Supplemental Digital Content is available in the text.
Keywords: endothelial cells; gene expression; hemodynamics; microscopy, fluorescence; RNA-seq; single-cell analysis; transcriptome
Objective:
Endothelial cells (ECs) that form the innermost layer of all vessels exhibit heterogeneous cell behaviors and responses to pro-angiogenic signals that are critical for vascular sprouting and angiogenesis. Once vessels form, remodeling and blood flow lead to EC quiescence, and homogeneity in cell behaviors and signaling responses. These changes are important for the function of mature vessels, but whether and at what level ECs regulate overall expression heterogeneity during this transition is poorly understood. Here, we profiled EC transcriptomic heterogeneity, and expression heterogeneity of selected proteins, under homeostatic laminar flow.
Approach and Results:
Single-cell RNA sequencing and fluorescence microscopy were used to characterize heterogeneity in RNA and protein gene expression levels of human ECs under homeostatic laminar flow compared to nonflow conditions. Analysis of transcriptome variance, Gini coefficient, and coefficient of variation showed that more genes increased RNA heterogeneity under laminar flow relative to genes whose expression became more homogeneous, although small subsets of cells did not follow this pattern. Analysis of a subset of genes for relative protein expression revealed little congruence between RNA and protein heterogeneity changes under flow. In contrast, the magnitude of expression level changes in RNA and protein was more coordinated among ECs in flow versus nonflow conditions.
Conclusions:
ECs exposed to homeostatic laminar flow showed overall increased heterogeneity in RNA expression levels, while expression heterogeneity of selected cognate proteins did not follow RNA heterogeneity changes closely. These findings suggest that EC homeostasis is imposed post-transcriptionally in response to laminar flow.
Highlights.
Endothelial cells exposed to homeostatic laminar flow showed overall increased heterogeneity in RNA expression levels via single-cell profiling.
Expression heterogeneity of selected cognate proteins did not follow RNA heterogeneity changes closely.
Results indicate that gene expression homeostasis in endothelial cells under flow is imposed post-transcriptionally.
Endothelial cells (ECs) drive new blood vessel formation. Once vessels form, EC line all conduits, where they form and maintain a barrier to solute leak, and participate in blood clotting and other homeostatic functions.1–3 The ability to modulate the relative heterogeneity of an EC cohort in terms of signaling and cell behaviors is critical to vascular function.4 For example, during sprouting angiogenesis, the formation of new blood vessels from preexisting vessels, it is important that some EC migrate while others proliferate to expand both the mass and complexity of the network.1 These differential cellular behaviors result from heterogeneous responses to incoming signals, accompanied by differential gene expression in selected signaling components within an EC cohort.5–7
In contrast, EC respond to remodeling cues, including mechanical input from laminar blood flow, to synchronize signaling responses, as they adopt similar cellular morphologies and become quiescent. This change promotes homeostatic functions such as barrier formation, and lesions that perturb EC responses to homeostatic inputs are often lethal and/or lead to diseases such as atherosclerosis.8,9 For example, as vessels mature, Notch signaling becomes arterial and relatively equalized in magnitude,10 while heterogeneous Notch signaling is a hallmark of early network sprouting.11 EC align along the direction of blood flow and morphologically appear more homogeneous under homeostatic laminar shear stress both in vivo and in vitro,8 suggesting a more homogeneous functional status of EC. However, this heterogeneity loss as EC transition from sprouting to homeostasis has not yet been quantified at the genome level.
Genome-wide expression heterogeneity of EC cohorts is not revealed by assays that average signals, such as bulk RNA or protein profiling; thus, we applied single-cell RNA sequencing (RNA-seq) to EC under homeostatic laminar flow to determine transcriptomic heterogeneity, and immunofluorescence with single-cell resolution to interrogate protein expression heterogeneity. Surprisingly, we found overall increased RNA expression heterogeneity under laminar flow, and that protein expression heterogeneity did not closely follow RNA heterogeneity, suggesting that flow-mediated EC homeostasis is imposed post-transcriptionally.
Materials and Methods
Data Availability
The single-cell RNA sequencing data supporting the findings of this study have been made publicly available at the GEO database under the accession number GSE151867.
Cell Culture and Laminar Flow
Human umbilical vein ECs (HUVEC, Lonza, C2519A) were cultured in endothelial growth medium (basal medium-2 (EBM-2, Lonza CC-3156) supplemented with Endothelial Cell Growth Medium (EGM)-2 Bullet Kit (CC-3162; Lonza). For flow, 5×105 passage 4 HUVECs were seeded onto µ channel slides (ibidi, 80176) coated with 5 µg/mL human plasma fibronectin (Roche, 11080938001) in growth medium overnight, then the medium was replaced with EBM-2 supplemented with 10% fetal bovine serum (FBS, Gibco, 26140-087). At 4- to 6-hour post medium change, static samples were collected for single-cell RNA-seq or fixed for immunofluorescence, and flow samples were exposed to 15 dyn/cm2 laminar shear stress for 72 hours (homeostatic laminar flow, ibidi system). After flow, ECs were either collected for single-cell RNA-seq or fixed for immunofluorescence. Mouse embryo fibroblasts (MEFs) were isolated as previously described12 from E13.5 CD1 mouse embryos, plated into 0.1% gelatin-coated dishes, and cultured in DMEM (Gibco, 11995-065) with 10% FBS. Antibiotics (Gibco, 15240-062) were used in all culture media.
Immunofluorescence
HUVEC in channel slides were fixed in 4% paraformaldehyde in PBS for 10 minutes, permeabilized in 0.1% Triton X-100 in PBS for 5 minutes, blocked in 5% donkey serum in PBS (blocking buffer) for 30 minutes, sequentially incubated with primary and secondary antibodies, or a mixed solution of DAPI and conjugated antibodies in blocking buffer for 1 hour each, then mounted with 80% glycerol in PBS. Cells were washed 3× with PBS for 5 minutes each between steps, and all incubations were at room temperature. Concentrations of primary and secondary antibodies used are in Major Resources Table. A control sample without primary antibody was included in each experiment. Z-stack images were acquired with an Olympus FV3000 confocal microscope (Olympus) at 60×, and total fluorescence intensity (sum of all slices in the Z stack) of each image was quantified using the Fiji version of ImageJ (NIH). For nuclear staining, only nuclear pixels were quantified by creating a mask using the DAPI channel. For each independent experiment, >100 cells from 5 images were quantified. Gini coefficient of total fluorescence intensity was calculated with the Gini function in the package ineq in R.
