Abstract
It has recently become clear that several human lineages coexisted with Homo sapiens during the late Middle and Late Pleistocene. Here, we report an archaic human fossil that throws new light on debates concerning the diversification of the Homo genus and the origin of H. sapiens. The fossil was recovered in Harbin city in northeastern China, with a minimum uranium-series age of 146 ka. This cranium is one of the best preserved Middle Pleistocene human fossils. Its massive size, with a large cranial capacity (∼1,420 mL) falling in the range of modern humans, is combined with a mosaic of primitive and derived characters. It differs from all the other named Homo species by presenting a combination of features, such as long and low cranial vault, a wide and low face, large and almost square orbits, gently curved but massively developed supraorbital torus, flat and low cheekbones with a shallow canine fossa, and a shallow palate with thick alveolar bone supporting very large molars. The excellent preservation of the Harbin cranium advances our understanding of several less-complete late Middle Pleistocene fossils from China, which have been interpreted as local evolutionary intermediates between the earlier species Homo erectus and later H. sapiens. Phylogenetic analyses based on parsimony criteria and Bayesian tip-dating suggest that the Harbin cranium and some other Middle Pleistocene human fossils from China, such as those from Dali and Xiahe, form a third East Asian lineage, which is a part of the sister group of the H. sapiens lineage. Our analyses of such morphologically distinctive archaic human lineages from Asia, Europe, and Africa suggest that the diversification of the Homo genus may have had a much deeper timescale than previously presumed. Sympatric isolation of small populations combined with stochastic long-distance dispersals is the best fitting biogeographical model for interpreting the evolution of the Homo genus.
Keywords: human phylogeny, human cranium fossil, human dispersal, human diversification
Graphical abstract
Public summary
-
•
More than 100,000 years ago, several human species coexisted in Asia, Europe, and Africa
-
•
A completely preserved fossil human cranium discovered in the Harbin area provides critical evidence for understanding the evolution of humans and the origin of our species
-
•
The Harbin cranium has a large cranial capacity (∼1,420 mL) falling in the range of modern humans, but is combined with a mosaic of primitive and derived characters
-
•
Our comprehensive phylogenetic analyses suggest that the Harbin cranium represents a new sister lineage for Homo sapiens
-
•
A multi-directional “shuttle dispersal model” is more likely to explain the complex phylogenetic connections among African and Eurasian Homo species/populations
Introduction
The origin of modern humans (Homo sapiens, our own species) has long been a controversial topic. During the late Middle and Late Pleistocene, several human lineages, evidently at species level, coexisted with H. sapiens across Africa and Eurasia. These extinct hominins include H. heidelbergensis/H. rhodesiensis, Homo naledi, Homo floresiensis, H. luzonensis, Denisovans, Neanderthals (Homo neanderthalensis), and Homo erectus.1, 2, 3, 4, 5 The phylogenetic relationship between these coexisting hominins and H. sapiens has long been debated. Before the appearance of undoubted modern humans in Asia, some archaic fossils, such as those from Narmada, Maba, Dali, Jinniushan, Xuchang, and Hualongdong show mosaic combinations of features present in H. erectus, H. heidelbergensis/H. rhodesiensis, Neanderthals, and H. sapiens. Therefore, it is widely believed that these Asian hominins are critical for studying the later evolution of the genus Homo and the origin of H. sapiens. The incomplete preservation of these fossils and the fact that they have largely been described by advocates of regional continuity have made it difficult to integrate them into the wider picture of human evolution. For example, Xuchang, Dali, and Hualongdong have all been described as transitional forms between Chinese H. erectus and H. sapiens, whose affinities can be understood in the context of a braided stream network model of gene flow.6, 7, 8, 9 Here, we report a fossil human cranium that is characterized by a combination of large cranial capacity, short face, and small check bones as in H. sapiens, but also a low vault, strong browridges, large molars, and alveolar prognathism as in most archaic humans. Through phylogenetic and biogeographic analyses, we argue that this fossil is the most complete representative of a distinct Middle Pleistocene lineage, with a separate evolutionary history in East Asia.
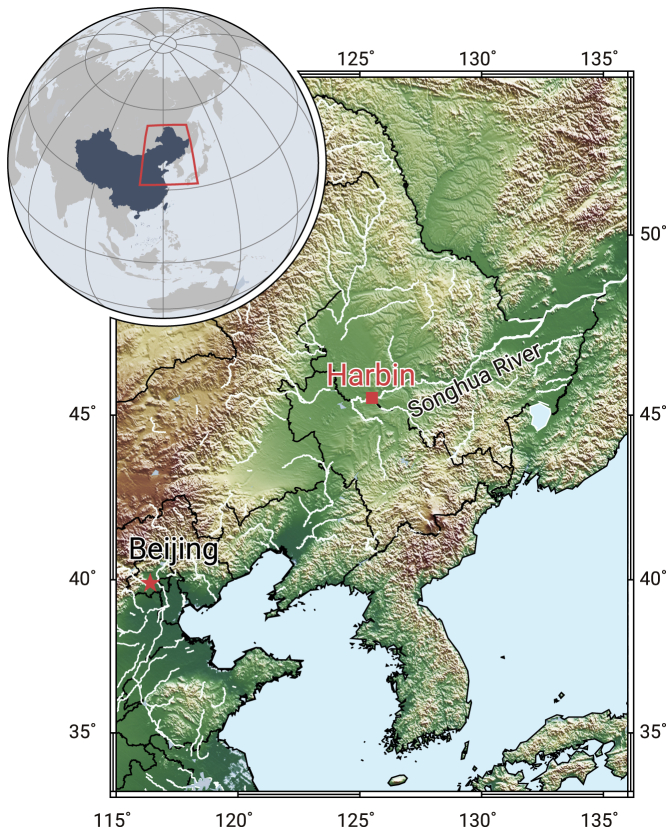
The Harbin human fossil is represented by a single cranium (HBSM2018-000018(A), housed in the Geoscience Museum of Hebei GEO University, Shijiazhuang, Hebei Province, China), which was reportedly discovered in 1933 during construction work when a bridge (Dongjiang Bridge) was built over the Songhua River in Harbin city (Figure 1). Because of a long and confused history since the discovery (see the supplemental information), the exact site of the find is uncertain. We tested the concentrations of rare earth elements (REEs) and the Sr isotopic composition of the human fossil and a range of mammalian fossils collected from deposits of the Songhua River near the supposed locality (Dongjiang Bridge), and used non-destructive X-ray fluorescence analyses to examine the element distributions of these human and mammalian fossils. The results of our experiments show that element distributions and REE concentrations of the Harbin cranium and the mammalian fossils found near Dongjiang Bridge have similar distribution patterns.10 The Sr isotopic composition of the Harbin cranium falls in the range of the local Middle Pleistocene-Early Holocene human and mammalian fossils.10 We also directly dated the Harbin fossil cranium by the uranium-series disequilibrium (U-series) method. The results suggest a minimum age for the cranium of ∼146 ka.10 While these results cannot pin the Harbin cranium to an exact site and layer, they are consistent with the conclusion that the cranium is from the late Middle Pleistocene of the Harbin area.10
Figure 1.
