., et al. (2015). Antimicrobials increase travelers’ risk of colonization by extended-spectrum betalactamase-producing Enterobacteriaceae. Clin. Infect. Dis. 60 837–846. 10.1093/cid/ciu957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartsev N. N., Fursova N. K., Pachkunov D. M., Bannov V. A., Eruslanov B. V., Svetoch E. A., et al. (2015). Molecular Characterization of Enterotoxin-Producing Escherichia coli Collected in 2011–2012, Russia. PLoS One 10:e0123357. 10.1371/journal.pone.0123357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik M., Khare N., Kumar S., Gulati P. (2019). High Prevalence of Antibiotic Resistance and Integrons in Escherichia coli Isolated from Urban River Water, India. Microb. Drug Resist. 25 359–370. 10.1089/mdr.2018.0194 [DOI] [PubMed] [Google Scholar]
- Kim J. S., Park J., Shin E., Kim S., Oh S. S., Yang H.-J., et al. (2017). Outbreak of CTX-M-15-Producing Enterotoxigenic Escherichia coli O159:H20 in the Republic of Korea in 2016. Antimicrobial. Agents Chemother. 61:17. 10.1128/AAC.00339-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti L. M., Bourgeois A. L., Walker C. L. F., Black R. E., Sack D. (2014). Estimating Diarrheal Illness and Deaths Attributable to Shigellae and Enterotoxigenic Escherichia coli among Older Children, Adolescents, and Adults in South Asia and Africa. PLoS Neglected Tropical Diseases 8:e2705. 10.1371/journal.pntd.0002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leray M., Knowlton N., Ho S.-L., Nguyen B. N., Machida R. J. (2019). GenBank is a reliable resource for 21st century biodiversity research. PNAS 116 22651–22656. 10.1073/pnas.1911714116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47 W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K. C., Leung A. A. M., Wong A. H. C., Hon K. L. (2019). Travelers’ Diarrhea: A Clinical Review. Recent Pat. Inflamm. Allergy Drug Discov. 13 38–48. 10.2174/1872213X13666190514105054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Reid C. J., Kudinha T., Jarocki V. M., Djordjevic S. P. (2020). Genomic analysis of trimethoprim-resistant extraintestinal pathogenic Escherichia coli and recurrent urinary tract infections. Microbial. Genomics 6:e000475. 10.1099/mgen.0.000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-J., Spychala C. N., Hu F., Sheng J.-F., Doi Y. (2015). Complete Nucleotide Sequences of blaCTX–M-Harboring IncF Plasmids from Community-Associated Escherichia coli Strains in the United States. Antimicrob. Agents Chemother. 59 3002–3007. 10.1128/AAC.04772-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenthal C., Elsinghorst E. A. (2001). Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69 52–57. 10.1128/IAI.69.1.52-57.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zheng D., Jin Q., Chen L., Yang J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47 D687–D692. 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.-P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microb. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mammarappallil J. G., Elsinghorst E. A. (2000). Epithelial Cell Adherence Mediated by the Enterotoxigenic Escherichia coli Tia Protein. Infect. Immun. 68 6595–6601. 10.1128/iai.68.12.6595-6601.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A. R., Johnson J. R. (2015). Reservoirs of Extraintestinal Pathogenic Escherichia coli. Microbiol. Spectr. 3:2012. 10.1128/microbiolspec.UTI-0006-2012 [DOI] [PubMed] [Google Scholar]
- Margulieux K. R., Srijan A., Ruekit S., Nobthai P., Poramathikul K., Pandey P., et al. (2018). Extended-spectrum β-lactamase prevalence and virulence factor characterization of enterotoxigenic Escherichia coli responsible for acute diarrhea in Nepal from 2001 to 2016. Antimicrobial. Res. Infect. Control 7:87. 10.1186/s13756-018-0377-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez Arancibia E., Pitart C., Ruiz J., Marco F., Gascón J., Vila J. (2009). Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveller’s diarrhoea. J. Antimicrob. Chemother. 64 343–347. 10.1093/jac/dkp178 [DOI] [PubMed] [Google Scholar]