Single-Cell RNA-Seq
For each condition, 2 biological replicates were collected in 2 independent experiments. HUVEC under flow or nonflow static conditions were digested with 0.05% trypsin-EDTA (Gibco, 25300-054) at 37 °C for 3 minutes, neutralized with 50% newborn calf serum (Gibco, 16010-159) in PBS, collected with a 1 mL syringe, pelleted by centrifugation at 100×g and 4 °C for 5 minutes, washed once with 0.04% BSA in PBS, filtered with a Flowmi 40 µm cell strainer (Sigma, BAH136800040), counted using Countess (Life Technology, AMQAX1000), diluted to 1000 cells/µL with 0.04% BSA in PBS, mixed with 10% MEF in 0.04% BSA/PBS, and loaded onto 10X Genomics Chromium Controller for droplet generation. MEFs were added to HUVEC to (1) determine multiplet rate via mixed-species design; (2) control for technical variability in post-MEF addition steps. HUVECs from 3 ibidi channel slides were combined for each static or flow sample. Viability of HUVEC was >95% via trypan blue staining. Four thousand cells (3600 HUVECs, 400 MEFs) per sample were loaded. Reverse transcription and library preparation were performed with the Single Cell 3′ Library & Gel Bead Kit v2 according to 10X Genomics’ standard protocol, except that 13 cycles were used for cDNA amplification. Libraries of 2 flow experiments (Flow1 and Flow2) and 2 nonflow experiments (Static1 and Static2) were combined and sequenced in one NextSeq500 run using the high output kit at the UNC Translational Genomics Lab (TGL) core. Each sample provided ≈1 to 1.5×108 150 bp paired-end reads. See experimental workflow in Figure IA in the Data Supplement.
Bioinformatic Analysis
Raw reads were mapped to the merged genome of mm10 (mouse) and hg38 (human) using Cell Ranger (10X Genomics). Reads containing the same unique molecular identifier were collapsed, then assigned to individual cells via barcodes and analyzed in R. The following quality control steps were implemented: (1) mouse-human multiplets containing both mouse and human reads were excluded (Figure IB in the Data Supplement). Total multiplet rate including cross-species multiplets and intraspecies multiplets were ≈0.34% based on %MEF and observed cross-species multiplets/sample (Figure IC in the Data Supplement). (2) Outliers with <200 detected genes/cell were excluded as low-quality or debris. (3) Outliers with low (<0.6%) or high (>10%) levels of mitochondria-encoded genes were excluded as cell debris (low) or dying cells (high). (4) Genes whose expression was undetectable in any HUVEC were also excluded, resulting in 20 722 genes in the dataset. A total of 5251 high-quality HUVEC passed all filters, with ≈10 000 unique molecular identifier counts/cell. Gene expression normalization was performed with the LogNormalize function in Seurat13 in R: unique molecular identifier counts were normalized to 10 000 total counts/cell before the logarithm was taken with an added pseudo-count. Principle component analysis (Figure ID and IE in the Data Supplement) and t-distributed Stochastic Neighbor Embedding (tSNE, Figure IF in the Data Supplement) analysis of MEF collected from all 4 samples was performed with Seurat13 in R, and cells were well mixed in the low-dimensional representation, suggesting minimal technical batch effect during droplet generation, library preparation, and sequencing.
Boxplots and violin plots of normalized gene expression were generated by R with the geom_boxplot function of ggplot2 and the VlnPlot function of Seurat, respectively. Curated gene expression level fold change between static and flow conditions was exported from Seurat for bar plots. Gene ontology analysis was performed with DAVID v6.8.14 Dimension reduction by Uniform Manifold Approximation and Projection and graph-based clustering of HUVEC were performed with Seurat15 in R. Data from the 2 replicates were integrated using the canonical correlation analysis by Seurat. Complete overlap of cells from the 2 replicates on Uniform Manifold Approximation and Projection indicates good reproducibility.
Statistical Analysis
Differential tests of Gini coefficient (Gini) and coefficient of variation (CV) were performed with DESCEND.16 DESCEND is a statistical framework specifically designed to recover the true cross-cell gene expression distribution in single-cell RNA-seq data. The integer-valued single-cell RNA-seq read counts do not follow a normal distribution. DESCEND models the complex gene-specific expression distribution with a point mass at zero and a nonzero component belonging to an exponential family of distributions, with the zero-inflated Poisson as a special case. This deconvolved expression distribution allows unbiased estimates of distributional properties such as dispersion (CV and Gini coefficient) and nonzero fraction. The noise model and estimation accuracy of DESCEND has been validated in the original DESCEND article with 9 previously published single-cell RNA-seq datasets including one side-by-side single-cell RNA-seq and RNA fluorescence in situ hybridization study. To confirm that DESCEND has proper type I error (false positive) control in our dataset, we randomly split cells in experiment 1 into 2 sets and ran DESCEND to compare gene expression heterogeneity in these 2 sets. DESCEND found no genes showing significantly different Gini or CV between the 2 sets of cells, confirming good type I error control and no false positives by DESCEND. To accurately recover the true gene expression distribution, genes with overall low expression were filtered out based on: fraction of nonzero counts <5%, number of nonzero counts <20, or mean unique molecular identifier <0.15. 5370 genes passed the filters, and their true expression distribution in each replicate was deconvolved by DESCEND. Permutation-based differential tests were then performed for Flow1 versus Static1 and Flow2 versus Static2, followed by multiple hypothesis testing correction with the Benjamini & Hochberg method in the R package stats to obtain FDR-adjusted P. In parallel, differential tests of Gini and CV between the flow and the static population as a whole were also performed by including batch as a covariate.
Differential tests of gene expression levels between static and flow conditions were performed with the FindMarkers function in Seurat using the Wilcoxon rank-sum test with default settings (log fold change threshold of 0.25 and Bonferroni corrected P threshold of 10−6). Protein expression data are representative of multiple independent experiments. Statistical tests were 1-way ANOVA followed by Tukey multiple comparison correction, paired t test and ratio paired t tests in Prism 8.0.1. Due to the small sample size, the normality and variance were not tested to determine whether the applied parametric tests were appropriate.