Geographic location of the Harbin cranium
The red square indicates the Dongjiang Bridge in Harbin city.
Results and discussion
Morphology
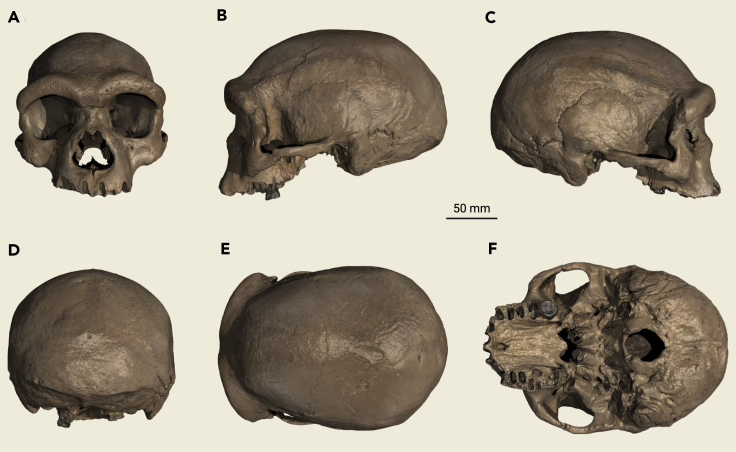
The Harbin cranium is undistorted and almost intact, with the main losses being all but one tooth (the left M2), and slight damage to the left zygomatic arch (Figure 2). It is massive in size, showing the largest values in our comparative fossil database (see the supplemental information) for measurements, such as maximum cranial length, nasio-occipital length, and supraorbital torus breadth, and the second largest values for measurements, such as biauricular breadth, frontal chord, zygomatic breadth, and biorbital breadth. Detailed morphological descriptions and comparisons of the cranium are given in the supplemental information, and are summarized below.
Figure 2.
The Harbin cranium in standard views
(A) Anterior view.
(B) Lateral view, left side.
(C) Lateral view, right side.
(D) Posterior view.
(E) Superior view.
(F) Inferior view. Scale bar, 50 mm.
The cranial vault is voluminous (∼1,420 mL capacity, measured using high-resolution computed tomography [CT] scanning and three-dimensional reconstruction of the endocranial cast). However, the braincase is clearly archaic, with a very wide supraorbital torus, base and palate, and a long and low shape in lateral view, with a receding frontal and evenly curved parietal contour. Nevertheless, it lacks both the angulated occipital with a strong transverse torus found in H. erectus and H. heidelbergensis/H. rhodesiensis crania, and the protruding occipital region with a central suprainiac fossa typical of Neanderthals. In posterior view the unkeeled cranium is widest in the supramastoid area, below which the well-developed mastoid processes slope inward. The temporals and parietals do not converge strongly as in H. erectus fossils, but there is no upper parietal expansion, as found in recent H. sapiens, nor the “en bombe” shape typical of Neanderthals. In lateral view the face is relatively low in height and retracted under the cranial vault, lacking the total anterior projection typical of H. erectus and H. heidelbergensis/H. rhodesiensis. The upper face and nasal aperture are very wide, but the zygomaxillary region is transversely flat and faces anteriorly, with a morphology like that of H. sapiens.
The combination of an archaic but large-brained cranial vault and a wide but H. sapiens-like face is striking, and is also found in the less-complete Middle Pleistocene Chinese fossils from Dali and Jinniushan, although they differ in details of morphology (see the supplemental information and Video S1. Virtual reconstruction of the Harbin cranium based on CT scanning, yaw movie, Video S2. Virtual reconstruction of the Harbin cranium based on CT scanning, pitch movie, Video S3. Virtual Reconstruction of the Harbin Cranium based on CT scanning, roll movie). The less-complete Hualongdong cranium resembles Dali more closely in several respects, and some of its differences may be due to its immaturity, while the Xuchang and Maba partial crania appear more distinct (see the supplemental information for more details and comparative data).
