Abstract
Functional gastrointestinal disorders (FGIDs), currently known as disorders of gut–brain interaction, are emerging microbiota–gut–brain abnormalities that are prevalent worldwide. The pathogenesis of FGIDs is heterogeneous and is intertwined with gut microbiota and its derived molecule‐modulated mechanisms, including gut dysmotility, visceral hypersensitivity, gut immune abnormalities, abnormal secretion, and impaired barrier function. There has been phenomenal progress in understanding the role of gut microbiota in FGIDs by underpinning the species alternations between healthy and pathological conditions such as FGIDs. However, the precise gut microbiota‐directed cellular and molecular pathogeneses of FGIDs are yet enigmatic. Determining the mechanistic link between the gut microbiota and gastrointestinal (GI) diseases has been difficult due to (i) the lack of robust animal models imitating the various aspects of human FGID pathophysiology; (ii) the absence of longitudinal human and/or animal studies to unveil the interaction of the gut microbiota with FGID‐relevant pathogenesis; (iii) uncertainty about connections between human and animal studies; and (iv) insufficient data supporting a holistic view of disease‐specific pathophysiological changes in FGID patients. These unidentified gaps open possibilities to explore pathological mechanisms directed through gut microbiota dysbiosis in FGIDs. The current treatment options for dysbiotic gut microbiota are limited; dietary interventions, antibiotics, probiotics, and fecal microbiota transplantation are the front‐line clinical options. Here, we review the contribution of gut microbiota and its derived molecules in gut homeostasis and explore the possible pathophysiological mechanisms involved in FGIDs leading to potential therapeutics options.
Keywords: disorders of gut–brain interaction, fecal microbiota transplantation, gut immune dysfunction, impaired intestinal barrier function, visceral hypersensitivity
This paper reviews the contribution of gut microbiota and its derived molecules in gut homeostasis and explore the possible pathophysiological mechanisms involved in FGIDs leading to potential therapeutics options. The current knowledge gaps warrant a modern approach for a holistic view to characterize patients based on the multiomic data from the gut microbiome, metabolome, transcriptome, host epigenome, and dietary profiles on longitudinal studies. The integration of these factors with physiological changes will enable us to develop targeted approaches to treat the patients, not only relieving symptoms but also restoring gut homeostasis.
Introduction
Functional gastrointestinal disorders (FGIDs), currently known as disorders of gut–brain interaction, including irritable bowel syndrome (IBS) and functional dyspepsia (FD), are common conditions in clinical practice and community.1 The prevalence of FGIDs is approximately 40% in the world based on a recent survey using the Rome IV diagnostic criteria.2 The spectrum of FGID symptoms includes abdominal pain/burning, bloating, nausea, fullness, vomiting, and altered bowel habit.3 The pathophysiology of FGIDs is complex, but mounting studies indicate that gut microbiota plays a central role in the development of FGID and modulation of the symptoms.4 Understanding of the pathogenesis of FGIDs is evolving with the elucidation of pathophysiological mechanisms at the cellular and molecular levels.5, 6 Not long ago, “functional” conditions, such as FGIDs, were explained as idiopathic, and the patients were suspected by physicians and researchers to be neurotic and/or healthy subjects with an imaginary illness.7 By far, with enhanced pathophysiological knowledge, the best approach to examining patients with FGIDs would be a holistic view of the patients' genetic predisposition, epigenetics, brain connectome, enteric nervous system (ENS), lifestyle and environmental factors, and their interface with gut microbiota.8, 9
Gut microbiota changes quite rapidly in humans because of variations in diets, medication, mode of birth, age, gender, and psychological state.10 The pathogenic potentials of gut microbiota have been well documented as up to 10% of IBS patients had gastroenteritis followed by gut microbiota dysbiosis, resulting in the development of IBS (postinfection IBS).11 The advent of modern technology broadened our understanding of the gut microbiome.12 Next‐generation sequencing has expanded our knowledge of characterizing gut microbial alternations using culture‐independent techniques and functional changes in patients with FGID, although the findings between studies are inconsistent.13, 14, 15 Notably, the most convincing evidence of the role of microbiota comes from the findings of alterations in gut transit, intestinal barrier function, and visceral sensation observed in germ‐free (GF) mice after fecal microbiota transplantation (FMT) from patients with FGIDs.13, 14 Extensive research identifies a knowledge gap regarding whether gut microbiota dysbiosis is a cause or consequence of gut diseases. However, to ensure significant progress and to effectively incorporate gut microbiome science in the clinic, robust longitudinal studies in humans and animal models are warranted to underpin the precise functional roles of microbiota and its derived molecules that link to the specific pathophysiological mechanisms of FGIDs.
This review aims to assess the impacts of gut microbiota and its derived molecules on the pathophysiological mechanisms of FGIDs, including gastrointestinal (GI) dysmotility, impaired barrier dysfunction, GI immune dysfunction, and visceral hypersensitivity. Besides, we summarize the potential therapeutic strategies for gut microbiota dysbiosis in FGIDs, highlighting the gaps and areas for further explorations.
Pathophysiological mechanisms of FGIDs with special emphasis on gut microbiota dysbiosis
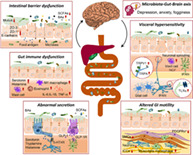
The human gut is home to a heterogeneous microbial system termed the gut microbiome, mediating a wide array of biochemical transformations that modulate host physiology and pathophysiology (Fig. 1).7 Disruption of gut microbial equilibrium can trigger mucosal immune activation, leading to breach in the epithelial barrier, which further results in visceral hypersensitivity and gut motility abnormalities, which are the hallmarks of FGIDs, reinforcing intestinal barrier dysfunction as the core pathophysiology.11, 16 Currently, the majority of gut microbiota‐mediated mechanistic studies related to FGIDs have been performed using animal models.13, 17 The implications of the animal studies enable us to extend the understanding of the crosstalk between the gut microbiota and the underlying pathogenies implicated in FGIDs.
Figure 1.
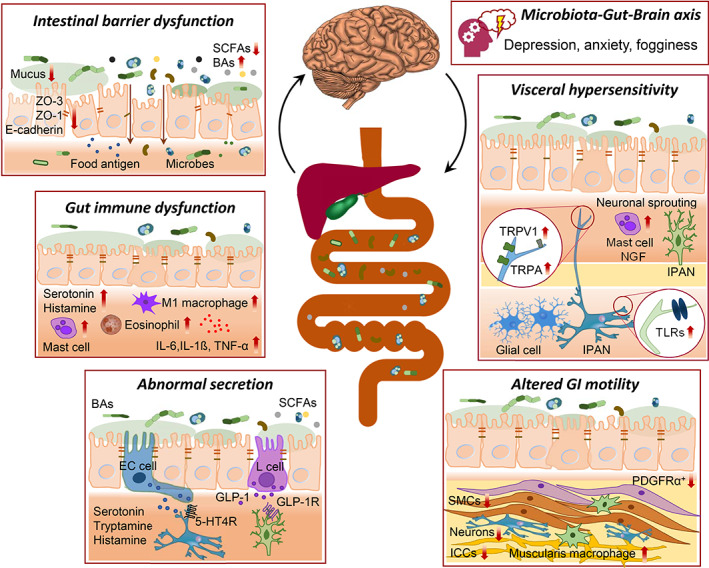
Gut microbiota‐directed pathophysiological mechanisms of functional gastrointestinal disorders (FGIDs), currently known as disorders of gut–brain interaction. 5‐HTR, 5‐hydroxytryptamine receptor; BAs, bile acids; E‐cadherin, epithelial cadherin; GLP‐1R, glucagon‐like peptide‐1 receptor; ICCs, interstitial cells of Cajal; IPAN, intrinsic primary afferent neuron; NGF, nerve growth factor; PDGFRα, platelet‐derived growth factor receptor alpha; SCFAs, short‐chain fatty acids; SMCs, smooth muscle cells; TLRs, toll‐like receptors; TNF‐α, tumor necrosis factor‐alpha; TRPA1, transient receptor potential ankyrin 1; TRPV1, transient receptor potential cation channel subfamily V member 1; ZO, zonula occludens.
Gut microbiota dysbiosis is an emerging pathophysiology of FGIDs
Gut microbiota alterations in the small bowel and colon have been evidenced extensively in patients with FGIDs (Fig. 2).12 Several studies showed that the gut microbiota profile of IBS patients significantly differs from that of healthy subjects. A recent meta‐analysis showed that IBS patients presented higher levels of pathogenic bacteria of the genera Enterobacter and lower commensal bacteria of the genera Bifidobacterium and Lactobacillus compared with the healthy controls.18 A microbiome signature from patients with IBS was found to be decrease in microbial diversity, and the severity of the symptoms was associated with Clostridiales species and/or methanogenic microbes.19 Another study demonstrated the overrepresentation of Methanobravibacter smithii in fecal samples from patients with functional constipation, and constipation‐predominant IBS (IBS‐C).19, 20 One study showed an increased ratio of Firmicutes to Bacteroidetes, increased Streptococcus and Ruminococcus, and decreased Lactobacillus and Bifidobacterium species in patients with IBS.21
Figure 2.