Results
ECs Have Increased Transcriptomic Heterogeneity Under Laminar Flow
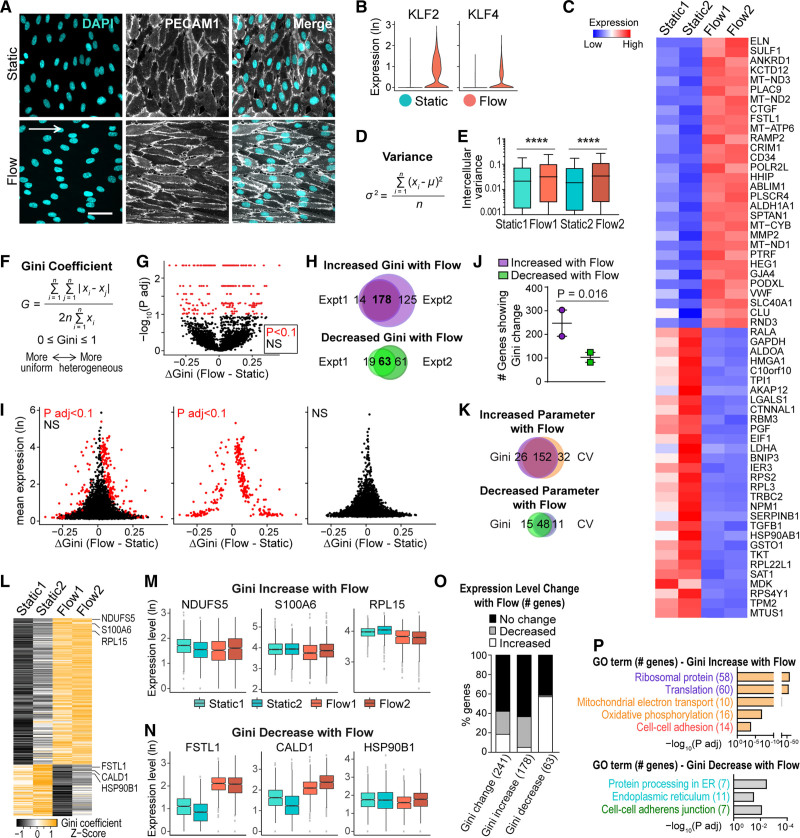
EC become more homogeneous when exposed to homeostatic laminar flow, such as is found in the larger arteries of the body, based on the morphological appearance of EC as they align to the shear stress gradient, and the fact that some signaling (ie, Notch) is less heterogeneous under laminar flow. We hypothesized that flow-stimulated EC morphological changes were accompanied by homogenization of the transcriptome. To examine the relative heterogeneity of EC at the RNA level with homeostatic laminar flow, we applied 15 dyn/cm2 laminar shear stress to HUVEC for 72 hours, which leads to strong EC alignment and morphological homogeneity (Figure 1A). We then performed single-cell RNA-seq on flowed HUVEC and nonflowed HUVEC controls (called static) using the 10x Genomics platform (Figure IA in the Data Supplement). Over 20 000 genes were detected in 5251 high-quality HUVEC, with a low doublet rate (<0.5%) and good sequencing depth (≈3000 detected genes/cell, Figure IB through IF and IIA in the Data Supplement, see Methods for quality control). Correlation analysis of pseudo-bulk samples assembled from single-cell RNA-seq data showed a much higher correlation between biological replicates than across different conditions, suggesting minimal batch effects and high reproducibility (Figure IIB and IIC in the Data Supplement). Significant expression of the EC markers PECAM1 and CDH5 (VE-cadherin) was detected, while expression of markers of several other lineages were low or undetectable, confirming the EC identity of the analyzed cells (Figure IID in the Data Supplement). Upregulation of 2 known flow-responsive genes, KLF2 and KLF4,17 was seen in HUVEC exposed to homeostatic laminar flow (Figure 1B), confirming appropriate transcriptional flow responses under these conditions. Additionally, 70% or more of the top 30 flow upregulated genes and flow downregulated genes in our dataset (Figure 1C) correlate with trends in at least one other published dataset of flow-regulated genes.18–22
Figure 1.
Single-cell RNA-seq shows endothelial cell (EC) transcriptomic heterogeneity under homeostatic laminar flow. A, Human umbilical vein ECs (HUVEC) under indicated conditions labeled for cell-cell borders (PECAM1 [platelet and endothelial cell adhesion molecule 1], white) and nuclei (DAPI, blue). White arrow, direction of flow; Scale bar, 50 µm. B–P, Single-cell RNA-seq of HUVEC under indicated conditions. B, Distribution of normalized gene expression of KLF2 (Kruppel-like factor 2) and KLF4 (Kruppel-like factor 4). C, Heatmap showing normalized expression of top 30 upregulated and downregulated genes (ranked by fold change) in flow vs static cells. D, Variance formula. E, Intercellular variance of HUVEC transcriptome under indicated conditions. Statistics, 1-way ANOVA then Tukey multiple comparisons test. ****P<0.0001. F, Gini coefficient formula. G–K, Gini was calculated for each gene with DESCEND (deconvolution of single-cell expression distribution),16 and differential test of Gini between conditions was performed. G, Each gene’s Gini change plotted against adjusted P. Red dots, significant genes (P adj <0.1); black dots, nonsignificant genes. H, Venn plots showing gene overlaps of significant Gini change in 2 independent experiments. I, Each gene’s Gini change plotted against mean expression. J, Number of genes showing Gini change in each experiment. Statistics, ratio paired t test. K, Venn plots showing overlap of genes with significant Gini (overlapping genes in H) vs coefficient of variation (overlapping genes in Figure IIIG in the Data Supplement) changes. L, Heatmap of Gini coefficient of genes showing significantly increased or decreased Gini (overlapping genes in H). M and N, Box plots of overall expression levels of representative genes with Gini increase (M) or decrease (N). Hinges correspond to first and third quartiles of expression levels. Gini is indicated by the width of box plots relative to y axis. Differential tests of gene expression levels performed with the Wilcoxon rank-sum test (P adj <10−6). O, Bar plot showing percentage and direction of gene expression level changes of genes showing Gini change. P, Representative gene ontology (GO) terms enriched in genes showing Gini increase/decrease with flow.