Overall, the Harbin cranium shows an individual combination of traits, and probably represents a distinct species of Homo from other designated Middle-Late Pleistocene human taxa, such as H. sapiens, H. neanderthalensis, and H. heidelbergensis/H. rhodesiensis. Its enormous overall size sets it apart from nearly every other fossil but, in terms of cranial vault proportions, the braincase clearly overlaps in shape with those of other large-sized late archaic Homo species. However, the face, despite its enormous breadth dimensions, is relatively low in height and has an H. sapiens and H. antecessor-like zygomaxillary shape that is also found in the Middle Pleistocene Chinese fossils from Dali and Jinniushan. It is also hafted onto the braincase with reduced prognathism, as in recent humans. In its combination of traits, the Harbin cranium is more like fossils attributed to early H. sapiens, such as Jebel Irhoud 1 and Eliye Springs, than to later members of our lineage. Finally, and perhaps significantly, the morphology and large size of the surviving Harbin M2 (Figure S1, mesiodistal length 13.6 mm and buccolingual width 16.6 mm) are matched most closely in the Late Pleistocene record by the permanent molars from Denisova Cave (Denisovan 4: M2/3, mesiodistal length 13.1 mm, and buccolingual width 14.7 mm; Denisovan 8: M3, mesiodistal length 14.3 mm, and buccolingual width 14.65 mm).11,12
Life reconstruction
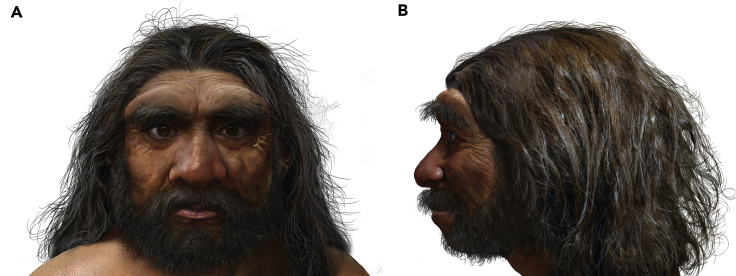
The overall size, robustness, thick and strong supraorbital tori, large mastoid processes, and salient temporal lines of the Harbin cranium suggest that it probably represents a male individual. The ectocranial sutures at the midlambdoid, lambdoid, obelion, anterior sagittal, superior sphenotemporal, incisive, anterior and posterior median palatine, and transverse palatine are all completely obliterated. The ectocranial sutures at bregma, midcoronal, pterion, sphenofrontal, and inferior sphenotemporal show significant closure. For the standard of H. sapiens, the ectocranial suture composite scores would suggest an old adult around 50 years old.11,12 However, the tooth wear seems to suggest a younger age. The only preserved M2 still has much enamel present, and dentine exposure is present on the protocone and paracone. The relatively complete ectocranial suture closure may be related to the robustness of the Harbin cranium. The large square eye sockets with strong supraorbital tori indicate deep eyes. The large and wide piriform aperture indicates a large and bulbous nose. The expanded paranasal region and relatively projecting middle face are matched with flat and short modern human-like cheek regions. Large incisor and canine tooth sockets indicate that the man probably had quite large front teeth and a broad mouth. The mandible of this individual is not known, but the phylogenetic analyses suggest that the Harbin cranium and the Xiahe mandible from Gansu Province of China form a sister group. The M2 size of the Harbin cranium matches the tooth size of the Xiahe mandible. It is reasonable to deduce that the Harbin cranium probably matches a mandible as robust as the Xiahe mandible and without a chin. It is hard to reconstruct the skin tone and hair color of the Harbin individual without genetic information, but available genetic data suggest that Neanderthals, Denisovans, and early H. sapiens generally had relatively dark skin, hair, and eye color. Considering the high latitude of the provenance of the Harbin cranium, we have chosen to give the reconstruction only a medium-dark skin color (Figure 3).
Figure 3.
Life reconstruction of the Harbin cranium
(A) Anterior view.
(B) Lateral view, left side.
Phylogenetic position of the Harbin cranium
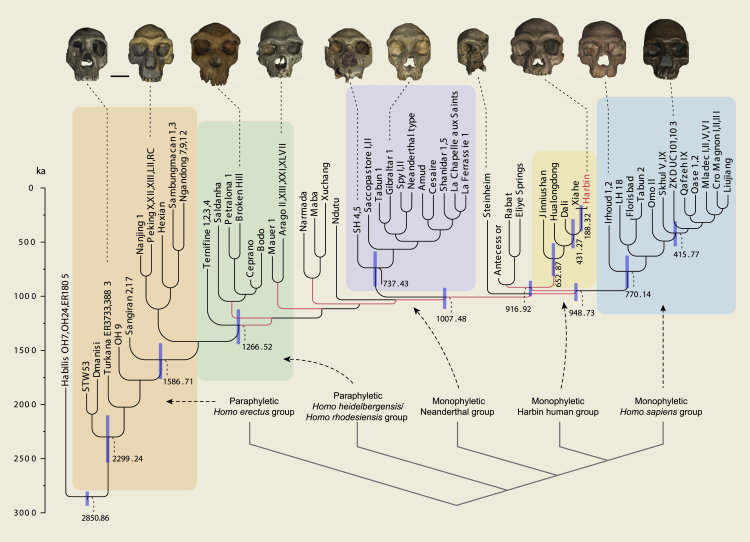
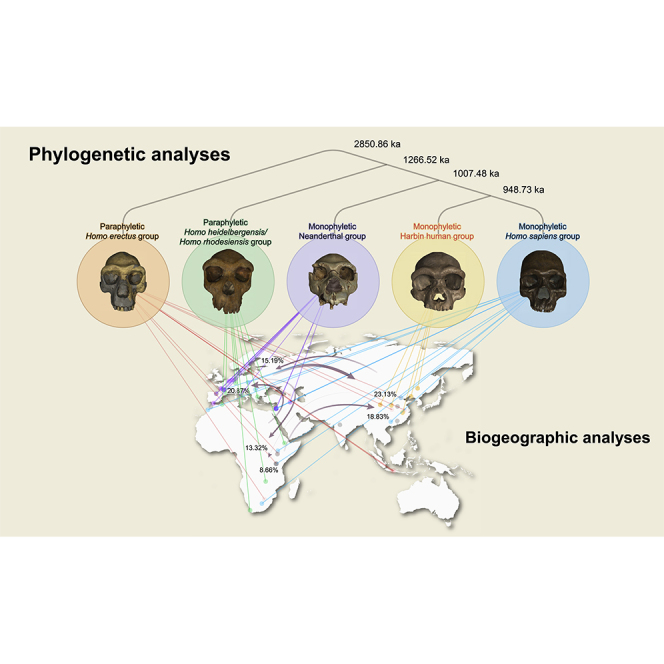
Our extensive phylogenetic analyses based on parsimony criteria13 and Bayesian inference14, 15, 16, 17 firstly support the monophyly of Neanderthals and the monophyly of H. sapiens (Figures 4 and S19–S23). The Irhoud fossils from Morocco form the most basal operational taxonomic unit (OTU) of the H. sapiens clade, and the Sima de los Huesos crania from Spain form the most basal OTU of the Neanderthal clade, in line with other current interpretations.18, 19, 20 The Harbin cranium and Xiahe mandible form a sister group, and they, plus the Dali, Hualongdong, Jinniushan specimens, the European H. antecessor partial cranium, the African Eliye Springs cranium, and Rabat palate, form a monophyletic group. This clade forms the sister group of the similarly monophyletic H. sapiens clade. The specimens traditionally grouped in H. heidelbergensis/H. rhodesiensis do not constitute a monophyletic group and the Asian and African H. erectus specimens similarly form a paraphyletic group. When backbone constraints are used to reflect the results from palaeoproteomic and ancient DNA research by forcing the Xiahe mandible as the sister group of Neanderthals21 and H. antecessor outside of the H. sapiens-Neanderthal clade,22 Chinese late Middle Pleistocene humans, including the Harbin cranium, form a monophyletic clade as the sister group of Neanderthals (Figure S20). Both most parsimonious and backbone partially constrained phylogenetic trees support the monophyly of the group, including Dali, Jinniushan, Hualongdong, Xiahe, and Harbin.
Figure 4.