- Meng X., Zhang Z., Li K., Wang Y., Xia X., Wang X., et al. (2017). Antibiotic Susceptibility and Molecular Screening of Class I Integron in Salmonella Isolates Recovered from Retail Raw Chicken Carcasses in China. Microb. Drug Resist. 23 230–235. 10.1089/mdr.2015.0359 [DOI] [PubMed] [Google Scholar]
- Michie K. A., Boysen A., Low H. H., Møller-Jensen J., Löwe J. (2014). LeoA, B and C from Enterotoxigenic Escherichia coli (ETEC) Are Bacterial Dynamins. PLoS One 9:107211. 10.1371/journal.pone.0107211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., et al. (2020). IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37 1530–1534. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirhoseini A., Amani J., Nazarian S. (2018). Review on pathogenicity mechanism of enterotoxigenic Escherichia coli and vaccines against it. Microbial. Pathog. 117 162–169. 10.1016/j.micpath.2018.02.032 [DOI] [PubMed] [Google Scholar]
- Mosquito S., Pons M. J., Riveros M., Ruiz J., Ochoa T. J. (2015). Diarrheagenic Escherichia coli Phylogroups Are Associated with Antibiotic Resistance and Duration of Diarrheal Episode. Sci. World J. 2015:610403. 10.1155/2015/610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada R. A., Shaheen H. I., Khalil S. B., Mansour A., El-Sayed N., Touni I., et al. (2011). Discovery and Phylogenetic Analysis of Novel Members of Class b Enterotoxigenic Escherichia coli Adhesive Fimbriae. J. Clin. Microb. 49 1403–1410. 10.1128/JCM.02006-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklasson M., Klena J., Rodas C., Bourgeois A. L., Torres O., Svennerholm A.-M., et al. (2010). Enterotoxigenic Escherichia coli Multilocus Sequence Types in Guatemala and Mexico. Emerg. Infect. Dis. 16 143–145. 10.3201/eid1601.090979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa Y. (1995). Heat-Stable Enterotoxin-Producing Escherichia coli O169:H41 in Japan. Emerg. Infect. Dis. 1 61–61. 10.3201/eid0102.950206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa Y., Helander A., Ogasawara J., Moyer N. P., Hanaoka M., Hase A., et al. (1998). Epidemiology and properties of heat-stable enterotoxin-producing Escherichia coli serotype O169:H41. Epidemiol. Infect. 121 31–42. 10.1017/s0950268898001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B. D., Treangen T. J., Melsted P., Mallonee A. B., Bergman N. H., Koren S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17:132. 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang-Latimer J., Jafri S., VanTassel A., Jiang Z.-D., Gurleen K., Rodriguez S., et al. (2011). In Vitro Antimicrobial Susceptibility of Bacterial Enteropathogens Isolated from International Travelers to Mexico, Guatemala, and India from 2006 to 2008. Antimicrob. Agents Chemother. 55 874–878. 10.1128/AAC.00739-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Kwong S. M., Firth N., Jensen S. O. (2018). Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 31:17. 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. K., Dotson J., Allen K. P., Fleckenstein J. M. (2004). Identification and Molecular Characterization of EatA, an Autotransporter Protein of Enterotoxigenic Escherichia coli. Infect. Immun. 72 1786–1794. 10.1128/IAI.72.3.1786-1794.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhani G. P., Chakraborty S., Fujihara K., Yamasaki S., Ghosh A., Nair G. B., et al. (2011). QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad, India. Indian J. Med. Res. 2011:10. [PMC free article] [PubMed] [Google Scholar]