We tested our hypothesis that laminar flow leads to reduced heterogeneity of RNA transcript levels by assessing EC transcriptomic heterogeneity using 3 different measurements: variance, CV, and Gini coefficient (Gini). We first calculated the intercellular variance (Figure 1D) of each gene in each sample. Surprisingly, a higher overall variance was observed for genes in both replicates of HUVEC under flow compared with static controls (Figure 1E), suggesting that EC have increased transcriptomic heterogeneity under homeostatic laminar flow. However, since variance can be influenced by gene expression levels, we additionally tested our hypothesis with CV (Figure IIIA in the Data Supplement) and Gini (Figure 1F), both of which are unitless and do not suffer from mean-variance dependence. Since Gini is more robust to extreme outliers than CV,23 we focused on Gini but also tested CV to determine congruence between the 2 measurements. We calculated Gini for each gene and performed differential tests with DESCEND,16 a statistical framework specifically designed for single-cell RNA-seq data (Figure 1G). We used 2 strategies to ensure that true biological variation was measured without batch effect as a confounding issue. We first tested each replicate separately and found highly consistent results between replicates, showing largely overlapping lists of genes with significant Gini changes between flow and static conditions (Figure 1H), suggesting good reproducibility of our data. The second strategy tested the replicates together but with batch as a covariate, and this comparison also led to highly consistent results (Figure IIIB and IIIC in the Data Supplement), suggesting that true equality (Gini) change of RNA expression was measured. We then plotted Gini change between flow and static HUVEC against mean gene expression levels or nonzero fraction; these comparisons suggested that significant Gini changes were not driven by large changes in gene expression or “dropout” effect of single-cell RNA-seq, since the number of genes with significant Gini change was not biased towards the top of the y axis (Figure 1I, Figure IIID in the Data Supplement). Similar to the variance measurement, a higher number of genes have more heterogeneous expression under flow (178 genes increased Gini) than genes that have more uniform expression under flow (63 genes decreased Gini, Figure 1H and 1J), also suggesting increased EC transcriptomic heterogeneity under homeostatic laminar flow. Finally, we calculated CV and performed a differential test as for Gini (Figure IIIE through IIIJ in the Data Supplement). The results showed that more genes exhibited increased transcriptomic heterogeneity under flow (184 genes increased CV) than decreased transcriptomic heterogeneity (59 genes decreased CV, Figure IIIG in the Data Supplement). Moreover, the lists of genes showing significant CV changes between flow and static conditions largely overlap with those showing Gini changes (Figure 1K, Figure IIIJ in the Data Supplement). In summary, results from 3 heterogeneity measurements consistently indicate an overall increased transcriptomic heterogeneity of EC under homeostatic laminar flow. These findings refute our hypothesis that loss of heterogeneity at the morphological level is accompanied by homogenization of the RNA transcriptome.
To further examine EC transcriptional heterogeneity in response to laminar flow, we ranked individual genes showing significant Gini changes under flow (Figure 1L). Top-ranked genes showing increased Gini under flow were individually plotted by experiment and condition to confirm Gini increase in flow experiments, as evidenced by increased spread along the y axis (Figure 1M). Likewise, top-ranked genes with decreased Gini showed decreased spread along the y axis in EC exposed to laminar flow (Figure 1N). We next calculated the percentage of genes whose Gini change are accompanied by expression level changes (Figure 1O). This analysis showed that a majority (58%) of the genes showing significant Gini changes had no overall expression changes. When we further broke down by the direction of Gini change, genes showing Gini increase with flow were less correlated with overall expression changes than genes showing Gini decrease, with 63% percent of Gini increase genes showing no significant expression change. Overall, this analysis indicates that the increased flow-induced transcriptomic variance in EC resulted from changes unlinked to overall expression level changes. Gene Ontology analysis of the genes showing significant Gini change under flow revealed that protein translation, oxidative metabolism, and cell-cell adhesion categories were enriched in the genes with upregulated Gini, while protein processing and cell-cell adherens junctions categories were enriched in genes with downregulated Gini (Figure 1P), suggesting that some categories of genes may harbor more Gini changes under flow, and that cell adhesion and adherens junctions may be differentially affected, although the overall limited number of genes in this category preclude drawing strong conclusions from this analysis.
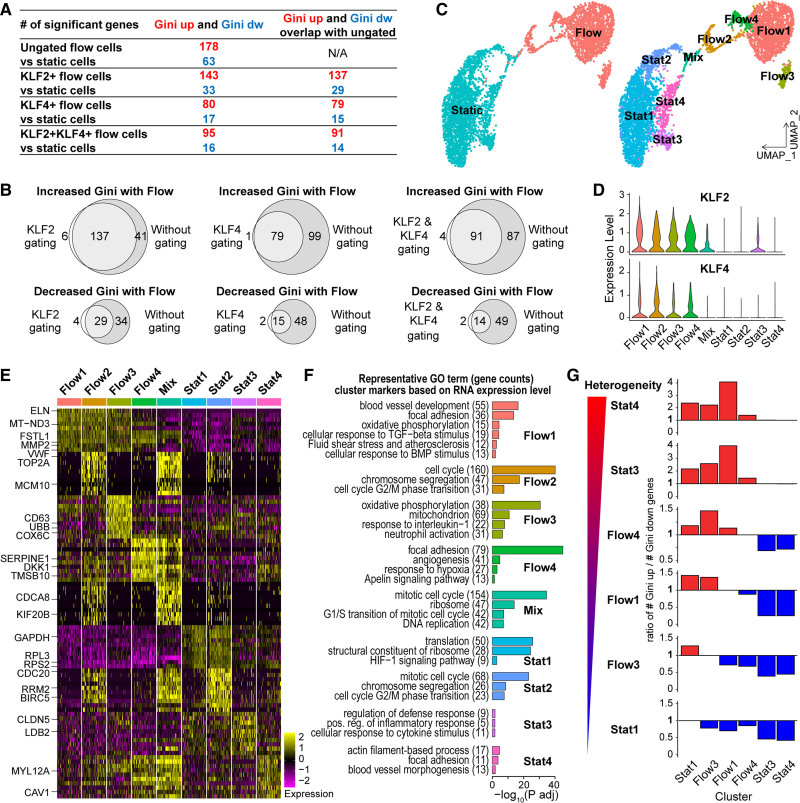
We next asked whether there are EC subpopulations whose gene expression heterogeneity differs in response to laminar shear stress. We first analyzed EC binned by flow-responsiveness via upregulation of KLF2, KLF4, or both genes. We compared gene expression heterogeneity of these subsets to EC under static conditions and found that more genes showed increased Gini than showed decreased Gini, regardless of the gating criteria (Figure 2A). The relationship is similar to the initial comparison of the entire populations, suggesting that EC flow-responsiveness does not significantly change the Gini relationships. The list of genes showing significant Gini changes in the flow-responsive subpopulation are also highly overlapping with the original gene lists ungated for flow-responsiveness (Figure 2B). Both of these relationships were reproduced when CV was analyzed (data not shown). These comparisons suggest that EC subpopulations with different flow responses do not have different Gini or CV relationships based on this distinction.