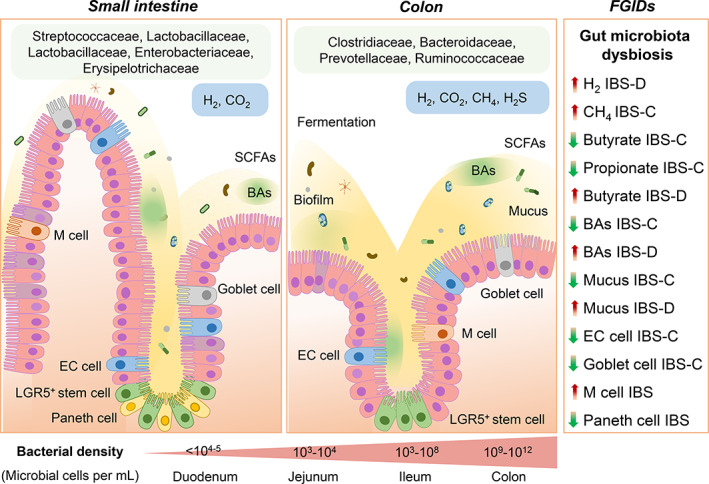
Pathophysiological mechanisms of functional gastrointestinal disorders (FGIDs), currently known as disorders of gut–brain interaction. BAs, bile acids; CH4, methane; EC cell, enterochromaffin cell; H2, hydrogen; H2S, hydrogen sulfide; IBS‐C, constipation‐predominant irritable bowel syndrome; IBS‐D; diarrhea‐predominant irritable bowel syndrome; LGR5+, leucine‐rich repeat‐containing G‐protein coupled receptor 5; M cell, microfold cell; SCFAs, short‐chain fatty acids.
Gut microbiota alterations in the small bowel include an increase in small intestinal bacteria on quantitative culture (≥105 colony‐forming units/mL of jejunal aspirate), defined as small intestinal bacterial overgrowth (SIBO), and low‐grade SIBO (≥103 but <105 colony‐forming units/mL of jejunal aspirate), which have clinical significance.22, 23, 24 Several studies found a strong association between the IBS and SIBO. A meta‐analysis showed a higher relative risk of the SIBO among IBS patients compared with controls.25 A recent study on patients presenting with bloating, abdominal pain, or diarrhea, thought to be suffering from SIBO, found alterations in small intestinal microbial composition.26 Another study showed a significant reduction of the anaerobic genera Veillonella, Prevotella, and Actinomyces in the duodenal mucosal biopsies from patients with FD compared with healthy controls.27 One study demonstrated the absence of Acidobacteria and an increased ratio of Bacteroidetes to Proteobacteria in gastric fluid samples from FD patients.28 However, skepticism about gut microbiota dysbiosis causing IBS still continues, partly because what constitutes a healthy microbiome is unclear. Taken together, this breakthrough evidence indicates that gut microbiota alteration is a common finding in FGIDs and has provided the impetus to whether reforming the gut microbiota composition might be a potential treatment for FGIDs.
Gut microbiota alterations modulating gut motility
Abnormal GI muscular movements driving fast or delayed gut transit is a major pathophysiological mechanism of FGIDs.12 Alterations in gut microbiota due to GI hypomotility can further perpetuate GI dysmotility resulting from gut dysbiosis.16 Gut microbes could alter gut transit; similarly, an irregular gut transit could modify the spatial organization and proportion of the microbiome by creating a luminal microenvironment for the growth of specific bacterial taxa or by affecting bacterial colonization.12 Patients with IBS‐C tend to present slow gut motility, while patients with diarrhea‐predominant IBS (IBS‐D) have relatively faster motility.12 One proof‐of‐concept study demonstrated that GF mice colonized with the fecal samples from IBS‐D patients had accelerated gut transit, impaired intestinal barrier function, gut immune dysfunction, and increased anxiety levels compared to those colonized with fecal samples from healthy controls.13 Another proof‐of‐concept study showed that GF mice colonized with the fecal samples from patients with IBS‐C had slowed gut transit, upregulated 5‐HT transporter, and reduced serotonin (5‐hydroxytryptamine, 5‐HT) levels in the colonic biopsy, which were supported by decreased relative abundance of gut microbiota such as Firmicutes and increased Bacteroidetes.29, 30, 31 These findings support the potential regulatory functions of gut microbiota dysbiosis in gut motility.
Gut microbiota‐derived molecules and gut motility
In addition, several studies showed that gut microbiota‐derived molecules directly regulate gut motility (Fig. 1). These molecules relay signals modulating the function of targeted host cells and subsequently alter the GI functions (Table 1).
Table 1.
Gut microbiota and its derived molecules in functional gastrointestinal disorders (FGIDs), currently known as disorders of gut–brain interaction FGIDs
| Gut microbiota derived molecules | Gut microbes | FGIDs/animal model | Key findings | Pathophysiological mechanism | References |
|---|---|---|---|---|---|
| Propionate and butyrate | Bifidobacterium adolescentis, Roseburia‐E.rectale | Constipation‐predominant irritable bowel syndrome (IBS) (IBS‐C) | Bifidobacterium adolescentis increased and Roseburia‐E.rectale decreased in fecal samples of IBS‐C, possibly regulating the levels of butyrate and ropionate, resulting in slowed gut transit | Altered gut motility | Sun et al.32 |
| Butyrate | Fecalibacterium prausnitzii | Diarrhea‐predominant IBS (IBS‐D) | Fecalibacterium prausnitzii increased in IBS‐D, which is a major producer of butyrate, resulting in accelerated gut transit. Butyrate might be considered a biomarker for IBS‐D | Altered gut motility | Sun et al.32 |
| Butyrate | Blautia obeum ATCC 29174 | IBS‐C | Blautia obeum ATCC 29174 were decreased in IBS‐C and correlated with butyrate levels | Altered gut motility | Bhattarai et al.17 |
| Tryptamine | Ruminococcus gnavus, Clostridium sporogenes | IBS, slow transit constipation (STC) |
Ruminococcus gnavus and Clostridium sporogenes identified as Tryptophan decarboxylase Tryptamine accelerates gut transit and increases colonic secretion by activating epithelial serotonin type 4 (5‐HT4) receptor Genetically engineered bacteria Bacteroides thetaiotaomicron produce tryptamine |
Altered gut motility Abnormal secretion |
Bhattarai et al.17 Williams et al.33 |
| BAs (CDCA, CA) | Blautia wexlerae DSM19850 | IBS‐C |
Reduced bile acid (BA) levels correlated with delayed gut transit Chenodeoxycholic acid (CDCA) was the most frequent associated metabolite of Blautia wexlerae DSM19850 |
Altered gut motility Abnormal secretion |
Mars et al.34 Camilleri et al.35 |
| BAs (CDCA, CA) | Blautia wexlerae DSM19850 | IBS‐D |
Increased BA levels correlated with bowel movements, colonic transit and visceral pain Elevated CDCA levels increased water content in stools from IBS‐D |
Altered gut motility | Mars et al.34 |
| Methane | Methanobravibacter smithii | IBS‐C, STC | Methane (CH4) in the depletes gut serotonin resulting in slowed gut transit and constipation |
Altered gut motility Visceral hypersensitivity |
Triantafyllou et al.36 |
| Hydrogen | Hydrogen‐producing bacteria | Small intestinal bacterial overgrowth (SIBO), IBS‐D | Hydrogen gas correlated with SIBO condition in IBS‐D patients |
Altered gut motility Gut immune dysfunction Visceral hypersensitivity Abdominal bloating and distension |
Ghoshal et al.37 Pimentel et al.38 |
| Hydrogen sulfide | Escherichia coli, Salmonella enterica, Clostridia, and Enterobacter aerogenes | Diarrhea, SIBO, IBS‐D? |
Probably due to formation of sulfuric acid in the cells? Hydrogen sulfide (H2S) might be considered a biomarker for SIBO |
Altered gut motility Abnormal secretion Visceral hypersensitivity Gut immune dysfunction |
Kalantar‐Zadeh et al.39 Pimentel et al.38 Banik et al.40 |
| Lactic acid | Lactobacillus, Bifidobacterium | SIBO, IBS‐D | Possible link between SIBO, brain fogginess, and D‐lactic acidosis. Patients with brain fogginess presented a higher SIBO prevalence with bloating, pain, and distension | Dysregulated microbiota‐gut‐brain axis | Rao et al.39 |
| Alcohol (Ethanol) | Klebsiella pneumonia | Need to explore in FGIDs and SIBO? | Alcohol abuse triggers gut microbiota dysbiosis, mucosal inflammation, and intestinal barrier derangement which corroborates liver damage | Impaired barrier dysfunction, gut immune dysfunction, dysregulated microbiota‐gut–brain axis | Meroni et al.41 |
Short‐chain fatty acids
Short‐chain fatty acids (SCFAs) are primary metabolites produced from gut microbial dietary fiber fermentation. SCFAs, particularly butyrate, acetate, and propionate, are versatile molecules modulating host physiology through various mechanisms, including interactions via specific G‐protein‐coupled receptors.42 SCFAs affect GI physiology, including contractility, visceral sensation, secretion, and intestinal barrier integrity (Figs 1, 2).43 These effects suggest considering the dynamic functions in FGIDs based on microbial activity.44, 45 One study demonstrated that antibiotic‐treated mice colonized with fecal samples of patients with slow‐transit constipation had altered SCFAs profiles and delayed gut transit.46 The authors further showed that butyrate supplementation reversed the delayed gut motility in mice colonized with fecal samples of patients with slow‐transit constipation (Table 1).46 A recent meta‐analysis compared the differences in SCFAs in patients with IBS and found that butyrate and propionate were decreased in patients with IBS‐C, whereas butyrate was increased in patients with IBS‐D compared to healthy controls.32 One study showed increased gene expression of tryptophan hydroxylase 1 (Tph1) and serotonin production in colon biopsy through stimulation of enterochromaffin (EC) cells, a specialized subtype of enteroendocrine cells, via butyrate.47 Taken together, these findings support the fact that SCFAs regulate gut motility.