- Poolman J. T., Wacker M. (2016). Extraintestinal Pathogenic Escherichia coli, a Common Human Pathogen: Challenges for Vaccine Development and Progress in the Field. J. Infect. Dis. 213 6–13. 10.1093/infdis/jiv429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puiprom O., Chataroj S., Gangnonngiw W., Okada K., Honda T., Taniguchi T., et al. (2010). Identification of colonization factors of enterotoxigenic Escherichia coli with PCR-based technique. Epidemiol. Infect. 138 519–524. 10.1017/s0950268809990720 [DOI] [PubMed] [Google Scholar]
- Qadri F., Svennerholm A.-M., Faruque A. S. G., Sack R. B. (2005). Enterotoxigenic Escherichia coli in Developing Countries: Epidemiology, Microbiology, Clinical Features, Treatment, and Prevention. CMR 18 465–483. 10.1128/CMR.18.3.465-483.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Rosovitz M. J., Myers G. S. A., Mongodin E. F., Fricke W. F., Gajer P., et al. (2008). The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J. Bacteriol. 190 6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regua-Mangia A. H., Guth B. C., Andrade J. R. D. C., Irino K., Pacheco A. B. F., Ferreira L. C. S., et al. (2004). Genotypic and phenotypic characterization of enterotoxigenic Escherichia coli (ETEC) strains isolated in Rio de Janeiro city, Brazil. FEMS Immunol. Med. Microbiol. 40 155–162. 10.1016/S0928-8244(03)00308-0 [DOI] [PubMed] [Google Scholar]
- Rivera F. P., Medina A. M., Aldasoro E., Sangil A., Gascon J., Ochoa T. J., et al. (2013). Genotypic Characterization of Enterotoxigenic Escherichia coli Strains Causing Traveler’s Diarrhea. J. Clin. Microbiol. 51 633–635. 10.1128/JCM.02572-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roer L., Tchesnokova V., Allesøe R., Muradova M., Chattopadhyay S., Ahrenfeldt J., et al. (2017). Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J. Clin. Microbiol. 55 2538–2543. 10.1128/JCM.00737-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K., Hamilton D., Allen K. P., Randolph M. P., Fleckenstein J. M. (2008). The EtpA Exoprotein of Enterotoxigenic Escherichia coli Promotes Intestinal Colonization and Is a Protective Antigen in an Experimental Model of Murine Infection. Infect. Immun. 76 2106–2112. 10.1128/IAI.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J. (2003). Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51 1109–1117. 10.1093/jac/dkg222 [DOI] [PubMed] [Google Scholar]
- Sahl J. W., Sistrunk J. R., Baby N. I., Begum Y., Luo Q., Sheikh A., et al. (2017). Insights into enterotoxigenic Escherichia coli diversity in Bangladesh utilizing genomic epidemiology. Sci. Rep. 7 1–12. 10.1038/s41598-017-03631-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer L., Singh B., Rupali P., Libman M. (2019). Emerging concepts in the diagnosis, treatment, and prevention of travelers’ diarrhea. Curr. Opin. Infect. Dis. 32 468–474. 10.1097/QCO.0000000000000581 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P., Lindsey R. L., Rowe L. A., Batra D., Stripling D., Garcia-Toledo L., et al. (2018). High-Quality Whole-Genome Sequences for 21 Enterotoxigenic Escherichia coli Strains Generated with PacBio Sequencing. Genome Announc. 6:17. 10.1128/genomeA.01311-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O. D., Ajiboye R. M., Riley L. W. (2006). Origin of Class 1 and 2 Integrons and Gene Cassettes in a Population-Based Sample of Uropathogenic Escherichia coli. J. Clin. Microbiol. 44 1347–1351. 10.1128/JCM.44.4.1347-1351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. N., Hamer D. H., Shlim D. R. (2017). Medications for the prevention and treatment of travellers’ diarrhea. J. Travel Med. 24 S17–S22. 10.1093/jtm/taw097 [DOI] [PubMed] [Google Scholar]