Figure 2.
Subpopulation analysis of endothelial cell (EC) under homeostatic laminar flow. A and B, Human umbilical vein ECs (HUVEC) flow samples gated based on expression of KLF2, KLF4, or both; differential Gini test performed between static and indicated flowed EC with DESCEND (deconvolution of single-cell expression distribution). A, Genes with significant Gini changes under flow by each gating criteria. B, Venn diagrams showing gene list overlap from gated vs ungated analysis (A). C–G, Subpopulation analyses. C, UMAP (Uniform Manifold Approximation and Projection) plots of HUVEC colored by condition (left) or subpopulations (right). D, Violin plots showing distribution of normalized KLF2 and KLF4 expression by subpopulation. E, Heatmap showing expression of top 10 cluster markers ranked by fold change. Left, representative genes for each cluster. F, Representative gene ontology terms for genes highly expressed in each cluster. G, Differential test of Gini performed between HUVEC subpopulations with DESCEND; ratio of genes showing increased Gini over genes showing decreased Gini for each pairing. Red bars, ratio >1. Blue bars, ratio <1.
Second, we performed dimension reduction and clustering with Seurat to interrogate flow-induced EC subpopulation heterogeneity. Dimension reduction with Uniform Manifold Approximation and Projection showed that homeostatic laminar flow induced significant transcriptome changes, and 9 clusters with distinct gene expression profiles were identified: Stat1-4, Flow1-4, and a cluster termed “Mix” that contained cells from both stat (nonflow) and flow conditions (Figure 2C, Figure IVA in the Data Supplement). KLF2 and KLF4 were highly expressed in all Flow clusters but not Stat clusters (Figure 2D), suggesting that all flowed subpopulations responded to shear stress. Stat1 and Flow1 contained the highest cell numbers, and highly expressed genes in these clusters are also highly expressed by other static or flow clusters (Figure 2E), suggesting that they are the “ground state” for nonflow (static) and flowed EC. Further analyses of cluster markers and enriched Gene Ontology terms (Figure 2F) suggest that Stat2, Flow2 and Mix clusters are likely proliferating and clustered as described because of this property. Differences in cell cycle activity are reported as primary drivers of transcriptional heterogeneity in published scRNA-seq studies24–26 and progression through the cell cycle has been found to correlate with total transcript level per cell.27 Therefore, to focus on flow-induced gene expression heterogeneity and not cell cycle-related heterogeneity, we excluded the 3 cell-cycle active clusters from further analysis.
We then performed the same DESCEND analysis used for the total populations to compare clusters: Stat1, 3, 4 and Flow1, 3, 4. Comparison via Gini (Figure 2G) revealed more genes with increased expression heterogeneity than with decreased heterogeneity when each Flow cluster was compared with ground state cluster Stat1. Stat1 contains the majority (≈70%) of nonflowed EC, supporting our overall conclusion that EC transcriptional heterogeneity increases under homeostatic laminar flow conditions. However, Stat3 and Stat4 clusters had higher gene expression heterogeneity than Stat1 and the analyzed Flow clusters, suggesting a higher level of inter-cluster heterogeneity among some nonflowed (static) EC, and increased Gini in these clusters relative to flow. Stat3 markers suggest that this cluster contains pro-inflammatory EC, while Stat4 EC have elevated expression of actin cytoskeleton-related and focal adhesion genes and are likely migratory. These findings suggest that structural and signaling changes in small subsets of nonflowed EC affect transcriptome expression heterogeneity in a context of overall decreased transcriptional heterogeneity in nonflow conditions. The Flow cluster Ginis were much closer to each other, and they were all more heterogeneous than Stat1. Flow 4 contained EC with a more activated “angiogenesis” profile and had a slightly more heterogeneous Gini compared with Flow1, while Flow 3 was characterized by oxidative metabolism and perhaps immune functions and had a slightly less heterogeneous Gini compared with Flow 1 (Figure 2F and 2G).
