Abstract
Soil salinity is a major environmental stress that restricts the growth and yield of crops. Understanding the physiological, metabolic, and biochemical responses of plants to salt stress and mining the salt tolerance-associated genetic resource in nature will be extremely important for us to cultivate salt-tolerant crops. In this review, we provide a comprehensive summary of the mechanisms of salt stress responses in plants, including salt stress-triggered physiological responses, oxidative stress, salt stress sensing and signaling pathways, organellar stress, ion homeostasis, hormonal and gene expression regulation, metabolic changes, as well as salt tolerance mechanisms in halophytes. Important questions regarding salt tolerance that need to be addressed in the future are discussed.
Keywords: salt stress, ion homeostasis, halophyte, hormones, oxidative stress, salt stress sensing, osmotic stress
Main Text
Introduction
Soil salinity is a worldwide problem that threatens the growth and yield of crops and prevents the sustainable development of modern agriculture. More than one-third of irrigated lands in the world are affected by salinization.1 The major causes of soil salinity are rising levels of groundwater with high salt content and poor-quality drainage and irrigation systems.2 All the major staple crops responsible for the bulk of calorie uptake by humans (e.g., rice, wheat, and corn) are glycophytes, which are unable to complete their life cycle when soil NaCl concentrations exceed 200 mM.3,4 Thus, improving salinity stress tolerance in crops is of paramount importance for global food security. To achieve this goal, it is necessary to understand how high salinity affects the morphological, physiological, biochemical, metabolic, and gene expression properties of plants.
The ability of plants to tolerate high salinity varies between and within species,4 which enables us to identify gene loci and natural variations that are critical for salt stress tolerance in plants. Fundamental studies in the model plant Arabidopsis have revealed many genes that are required for salt stress tolerance, and applications of some of these genes to crops increase their salt stress tolerance. The discovery of the salt overly sensitive (SOS) signaling pathway,5,6 a major mechanism behind the exclusion of Na+ from the cytosol to the outside of cells, was a milestone in our understanding of how plants deal with salt load. Recent work showed that glycosyl inositol phosphorylceramide (GIPC) sphingolipids may function as salt stress sensors in plants.7 Research on halophytic plants, which usually reside in high salinity environments, have also been important for us to understand salt tolerance mechanisms in plants. Some halophytes have developed special structures, such as epidermal bladder cells that accumulate excessive Na+ in their vacuoles, which enable them to adapt to high salinity. Recent progress in understanding the mechanisms underlying the salt stress tolerance of halophytes will facilitate the breeding of salt-tolerant crops. This review provides a synopsis of salt stress responses and adaptation in plants and considers the newest developments in the field. These developments include adaptative physiological responses, sensing and signaling pathways, salinity-induced stress on specific organelles, hormonal regulation, ion homeostasis, metabolic changes, and salt tolerance mechanisms of halophytes.
Physiological Responses to Salt Stress
Salinity stress inhibits plant growth and development by imposing several major constraints. The first constraint is an osmotic stress (the lowering of the external water potential) that compromises a plant's ability to take up water. This process triggers several major events in plant tissues. At the macroscopic level, expansion of both root8 and shoot9 cells is immediately arrested as a result of a decreased turgor pressure. To deal with this issue, plants must adjust osmotically. Most of the cell turgor in the root is regained within 40–60 min10 by an increase in the uptake of inorganic ions, and growth is resumed, although at a reduced rate. The latter fact is most likely explained by modification of the composition of the cell wall resulting from the binding of Na+ to cell wall components.11 Osmotic stress also results in rapid closure of stomata, which reduces the plant's ability to assimilate CO2. Rapid closure of stomata can be explained by the rapid drop in xylem pressure (e.g., by 0.05 MPa in maize roots exposed to 100 mM NaCl)12 that accompanies salinity stress. This drop in xylem pressure occurs within minutes upon stress onset, and the hydraulic signals sensed by roots move at the speed of sound13 and are transduced almost instantaneously to the shoot, where they are decoded and alter shoot metabolism.14 Because stomatal guard cells possess a range of mechano-sensitive (stretch-activated) ion channels,15,16 they could potentially transduce a change in the xylem pressure caused by salinity into altered stomatal apertures.