Serotonin and tryptamine
A study reported that indigenous spore‐forming bacteria from human and mouse promote biosynthesis of serotonin from colonic mucosal EC cells.48 The host as well as the microbes appear benefit from serotonin signaling. A state‐of‐the‐art study suggested that increased levels of gut luminal serotonin increased the relative abundance of spore‐forming members of the gut microbiota. For instance, Turicibacter sanguinis expresses a eukaryotic neurotransmitter sodium symporter‐related protein‐sharing sequence and structure with the mammalian serotonin transporter.49 Another group of researchers detected the mechanism of crosstalk between enteric neurons and the gut microbiota as the stimulus signal for serotonin release and the subsequently activated serotonin type 4 (5‐HT4) receptor, which regulates smooth muscle contraction and relaxation.50 This study demonstrates direct crosstalk between the microbiota and the ENS mediated via serotonin and indicates a potential mechanism linking microbial dysbiosis to FGIDs.
Tryptamine is produced by gut bacteria, particularly Clostridium sporogenes and Ruminococcus gnavus from typtophan.33 One study examined whether tryptamine in the gut can function by facilitating the 5‐HT4 receptor.17 The authors showed that tryptamine increased the ionic flux across the colonic epithelium, as well as fluid secretions in colonoids from GF and humanized mice, validating the physiological function of tryptamine for gut secretion. Furthermore, improved gut motility was observed in GF mice that were colonized with Bacteroides thetaiotaomicron microbes, which were genetically engineered to produce tryptamine.17
Bile acids
Bile acid (BA) pool alternations induced by microbial dysbiosis have been proposed in FGID pathogenesis (Fig. 2).51, 52 In humans, the primary BAs cholic acid (CA) and chenodeoxycholic acid (CDCA) are deconjugated from their glycine or taurine conjugate by microbial bile salt hydrolases through the microbial biotransformation process.53 Microbiota‐ mediated effects on intestinal fluid secretion are the consequences of differential BA biotransformation.11 BA malabsorption is known to be involved in IBS‐D.7 BA malabsorption is observed more in patients with IBS‐D than those with IBS‐C.54 A meta‐analysis demonstrated that approximately 10% patients were IBS‐D had severe BAs malabsorption (425 patients in 5 studies, SeHCAT retention <5% of baseline), 32% patients presented moderate BA malabsorption (1,073 patients in 17 studies, SeHCAT retention <10%), and 26% patients showed mild BA malabsorption (618 patients in 7 studies, SeHCAT retention <15%).55 One study showed that the quantity of BAs (CDCA and CA) were noticeably higher in stool samples from IBS‐D patients compared to IBS‐C patients.34 The authors observed variation in BA signature associated with IBS, with a significantly elevated level of unconjugated primary BAs in stool samples from IBS‐D patients and lower amounts in IBS‐C patients. Another study showed that patients with IBS‐D had elevated BA levels, and it correlated with bowel movements, colonic transit, and visceral pain.35 One study on patients with FD showed reduced duodenal BA concentrations, which was associated with the impaired intestinal barrier function.52, 56
Gut microbiota alterations modulate intestinal gas profile
Carbohydrate fermentation leads to the formation of hydrogen (H2) and carbon dioxide (CO2), which constitute intestinal gases produced by gut microbiota (Fig. 2).38, 39 Variations in the intestinal gas profile have been consistently associated with FGIDs.57 A few studies, using computed tomography scans, showed increased luminal gas in patients with FGIDs during a flare of abdominal distension that might or might not result from consumption of a “high‐flatulence” diet.58 Numerous studies highlighted the contribution of gut microbiota to the alterations of intestinal gases.41, 59
Gut microbiota dysbiosis, which includes a quantitative alteration called SIBO and qualitative variations, is common among FGID patients and is known to be associated with abdominal distension, flatulence and bloating.60 Dysbiotic gut microbiota may cause these symptoms due to carbohydrate fermentation followed by substantial gas production.60 The intestinal gases diffuse into the systemic circulation and are exhaled in the breath, such as H2 and methane (CH4) (Table 1).7, 39 An increase of ≥20 parts per million (PPM) of H2 or >10 PPM of CH4 above the baseline after glucose and within 90 min after lactulose ingestion on breath tests suggest the presence of SIBO, are associated with GI symptoms, for instance, abdominal bloating and/or distension.7, 61 A few studies showed an association between high breath CH4 and constipation.36 Methanogenic microbes in the colon produce CH4, which depletes serotonin, resulting in slowed gut transit and constipation.36 Another study showed the association of H2 gas mainly with IBS‐D.37 Some gut microbes such as Escherichia coli, Salmonella enterica, Clostridia, and Enterobacter aerogenes produce hydrogen sulfide (H2S) in the colon.62 H2S is quickly absorbed in the colonocytes and forms sulfuric acid, which is highly toxic to the cells, resulting in altered gut motility, abnormal secretion, visceral hypersensitivity, and gut immune dysfunction (Table 1).38, 40 One recent study showed that there was “brain fogginess” due to D‐lactic acidosis in patients with SIBO.59 Another study showed that gut bacteria such as Klebsiella pneumonia produces alcohol, which might modulate brain and intestinal barrier functions.41
Thus, the aforementioned findings suggested that gut microbes and their derived molecules are essential to maintain proper GI function. These signaling molecules further enlighten the understanding of host and gut microbiota crosstalk, alluding to potential therapeutic options for FGIDs.
Gut microbiota alterations modulating gut immune function
Gut immune dysregulation has been implicated in the pathogenesis of subsets of patients with FGIDs.3 A few studies showed an increased number of gut immune cells such as macrophages, mast cells, eosinophils, and T cells in patients with FGIDs.63 Mast cells, upon activation, release tryptase, histamine, cytokines, and prostaglandins, which have been implicated in altering nociceptive pathways and intestinal barrier functions in FGIDs.64 A breach of the intestinal barrier allows infiltration of luminal antigens, activating the immune response, and contributes to the severity of the symptom in FGIDs through neuroimmune dysregulation.65
There is a plethora of evidence supporting gut immune system activation triggered by the gut microbiota.11 Several studies showed increased expression of proinflammatory cytokines following interactions between bacterial components and pattern recognition receptors, including toll‐like receptors (TLRs), which play essential roles in the innate immune system's ability to recognize structurally conserved molecules derived from microbes.66 One study on patients with IBS showed that the expression levels of proinflammatory cytokine IL‐6, TLR4, TLR5, C‐X‐C motif chemokine ligand‐11, and CXC‐3 receptor (CXCR) were elevated in colonic biopsies of patients compared to controls; on the contrary, the expression levels of anti‐inflammatory cytokine IL‐10 were downregulated.67 The authors confirmed a positive correlation between the number of gene copies of the commensal bacteria such as Bifidobacteria, Lactobacillus, and IL‐10.67 This study further demonstrated a positive correlation between weekly stool frequency and the expression levels of TLR4 and CXCR‐3; in contrast, it was negatively correlated with IL‐10.67 Another study showed that IBS patients with SIBO presented higher mucosal IL‐1 α and β levels than those without SIBO, and the elevated IL‐1 β levels were associated with abdominal bloating and loose stool.68 Studies on mucosal T cell alterations in FGIDs are scant. One study showed reduced expression of lymphocyte marker, Fas cell surface death receptor (FAS), and human leukocyte antigen‐DR isotype (HLA‐DR) in patients with FD, while duodenal eosinophilia has been implicated in dyspeptic symptom generation.69 Another study showed increased small bowel‐homing CCR9 T lymphocytes and α4β7 integrin, which correlated with the symptom severity in patients with FD.70 This evidence strongly suggests that patients with FGIDs experience gut immune activation influenced by microbiota alternations, reinforcing low‐grade inflammation as a potential mechanism of FGIDs.