Phylogeny of the 55 selected fossils from the genus Homo
The topology of the tree was inferred from a Bayesian tip-dating analysis in MrBayes 3.223 and summarized as the all-compatible tree. To reduce the polytomy at some clades, the strict consensus of the most parsimonious trees from the parsimony analysis in TNT13 was used as a reference. The branches in red indicate the backbone constraints based on the most parsimonious trees. Branch lengths are proportional to the division age in thousands of years. Numbers at the internal nodes are the median ages, and the blue bars indicate the 95% highest posterior density interval of the node ages. Color shadows indicate the monophyletic H. sapiens group, Neanderthal group and Harbin human group, and the paraphyletic H. heidelbergensis/H. rhodesiensis group and H. erectus group. A simplified phylogenetic relationship of the five groups is shown on the lower right. Human crania images are aligned to the Frankfurt horizontal plane. Scale bar, 50 mm (between the Turkana and Peking crania).
Some researchers have proposed that all Middle Pleistocene hominins belong to a single lineage leading to modern humans, with Asian Middle Pleistocene hominins, such as Dali and Hualongdong, suggested as transitional forms between Asian H. erectus and Asian H. sapiens specimens.6,9,24 Some other researchers have recognized these Asian hominins as part of the H. heidelbergensis/H. rhodesiensis hypodigm.25, 26, 27 A previous analysis based on overall similarity showed differences between Dali-Maba and the H. heidelbergensis hypodigm, and the potential connection between Dali-Maba and African early H. sapiens.28 Our analyses suggest that the Harbin cranium, together with Dali, Jinniushan, Hualongdong, and Xiahe, is not a part of the African and European H. heidelbergensis/H. rhodesiensis clade, but is the sister group of H. sapiens (see also the backbone partially constrained parsimony analysis in the supplemental information). The sister relationship between Harbin and Xiahe, as identified by Bayesian inference (but not parsimony analysis, see the supplemental information), is particularly interesting. The Xiahe mandible shows some proteomic features of the Denisovans,21 who were informally called “Homo sapiens altaiensis” or “Homo altaiensis,”12,29 and sediments from Baishiya Cave have yielded Denisovan mtDNA.30 The Harbin M2 also matches the known permanent Denisovan molars in size and root morphology, and, ever since the discovery of Denisovans, Asian Middle Pleistocene hominins, such as Dali, Jinniushan, and Xujiayao, have been suspected to represent an East Asian population of the Denisovans.31 More mandibular specimens for the Harbin population or cranial specimens corresponding to the Xiahe mandible will test how close the Harbin and Xiahe humans are morphologically, while new genetic material will test the relationship of these populations to each other and to the Denisovans.
The results of the Bayesian tip-dating analyses suggest that the Harbin and Xiahe fossils shared a common ancestor ∼188 ka (397–155 ka), and the clade, including the Harbin cranium and H. sapiens shared a common ancestor at ∼949 ka (1,041.41–875.25 ka). The Neanderthal-H. sapiens divergence time in our analysis was ∼1,007 ka (1,114–919 ka). This estimation falls in the range based on mtDNAs for the split between the basal Neanderthal (Sima de los Huesos) and the H. sapiens lineage,20 but is much older than the estimation based on nuclear DNAs for the splits between the Neanderthal and H. sapiens lineages.32, 33, 34 However, it is possible that this younger estimated divergence date is an artifact of statistical averaging between “super-archaic” and “recent gene flow” events.35 The common ancestor of the H. sapiens OTUs included in our analysis is as old as ∼770 ka (922–622 ka), suggesting that the H. sapiens clade has a much deeper origin time than previously estimated. The Eurasian H. sapiens OTUs share a common ancestor ∼416 ka (534–305 ka) old. Outside of Africa, however, the earliest known H. sapiens fossil is only ∼210 ka.36
There is a large time gap between the hypothetical common ancestor of Eurasian H. sapiens and the actual fossil record, from the Bayesian tip-dating analysis. One plausible hypothesis is that the ancestral population of Eurasian H. sapiens may have diversified in Africa for many millennia before they dispersed into Eurasia. Genetic studies on ancient DNA suggest that the initial genetic exchanges between Neanderthals and H. sapiens occurred between 468 and 219 ka,33 or between ∼370 and 100 ka,34 and the introgression may have originated through gene flow from an African source.19,33 Interestingly, not only does the estimated time of the introgression event between Neanderthals and H. sapiens roughly overlap our prediction for the age of the common ancestor of Eurasian H. sapiens, but the African origin of the introgression is also consistent with our African ancestral population hypothesis.
Biogeography of the Homo species/populations
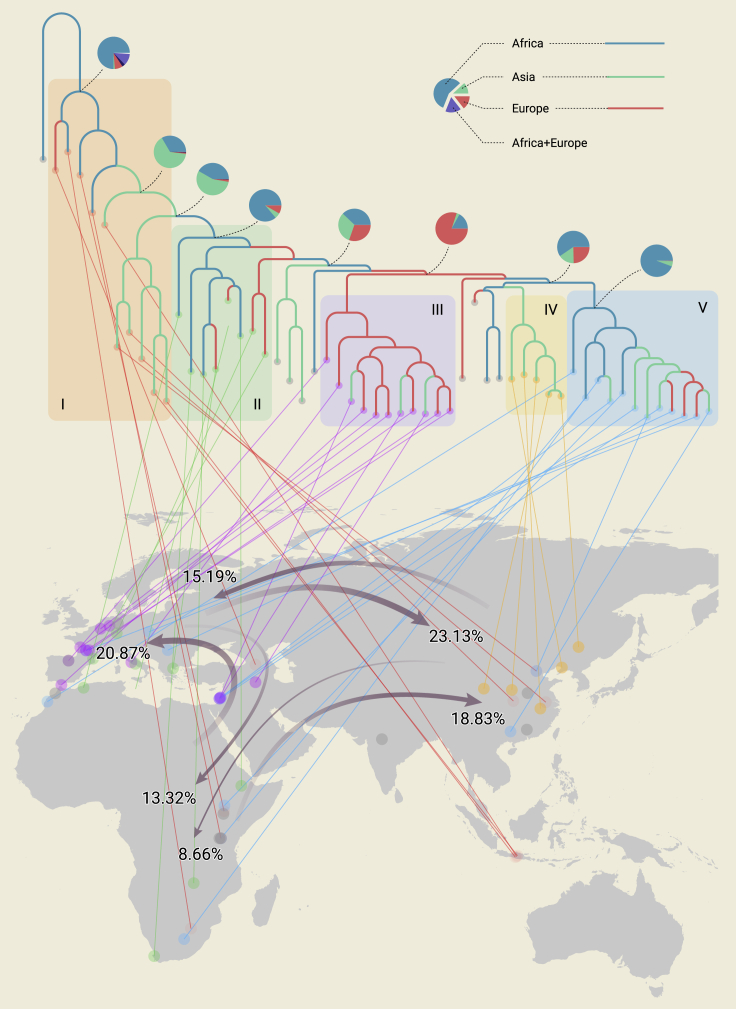
We conducted maximum likelihood analysis under 18 different biogeographical models and estimated the number and type of biogeographical events using biogeographical stochastic mapping (BSM). The Akaike information criterion (AIC) model selection strongly supported dispersal-extinction cladogenesis37,38 with the founder-event dispersal (“jump dispersal”)39,40 model (DEC + j) as the best fit and the most probable biogeographical model (Tables S12–S14). Under this best fitting model (Figure 5), the ancestral distribution range of the Harbin, Dali, Jinniushan, Xiahe and Hualongdong group is most probably in Asia. The ancestral area for the Harbin-H. sapiens clade is most probably from Africa, supporting the idea that Africa is the center of origin of the H. sapiens clade. The ancestral distribution of the group bracketing Neanderthal, H. sapiens, and Harbin is from Africa or Europe.