The second constraint imposed by salinity is ionic imbalance (often called “ionic stress” or “ion toxicity” in the literature). In most cases, this constraint is associated with an excessive accumulation of Na+ and Cl− in metabolically active intracellular compartments. Surprisingly, the mechanistic basis of such toxicity is poorly understood. Although it is well known that Na+ can harm plant metabolism and can potentially kill the plant,17 the target(s) of Na+ in the plant is unknown. The most common explanation for Na+ toxicity is that it has an inhibitory effect on the activities of enzymes. The cytosolic compartment, for example, contains many enzymes involved in primary metabolism, the Calvin cycle, the phenylpropanoid pathway, glycolysis, and polyamine and starch synthesis. Many of these enzymes are controlled by K+.18 Given the close similarity between Na+ and K+,19 it is usually accepted that the Na+ tends to replace K+ in those enzymatic reactions, but with much less efficiency.14,18,20 In addition Cheeseman,17 noted that biomacromolecules occupy 20%–30% of the cytoplasmic volume. Because biomacromolecules are complexed and charged 3D structures, their operation is strongly affected by electrostatic interactions and the ionic strength of the solution, as well as by the presence of both screened and unscreened electrostatic forces.21 As a result, small hydrated Na+ ions tend to accumulate in the regions of greater density, while larger K+ ions tend to be found in the less dense regions. This differential intracellular ion distribution will have an impact on cell operation, as the fixed charge and ionic conditions of the cytosol will inevitably determine the local water relations relevant to the cytoskeleton and proteins.17
A related issue is chloride (Cl−) toxicity. The current notion is that Cl− exclusion from the shoot is crucial for salt tolerance.22 This inference is supported by the findings in certain salt-sensitive species that high shoot Cl− levels are positively correlated with severe physiological dysfunctions.23,24 However, the negative correlation between shoot Cl− concentration and plant biomass recorded for some salt-grown non-halophytes does not hold for halophytes,23 some of which are capable of accumulating Cl− at a concentration >1 M without experiencing a major negative effect on plant performance.25 Researchers have argued that the detrimental effects of Cl− on plant performance may be not a result of toxicity per se but rather from a Cl−-induced deficiency of key macro-nutrients (e.g., N and S), as uptake of NO3− and SO42− are mediated by the same (non-selective) anion transporters as Cl−.23 It therefore appears that the negative effect caused by either Na+ or Cl− is not a nutrient toxicity per se but instead results from interference with the uptake or metabolism of other essential ions. For this reason, the use of the term ionic imbalance is more suitable and should be used instead of the more popular specific ion toxicity.
Another widely held misconception is related to the timescale imposed by these two constraints. The traditional view is that ionic stress has a slower speed of onset than osmotic stress and operates at a timescale of days if not weeks.26,27 Although this view does apply to shoot tissues, it does not apply to the plant as a whole. In response to ionic stress, various PCD (programmed cell death) events are observed in roots at a much more rapid timescale. Apoptotic events, such as DNA laddering or cytochrome c release, are observed in plant roots within hours of salinity exposure.28, 29, 30 In Arabidopsis roots, the level of autophagy (another form of PCD) peaks within 30 min of salt stress.31 Importantly, these salinity-induced PCD events are Na+ specific and not related to the osmotic component of salt stress.32,33 The salinity-induced PCD events in plant roots are significantly reduced or prevented in Arabidopsis mutants lacking a gated outwardly rectifying K+ channel (GORK),34 suggesting a causal link between Na+ entry into the cytosol, resulting in membrane depolarization and accompanied by K+ efflux, and activation of caspase-like proteases and endonucleases that execute PCD.35,36
Salinity and Oxidative Stress
Reactive oxygen species (ROS), which function as versatile signals, are rapidly induced by a variety of environmental stresses, including pathogen infection, high salinity, drought, and heat stress.37 The major ROS in plants include hydrogen peroxide (H2O2), superoxide anion (O2·−), singlet oxygen (1O2), and hydroxyl radical (OH·). These ROS are mainly produced in the apoplast, chloroplasts, mitochondria, and peroxisomes.37 Production of ROS in the apoplast is mediated by plasma membrane-localized respiratory burst oxidase homologs (such as AtRbohD and AtRbohF), apoplastic diamine oxidase, peroxidase, and polyamine oxidases.37 AtRbohD and AtRbohF genes are both upregulated under salt stress, and simultaneous mutations of these two genes result in hypersensitivity to salt stress.38 The mechanisms underlying the positive roles of AtRbohD/AtRbohF in salt stress tolerance have been revealed by several studies. Salt-induced production of ROS by AtRbohD/AtRbohF promotes the movement of K+ into the cytosol and thus reduces Na+/K+ ratios.38 AtRbohF is able to restrict the distribution of Na+ in xylem sap, and thereby reduces the delivery of Na+ from roots to shoots via transpiration.39 AtRbohD mediates the propagation of long-distance signals triggered by a variety of environmental stimuli, including high salinity, wounding, heat, cold, and high-intensity light,40 suggesting that ROS are required for systemic signaling in plants. AtRbohD/F-mediated production of H2O2 at the early stage of salt stress could be a signal that triggers an antioxidative response that reduces the oxidative damage to cells.41 ROS production in the apoplast contributes to lignin formation under a saline environment.42