Gut microbiota alterations modulating intestinal barrier function
The gut luminal–mucosal interface represents the place where food particles, immunogenic and toxic molecules, and gut bacteria and their molecules challenge the scrutiny of the gut mucosa‐associated immune system.7 As a core pathophysiological mechanism, intestinal barrier dysfunction leads to mucosal immune response activation, which modulates GI motility in relation to the severity of symptoms of FGIDs.64 The gut epithelium is a single layer of cells that acts as a selective barrier with astonishing dynamics, preventing the passage of detrimental luminal contents while allowing the absorption of water, electrolytes, and dietary nutrients.71
The mucus layer overlaying the gut epithelial cells serves as a primary factor in protecting the host by providing a strong immunologic and physical barrier against pathogens.71 The mucus layer is a reservoir of antimicrobial immunoglobulins and peptides and is enriched in polysaccharides, a major nutrient source for different populations of bacteria.72 Furthermore, the intestinal barrier is protected by the epithelial tight junction proteins, which are multifunctional complexes that form a seal between adjacent intestinal epithelial cells near the apical surface and seal the paracellular space, thus preventing gut luminal contents from entering the subepithelial space.71 The starvation of microbiota, for instance, with decreased fiber consumption might increase the reliance of microbiota on the mucus polysaccharides, leading to mucus layer degradation and increasing pathogen susceptibility, which further leads to immune activation.73 A few studies showed that alterations in mucosa‐associated bacteria, for instance, Ruminococcus gnavus, Akkermansia muciniphila, and Ruminococcus torques, were associated with mucus integrity and intestinal secretion.74 Furthermore, in susceptible conditions, a harmful pathogen or the consumption of certain foods impairs intestinal barrier function by altering tight junctions, resulting from proteasome‐mediated degradation triggered by inflammatory mediators including proteases, eicosanoids, and histamine.75 Several studies showed that the expression of adhesion proteins, zonula occludens‐1 and occludin, was significantly lower in duodenal biopsy samples with decreased transepithelial resistance and impaired intestinal barrier function in FGID patients compared to controls.76, 77 Recently, a new endoscopic technology, confocal laser endomicroscopy and endocytoscopy, enables us to carry out a high‐resolution assessment of GI mucosal histology at cellular and subcellular levels and obtain “optical biopsies” of the endoluminal surface.78 Exploiting the endomicroscopy, one study reported duodenal mucosal epithelial breaks and increased intervillous spaces when the mucosa was challenged with foods in patients with IBS.79 In the setting of impaired barrier function, the pathogens or food antigens cross the intestinal barrier and trigger a T helper 2 (TH2) immune response that results in the activation of immune responses mediated via eosinophils and mast cells.64 Mast cells and eosinophils act as antigen‐presenting cells to TH2 lymphocytes, which leads to eosinophil and mast cell degranulation close to nerve fibers, which is responsible for muscle contraction and abdominal pain.80 TH2 cells also trigger immunoglobulin (Ig) class switching in B cells and elevate IgE antibody expression, suggesting an increased number of IgE+ B cells in conditions associated with leaky gut, including FGIDs.81 A recent study showed an increased number of IgE+ mast cells in colonic tissue positively correlated with visceral hypersensitivity and intestinal barrier dysfunction in IBS patients.80 Thus, a healthy gut barrier function may play a fundamental role in gut homeostasis, microbial colonization resistance, community resilience, host defense, drug modifications, and food digestion.
Gut microbiota alterations modulating visceral sensation
The “taste” and “smell” of food particles, the gut bacteria, and their derived molecules could be sensed and transduced through the enteroendocrine cells and intrinsic nerves via specific receptors.82 Abnormal activation of a mucosal immune response affects visceral sensitivity in relation to the degree of inflammatory response accompanied by the proximity of immune cells and sensory neurons.83 Visceral hypersensitivity is a critical contributor to the occurrence and severity of symptoms in FGIDs.3 It manifests as an amplified perception of mechanochemical stimuli applied to the gut, which results in the exaggerated sensation of pain and burning.7 The modulators of visceral sensation include transient receptor potential vanilloid subtype 1 (TRPV1) and the transient receptor potential ankyrin 1 channel (TRPA1), serotonin, histamine, tachykinin, cannabinoid, acid‐sensing ion channel, protease activated receptors, and voltage‐gated calcium and sodium channels.84, 85 Activation of TRPV1 by capsaicin, acidic pH, thermal stimulus, nerve growth factor (NGF), prostaglandins, inflammatory mediators, and microbes triggers the release of neuropeptides, such as substance P and calcitonin gene‐related peptide‐1.84
Several studies showed that the upregulated expression of TRPV1 in colonic biopsies was positively correlated with abdominal pain in IBS‐D patients.86, 87 One recent study showed a positive correlation between an increase in Escherichia coli abundance and visceral hypersensitivity followed by induction of hypersensitivity in response to Escherichia coli gavage in Paneth cell‐defect mice.88 One state‐of‐the‐art study showed that EC cells were activated via the stimuli through TRPA1 and led to voltage‐gated calcium channel‐dependent serotonin release, which might be a potential target for the management of FGIDs.85 The gut epithelial cells are observed to provide a barrier between the nerve fibers and lumen, suggesting that TRPA1 serves as the primary detector of luminal irritants in the EC cells prior to direct submucosal damage. Further studies explored a mechanistic link contributing to visceral hypersensitivity by demonstrating the induction of epithelial cannabinoid and μ‐opioid receptors after administration of Lactobacillus strains; regulation of peripheral and central neuronal pathways and antinociceptive effects through the inhibition of TRPV1 after treatment with Lactobacillus reuteri have been shown.89, 90
Gut microbiota modulates neuroimmunoendocrine interface
The neuron, immune, and endocrine regulations are anatomically and physiologically interconnected in the gut.91 Microbiota‐derived molecules could be detected by TLRs and expressed in host cells, particularly neurons, glial cells, macrophages and dendritic cells, and enteroendocrine cells, which specifically recognize microbe‐associated molecular patterns.66 For instance, TLR4 recognizes the lipopolysaccharide of Gram‐negative bacteria regulating ENS integrity, as well as neuromuscular function.92 TLR2 detects lipoteichoic acid from Gram‐positive bacteria and promoted neurogenesis after antibiotic treatment.