Figure 5.
Maximum likelihood ancestral range estimations and dispersal events for the Pleistocene Homo species/populations
R41 package BioGeoBEARS39,40 was used to estimate ancestral range probabilities and the number of dispersals. Topology of the phylogenetic tree is the same as that in Figure 4. The branch colors (red, blue, and green) indicate the geographical occurrences of the Homo fossils and the maximum likelihood ancestral range estimations for Homo under the best DEC + j model (dispersal-extinction cladogenesis38 with the founder-event dispersal39,40 model). The pie diagrams at the nodes show the relative probability of all possible ancestral distribution (areas or combinations of areas). Color shadows behind the phylogenetic tree indicate: I, H. erectus group; II, H. heidelbergensis/H. rhodesiensis group; III, Neanderthal group; IV, Harbin human group; V, H. sapiens group. Terminal taxa are linked with their geographical distributions. Grey arrows indicate the dispersal events between Africa, Asia, and Europe. Numbers near the arrowheads show the percentages of the means for the count of dispersal events between each pair of regions. The means are calculated from the event counts in each of 100 biogeographical stochastic maps. The common ancestor of the H. sapiens group and the common ancestor of the H. sapiens group, Harbin human group, and Neanderthal group are from Africa. However, the monophyletic clade embraced between the H. sapiens group and Asian H. erectus has an ancestral distribution in Asia. Asia received more dispersals from the other two continents. Africa received fewer dispersal from the other two continents.
Our simulation of the biogeographical history of Homo species/populations identified sympatry diversification (∼57%) and founder-event dispersal (∼42%) as the common types of biogeographical modes across the phylogenetic tree of Homo (Table S15). Because all the OTUs are at the population level from a single locality, it is reasonable to find that no range expansion or range contraction event is detected from the BSM simulations. Founder-event dispersal usually involves a small number of individuals that dispersed to a new locality through a long dispersal distance and established a new isolated founder population.42, 43, 44 The changes in distribution range occurred at a lineage-splitting node, resulting in one daughter lineage dispersal into a new range, and the other daughter lineage remaining in the ancestral range. Sympatric diversification and founder-event dispersal being the most dominant biogeographical modes reflects the fact that multi-lineages of Homo coexisted in Africa, Europe, and Asia during the Middle and Late Pleistocene. These Homo lineages probably had a strong capability of dispersing for long distances, but remained in relatively small and isolated populations.
BSMs indicate that the directionality of the dispersals between Africa, Asia, and Europe is asymmetric (Figure 5; Table S15). Asia is a sink of Homo species/populations that receives more dispersals from Africa and Europe than it gives dispersals to Africa and Europe. In total, Asia receives ∼42% of the total dispersal events and only provides ∼24% dispersals to other continents (Figure 5; Table S16). Africa is the major source of Homo dispersals. In total, ∼40% of all the dispersals are from Africa, while Africa also receives ∼22% dispersals from Asia and Europe. Instead of a unidirectional “out of Africa” model, a multi-directional “shuttle dispersal model” is more likely to explain the complex phylogenetic connections among African and Eurasian Homo species/populations.
Conclusions
The Harbin cranium is one of the best preserved of all archaic human fossils and its estimated late Middle Pleistocene age places it as an Asian contemporary of the evolving H. sapiens, H. neanderthalensis, and Denisovan lineages. It is huge in size, and its distinctive combination of traits in the cranial vault and face differentiate it from H. sapiens and H. neanderthalensis, as well as from the earlier species H. heidelbergensis/H. rhodesiensis. Instead it shows the greatest resemblances to Middle Pleistocene Chinese fossils, such as Hualongdong, Dali, and Jinniushan. This is confirmed by phylogenetic analyses using parsimony and Bayesian methods, which place these Chinese fossils with Harbin as a part of the sister group to H. sapiens, based on synapomorphies, such as a moderate post-toral sulcus, gently arched zygomaticoalveolar crest, presence of inferior orbital torus, strong malar tubercle, and thick mastoid processes. Our analyses also suggest a potential link between the Harbin cranium and the Xiahe mandible, a fossil attributed to the Denisovan lineage. The northerly location of the Harbin site also has implications for Middle Pleistocene human adaptive capabilities, since, even in the present interglacial, this region has winter temperatures averaging more than 16°C below zero. The very large size of the Harbin individual (as judged from the size of the cranium) may indicate physical adaptation to such conditions.45 The coexistence of several human lineages during the late Middle and Late Pleistocene of Asia is probably related to its diverse palaeoenvironments (ranging from the Gobi Desert to rainforest, and from coastal plains to the Qinghai-Tibet Plateau), which produced a varied biogeographic sink for human evolution.
Material and methods
Morphological studies
We scored and measured morphological characters from 95 cranial, mandibular, or dental specimens of the Homo genus (Table S1). All the specimens and replicas used in this research are under the oversight of the institutional review board of the Hebei GEO University, the Institute of Vertebrate Paleontology or the Natural History Museum, London. We used the high-resolution CT facilities and the surface scanner at the Key Laboratory of Vertebrate Evolution and Human Origins of the Chinese Academy of Sciences and the Natural History Museum, London, to CT scan or surface scan all the Homo fossils and casts included in this study. We used VG Studio Max 3.2 to build three-dimensional models. All measurements were taken from the digital three-dimensional models.