In shoots, both osmotic stress-induced stomatal closure and accumulation of high levels of Na+ in the cytosol impair the photosynthetic machinery. As a result, the amount of absorbed light exceeds the demand for photosynthesis, which leads to the formation of ROS in green tissues.43 There are three major sites of ROS production in chloroplasts: (1) the Mehler reaction in the PSI; (2) 1O2 production by photosystem II (PSII) in the thylakoid membrane resulting from limitation of the electron transport chain between photosystems; and (3) H2O2 production at the electron donor side of PSII via incomplete oxidation of water due to the inhibition of the water-splitting manganese complex.44,45 Another major source of ROS production in salt-affected plants is mitochondrial respiration. Over-reduction of the ubiquinone pool during salt stress allows electrons to leak from complexes I and III of the mitochondrial electron transport chain to molecular oxygen, which results in O2·− production.37 The peroxisome is a major site for the production of intracellular H2O2.46 The reduced CO2/O2 ratio in plant cells under salt stress promotes H2O2 production in peroxisomes through enhanced photorespiration.47
At low concentrations, ROS act as signal molecules to regulate many biological processes, including plant growth and responses to a variety of biotic and abiotic stresses. An excessive accumulation of ROS under saline conditions, however, has detrimental effects on plant tissues. The detrimental effects of ROS are traditionally attributed to their ability to damage key cellular structures, including lipid peroxidation in cellular membranes, DNA damage, protein denaturation, carbohydrate oxidation, pigment breakdown, and an impairment of enzymatic activities.48 Before this damage to key cellular structures occurs, however, stress-induced accumulation of ROS disturbs ionic homeostasis in the cells by activating many different types of ROS-sensitive ion channels. H2O2-sensitive Ca2+-permeable ion channels have been found in both root epidermal cells49 and stomatal guard cells.50 Constitutively expressed inward-rectifying K+ channels in stomata are also inhibited by H2O.51 OH·, which is generated upon H2O2 reduction in cell walls, has a much wider action spectrum and can activate a broad range of non-selective (thus, Na+-permeable) cation channels52,53 as well as GORK-like K+ efflux channels,34,54 annexin-mediated conductance,55 and a Ca2+-pump.54 ROS-regulated ion channels are also present at organellar membranes (e.g., chloroplasts,56 vacuoles57) and alter the operation of these organelles. For example, the salt stress-induced decrease in the photosynthetic performance of chloroplasts is associated with the swelling of thylakoids58 that results from increased ion fluxes across the thylakoid membrane via H2O2-activated ion channels.56
To reduce the oxidative stress caused by the accumulation of ROS under high salinity, plants rely on activation of ROS-scavenging machineries. The enhanced tolerance of halophytes to high salinity is to some extent due to an enhanced capacity to maintain ROS homeostasis.59 The scavenging of excessive ROS under high salinity may be attributed to non-enzymatic antioxidant metabolites, including ascorbate, glutathione, and tocopherols, and to enzymatic agents, such as catalases (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), and glutathione reductase (GR).60 Enhancement of these antioxidant systems increases salt stress tolerance in plants. For example, the increased accumulation of total glutathione in peroxisomes, chloroplasts, and mitochondria conferred increased tolerance to salt stress,59,61 and a reduction in the ascorbate content in the vtc2-1 mutant is correlated with a reduction in salt stress tolerance.62 In chloroplasts, SOD catalyzes the transfer of O2·− to H2O2, and APX is required for the conversion of H2O2 to H2O.43 Plants that overexpress the APX gene show enhanced tolerance to salt and drought stresses.63 NCA1, a chaperone protein that is required for the maintenance of the functional state of CAT, positively regulates salt stress tolerance.64 In mitochondria, alternative oxidase (AOX) and manganese SOD (Mn-SOD) are the major enzymes involved in the detoxification of ROS. Plants with higher activities of mitochondrial AOX1 and Mn-SOD exhibit increased tolerance to salt and drought stress.65,66
Salt Stress Sensors
To avoid the damage caused by the high concentrations of salts in soil, plants must have evolved the ability to sense salt stress, to transduce signals to cell interiors, and to adjust cellular traits. Identification of salt stress sensor(s) has been challenging, perhaps because of the functional redundancy of sensors, technical difficulties, or lethality when salt stress sensors are knocked out. Many abiotic stresses, including high salinity, drought, and cold, trigger increases in the cytosolic Ca2+ concentration within seconds to minutes.67,68 For this reason, identification of proteins or other components that are required for the rapid influx of Ca2+ under stress conditions is considered to be a good way to discover stress sensors. Based on this possibility, researchers have conducted genetic screens for mutants that are defective in the activation of Ca2+ signaling under a variety of environmental stresses.
Reduced hyperosmolality-induced [Ca2+]iincrease1 (OSCA1), which encodes a hyperosmolality-gated calcium-permeable channel, was identified as an osmotic stress sensor.69 Mutation in the OSCA1 gene impairs osmotic stress-induced Ca2+ signaling in guard cells and root cells, which results in reduced stomatal closure and root growth in response to osmotic stress. The osca1 mutant does not show altered phenotypes under high salinity, which raises a question about the role of OSCA1 in the sense of high salinity-triggered osmotic stress. Using a similar screening strategy, researchers recently identified monocation-induced [Ca2+]i increases 1 (MOCA1), which encodes a glucuronosyltransferase that is involved in the biosynthesis of glycosyl inositol phosphorylceramide (GIPC) sphingolipids and that is specifically required for spikes in cytosolic Ca2+ in response to ionic stress but not in response to osmotic stress.7 Mutation in the MOCA1 gene leads to reduced weight and reduced survival under salt stress. GIPCs were shown to directly bind to Na+ and regulate the entry of Ca2+ into the cytosol.7 However, which Ca2+ channels are involved in this process and how binding of Na+ to GIPCs activates Ca2+ channels remain unknown.