EC Protein Expression Becomes More Homogeneous Under Flow
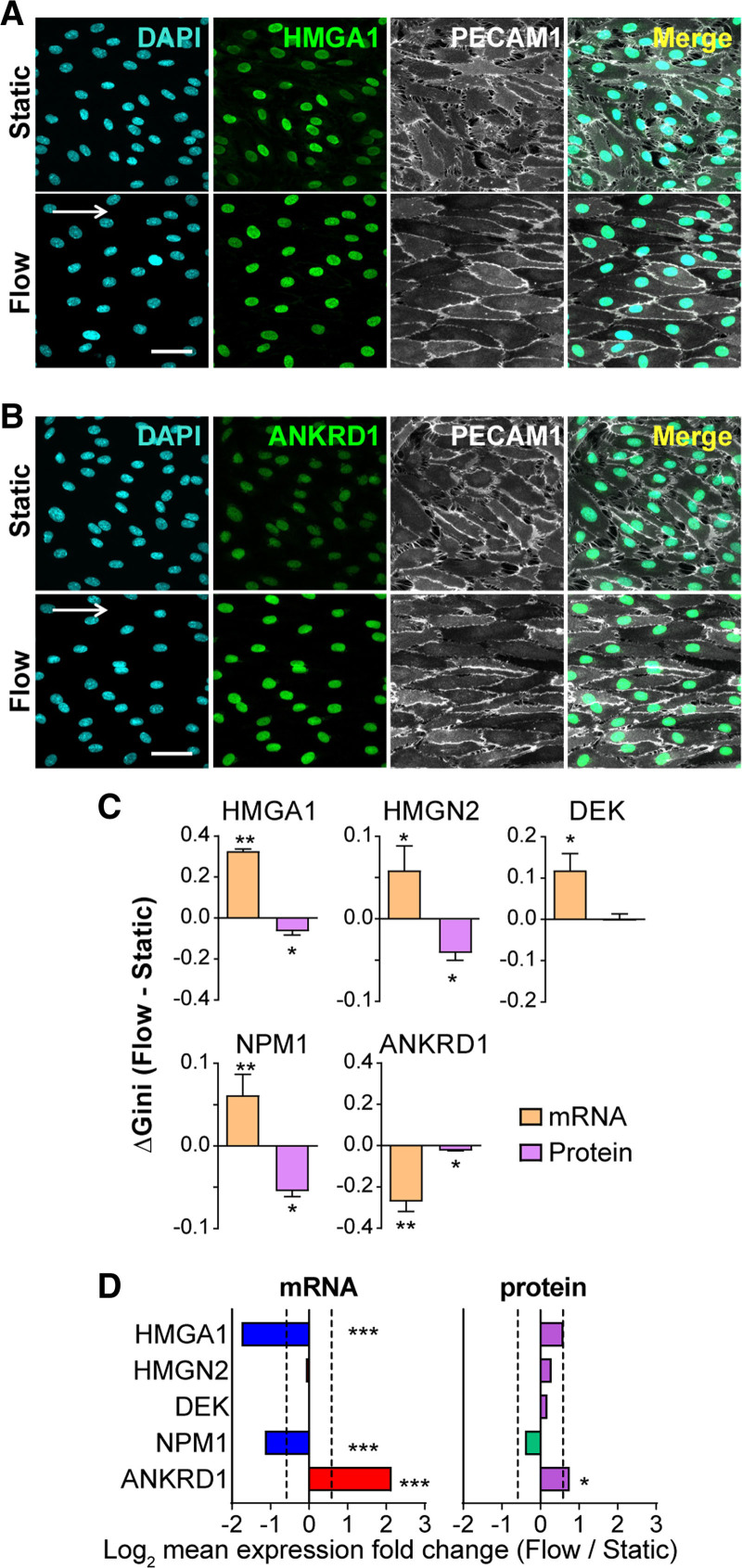
To investigate EC protein expression heterogeneity changes with homeostatic laminar flow, we performed antibody staining for selected EC proteins at single-cell resolution. We tested nuclear-localized proteins to provide more accurate quantification on a per-cell basis, and the selected genes included some with significant heterogeneity (Gini) change at the RNA level with flow, and others whose Gini was not significant. Representative images showed that some genes exhibited more uniform fluorescence intensity via antibody staining under flow compared with static controls (Figure 3A and 3B), suggesting decreased protein expression heterogeneity under flow. Quantification of total fluorescence intensity/nucleus, followed by a Gini coefficient calculation, showed decreased protein expression heterogeneity under flow for 4/5 genes with significant RNA expression heterogeneity, while 1/5 genes showed unchanged protein heterogeneity under flow (Figure 3C). Genes whose RNA Gini was not significant but trended downward (3/5) had little protein Gini, while one tested gene had decreased RNA Gini and significantly decreased protein Gini (Figure IVB in the Data Supplement), suggesting that EC protein heterogeneity under flow is uniform regardless of the RNA Gini. In contrast, the overall expression level changes between static and flow were more concordant for RNA and protein (Figure 3D, Figure IVC in the Data Supplement), although the sample size for protein data was small and some of the protein level changes were not significant.
Figure 3.
Endothelial cell (EC) transcriptomic heterogeneity compared with EC protein expression heterogeneity under homeostatic laminar flow. Human umbilical vein ECs (HUVEC) immunostained for genes showing significant Gini change under flow vs static in single-cell RNA-seq data. Representative images of HMGA1 (high mobility group AT-hook 1; A), ANKRD1 (ankyrin repeat domain 1; B) staining, with DAPI (blue, nucleus), and PECAM1 (platelet endothelial cell adhesion molecule 1; gray, cell borders). White arrow, direction of flow; scale bar, 50 µm. C, Gini change for tested nuclear genes at the RNA (single-cell RNA-seq) and protein level (immunostaining). Error bars, mean±SEM from multiple replicates. Statistics for mRNA, DESCEND (deconvolution of single-cell expression distribution), **P<0.01, *P<0.05. Statistics for protein, paired t test, *P<0.05. D, Mean expression level fold change for tested genes at the RNA (single-cell RNA-seq) and protein level (mean fluorescence intensity from 3 independent experiments). Dotted lines, Fold change=1.5. Statistics for mRNA, Wilcoxon rank-sum test, ***P<0.001. Statistics for protein, ratio paired t test, *P<0.05.
Discussion
The responses of ECs to laminar flow are crucial to establishing and maintaining vascular homeostasis and quiescence; yet, whether the homogenization of morphology and signaling is reflected in global homogenization of transcription profiles was not known. Here, we use genome-wide single-cell RNA-seq analysis to show that a subset of RNA transcripts becomes more heterogeneous in expression among a cohort of EC undergoing morphological homogenization in response to flow, while a smaller subset becomes less transcriptionally heterogeneous under flow. In contrast, a sampling of heterogeneity in protein expression levels indicates that protein expression tends to become less heterogeneous under homeostatic laminar flow, regardless of the RNA expression heterogeneity. Taken together, these findings suggest that EC expression heterogeneity under flow is complex, and that RNA expression heterogeneity does not always follow morphological homogenization as EC respond to homeostatic laminar flow.
Differential gene expression is well-documented between different cell types, but intercellular gene expression heterogeneity within a cell type is relatively understudied due to technical challenges. The recent development of single-cell OMICS and other single-cell techniques have enabled interrogation of global regulation of intercellular heterogeneity, although most studies query expression level changes. Among the few studies focusing on gene expression variation, one provided evidence that regulated heterogeneity but not gene expression levels correlated with epithelial cell differentiation to fiber cells during ocular lens development.28 DNA methylation heterogeneity across cells was linked to splicing variability during differentiation of induced pluripotent stem cells in another study.29 Multiplexed RNA in situ hybridization revealed high spatial heterogeneity of biomarker gene expression in tumor tissues, suggesting the potential application of spatial heterogeneity as a complementary approach for breast cancer subtype differentiation.30 Thus intercellular/ interregional gene expression heterogeneity has functional consequences and clinical applications, underscoring its importance.