Phylogenetic analyses
Although it is debatable how phenomic features are correlated to each other and whether some characters are more important than others for phylogeny reconstruction, phylogenetic analysis based on phenomic characters has long been practiced to generate phylogenetic frameworks for hominins (e.g., Wood and other workers25,46, 47, 48, 49). We built a phenomic character data matrix (232 discrete characters and 400 continuous characters) using MorphoBank.50 Most of the 234 discrete characters are widely used and discussed in paleoanthropological research (see the supplemental information). The continuous characters include 184 linear measurements, 22 angles, and 194 ratios. The linear and angular measurements were taken following the standards defined by Martin and Saller51 and Howells.52 In total, 1,379 annotated images and 9,618 labels were used in MorphoBank (MorphoBank: Project 3385) to illustrate the phenomic homologies. To remove the effect of body size, the linear measurements of the crania and the upper dentitions of a scored specimen were divided by the 1/3 power of the cranial capacity of this specimen. The linear measurements of the mandibles and lower dentitions of a scored specimen were divided by the biramus breadth at the alveolar margin of this specimen. We consciously avoided redundant and potentially correlated discrete characters. All the continuous characters were normalized to have a range between 0 and 1. Normalization of the continuous characters can significantly reduce the potential correlations among different characters.
For most of the species/populations of Homo, palaeoproteomic or ancient DNA data are unknown. Thus, phenomic data form the base of evidence for setting taxonomic boundaries and/or phylogenetic relationships. It has been shown that hybridization does not cause significant taxonomic problems in most analyses,53,54 and we assume that any interbreeding between the OTUs did not affect the distribution or expression of characters for parsimony or Bayesian tip-dating analyses.
To reflect intra- and inter-species morphological variation, specimens that were from the same locality and generally accepted as the same species/population were grouped into one OTU. After combination, 55 OTUs were used as terminal taxa for the phylogenetic and biogeographic analyses. The OTUs cover most of the major clades or groups of the Homo genus. For each terminal taxon, we use the most recently published dating results (Table S1). The Hualongdong skull, H. antecessor ATD6-69, and the Turkana KNM-WT 15000 fossil are adolescent individuals. The main effects of their young ages will be in the final stages of cranial growth and the full development of face and mandible size, and cranial superstructures. When scoring these young specimens, we chose characters and character states that are little affected by their immaturity.
Parsimony analysis of the data matrix, including discrete and continuous characters was undertaken by using TNT, Tree analysis using New Technology, a parsimony analysis program subsidized by the Willi Hennig Society.13 We used the parallel version of TNT on 100 CPU cores. In total, one million replications were performed (10,000 replications on each core). The 234 discrete characters were all equally weighted. Forty-six multi-state characters were set as “ordered.” The merged cells with multiple states were set to polymorphism. To separately reflect recent results from palaeoproteomic and ancient DNA research,5,21,22,32 partial backbone constraints were used to force the Xiahe mandible as the sister group of Neanderthals and to force H. antecessor outside of the Neanderthal-Xiahe-H. sapiens clade (see the supplemental information). We used Bremer supports55 calculated in TNT to describe the stability of the phylogenetic results.
Estimating the split time of ancestral species/populations of Homo should be treated with caution, because all estimations must be based on particular models. The divergence times between Neanderthal, Denisovan, and fossil H. sapiens populations, as reflected by ancient DNA sequences and favored by one of the present authors (C.S. in Bergström et al.35), rely on a fixed human DNA sequence mutation rate.19,20,32,33,56 However, in our Bayesian tip-dating analysis we included fossil ages for all the OTUs to inform the divergence times of all the Homo clades, and co-estimated the clock rate together with the divergence times (instead of using a fixed mutation rate, see the supplemental information). We used the Bayesian tip-dating approach14, 15, 16, 17 implemented in MrBayes 3.2.723 to infer the timetree and evolutionary rates. This method integrates both the fossil ages and the morphological data while accounting for their uncertainties in a coherent analysis. Since MrBayes 3.2.7 cannot handle continuous characters directly and can only deal with ordered characters up to six states, all the continuous characters were discretized into six states. We executed four independent runs and eight chains per run (one cold and seven hot chains with temperature 0.05) in the Markov chain Monte Carlo simulation. Each run was executed with 100 million iterations, and sampled every 2,000 iterations.
To test whether different age estimates for the Harbin cranium would change its phylogenetic position in the Bayesian tip-dating analyses, we also used 296 ± 8 and 59–304 ka (the maximum U-series age and the maximum U-series age range, see the supplemental information) as the tip ages for the Harbin cranium. Different tip age estimates have very minor influence on the topology and the divergence age estimation of the whole tree (Figures S22 and S23).
Biogeographical analyses
We used the R41 package BioGeoBEARS39,40 to compare biogeographical models and estimate ancestral range probabilities of Homo species/populations. The same R package was also used to estimate the number of dispersal, vicariance, and sympatry events with BSM40 using the same R package. The Bayesian tip-dating all-compatible consensus tree was used for biogeographical analyses. We tested 18 biogeographical models, including 3 hypotheses of dispersal routes from Africa to Asia. AIC was used to select the best fitting model.37 We ran 1,000 BSMs under the best fitting biogeographical model, and calculated the means and standard deviations of biogeographical events across the 100 mapping processes.
Data availability
The phenomic data matrix, including scoring, metrical measurements, illustrations, and labels will be released on MorphoBank (project 3385) after publication. Full description of the methods and the scripts for computational analyses are given in the supplemental information. Other data will be available by request to X.N. and Q.J.
Acknowledgments
We thank Messrs Yemao Hou, Wei Zhang, Ning Ma, Ruiping Tang, and Ms Fang Zheng for CT scanning, surface model scanning, geological imaging, and Anthropology staff, especially Dr. Rachel Ives, at the Natural History Museum London for the provision of comparative material. We are grateful for the support of Drs. Fengming Wang, Chun Li, Xiujie Wu, Dongju Zhang, Lucile Crété, Tao Deng, and Tao Zhan. Heilongjiang Academy of Geological Sciences helped with core-drilling and sample collections. Dr. Tao Yang from the Nanjing University helped with the REE and Sr isotopic analyses. Drs. Lei Zhang, Jinhai Zhang, and Wei Yang from the Innovation Academy for Earth Science of Chinese Academy of Sciences helped with geographic imaging. We also thank Jinhai Zhang and Wei Yang for helpful discussions. This project has been supported by the National Natural Science Foundation of China (41842039, 41625005, 41888101, 41988101, 41877430, 41977380), the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB26030300, XDA20070203, XDA19050100), the People's Government of Hebei Province (Z20177187), the China Geological Survey (DD20190601), the Science Foundation of Hebei GEO University (TS2017-001), and the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0705). C.S.'s research is supported by the Calleva Foundation and the Human Origins Research Fund. Mr. Chuang Zhao produced the artist’s illustration of the life reconstruction. We thank the reviewers for their help in improving the paper.