Other proteins that mediate salt-induced Ca2+ signaling have also been reported. These proteins include FERONIA (FER), annexin1 (ANN1), and plastid K+ exchange antiporters (KEAs).70, 71, 72 FER, a member of the Catharanthus roseus receptor-like kinase 1-like (CrRLK1L) protein family, directly binds to pectin. Loss of function of FER results in reduced salt-induced Ca2+ signaling and increased sensitivity to high salinity.70 ANN1 mediates ROS-activated Ca2+ influx in response to increased accumulation of extracellular Na+. Mutation in the ANN1 gene leads to increased Na+ influx and impaired salt-induced transcriptional and growth adaptation.71 The KEA1/2 and KEA3 transporters are required for osmotic stress-induced Ca2+ response, suggesting that KEA1/2 and KEA3 may function as sensors of osmotic stress.72
In addition to sensing salt stress via Ca2+ signaling, plants may also sense salt stress by recognizing salinity-induced changes in cellular structures. High salinity rapidly reduces turgor pressure, which is the consequence of osmotic stress-mediated water loss. The reduced turgor pressure can be perceived by mechanosensitive sensors, such as MscS-like (MSL), Mid1-complementing activity (MCA), and two-pore potassium (TPK) family proteins.73 MSL8 is required for the survival of pollens subjected to the hyperosmotic shock of rehydration,74 and MSL2 and MSL3 are required for the adaptation of plastids to hyperosmotic stress.75 It is well known that excessive accumulation of ions in the apoplast affects the properties of cell wall components, which are perceived by cell wall-localized glycoproteins or plasma membrane-localized receptor-like kinases.76 Proteins potentially involved in sensing cell wall changes include hydroxyproline-rich glycoproteins (HRGPs), wall-associated kinases (WAKs), and CrRLK1L family proteins.76, 77, 78, 79 WAKs are able to bind to pectin and Ca2+, and excessive Na+ may affect these interactions and thus trigger stress signaling.80 Recent studies indicate that the cell wall-localized leucine-rich repeat extensins LRX3, LRX4, and LRX5, together with secretory peptides RALFs and the receptor-like kinase FER, are involved in sensing and relaying salt stress signals by monitoring the status of cell wall integrity, although the initial sensing of salinity-triggered cell wall changes by the LRXs is still not understood.79 FER inhibits the proton transport activity of plasma membrane H+-ATPase (AHA2) and thus regulates pH in the apoplast.81 FER is also required for the activation of cell wall repair pathways to maintain cell wall integrity under high salinity.70 FEI1 and FEI2, two leucine-rich repeat receptor kinases (LRR-RKs), are also associated with cell wall integrity sensing. Loss of function of FEI1 and FEI2 results in hypersensitivity to high sucrose and high salt.82,83
Ionic Stress Signaling
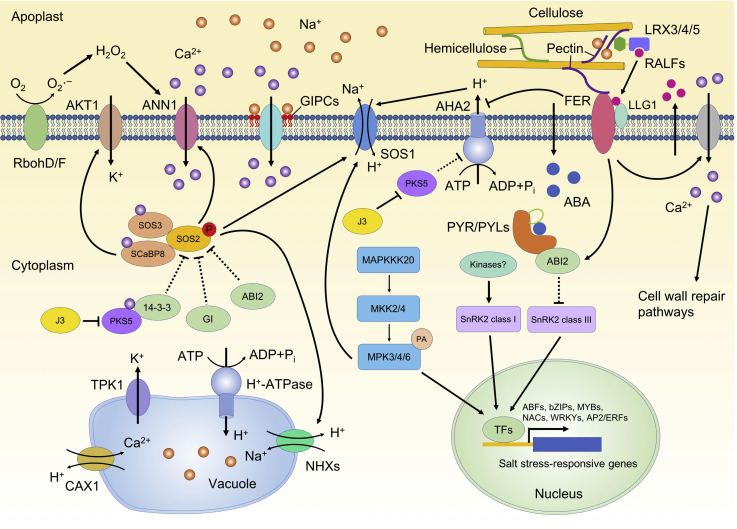
The environmental stimuli-triggered Ca2+ influx signal into the cytoplasm can be decoded by diverse Ca2+-dependent proteins, such as calcium-dependent protein kinases (CDPKs), calcineurin B-like proteins (CBLs)/SOS3-like calcium-binding proteins (SCaBPs), CBL-interacting protein kinases (CIPKs)/protein kinases of the SOS2 family (PKS).67 In Arabidopsis, the SOS signaling pathway, which consists of SOS3 and SCaBP8, SOS2, and SOS1, plays a critical role in the regulation of Na+/K+ ion homeostasis5,6,84 (Figure 1). SOS3 and SCaBP8 relay salt-induced Ca2+ signals to SOS2 kinase.84, 85, 86 SOS3 activates and recruits SOS2 to the plasma membrane via its interaction with the regulatory domain of SOS2.5 SOS2 is a serine/threonine protein kinase