ECs are highly specialized in structure and function and remarkably heterogeneous in different tissues and organs.4 EC transcriptomes and proteomes also vary dramatically across vascular beds.31,32 Single-cell RNA-seq has revealed new EC subpopulations defined by distinct gene expression patterns from a variety of tissues.32–34 Our cluster analysis of EC exposed to homeostatic laminar flow compared with nonflowed controls revealed that each condition had 4 clusters plus one that was shared between stat and flow EC. This indicates that while homeostatic laminar flow imposes morphological homogeneity on EC, there exist different subtypes of EC exposed to flow, as defined by expression level profiling of gene sets. This is consistent with other studies that analyzed freshly isolated aortic EC.34,35 Most of our top significant changes in EC gene expression under flow are shared with published data,18–22 suggesting that EC flow responses are similar when flow parameters are similar, and that species differences or use of primary EC from a particular source (ie, arterial versus venous) in culture do not affect the outcomes. Gene ontology analysis of transcriptome expression heterogeneity changes with flow revealed that protein translation and oxidative metabolism pathways are associated with elevated variance while protein processing is associated with reduced variance, and cell-cell interactions are found in both categories, suggesting complexity in these categories in response to flow.
Our findings reveal that the relationship between RNA and protein expression heterogeneity in response to flow is complex. With the caveat that the protein data were limited in scope relative to the RNA data, we asked whether a subset of genes with significant Gini change at the RNA level showed a similar trend in protein and found little correlation, suggesting divergence in RNA and protein expression heterogeneity regulation. However, similar analysis of another small gene subset not chosen on the basis of Gini change showed little overall change in RNA Gini and a trend in the same direction for overall RNA and protein levels. Thus, we conclude that RNA expression heterogeneity does not strongly correlate with protein expression heterogeneity changes under flow. In contrast, overall expression levels of the same genes were more correlated, suggesting that regulation of expression heterogeneity may be distinct from regulation of expression levels under flow. Although most single-cell EC profiling bins by gene expression levels, one recent report revealed the variability of VCAM1 gene expression at both RNA and protein levels in HUVEC after TNFα stimulation,36 indicating that transcriptional heterogeneity reflects protein heterogeneity under these conditions. This heterogeneity was linked to preexisting heterogeneity of VCAM1 promoter methylation rather than stochastic TNFα signaling or histone modification. von Willebrand factor is also expressed heterogeneously at both the RNA and protein levels, and promoter methylation was linked to control of heterogeneity in VWF expression.37
Here, we interrogated the entire transcriptome of a cohort using 3 measures of variance as EC responded to homeostatic laminar flow, and we found that overall transcriptional heterogeneity increased, and was unlinked to morphological homogenization. However, when transcriptional heterogeneity was assessed in different EC subtypes within the nonflow and flow populations, a large cluster in each condition mirrored the total set for that condition in Gini relationships, while some nonflowed clusters with smaller cell numbers had higher heterogeneity than the flow clusters. The association of these clusters with inflammation and migration suggests that activation of these pathways may be a stimulus for increased transcriptional heterogeneity. Several smaller flow clusters also varied slightly from the main flow cluster in RNA expression heterogeneity, suggesting that EC RNA Gini responses to flow vary within a small range.
These findings have implications for EC dysfunction and disease. For example, atherosclerotic lesions form preferentially in areas of vessels that experience disturbed rather than laminar flow, such as vessel branch points, and EC in these areas do not exhibit the morphological homogeneity of EC in laminar areas.8,38 Based on our findings, we predict that important regulation of EC responses to homeostatic laminar flow may include homogenization of expression post-transcriptionally for some genes. Thus, therapies that target pathways post-transcriptionally may be more effective in mitigating the effects of disturbed flow on EC function.
Acknowledgments
We thank the UNC TGL Core for technical support for single-cell RNA-seq experiments. We thank members of the Bautch laboratory for constructive discussion and comments on the article.
Sources of Funding
This work was supported by US National Institutes of Health grants R35 HL139950 (to V.L. Bautch), R01 GM129074 (to V.L. Bautch), and R35 GM138342 (to Y. Jiang).
Disclosures
None.
Supplemental Materials
Data Supplement Major Resources Table
Data Supplement Figures I–IV
Supplementary Material
Nonstandard Abbreviations and Acronyms
- EC
- endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- RNA-seq
- RNA sequencing
- CV
- coefficient of variation
- MEF
- mouse embryo fibroblast
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.121.316797.
For Sources of Funding and Disclosures, see page 2583.
Contributor Information
Ziqing Liu, Email: ziqing@email.unc.edu.
Dana L. Ruter, Email: dlynn313@gmail.com.
Kaitlyn Quigley, Email: kmq97@live.unc.edu.
Natalie T. Tanke, Email: ntanke22@live.unc.edu.
Yuchao Jiang, Email: yuchaoj@email.unc.edu.