Author contributions
Q.J. obtained the Harbin cranium, organized the project, and edited the manuscript. Q.S. performed U-series dating, REE and Sr isotopic analyses, analyzed the U-series dating data, and edited the manuscript. J.G. analyzed the REE and Sr isotopic data, and edited the manuscript. J.L. performed XRF analyses, and edited the manuscript. C.Z. performed the Bayesian tip-dating analysis, and edited the manuscript. R.G. analyzed the U-series dating data, and edited the manuscript. Q.L. collected the mammalian fossils, revised the phylogenetic data matrix, and edited the manuscript. W.W., Y.J., Z.G., and L.L. collected data, drilled the core, and measured sections. C.S. described and compared the fossils, revised the phylogenetic data matrix, and wrote the manuscript. X.N. organized the project, developed the phylogenetic data matrix, described and compared the fossils, performed phylogenetic and biogeographical analyses, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2021.100130.
Contributor Information
Xijun Ni, Email: nixijun@hgu.edu.cn.
Qiang Ji, Email: jiqiang@hgu.edu.cn.
Chris Stringer, Email: c.stringer@nhm.ac.uk.
Supplemental information
References
- 1.Grün R., Pike A., McDermott F., et al. Dating the skull from Broken Hill, Zambia, and its position in human evolution. Nature. 2020 doi: 10.1038/s41586-020-2165-4. [DOI] [PubMed] [Google Scholar]
- 2.Berger L.R., Hawks J., de Ruiter D.J., et al. Homo naledi, a new species of the genus Homo from the Dinaledi Chamber, South Africa. eLife. 2015;4:e09560. doi: 10.7554/eLife.09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hublin J.-J., Ben-Ncer A., Bailey S.E., et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 2017;546 doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 4.Wood B., Baker J. Evolution in the genus Homo. Ann. Rev. Ecol. Evol. Syst. 2011;42:47–69. doi: 10.1146/annurev-ecolsys-102209-144653. [DOI] [Google Scholar]
- 5.Reich D., Green R.E., Kircher M., et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X., Athreya S. A description of the geological context, discrete traits, and linear morphometrics of the middle Pleistocene hominin from Dali, Shaanxi Province, China. Am. J. Phys. Anthropol. 2013;150:141–157. doi: 10.1002/ajpa.22188. [DOI] [PubMed] [Google Scholar]
- 7.Athreya S., Wu X. A multivariate assessment of the Dali hominin cranium from China: morphological affinities and implications for Pleistocene evolution in East Asia. Am. J. Phys. Anthropol. 2017;164:679–701. doi: 10.1002/ajpa.23305. [DOI] [PubMed] [Google Scholar]
- 8.Li Z.-Y., Wu X.-J., Zhou L.-P., et al. Late Pleistocene archaic human crania from Xuchang, China. Science. 2017;355:969–972. doi: 10.1126/science.aal2482. [DOI] [PubMed] [Google Scholar]
- 9.Wu X.-J., Pei S.-W., Cai Y.-J., et al. Archaic human remains from Hualongdong, China, and Middle Pleistocene human continuity and variation. Proc. Natl. Acad. Sci. U.S.A. 2019;116:9820–9824. doi: 10.1073/pnas.1902396116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Q., Ge J., Ji Q., et al. Geochemical locating and direct dating of the Harbin archaic human cranium. The Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100131. In this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawyer S., Renaud G., Viola B., et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15696–15700. doi: 10.1073/pnas.1519905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zubova A.V., Chikisheva T.A., Shunkov M.V. The morphology of permanent molars from the Paleolithic layers of Denisova Cave. Archaeol. Ethnol. Anthrop. Euras. 2017;45:121–134. doi: 10.17746/1563-0110.2017.45.1.121-134. [DOI] [Google Scholar]
- 13.Goloboff P.A., Farris J.S., Nixon K.C. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- 14.Ronquist F., Klopfstein S., Vilhelmsen L., et al. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 2012;61:973–999. doi: 10.1093/sysbio/sys058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavryushkina A., Heath T.A., Ksepka D.T., et al. Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 2017;66:57–73. doi: 10.1093/sysbio/syw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C., Stadler T., Klopfstein S., et al. Total-evidence dating under the fossilized birth-death process. Syst. Biol. 2016;65:228–249. doi: 10.1093/sysbio/syv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C., Wang M. Bayesian tip dating reveals heterogeneous morphological clocks in Mesozoic birds. R. Soc. Open Sci. 2019;6:182062. doi: 10.1098/rsos.182062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arsuaga J.L., Martínez I., Arnold L.J., et al. Neandertal roots: cranial and chronological evidence from Sima de los Huesos. Science. 2014;344:1358–1363. doi: 10.1126/science.1253958. [DOI] [PubMed] [Google Scholar]
- 19.Meyer M., Arsuaga J.-L., de Filippo C., et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016;531:504–507. doi: 10.1038/nature17405. [DOI] [PubMed] [Google Scholar]
- 20.Meyer M., Fu Q., Aximu-Petri A. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505:403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 21.Chen F., Welker F., Shen C.-C., et al. A late Middle Pleistocene Denisovan mandible from the Tibetan plateau. Nature. 2019;569:409–412. doi: 10.1038/s41586-019-1139-x. [DOI] [PubMed] [Google Scholar]
- 22.Welker F., Ramos-Madrigal J., Gutenbrunner P., et al. The dental proteome of Homo antecessor. Nature. 2020;580:235–238. doi: 10.1038/s41586-020-2153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F., Teslenko M., van der Mark P., et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etler D.A. Homo erectus in East Asia: human ancestor or evolutionary dead-end? Athena Rev. 2006;4:37–50. [Google Scholar]