belonging to the sucrose non-fermenting 1 (SNF1)/AMPK family.5 Under normal conditions, the kinase activity of SOS2 is inhibited by 14-3-3 and GIGANTEA (GI) proteins. Salt stress promotes the degradation of 14-3-3 and GI, resulting in the release of SOS2 from SOS2-GI/14-3-3 complexes and consequently the activation of SOS2 by SOS3.87,88 A recent report indicated that PKS5 inhibits the activity of SOS2 by promoting interaction between SOS2 and 14-3-3 proteins; the salt stress-induced Ca2+ signal induces the interaction between PKS5 and 14-3-3 and thus releases the inhibition of SOS2.89 The kinase activity of SOS2 is also regulated by the protein phosphatase 2C ABI2.90 SOS1 is a plasma membrane Na+/H+ antiporter that is required for the extrusion of the excess of Na+ out of the cells (i.e., into the rhizosphere via root epidermal cells, or into the xylem via xylem parenchyma cells) and therefore for the alleviation of ionic stress. SOS1 is autoinhibited under normal conditions, and the inhibition is released by the phosphorylation of Ser1044 at the C-terminal domain of SOS1 by SOS2 under salt stress.91 Notably, sos mutants only show a hypersensitive phenotype under high salinity but grow normally under osmotic stress imposed by mannitol or PEG (polyethylene glycol), indicating that the SOS signaling pathway is specifically involved in the response to ionic stress. SCaBP8 also mediates the regulation of Na+/K+ ion homeostasis by modulating the activity of the K+ channel AKT1, a process that requires SOS2-mediated phosphorylation of SCaBP8.92,93
Figure 1.
Salt Stress Signaling Pathways
The SOS signaling pathway, consisting of SOS3/SOS3-like calcium-binding protein 8 (SCaBP8), SOS2, and SOS1, is important for sensing salt-induced Ca2+ signals and in the regulation of ion homeostasis by extruding excessive Na+ out of cells. 14-3-3, GI, and ABI2 negatively regulate the kinase activity of SOS2. Ca2+-mediated binding of PKS5 with 14-3-3 releases the inhibition on SOS2. GIPCs act as putative salt stress sensors that directly bind to Na+ and trigger Ca2+ influx via an unknown Ca2+ channel. GIPCs-mediated Ca2+ influx is required for the activation of the SOS signaling pathway. RbohD/F are involved in the production of ROS at the plasma membrane, and ROS can activate the ANN1-mediated Ca2+ signaling pathway. AKT1, which is regulated by SCaBP8, mediates the influx of K+ to the cytosol under salt stress. MAP kinase cascades, including MAPKKK20, MKK2, MKK4, MPK3, MPK4, and MPK6, are involved in the relay of salt stress signals. Salt stress-induced accumulation of ABA activates subclass III SNF1-related protein kinase 2s (SnRK2s) via the PYR/PYLs-PP2Cs-mediated regulatory module. Subclass I SnRK2s are activated via an ABA-independent pathway under osmotic stress. Activated MPKs and SnRK2s transduce signals to downstream transcription factors, including ABFs, zips, MYBs, NACs, WRKYs, and AP2/ERFs, in the nucleus to induce the expression of stress-responsive genes. In the apoplast, cell wall-localized leucine-rich repeat extensins LRX3, LRX4, and LRX5, together with secreted peptides RALF22/23 and receptor-like kinase FER, function as a module to sense salt stress-induced cell wall changes. FER, RALFs, and LLG1 form a complex at the plasma membrane to trigger Ca2+ signaling and consequently activate the cell wall repair pathway. FER also inhibits the activity of AHA2 to regulate apoplastic pH. In the vacuole, NHXs, CAX1, TPK1, and H+-ATPase are involved in the regulation of ion homeostasis under high salinity. The dashed lines indicate that the negative regulatory roles are released under salt stress.