A recent study demonstrated that gut microbiota potentially affects embryonic neuronal development. This study showed that GF mice display an altered prenatal transcriptomic profile and activation of brain microglial cells, which are critical for neuronal development.93 Patients with FGIDs showed increased numbers of mast cells and lamina propria T lymphocytes in close proximity to submucosal neuronal plexus, with decreased responsiveness of the neurons.94 Mast cell infiltration in the colonic mucosa in IBS patients triggers neuronal sprouting, resulting in the release of NGF, which promotes topical neoinnervation and angiogenesis.94, 95 Duodenal fine nerve fibers were observed more often in IBS patients compared to healthy controls, and the degree of mast cell activation positively correlated with the grade of nerve fiber sprouting.95
Microfold (M) cells are presented in the epithelial lining of mucosa‐associated lymphoid tissues, for instance, Peyer's patches (PPs) of the gut. Luminal antigens are actively transported through M cells to the underlying lymphoid follicles to propagate an immune response.96 This study demonstrated that TRPV1+ neurons promoted gut immune regulation against the pathogenic bacteria Salmonella typhimurium. Another study suggested that the TRPV1+ nociceptors regulated the density of M cells in ileum PP follicle‐associated epithelia to limit entry points for pathogenic bacteria.96
Enteroendocrine cells represent less than 1% of the gut epithelium, and they are critical to maintain gut hemostasis.97 Accumulating evidence shows that interplay between enteroendocrine cells and enteric neurons modulate gut physiology via an extended axon‐like structure termed “neuropod” that directly crosstalks with neurons through modified synapses.98 EC cells secrete serotonin in response to a wide variety of microbiota‐derived molecules in the lumen.85 EC cells were found to be significantly increased in rectal biopsies from patients with IBS.99 One recent study showed that TLR9 colocalized with the satiety hormone cholecystokinin (CCK) in the enteroendocrine cells of the small intestine.100 The authors found that pathogenic bacteria, such as Shigella flexneri, Escherichia coli, and Salmonella typhimurium, evoke CCK secretion in TLR9‐expressing enteroendocrine cells.100
Gut microbiota modulates gut–brain axis
Dysregulation of the gut–brain axis is one of the primary pathophysiological mechanisms in a subset of patients with FGIDs.4, 101 Gut microbiota‐regulated alterations in epithelial barrier function triggered by immune system activation resulting in abnormal gut–brain communications.102 Stress‐related corticotropin‐releasing hormones have an important role in intestinal barrier function.103 In addition, eosinophils release corticotropin‐releasing hormone and substance P, which might result in the activation of mast cells and, subsequently, dysfunction of the intestinal epithelial barrier.64 Magnetic resonance imaging studies on FGIDs patients demonstrated that abnormal structural and functional connectivity in the brain were responsible for processing visceral afferent transduction.8, 104 Gut microbial alterations could modulate the function of neurotransmitters, such as dopamine, serotonin, γ‐aminobutyric acid, and acetylcholine, either by synthesis or consumption, resulting in changes in emotional state and behavior.105 Furthermore, BAs and their receptors have been identified in the brain, reinforcing a potential mechanistic role in the gut–brain axis.106
Psychological comorbid conditions, including stress, anxiety, or depression, are associated with FGIDs and contribute to the pathophysiology as part of an integrated biopsychosocial model.107 Within this construct, the bidirectional communication of the gut–brain axis is critical in FGIDs. The brain alters gut physiology, such as visceral sensitivity and motility, mediating symptoms of FGIDs.108 Meanwhile, changes in the gut provide feedback to the brain, influencing psychological well‐being.108 A meta‐analysis showed high prevalence of both depressive disorders and anxiety (23%), anxiety (39%), and depressive (29%) symptoms in IBS patients.109 Psychological comorbidity could be a consequence of chronic GI disease burden and reduced quality of life, and its role in the pathophysiology of FGIDs is still unequivocally broader.
Microbiota‐directed therapeutic interventions for FGIDs
The current treatment strategies to manipulate gut microbiota dysbiosis include probiotics, antibiotics, fecal microbiota transplantation (FMT), and dietary interventions.
Probiotics and antibiotics
The efficacy of probiotic treatment for FGIDs symptoms has been documented but lacks consistency in symptom improvement. Numerous studies suggested the benefit of specific probiotic strains, such as Bifidobacterium animalis DN‐173010154, Bifidobacterium lactis DN‐173, and multispecies probiotics in patients with IBS.110 One study showed that treatment with Lactobacillus gasseri OLL2716 shifted the microbial community composition in gastric fluid as found in healthy subjects.111 Other studies demonstrated that treatment with a probiotic containing Bifidobacterium lactis accelerated gut transit and improved symptoms in IBS‐C patients.110
Interventional studies reported a decrease in breath CH4 through treatment with either rifaximin and neomycin or rifaximin alone, which led to improvement in constipation in IBS‐C and functional constipation patients.112, 113 Randomized controlled trials on FD patients showed that rifaximin treatment was associated with improvements in dyspeptic symptoms, including abdominal bloating/fullness and belching.114 Furthermore, systematic reviews and meta‐analysis of rifaximin and other antibiotics showed improvement in SIBO.20, 115 These studies reinforce the fundamental role of gut microbiota in FGIDs.
Fecal microbiota transplantation
FMT is considered to be beneficial in managing microbial dysbiosis due to its ability to restore “healthy” microbes in patients.116 However, whether FMT as a treatment for FGIDs is a panacea or placebo is still controversial and warrants a precise definition of gut microbial dysbiosis and the constitution of healthy microbes.117, 118 A recent meta‐analysis of randomized controlled trials showed no significant symptom improvement in IBS patients after FMT compared to placebo.119 In contrast, a double‐blind controlled trial in IBS patients showed improvement in symptom severity after FMT compared to placebo.120 However, more rigorous and multicentric larger trials are warranted on this issue.
Dietary intervention
The safest treatment intended to resume the healthy gut microbiota would be dietary intervention.121, 122 A few randomized controlled trials showed that diets low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) are helpful to treat IBS.123, 124 A few studies showed that vegetarian diets rich in fiber would increase SCFA production, which further inhibits pathogenic bacterial colonization in the gut.125, 126
Future treatment strategies
As the field moves toward modern science utilizing a rigorous, robust design, large and multicentric longitudinal human studies might be exploited to integrate multiomic data including physiology and pathophysiological mechanisms to explore unique features of symptom response for individual treatment. Such insight would lead to better stratification of therapeutic approaches, leading to the targeted restoration of altered physiological functions in a subgroup of patients with FGIDs. Gut microbiota and its derived molecules might be key drivers of the FGID pathogenesis. Through the elucidation of the steps involved in the synthesis, the release and consumption of these molecules might explore novel therapeutics avenues. The therapeutic intervention of the gut microbiome might include (i) interventions of replacing an entire gut microbial community with an optimized healthy community127; (ii) introducing single specific microbes that might restore normal gut functions by producing critical gut microbiota‐derived molecules for specific pathophysiology, such as genetically engineered bacteria Bacteroides thetaiotaomicron that produce tryptamine, which results in increased intestinal secretion and accelerate gut transit17; and (iii) inhibiting and/or removing specific microbes or microbial processes through inhibitors of enzymatic pathways that are specific to the gut microbiota.128 Furthermore, the prior screening for SIBO/gut microbiota dysbiosis in FGID patients before antibiotic treatment may lead to better therapeutic response. Unselected patients with IBS treated with antibiotics showed a response rate of 40%.129 In contrast, according to a novel Indian study, IBS patients with SIBO on upper gut aspirate culture selected for antibiotics treatment showed a response rate as high as 87%.130 These improved antibiotics responses were able to be reproduced in subsequent studies from China and the United States using the lactulose H2 breath test, although studies with a larger sample size are needed on this issue.131, 132
Conclusion and further directions
State‐of‐the‐art studies showed the importance of gut microbiota and its derived molecules in the modulation of major pathophysiological mechanisms in FGIDs. There is substantial progress in characterizing the gut microbiota species in healthy and patients with FGIDs. However, the translational approach of gut microbiome research warrants robust and multicentric longitudinal studies in animals and humans to elucidate the precise role of gut microbes that can be therapeutically targeted to underpin each pathophysiology in FGIDs.15 Restoring healthy gut microbiota would maintain the microbiota–gut–brain axis in homeostasis from motility to mood, but it is important to keep in mind that clinical outcomes are more important compared to mechanistic parameters alone.117
Furthermore, the biggest challenge for gut microbiome‐based treatment is the use of broad‐spectrum antibiotics, leading to the scorched earth effect on the gut microbiome, which has faced challenges due to skepticism related to antibiotic resistance, though, hitherto unproven.133 Therefore, alternative interventions for antibiotics might be considered, such as optimized healthy gut microbiota and/or its metabolites, which may benefit the patients by hampering the pathogenic bacteria. Traditional approaches to treat patients with FGIDs have beneficial effects in general. Due to the enormous heterogeneous nature of FGIDs, without consideration of the focus on the underlying pathophysiological mechanisms mediated through gut microbiota alterations, it would likely dilute the impact of current therapeutic strategies. The current knowledge gaps warrant a modern approach for a holistic view to characterize patients based on the multiomics data from the gut microbiome, metabolome, transcriptome, host epigenome, and dietary profiles based on longitudinal studies. The integration of these factors with physiological changes will enable us to develop targeted approaches to treat the patients, not only relieving symptoms but also restoring gut homeostasis.
Lai Wei and Rajan Singh equally contributed to this work.
Declaration of conflict interest: None
Contributor Information
Seungil Ro, Email: sro@med.unr.edu.
Uday C Ghoshal, Email: udayghoshal@gmail.com.