References
- 1.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183 [DOI] [PubMed] [Google Scholar]
- 2.Michiels C. Endothelial cell functions. J Cell Physiol. 2003;196:430–443. doi: 10.1002/jcp.10333 [DOI] [PubMed] [Google Scholar]
- 3.Krüger-Genge A, Blocki A, Franke RP, Jung F. Vascular endothelial cell biology: an update. Int J Mol Sci. 2019;20:E4411. doi: 10.3390/ijms20184411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2:a006429. doi: 10.1101/cshperspect.a006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mouillesseaux KP, Wiley DS, Saunders LM, Wylie LA, Kushner EJ, Chong DC, Citrin KM, Barber AT, Park Y, Kim JD, et al. Notch regulates BMP responsiveness and lateral branching in vessel networks via SMAD6. Nat Commun. 2016;7:13247. doi: 10.1038/ncomms13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 7.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baeyens N, Bandyopadhyay C, Coon BG, Yun S, Schwartz MA. Endothelial fluid shear stress sensing in vascular health and disease. J Clin Invest. 2016;126:821–828. doi: 10.1172/JCI83083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistiakov DA, Orekhov AN, Bobryshev YV. Effects of shear stress on endothelial cells: go with the flow. Acta Physiol (Oxf). 2017;219:382–408. doi: 10.1111/apha.12725 [DOI] [PubMed] [Google Scholar]
- 10.Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571 [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Chen O, Zheng M, Wang L, Zhou Y, Yin C, Liu J, Qian L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 15.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Huang M, Torre E, Dueck H, Shaffer S, Murray J, Raj A, Li M, Zhang NR. Gene expression distribution deconvolution in single-cell RNA sequencing. Proc Natl Acad Sci USA. 2018;115:E6437–E6446. doi: 10.1073/pnas.1721085115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ajami NE, Gupta S, Maurya MR, Nguyen P, Li JY, Shyy JY, Chen Z, Chien S, Subramaniam S. Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc Natl Acad Sci USA. 2017;114:10990–10995. doi: 10.1073/pnas.1707517114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis. 2016;19:9–24. doi: 10.1007/s10456-015-9485-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S, Xu Y, Yin M, Zhang S, Liu P, Koroleva M, Si S, Little PJ, Pelisek J, Jin ZG. Flow-dependent epigenetic regulation of IGFBP5 expression by H3K27me3 contributes to endothelial anti-inflammatory effects. Theranostics. 2018;8:3007–3021. doi: 10.7150/thno.21966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conway DE, Williams MR, Eskin SG, McIntire LV. Endothelial cell responses to atheroprone flow are driven by two separate flow components: low time-average shear stress and fluid flow reversal. Am J Physiol Heart Circ Physiol. 2010;298:H367–H374. doi: 10.1152/ajpheart.00565.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruter DL, Liu Z, Ngo KM, X S, Marvin A, Buglak DB, Kidder EJ, Bautch VL. SMAD6 transduces endothelial cell flow responses required for blood vessel homeostasis. Angiogenesis. 2021;24:387–398. doi: 10.1007/s10456-021-09777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurya MR, Gupta S, Li JY, Ajami NE, Chen ZB, Shyy JY, Chien S, Subramaniam S. Longitudinal shear stress response in human endothelial cells to atheroprone and atheroprotective conditions. Proc Natl Acad Sci USA. 2021;118:e2023236118. doi: 10.1073/pnas.2023236118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Maio FG. Income inequality measures. J Epidemiol Community Health. 2007;61:849–852. doi: 10.1136/jech.2006.052969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barron M, Li J. Identifying and removing the cell-cycle effect from single-cell RNA-sequencing data. Sci Rep. 2016;6:33892. doi: 10.1038/srep33892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauridsen FKB, Jensen TL, Rapin N, Aslan D, Wilhelmson AS, Pundhir S, Rehn M, Paul F, Giladi A, Hasemann MS, et al. Differences in cell cycle status underlie transcriptional heterogeneity in the HSC compartment. Cell Rep. 2018;24:766–780. doi: 10.1016/j.celrep.2018.06.057 [DOI] [PubMed] [Google Scholar]
- 26.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746. doi: 10.15252/msb.20188746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolatabadi S, Candia J, Akrap N, Vannas C, Tesan Tomic T, Losert W, Landberg G, Åman P, Ståhlberg A. Cell cycle and cell size dependent gene expression reveals distinct subpopulations at single-cell level. Front Genet. 2017;8:1. doi: 10.3389/fgene.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangalum RK, Kim D, Kashyap RK, Mangul S, Zhou X, Elashoff D, Bhat SP. Spatial analysis of single fiber cells of the developing ocular lens reveals regulated heterogeneity of gene expression. iScience. 2018;10:66–79. doi: 10.1016/j.isci.2018.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linker SM, Urban L, Clark SJ, Chhatriwala M, Amatya S, McCarthy DJ, Ebersberger I, Vallier L, Reik W, Stegle O, et al. Combined single-cell profiling of expression and DNA methylation reveals splicing regulation and heterogeneity. Genome Biol. 2019;20:30. doi: 10.1186/s13059-019-1644-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voith von Voithenberg L, Fomitcheva Khartchenko A, Huber D, Schraml P, Kaigala GV. Spatially multiplexed RNA in situ hybridization to reveal tumor heterogeneity. Nucleic Acids Res. 2020;48:e17. doi: 10.1093/nar/gkz1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleuren ACA, van der Ent MA, Jiang H, Hunker KL, Yee A, Siemieniak DR, Molema G, Aird WC, Ganesh SK, Ginsburg D. The in vivo endothelial cell translatome is highly heterogeneous across vascular beds. Proc Natl Acad Sci U S A. 2019;116:23618–23624. doi: 10.1073/pnas.1912409116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 33.Feng W, Chen L, Nguyen PK, Wu SM, Li G. Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front Cardiovasc Med. 2019;6:165. doi: 10.3389/fcvm.2019.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, Regev A, Kathiresan S, Gupta RM. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation. 2019;140:147–163. doi: 10.1161/CIRCULATIONAHA.118.038362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald AI, Shirali AS, Aragón R, Ma F, Hernandez G, Vaughn DA, Mack JJ, Lim TY, Sunshine H, Zhao P, et al. Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell. 2018;23:210–225.e6. doi: 10.1016/j.stem.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turgeon PJ, Chan GC, Chen L, Jamal AN, Yan MS, Ho JJD, Yuan L, Ibeh N, Ku KH, Cybulsky MI, et al. Epigenetic heterogeneity and mitotic heritability prime endothelial cell gene induction. J Immunol. 2020;204:1173–1187. doi: 10.4049/jimmunol.1900744 [DOI] [PubMed] [Google Scholar]
- 37.Yuan L, Chan GC, Beeler D, Janes L, Spokes KC, Dharaneeswaran H, Mojiri A, Adams WJ, Sciuto T, Garcia-Cardeña G, et al. A role of stochastic phenotype switching in generating mosaic endothelial cell heterogeneity. Nat Commun. 2016;7:10160. doi: 10.1038/ncomms10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warboys CM, Amini N, de Luca A, Evans PC. The role of blood flow in determining the sites of atherosclerotic plaques. F1000 Med Rep. 2011;3:5. doi: 10.3410/M3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell RNA sequencing data supporting the findings of this study have been made publicly available at the GEO database under the accession number GSE151867.