- 25.Wood B., Lonergan N. The hominin fossil record: taxa, grades and clades. J. Anat. 2008;212:354–376. doi: 10.1111/j.1469-7580.2008.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tattersall I., Schwartz J.H. Evolution of the genus Homo. Ann. Rev. Ear. Plan. Sci. 2009;37:67–92. doi: 10.1146/annurev.earth.031208.100202. [DOI] [Google Scholar]
- 27.Tattersall I. In: Continuity and Discontinuity in the Peopling of Europe: One Hundred Fifty Years of Neanderthal Study. Condemi S., Weniger G.-C., editors. Springer; 2011. pp. 47–53. [Google Scholar]
- 28.Stringer C.B. Reconstructing recent human evolution. Phil. Trans. R. Soc. B. 1992;337:217–224. doi: 10.1098/rstb.1992.0099. [DOI] [PubMed] [Google Scholar]
- 29.Derevianko A.P. The origin of anatomically modern humans and their behavior in Africa and Eurasia. Archaeol. Ethnol. Anthrop. Euras. 2011;39:2–31. doi: 10.1016/j.aeae.2011.09.001. [DOI] [Google Scholar]
- 30.Zhang D., Xia H., Chen F., et al. Denisovan DNA in late Pleistocene sediments from Baishiya Karst Cave on the Tibetan plateau. Science. 2020;370:584–587. doi: 10.1126/science.abb6320. [DOI] [PubMed] [Google Scholar]
- 31.Stringer C. The status of Homo heidelbergensis (Schoetensack 1908) Evol. Anthropol. 2012;21:101–107. doi: 10.1002/evan.21311. [DOI] [PubMed] [Google Scholar]
- 32.Prüfer K., Racimo F., Patterson N., et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posth C., Wißing C., Kitagawa K., et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Comm. 2017;8:16046. doi: 10.1038/ncomms16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petr M., Hajdinjak M., Fu Q., et al. The evolutionary history of Neanderthal and Denisovan Y chromosomes. Science. 2020;369:1653–1656. doi: 10.1126/science.abb6460. [DOI] [PubMed] [Google Scholar]
- 35.Bergström A., Stringer C., Hajdinjak M., et al. Origins of modern human ancestry. Nature. 2021;590:229–237. doi: 10.1038/s41586-021-03244-5. [DOI] [PubMed] [Google Scholar]
- 36.Harvati K., Röding C., Bosman A.M., et al. Apidima Cave fossils provide earliest evidence of Homo sapiens in Eurasia. Nature. 2019;571:500–504. doi: 10.1038/s41586-019-1376-z. [DOI] [PubMed] [Google Scholar]
- 37.Burnham K.P., Anderson D.R. Second edition. Springer; 2002. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; p. 488. [Google Scholar]
- 38.Ree R.H., Smith S.A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 39.Matzke N.J. Wikidot.com; 2016. Stochastic Mapping under Biogeographical Models.http://phylo.wikidot.com/biogeobears#stochastic_mapping [Google Scholar]
- 40.Matzke N.J. GitHub; 2018. BioGeoBEARS: bioGeography with Bayesian (and Likelihood) Evolutionary Analysis with R Scripts.https://github.com/nmatzke/BioGeoBEARS [Google Scholar]
- 41.R Core Team . The R Foundation; 2018. R: A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- 42.Matzke N.J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst. Biol. 2014;63:951–970. doi: 10.1093/sysbio/syu056. [DOI] [PubMed] [Google Scholar]
- 43.Soto-Trejo F., Matzke N.J., Schilling E.E., et al. Historical biogeography of Florestina (Asteraceae: Bahieae) of dry environments in Mexico: evaluating models and uncertainty in low-diversity clades. Bot. J. Linn. Soc. 2017;185:497–510. doi: 10.1093/botlinnean/box069. [DOI] [Google Scholar]
- 44.Dupin J., Matzke N.J., Särkinen T., et al. Bayesian estimation of the global biogeographical history of the Solanaceae. J. Biogeogr. 2017;44:887–899. doi: 10.1111/jbi.12898. [DOI] [Google Scholar]
- 45.Ruff C. Variation in human body size and shape. Ann. Rev. Anthropol. 2002;31:211–232. doi: 10.1146/annurev.anthro.31.040402.085407. [DOI] [Google Scholar]
- 46.Strait D.S., Grine F.E. Inferring hominoid and early hominid phylogeny using craniodental characters: the role of fossil taxa. J. Hum. Evol. 2004;47:399–452. doi: 10.1016/j.jhevol.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 47.González-José R., Escapa I., Neves W.A., et al. Cladistic analysis of continuous modularized traits provides phylogenetic signals in Homo evolution. Nature. 2008;453:775–778. doi: 10.1038/nature06891. [DOI] [PubMed] [Google Scholar]
- 48.Argue D., Morwood M., Sutikna T., et al. Homo floresiensis: a cladistic analysis. J. Hum. Evol. 2009;57:623–639. doi: 10.1016/j.jhevol.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Mounier A., Balzeau A., Caparros M., et al. Brain, calvarium, cladistics: a new approach to an old question, who are modern humans and Neandertals? J. Hum. Evol. 2016;92:22–36. doi: 10.1016/j.jhevol.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 50.O’Leary M.A., Kaufman S. MorphoBank: phylophenomics in the “cloud”. Cladistics. 2011;27:529–537. doi: 10.1111/j.1096-0031.2011.00355.x. [DOI] [PubMed] [Google Scholar]
- 51.Martin R., Saller K. Systematischer Darstellung. Lieferung 3. Systematische Anthropologie. Gustav Fischer Verlag; 1956. Lehrbuch der Anthropologie; pp. 273–518. [Google Scholar]
- 52.Howells W.W. Cranial variation in man. A study by multivariate analysis of patterns of differences among recent human populations. Pap. Peab. Mus. Archaeol. Ethnol. 1973;67:1–190. [Google Scholar]
- 53.McDade L.A. Species concepts and problems in practice: insight from botanical monographs. Syst. Bot. 1995;20:606–622. doi: 10.2307/2419813. [DOI] [Google Scholar]
- 54.Rieseberg L.H., Wood T.E., Baack E.J. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- 56.Bokelmann L., Hajdinjak M., Peyrégne S., et al. A genetic analysis of the Gibraltar Neanderthals. Proc. Natl. Acad. Sci. U.S.A. 2019;116:15610–15615. doi: 10.1073/pnas.1903984116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phenomic data matrix, including scoring, metrical measurements, illustrations, and labels will be released on MorphoBank (project 3385) after publication. Full description of the methods and the scripts for computational analyses are given in the supplemental information. Other data will be available by request to X.N. and Q.J.