References
- 1.Ghoshal UC, Singh R. Frequency and risk factors of functional gastro‐intestinal disorders in a rural Indian population. J. Gastroenterol. Hepatol. 2017; 32: 378–87. [DOI] [PubMed] [Google Scholar]
- 2.Sperber AD, Bangdiwala SI, Drossman DAet al. Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome Foundation Global Study. Gastroenterology. 2021; 160: 99–114 e3. [DOI] [PubMed] [Google Scholar]
- 3.Black CJ, Drossman DA, Talley NJ, Ruddy J, Ford AC. Functional gastrointestinal disorders: advances in understanding and management. Lancet. 2020; 396: 1664–74. [DOI] [PubMed] [Google Scholar]
- 4.Ghoshal UC. Gut microbiota‐brain axis modulation by a healthier microbiological microenvironment: facts and fictions. J. Neurogastroenterol. Motil. 2018; 24: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R, Ha SE, Wei Let al. miR‐10b‐5p rescues diabetes and gastrointestinal dysmotility. Gastroenterology. 2021. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei L, Singh R, Ha SEet al. Serotonin deficiency is associated with delayed gastric emptying. Gastroenterology. 2021. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghoshal UC. Marshall and Warren Lecture 2019: a paradigm shift in pathophysiological basis of irritable bowel syndrome and its implication on treatment. J. Gastroenterol. Hepatol. 2020; 35: 712–21. [DOI] [PubMed] [Google Scholar]
- 8.Ford AC, Mahadeva S, Carbone MF, Lacy BE, Talley NJ. Functional dyspepsia. Lancet. 2020; 396: 1689–702. [DOI] [PubMed] [Google Scholar]
- 9.Ford AC, Sperber AD, Corsetti M, Camilleri M. Irritable bowel syndrome. Lancet. 2020; 396: 1675–88. [DOI] [PubMed] [Google Scholar]
- 10.Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017; 171: 1481–93. [DOI] [PubMed] [Google Scholar]
- 11.Barbara G, Feinle‐Bisset C, Ghoshal UCet al. The intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology. 2016; 150: 1305–1318.e8. [DOI] [PubMed] [Google Scholar]
- 12.Shin A, Preidis GA, Shulman R, Kashyap PC. The gut microbiome in adult and pediatric functional gastrointestinal disorders. Clin. Gastroenterol. Hepatol. 2019; 17: 256–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Palma G, Lynch MD, Lu Jet al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017; 9: eaaf6397. [DOI] [PubMed] [Google Scholar]
- 14.Edogawa S, Edwinson AL, Peters SAet al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut. 2020; 69: 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mars RAT, Frith M, Kashyap PC. Functional gastrointestinal disorders and the microbiome‐what is the best strategy for moving microbiome‐based therapies for functional gastrointestinal disorders into the clinic? Gastroenterology. 2021; 160: 538–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Zogg H, Wei Let al. Gut microbial dysbiosis in the pathogenesis of gastrointestinal dysmotility and metabolic disorders. J. Neurogastroenterol. Motil. 2021; 27: 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattarai Y, Williams BB, Battaglioli EJet al. Gut microbiota‐produced tryptamine activates an epithelial G‐protein‐coupled receptor to increase colonic secretion. Cell Host Microbe. 2018; 23: 775–785.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Alammar N, Singh Ret al. Gut microbial dysbiosis in the irritable bowel syndrome: a systematic review and meta‐analysis of case‐control studies. J. Acad. Nutr. Diet. 2020; 120: 565–86. [DOI] [PubMed] [Google Scholar]
- 19.Ghoshal U, Shukla R, Srivastava D, Ghoshal UC. Irritable bowel syndrome, particularly the constipation‐predominant form, involves an increase in Methanobrevibacter smithii, which is associated with higher methane production. Gut Liver. 2016; 10: 932–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoshal UC, Srivastava D, Misra A. A randomized double‐blind placebo‐controlled trial showing rifaximin to improve constipation by reducing methane production and accelerating colon transit: a pilot study. Indian J. Gastroenterol. 2018; 37: 416–23. [DOI] [PubMed] [Google Scholar]
- 21.Rodino‐Janeiro BK, Vicario M, Alonso‐Cotoner Cet al. A review of microbiota and irritable bowel syndrome: future in therapies. Adv. Ther. 2018; 35: 289–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghoshal UC, Ghoshal U. Small intestinal bacterial overgrowth and other intestinal disorders. Gastroenterol. Clin. North Am. 2017; 46: 103–20. [DOI] [PubMed] [Google Scholar]
- 23.Ghoshal UC, Goel A, Quigley EMM. Gut microbiota abnormalities, small intestinal bacterial overgrowth, and non‐alcoholic fatty liver disease: an emerging paradigm. Indian J. Gastroenterol. 2020; 39: 9–21. [DOI] [PubMed] [Google Scholar]
- 24.Ghoshal UC, Baba CS, Ghoshal Uet al. Low‐grade small intestinal bacterial overgrowth is common in patients with non‐alcoholic steatohepatitis on quantitative jejunal aspirate culture. Indian J. Gastroenterol. 2017; 36: 390–9. [DOI] [PubMed] [Google Scholar]
- 25.Ghoshal UC, Nehra A, Mathur A, Rai S. A meta‐analysis on small intestinal bacterial overgrowth in patients with different subtypes of irritable bowel syndrome. J. Gastroenterol. Hepatol. 2020; 35: 922–31. [DOI] [PubMed] [Google Scholar]
- 26.Rao SSC, Bhagatwala J. Small intestinal bacterial overgrowth: clinical features and therapeutic management. Clin. Transl. Gastroenterol. 2019; 10: e00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong L, Shanahan ER, Raj Aet al. Dyspepsia and the microbiome: time to focus on the small intestine. Gut. 2017; 66: 1168–9. [DOI] [PubMed] [Google Scholar]
- 28.Igarashi M, Nakae H, Matsuoka Tet al. Alteration in the gastric microbiota and its restoration by probiotics in patients with functional dyspepsia. BMJ Open Gastroenterol. 2017; 4: e000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, Liu X, An Yet al. Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci. Rep. 2017; 7: 10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang H, Liu Y, Chen Zet al. miR‐222 regulates cell growth, apoptosis, and autophagy of interstitial cells of Cajal isolated from slow transit constipation rats by targeting c‐kit. Indian J. Gastroenterol. 2021. (in press). [DOI] [PubMed] [Google Scholar]
- 31.Singh R, Wei L, Ghoshal UC. Micro‐organic basis of functional gastrointestinal (GI) disorders: role of microRNAs in GI pacemaking cells. Indian J. Gastroenterol. 2021. (in press). [DOI] [PubMed] [Google Scholar]
- 32.Sun Q, Jia Q, Song L, Duan L. Alterations in fecal short‐chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta‐analysis. Medicine (Baltimore). 2019; 98: e14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams BB, Van Benschoten AH, Cimermancic Pet al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014; 16: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mars RAT, Yang Y, Ward Tet al. Longitudinal multi‐omics reveals subset‐specific mechanisms underlying irritable bowel syndrome. Cell. 2020; 182: 1460–1473.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camilleri M, Oduyebo I, Halawi H. Chemical and molecular factors in irritable bowel syndrome: current knowledge, challenges, and unanswered questions. Am. J. Physiol. Gastrointest. Liver Physiol. 2016; 311: G777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J. Neurogastroenterol. Motil. 2014; 20: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoshal UC, Park H, Gwee KA. Bugs and irritable bowel syndrome: the good, the bad and the ugly. J. Gastroenterol. Hepatol. 2010; 25: 244–51. [DOI] [PubMed] [Google Scholar]
- 38.Pimentel M, Mathur R, Chang C. Gas and the microbiome. Curr. Gastroenterol. Rep. 2013; 15: 356. [DOI] [PubMed] [Google Scholar]
- 39.Kalantar‐Zadeh K, Berean KJ, Burgell RE, Muir JG, Gibson PR. Intestinal gases: influence on gut disorders and the role of dietary manipulations. Nat. Rev. Gastroenterol. Hepatol. 2019; 16: 733–47. [DOI] [PubMed] [Google Scholar]
- 40.Banik GD, De A, Som Set al. Hydrogen sulphide in exhaled breath: a potential biomarker for small intestinal bacterial overgrowth in IBS. J. Breath Res. 2016; 10: 026010. [DOI] [PubMed] [Google Scholar]
- 41.Meroni M, Longo M, Dongiovanni P. Alcohol or gut microbiota: who is the guilty? Int. J. Mol. Sci. 2019; 20: 4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priyadarshini M, Kotlo KU, Dudeja PK, Layden BT. Role of short chain fatty acid receptors in intestinal physiology and pathophysiology. Compr. Physiol. 2018; 8: 1091–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalile B, Van Oudenhove L, Vervliet Bet al. The role of short‐chain fatty acids in microbiota‐gut‐brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019; 16: 461–78. [DOI] [PubMed] [Google Scholar]