MAPK cascades94, 95, 96 and phosphatidic acid (PA)97 are also involved in the regulation of the salt stress signaling pathway. Salt stress activates MPK3, MPK4, and MPK6, which contribute to salt stress tolerance in Arabidopsis.94,95,97 The activities of MPK4 and MPK6 under salt stress are induced by MKK2. Phenotypic analysis revealed that plants overexpressing MKK2 exhibit increased salt stress tolerance, while mkk2 null mutants are hypersensitive to salt stress.94 In rice, the osmkk1-knockout mutant is hypersensitive to salt stress, which is caused by the impaired activation of OsMPK4.95 Conversely, activation of MKK9 increases sensitivity to salt stress in Arabidopsis,96 suggesting a complex regulatory network among distinct MAP cascades in response to salt stress. Salt stress triggers phospholipase D (PLDα)-mediated production of PA, which in turn regulates the salt stress response by activating MPK6. Activated MPK6 phosphorylates the C-terminal fragment of SOS1 and promotes salt stress tolerance.97 Mutations in PLDα1, PLDδ, or PLDα3 lead to hypersensitivity to salt stress.98, 99, 100 PLD-mediated production of PA is also required for the activation of H+-ATPase101 and root hair formation under high salinity.102
Osmotic Stress Signaling
Whether cellular responses are induced by salinity-induced ion stress or osmotic stress can be determined by comparing plants exposed to high salinity with those exposed to isotonic non-ionic solutions, such as mannitol and PEG. Osmotic stress triggers signaling pathways that promote the biosynthesis and accumulation of compatible osmolytes, which is important for both short-term and long-term osmotic stress tolerance in plants. The increased levels of compatible osmolytes in the cytosol reduce water loss and enhance turgor pressure and cell expansion.103 Both salt stress and osmotic stress can activate SNF1-related protein kinase 2s (SnRK2s).104,105 Osmotic stress triggers the activation of SnRK2s in both ABA-dependent and -independent manners. SnRK2 kinases are divided into three groups: subclasses I, II, and III.104,105 Subclass III SnRK2 kinases, including SnRK2.2, SnRK2.3, and SnRK2.6, play an important role in the transduction of ABA signals and in the regulation of downstream gene expression responses by activating AREB/ABF (ABA-responsive element binding factor) transcription factors.106, 107, 108 Subclass I SnRK2s, including SnRK2.1, SnRK2.4, SnRK2.5, SnRK2.9, and SnRK2.10, are specifically responsive to osmotic stress, but not to ABA.104,109,110 The molecular mechanisms underlying the specificity of subclass I SnRK2s to osmotic stress are still unclear. It is likely that some early signaling components, such as kinases, can be activated by osmotic stress to trigger the activation of subclass I SnRK2s. Abiotic stress-responsive Raf-like kinases mediate the activation of SnRK2s under osmotic stress in the moss Physcomitrella patens,110 and whether this is also the case in higher plants, such as Arabidopsis, needs to be investigated. Phenotypic analysis indicates that the simultaneous disruption of ten SnRK2s increases growth inhibition and leaf chlorosis under osmotic stress.111 Among these SnRK2s, SnRK2.4 and SnRK2.10 are important in the maintenance of root growth and architecture under saline conditions.105,112 The MKK4-MPK3 and MAPKKK20-MPK6 cascades are required for osmotic stress responses in Arabidopsis.113,114 Loss of function of MKK4 increases water loss and ROS accumulation under dehydration conditions, and salt-induced activation of MKK4 regulates the activity of MPK3 and increases the expression of abiotic stress-responsive genes NCED3 and RD29A.114 MAPKKK20 is required for the activation of MPK6 under various abiotic stresses. The plants overexpressing MAPKKK20 exhibit enhanced tolerance to salt stress.113
Salt-Induced Organellar Stresses
Proper functioning of cells requires coordination among different organelles and cellular compartments. The cellular damage caused by abiotic stresses, including high salinity, drought, cold, and heat, can cause stress on various organelles.115 Endoplasmic reticulum (ER) stress has been widely considered as an important cellular response to stress conditions, and significant advances have recently been made to understand the role of cell wall stress in salt stress tolerance. To attenuate stress-induced organellar stresses, each affected organelle must be able to perceive the stress signals and also to relay the signals to the nucleus.
Cell Wall Stress
The plant cell wall consists of cellulose, hemicellulose, pectins, and many glycoproteins. Cell wall integrity is an important factor that determines plant growth and salt stress tolerance.70,79 Several mutants that are defective in cell wall integrity are hypersensitive to salt stress. For example, CESA6, which is a core component of the cellulose synthase complex, is required for normal root elongation under salt stress.116 SOS5, encoding a fasciclin-like arabinogalactan protein (AGP),117 and SOS6, encoding a cellulose synthase-like protein, are required for root elongation under high salinity.118 CC1 and CC2 were discovered as companions of CESAs, and both are required for hypocotyl growth under high salinity.119 MUR4 is a Golgi-localized enzyme involved in the biosynthesis of UDP-arabinose (UDP-Ara). UDP-Ara is involved in the modification of diverse polysaccharides and glycoproteins, which are exported to the apoplast to maintain cell wall integrity. Mutation in MUR4 leads to reduced root elongation and defective cell-cell adhesion under high salinity,120 indicating that arabinose modifications are important for the regulation of cell wall integrity in roots under salt stress.