- Tobias J., Von Mentzer A., Loayza Frykberg P., Aslett M., Page A. J., Sjöling Å, et al. (2016). Stability of the Encoding Plasmids and Surface Expression of CS6 Differs in Enterotoxigenic Escherichia coli (ETEC) Encoding Different Heat-Stable (ST) Enterotoxins (STh and STp). PLoS One 11:152899. 10.1371/journal.pone.0152899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin-Hill G., MacAlasdair N., Ruis C., Weimann A., Horesh G., Lees J. A., et al. (2020). Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21:180. 10.1186/s13059-020-02090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Ondov B. D., Koren S., Phillippy A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biology 15 524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble D. R. (2017). Resistant pathogens as causes of traveller’s diarrhea globally and impact(s) on treatment failure and recommendations. J. Travel Med. 24 S6–S12. 10.1093/jtm/taw090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilkman K., Lääveri T., Pakkanen S. H., Kantele A. (2019). Stand-by antibiotics encourage unwarranted use of antibiotics for travelers’ diarrhea: A prospective study. Travel. Med. Infect. Dis. 27 64–71. 10.1016/j.tmaid.2018.06.007 [DOI] [PubMed] [Google Scholar]
- von Mentzer A., Connor T. R., Wieler L. H., Semmler T., Iguchi A., Thomson N. R., et al. (2014). Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nature Genetics 46 1321–1326. 10.1038/ng.3145 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhong Z., Luo Y., Cox E., Devriendt B. (2019). Heat-Stable Enterotoxins of Enterotoxigenic Escherichia coli and Their Impact on Host Immunity. Toxins 11:11010024. 10.3390/toxins11010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. A., McIver C. J., Deng Y.-M., Rawlinson W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182 265–269. 10.1111/j.1574-6968.2000.tb08906.x [DOI] [PubMed] [Google Scholar]
- Woerther P.-L., Andremont A., Kantele A. (2017). Travel-acquired ESBL-producing Enterobacteriaceae: impact of colonization at individual and community level. J. Travel. Med. 24 S29–S34. 10.1093/jtm/taw101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrsch E. R., Chowdhury P. R., Jarocki V. M., Brandis K. J., Djordjevic S. P. (2020). Duplication and diversification of a unique chromosomal virulence island hosting the subtilase cytotoxin in Escherichia coli ST58. Microbial. Genom. 2020:387. 10.1099/mgen.0.000387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W., Xu T., Qin T., Li P., Liu Y., Kang H., et al. (2016). Characterization of integrons and novel cassette arrays in bacteria from clinical isloates in China, 2000-2014. J. Biomed. Res. 30 292–303. 10.7555/JBR.30.20150153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Echeverria P. (1996). Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64 1441–1445. 10.1128/iai.64.4.1441-1445.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. S., Lee J. C., Kang H. Y., Jeong Y. S., Lee E. Y., Choi C. H., et al. (2004). Prevalence of dfr genes associated with integrons and dissemination of dfrA17 among urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 53 445–450. 10.1093/jac/dkh097 [DOI] [PubMed] [Google Scholar]
- Zankari E., Allesøe R., Joensen K. G., Cavaco L. M., Lund O., Aarestrup F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72 2764–2768. 10.1093/jac/dkx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeinalzadeh N., Salmanian A. H., Goujani G., Amani J., Ahangari G., Akhavian A., et al. (2017). A Chimeric protein of CFA/I, CS6 subunits and LTB/STa toxoid protects immunized mice against enterotoxigenic Escherichia coli. Microbiol. Immunol. 61 272–279. 10.1111/1348-0421.12491 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 39 W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Alikhan N.-F., Mohamed K., Fan Y., Group A. S., Achtman M. (2019). The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. gr. 251678 119. 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberberg M. D., Shorr A. F. (2013). Secular trends in gram-negative resistance among urinary tract infection hospitalizations in the United States, 2000-2009. Infect. Control Hosp. Epidemiol. 34 940–946. 10.1086/671740 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/639663, SAMN15244685 – SAMN15244730.