- 44.Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018; 362: eaat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurst NR, Kendig DM, Murthy KS, Grider JR. The short chain fatty acids, butyrate and propionate, have differential effects on the motility of the guinea pig colon. Neurogastroenterol. Motil. 2014; 26: 1586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge X, Zhao W, Ding Cet al. Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci. Rep. 2017; 7: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reigstad CS, Salmonson CE, Rainey JF 3rdet al. Gut microbes promote colonic serotonin production through an effect of short‐chain fatty acids on enterochromaffin cells. FASEB J. 2015; 29: 1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yano JM, Yu K, Donaldson GPet al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015; 161: 264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fung TC, Vuong HE, Luna CDGet al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 2019; 4: 2064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Vadder F, Grasset E, Manneras Holm Let al. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. U.S.A. 2018; 115: 6458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishima Y, Ishihara S. Molecular mechanisms of microbiota‐mediated pathology in irritable bowel syndrome. Int. J. Mol. Sci. 2020; 21: 8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beeckmans D, Riethorst D, Augustijns Pet al. Altered duodenal bile salt concentration and receptor expression in functional dyspepsia. United European Gastroenterol. J. 2018; 6: 1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016; 7: 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegyi P, Maleth J, Walters JRet al. Guts and gall: bile acids in regulation of intestinal epithelial function in health and disease. Physiol. Rev. 2018; 98: 1983–2023. [DOI] [PubMed] [Google Scholar]
- 55.Wedlake L, A'Hern R, Russell Det al. Systematic review: the prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea‐predominant irritable bowel syndrome. Aliment. Pharmacol. Ther. 2009; 30: 707–17. [DOI] [PubMed] [Google Scholar]
- 56.Beeckmans D, Farre R, Riethorst Det al. Relationship between bile salts, bacterial translocation, and duodenal mucosal integrity in functional dyspepsia. Neurogastroenterol. Motil. 2020; 32: e13788. [DOI] [PubMed] [Google Scholar]
- 57.Lacy BE, Cangemi D, Vazquez‐Roque M. Management of chronic abdominal distension and bloating. Clin. Gastroenterol. Hepatol. 2021; 19: 219–31 e1. [DOI] [PubMed] [Google Scholar]
- 58.Bendezu RA, Barba E, Burri Eet al. Intestinal gas content and distribution in health and in patients with functional gut symptoms. Neurogastroenterol. Motil. 2015; 27: 1249–57. [DOI] [PubMed] [Google Scholar]
- 59.Rao SSC, Rehman A, Yu Set al. Brain fogginess, gas and bloating: a link between SIBO, probiotics and metabolic acidosis. Clin. Transl. Gastroenterol. 2018; 9: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takakura W, Pimentel M. Small intestinal bacterial overgrowth and irritable bowel syndrome – an update. Front. Psych. 2020; 11: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rezaie A, Buresi M, Lembo Aet al. Hydrogen and methane‐based breath testing in gastrointestinal disorders: the North American Consensus. Am. J. Gastroenterol. 2017; 112: 775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh SB, Lin HC. Hydrogen sulfide in physiology and diseases of the digestive tract. Microorganisms. 2015; 3: 866–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matricon J, Meleine M, Gelot Aet al. Review article: associations between immune activation, intestinal permeability and the irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012; 36: 1009–31. [DOI] [PubMed] [Google Scholar]
- 64.Wouters MM, Vicario M, Santos J. The role of mast cells in functional GI disorders. Gut. 2016; 65: 155–68. [DOI] [PubMed] [Google Scholar]
- 65.Camilleri M, Madsen K, Spiller R, van Meerveld BG, Verne GN. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol. Motil. 2012; 24: 503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007; 449: 819–26. [DOI] [PubMed] [Google Scholar]
- 67.Shukla R, Ghoshal U, Ranjan P, Ghoshal UC. Expression of toll‐like receptors, pro‐, and anti‐inflammatory cytokines in relation to gut microbiota in irritable bowel syndrome: the evidence for its micro‐organic basis. J. Neurogastroenterol. Motil. 2018; 24: 628–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srivastava D, Ghoshal U, Mittal RD, Ghoshal UC. Associations between IL‐1RA polymorphisms and small intestinal bacterial overgrowth among patients with irritable bowel syndrome from India. Neurogastroenterol. Motil. 2014; 26: 1408–16. [DOI] [PubMed] [Google Scholar]
- 69.Gargala G, Lecleire S, Francois Aet al. Duodenal intraepithelial T lymphocytes in patients with functional dyspepsia. World J. Gastroenterol. 2007; 13: 2333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liebregts T, Adam B, Bredack Cet al. Small bowel homing T cells are associated with symptoms and delayed gastric emptying in functional dyspepsia. Am. J. Gastroenterol. 2011; 106: 1089–98. [DOI] [PubMed] [Google Scholar]
- 71.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019; 68: 1516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Motta JP, Wallace JL, Buret AGet al. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021. (in press). [DOI] [PubMed] [Google Scholar]
- 73.Desai MS, Seekatz AM, Koropatkin NMet al. A dietary fiber‐deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016; 167: 1339–1353.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020; 69: 2232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bischoff SC, Barbara G, Buurman Wet al. Intestinal permeability – a new target for disease prevention and therapy. BMC Gastroenterol. 2014; 14: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JY, Kim N, Choi YJet al. Expression of tight junction proteins according to functional dyspepsia subtype and sex. J. Neurogastroenterol. Motil. 2020; 26: 248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertiaux‐Vandaele N, Youmba SB, Belmonte Let al. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am. J. Gastroenterol. 2011; 106: 2165–73. [DOI] [PubMed] [Google Scholar]
- 78.Turcotte JF, Kao D, Mah SJet al. Breaks in the wall: increased gaps in the intestinal epithelium of irritable bowel syndrome patients identified by confocal laser endomicroscopy (with videos). Gastrointest. Endosc. 2013; 77: 624–30. [DOI] [PubMed] [Google Scholar]
- 79.Fritscher‐Ravens A, Schuppan D, Ellrichmann Met al. Confocal endomicroscopy shows food‐associated changes in the intestinal mucosa of patients with irritable bowel syndrome. Gastroenterology. 2014; 147: 1012–20 e4. [DOI] [PubMed] [Google Scholar]
- 80.Aguilera‐Lizarraga J, Florens MV, Viola MFet al. Local immune response to food antigens drives meal‐induced abdominal pain. Nature. 2021; 590: 151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 2003; 3: 331–41. [DOI] [PubMed] [Google Scholar]
- 82.Sternini C, Anselmi L, Rozengurt E. Enteroendocrine cells: a site of 'taste' in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008; 15: 73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barbara G, Stanghellini V, De Giorgio Ret al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004; 126: 693–702. [DOI] [PubMed] [Google Scholar]
- 84.Farzaei MH, Bahramsoltani R, Abdollahi M, Rahimi R. The role of visceral hypersensitivity in irritable bowel syndrome: pharmacological targets and novel treatments. J. Neurogastroenterol. Motil. 2016; 22: 558–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bellono NW, Bayrer JR, Leitch DBet al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell. 2017; 170: 185–198.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akbar A, Yiangou Y, Facer P, Walters JRF, Anand P, Ghosh S. Increased capsaicin receptor TRPV1‐expressing sensory fibres in irritable bowel syndrome and their correlation with abdominal pain. Gut. 2008; 57: 923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes PA, Brierley SM, Martin CMet al. TRPV1‐expressing sensory fibres and IBS: links with immune function. Gut. 2009; 58: 465–6. [DOI] [PubMed] [Google Scholar]
- 88.Riba A, Olier M, Lacroix‐Lamande Set al. Paneth cell defects induce microbiota dysbiosis in mice and promote visceral hypersensitivity. Gastroenterology. 2017; 153: 1594–1606.e2. [DOI] [PubMed] [Google Scholar]
- 89.Rousseaux C, Thuru X, Gelot Aet al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 2007; 13: 35–7. [DOI] [PubMed] [Google Scholar]
- 90.Wouters MM, Balemans D, Van Wanrooy Set al. Histamine receptor H1‐mediated sensitization of TRPV1 mediates visceral hypersensitivity and symptoms in patients with irritable bowel syndrome. Gastroenterology. 2016; 150: 875–87 e9. [DOI] [PubMed] [Google Scholar]