Na+ accumulated in the apoplast may directly bind to cell wall components and affect their chemical properties.121 Because detecting the changes in cell wall composition is challenging, our understanding of the mechanisms underlying the modifications of cell walls upon exposure to high salinity is limited. Cell wall polymers are negatively charged and are therefore able to reversibly bind to cations.121,122 Pectin, a major component of the cell wall, is composed of homogalacturonan, rhamnogalacturonan I and II (RGI and RGII), and xylogalacturonan.123 Boron is required for the cross-linking of RGII,124 and the cross-linking is coordinated by Ca2+.125 When present at high levels in the apoplast, Na+ may replace Ca2+ in binding to pectins and may thereby interfere with pectin cross-linking,11 leading to reduced cell expansion.126 Pectins are synthesized and secreted into the cell wall in a methylesterified form. Pectin methyl esterase (PME)-mediated demethylesterification is an important form of pectin modification.126 Binding of Na+ to the substrate sites of PMEs affects the demethylesterification of pectins and thus inhibits cell growth.11,127 AGPs function as a reservoir of extracellular Ca2+. Excessive Na+ may free the Ca2+ bound in the AGPs and initiate influx of Ca2+ into the cytosol.128 Plants that are defective in the production of AGPs are hypersensitive to high salinity.129,130
Apoplastic acidification promotes cell expansion. High salinity causes the alkalinization of the apoplast131 and thus inhibits cell growth. RALF peptides were identified that can cause alkalinization of the apoplast by regulating H+-ATPases at the plasma membrane.81 High salinity can induce the production of mature RALF peptides,79 suggesting that salt stress-induced alkalinization of the apoplast is probably mediated by RALFs. The effect of apoplastic pH on cell growth is mainly mediated by the regulation of expansin activities.11 Expansins facilitate cell expansion by loosening cell walls under a variety of environmental stresses, including high salinity and drought stress.124
Chloroplast Stress
In addition to functioning in photosynthesis, chloroplasts also contribute to the biosynthesis of amino acids, vitamins, isoprenoids, fatty acids, and lipids.132 Dysfunction of chloroplasts caused by environmental stresses can have harmful effects on the physiological, biochemical, and metabolic properties of plant cells. High salinity has multiple effects on chloroplasts, including reduced CO2 uptake due to stomatal closure, reduced photosynthetic efficiency, thylakoid membrane damage, oxidative stress, impaired osmotic and ionic homeostasis, and disrupted protein synthesis and turnover.133 The reduced efficiency of photosynthesis is a major reason for the growth inhibition that occurs under high salinity.134,135 K+, as an essential nutrient for plants, is required for the regulation of pH, volume, and electron transport in chloroplast.56,136 Excessive accumulation of Na+ and Cl− results in reduced K+ influx in chloroplasts and thus causes ionic, osmotic, and oxidative stresses (see more details in other sections). Transcriptomic profiling has indicated that 53 salt-responsive genes encode chloroplast-localized proteins, many of which are important for salt stress tolerance.133 Most steps of ABA biosynthesis occur in the chloroplast, and ABA biosynthesis-associated proteins, such as ABA1, ABA4, and NCED3, are localized in chloroplasts and are required for salt stress-induced accumulation of ABA.137,138 MsK4, a novel Medicago sativa GSK-3-like kinase localized in plastid, positively regulates the salt stress response by modulating sugar metabolism.139 Fad6 and GPAT, two plastid-localized enzymes, facilitate thylakoid membrane fluidity and thereby increase salt stress tolerance by modulating fatty acid metabolism.140,141 Some genes encoded in chloroplasts, such as RUB and RCI, are associated with the maintenance of PSII activity under high salinity.142,143
The signals caused by chloroplast stress can be transduced to the nucleus via retrograde signaling pathways.132 High salinity-induced production of 1O2, which causes photo-oxidative damage of PSII, acts as one of the retrograde signals.144 1O2 can be sensed by EXECUTER (EX1), a nuclear-encoded protein localized in the thylakoid membrane of chloroplasts.145 The stress-induced release of 1O2 promotes the degradation of EX1, the process of which depends on oxidative post-translational modification at the Trp643 residue in the DUF3506 domain of EX1.144 Besides the EX1-mediated pathway, 1O2 can trigger chloroplast-to-nucleus retrograde signaling via oxidative products of beta carotene.146 Methylerythritol cyclodiphosphate (MEcPP), a precursor of isoprenoids produced by the plastidial methylerythritol phosphate (MEP) pathway,147 and phosphonucleotide (3′-phosphoadenosine 5′-phosphate [PAP]),148 are another two retrograde signaling metabolites involved in the transduction of signals from chloroplast to the nucleus to regulate stress-responsive gene expression.