- 91.Furness JB, Kunze WA, Clerc N. Nutrient tasting and signaling mechanisms in the gut. II. The intestine as a sensory organ: neural, endocrine, and immune responses. Am. J. Physiol. 1999; 277: G922–8. [DOI] [PubMed] [Google Scholar]
- 92.Yarandi SS, Kulkarni S, Saha Met al. Intestinal bacteria maintain adult enteric nervous system and nitrergic neurons via toll‐like receptor 2‐induced neurogenesis in mice. Gastroenterology. 2020; 159: 200–213 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joly A, Leulier F, De Vadder F. Microbial modulation of the development and physiology of the enteric nervous system. Trends Microbiol. 2020. (in press). [DOI] [PubMed] [Google Scholar]
- 94.Barbara G, Wang B, Stanghellini Vet al. Mast cell‐dependent excitation of visceral‐nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 2007; 132: 26–37. [DOI] [PubMed] [Google Scholar]
- 95.Buhner S, Li Q, Vignali Set al. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology. 2009; 137: 1425–34. [DOI] [PubMed] [Google Scholar]
- 96.Lai NY, Musser MA, Pinho‐Ribeiro FAet al. Gut‐innervating nociceptor neurons regulate Peyer's patch microfold cells and SFB levels to mediate salmonella host defense. Cell. 2020; 180: 33–49 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Worthington JJ, Reimann F, Gribble FM. Enteroendocrine cells‐sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2018; 11: 3–20. [DOI] [PubMed] [Google Scholar]
- 98.Kaelberer MM, Rupprecht LE, Liu WW, Weng P, Bohórquez DV. Neuropod cells: the emerging biology of gut‐brain sensory transduction. Annu. Rev. Neurosci. 2020; 43: 337–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaelberer MM, Buchanan KL, Klein MEet al. A gut‐brain neural circuit for nutrient sensory transduction. Science. 2018; 361: eaat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Daly K, Burdyga G, Al‐Rammahi Met al. Toll‐like receptor 9 expressed in proximal intestinal enteroendocrine cells detects bacteria resulting in secretion of cholecystokinin. Biochem. Biophys. Res. Commun. 2020; 525: 936–40. [DOI] [PubMed] [Google Scholar]
- 101.Margolis KG, Cryan JF, Mayer EA. The microbiota‐gut‐brain axis: from motility to mood. Gastroenterology. 2021. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut‐brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015; 28: 203–9. [PMC free article] [PubMed] [Google Scholar]
- 103.Rodino‐Janeiro BK, Alonso‐Cotoner C, Pigrau Met al. Role of corticotropin‐releasing factor in gastrointestinal permeability. J. Neurogastroenterol. Motil. 2015; 21: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guleria A, Karyampudi A, Singh Ret al. Mapping of brain activations to rectal balloon distension stimuli in male patients with irritable bowel syndrome using functional magnetic resonance imaging. J. Neurogastroenterol. Motil. 2017; 23: 415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Needham BD, Kaddurah‐Daouk R, Mazmanian SK. Gut microbial molecules in behavioural and neurodegenerative conditions. Nat. Rev. Neurosci. 2020; 21: 717–31. [DOI] [PubMed] [Google Scholar]
- 106.McMillin M, DeMorrow S. Effects of bile acids on neurological function and disease. FASEB J. 2016; 30: 3658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Oudenhove L, Crowell MD, Drossman DAet al. Biopsychosocial aspects of functional gastrointestinal disorders. Gastroenterology. 2016; 150: 1355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quigley EMM. Microbiota‐brain‐gut axis and neurodegenerative diseases. Curr. Neurol. Neurosci. Rep. 2017; 17: 94. [DOI] [PubMed] [Google Scholar]
- 109.Zamani M, Alizadeh‐Tabari S, Zamani V. Systematic review with meta‐analysis: the prevalence of anxiety and depression in patients with irritable bowel syndrome. Aliment. Pharmacol. Ther. 2019; 50: 132–43. [DOI] [PubMed] [Google Scholar]
- 110.Aragon G, Graham DB, Borum M, Doman DB. Probiotic therapy for irritable bowel syndrome. Gastroenterol. Hepatol. (NY). 2010; 6: 39–44. [PMC free article] [PubMed] [Google Scholar]
- 111.Koga Y, Ohtsu T, Kimura K, Asami Y. Probiotic L. gasseri strain (LG21) for the upper gastrointestinal tract acting through improvement of indigenous microbiota. BMJ Open Gastroenterol. 2019; 6: e000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Low K, Hwang L, Hua J, Zhu A, Morales W, Pimentel M. A combination of rifaximin and neomycin is most effective in treating irritable bowel syndrome patients with methane on lactulose breath test. J. Clin. Gastroenterol. 2010; 44: 547–50. [DOI] [PubMed] [Google Scholar]
- 113.Pimentel M, Chatterjee S, Chow EJ, Park S, Kong Y. Neomycin improves constipation‐predominant irritable bowel syndrome in a fashion that is dependent on the presence of methane gas: subanalysis of a double‐blind randomized controlled study. Dig. Dis. Sci. 2006; 51: 1297–301. [DOI] [PubMed] [Google Scholar]
- 114.Iovino P, Bucci C, Tremolaterra F, Santonicola A, Chiarioni G. Bloating and functional gastro‐intestinal disorders: where are we and where are we going? World J. Gastroenterol. 2014; 20: 14407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gatta L, Scarpignato C. Systematic review with meta‐analysis: rifaximin is effective and safe for the treatment of small intestine bacterial overgrowth. Aliment. Pharmacol. Ther. 2017; 45: 604–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016; 13: 508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shanahan F, Ghosh TS, O'Toole PW. The healthy microbiome‐what is the definition of a healthy gut microbiome? Gastroenterology. 2021; 160: 483–94. [DOI] [PubMed] [Google Scholar]
- 118.Pulipati P, Sarkar P, Jakkampudi Aet al. The Indian gut microbiota‐Is it unique? Indian J. Gastroenterol. 2020; 39: 133–40. [DOI] [PubMed] [Google Scholar]
- 119.Ianiro G, Eusebi LH, Black CJ, Gasbarrini A, Cammarota G, Ford AC. Systematic review with meta‐analysis: efficacy of faecal microbiota transplantation for the treatment of irritable bowel syndrome. Aliment. Pharmacol. Ther. 2019; 50: 240–8. [DOI] [PubMed] [Google Scholar]
- 120.El‐Salhy M, Hatlebakk JG, Gilja OHet al. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double‐blind, placebo‐controlled study. Gut. 2020; 69: 859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delzenne NM, Bindels LB. Food for thought about manipulating gut bacteria. Nature. 2020; 577: 32–4. [DOI] [PubMed] [Google Scholar]
- 122.Walker AW, Ince J, Duncan SHet al. Dominant and diet‐responsive groups of bacteria within the human colonic microbiota. ISME J. 2011; 5: 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mehtab W, Agarwal A, Singh N, Malhotra A, Makharia GK. All that a physician should know about FODMAPs. Indian J. Gastroenterol. 2019; 38: 378–90. [DOI] [PubMed] [Google Scholar]
- 124.Altobelli E, Del Negro V, Angeletti PMet al. Low‐FODMAP diet improves irritable bowel syndrome symptoms: a meta‐analysis. Nutrients. 2017; 9: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tomova A, Bukovsky I, Rembert Eet al. The effects of vegetarian and vegan diets on gut microbiota. Front. Nutr. 2019; 6: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018; 23: 705–15. [DOI] [PubMed] [Google Scholar]
- 127.Zhao L, Zhang F, Ding Xet al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018; 359: 1151–6. [DOI] [PubMed] [Google Scholar]
- 128.Roberts AB, Gu X, Buffa JAet al. Development of a gut microbe‐targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018; 24: 1407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pimentel M, Lembo A, Chey WDet al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N. Engl. J. Med. 2011; 364: 22–32. [DOI] [PubMed] [Google Scholar]
- 130.Ghoshal UC, Srivastava D, Misra A, Ghoshal U. A proof‐of‐concept study showing antibiotics to be more effective in irritable bowel syndrome with than without small‐intestinal bacterial overgrowth: a randomized, double‐blind, placebo‐controlled trial. Eur. J. Gastroenterol. Hepatol. 2016; 28: 281–9. [DOI] [PubMed] [Google Scholar]
- 131.Rezaie A, Heimanson Z, McCallum R, Pimentel M. Lactulose breath testing as a predictor of response to rifaximin in patients with irritable bowel syndrome with diarrhea. Am. J. Gastroenterol. 2019; 114: 1886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhuang X, Tian Z, Luo M, Xiong L. Short‐course rifaximin therapy efficacy and lactulose hydrogen breath test in Chinese patients with diarrhea‐predominant irritable bowel syndrome. BMC Gastroenterol. 2020; 20: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Le Bastard Q, Vangay P, Batard Eet al. US immigration is associated with rapid and persistent acquisition of antibiotic resistance genes in the gut. Clin. Infect. Dis. 2020; 71: 419–21. [DOI] [PubMed] [Google Scholar]