ER Stress
Biotic and abiotic stresses can cause the accumulation of unfolded or misfolded proteins in the ER, resulting in ER stress. The misfolded proteins can be recognized by a protein quality control system in the ER, which induces the expression of chaperone genes that are important for protein folding and triggers ER-associated protein degradation (ERAD) and autophagy.149 Upon exposure to salt stress, the ubiquitinated proteins increase in the ER, and this accumulation activates an ER stress response. The positive role of the ER stress response in salt stress tolerance is supported by the finding that a defect in the HRD3A of the HRD1/HRD3 complex, which is required for the unfolded protein response in the ERAD pathway, confers hypersensitivity to salt stress, and Ca2+ and ROS are required for the ERAD-mediated response to salt stress.150 UBC32, which encodes an E2 ubiquitin-conjugating enzyme, is an active component of the plant ERAD compartment. UBC32 gene expression is highly induced by drought and salt stress, and loss of function of UBC32 enhances salt stress tolerance via a BR-dependent pathway.151 ER stress can trigger regulated intramembrane proteolysis under stress conditions. For example, salt stress induces subtilisin-like serine protease (AtS1P)-mediated cleavage of a membrane-localized bZIP transcription factor, AtbZIP17, in the ER. The activated AtbZIP17 translocates to the nucleus where it upregulates the expression of many salt stress responsive genes.152 S2P stimulates the nuclear localization of bZIP17 and bZIP28 via a process of cleavage. In the nucleus, these two transcription factors induce the expression of chaperone genes and the activation of BR signaling, and finally confer salt stress tolerance.153
Mitochondrial Stress
The mitochondrion is an energy-producing organelle that is important for the survival of plants under stress conditions.154 Abiotic stress-induced perturbation of mitochondrial functions can activate the expression of stress-responsive genes via a mitochondrial retrograde regulation (MRR) or retrograde signaling pathway.154,155 AOX is a well-studied mitochondrion-localized protein that is responsive to environmental stresses, and AOX induction has been used as a marker for MRR in response to stress.154 In Arabidopsis, influx of Ca2+ into mitochondrial is required for the induction of AOX1a gene expression under salt stress,156 and ABA INSENSITIVE 4 (ABI4) acts as a negative regulator of AOX1a gene expression. Cyclin-dependent kinase E1 (CDKE1), a component of the mediator complex, regulates AOX1a gene expression by interacting with SNF1-related kinase 1 (SnRK1/KIN10).157
Hormonal Regulation during Salt Stress
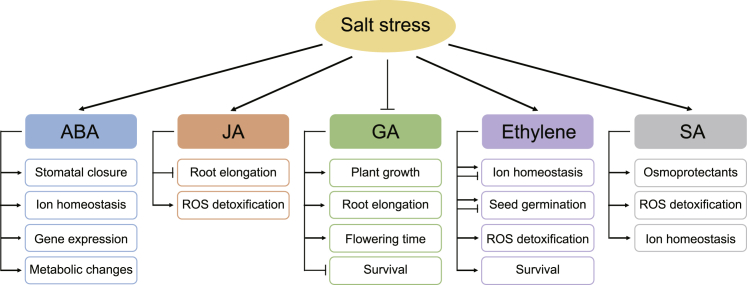
Response and adaptation to salt stress require the integration and coordination of multiple phytohormones, including ABA, jasmonic acid (JA), gibberellic acid (GA), ethylene, and salicylic acid (SA) (Figure 2). Among these hormones, ABA is the most involved in the response to diverse abiotic stresses. Osmotic stress imposed on roots results in a very rapid (within several minutes) and massive increase in ABA concentration in both root and leaf tissues.9,158 ABA is one of the key signaling molecules known to cause stomatal closure.159 This process involves binding of ABA to PYR/PYL/RCAR receptors.160,161 Once the receptors bind to ABA, they interact with PP2C phosphatases and inhibit their activity, thus releasing SnRK2s from repression.160,161 The SnRK2s then activate a range of anion efflux channels162,163 resulting in a loss of turgor pressure and stomatal closure. Although the role of ABA in salinity-induced stomatal closure is beyond any doubt, the origin of the ABA signal is still debated. For many years, it was thought that ABA is generated in osmotically stressed roots and is then rapidly transported to the shoot with the transpiration stream.164,165 More recent studies, however, showed that root-to-shoot ABA delivery may be not be required for stress-induced stomatal closure.166,167 The NCED3 gene, which encodes the first step of ABA biosynthesis, is expressed predominantly in the vascular parenchyma of leaves168 and is rapidly upregulated by osmotic stress, and many experiments using reciprocal grafting of ABA-deficient mutants suggest that drought resistance was conferred by the genotype of the scion and not of the rootstock (reviewed in Buckley166). In addition, ABA synthesis in roots may require precursors transported from leaves.169 On the other hand, the stress-induced increase in ABA concentration is several fold higher in roots than in leaves,158 but the role of ABA production in roots in the plant response to salt stress remains unclear.
Figure 2.
The Biological Functions of Phytohormones in the Regulation of Salt Stress Response in Plants
ABA is a major hormone involved in the regulation of salt stress response, including the regulation of stomatal closure, ion homeostasis, salt stress-responsive gene expression, and metabolic changes. JA is required for the inhibition of root elongation and activation of antioxidative enzymes upon exposure to high salinity. Salt stress reduces the accumulation of endogenous bioactive GAs, leading to inhibition of plant growth and root elongation, delay of flowering, and promotion of survival under high salinity. The effect of ethylene on salt stress tolerance acts in a species- or gene-specific manner. The components involved in ethylene biosynthesis or signaling transduction either positively or negatively regulate ion homeostasis and seed germination, but ethylene induces detoxifying machineries and promotes survival under salt stress. SA participates in the accumulation of osmoprotectants, induction of antioxidative enzymes, and improvement of ion homeostasis under salt stress.
A clue to how ABA production in roots may contribute to the salt stress response is provided by the fact that ABA interacts with H2O2 in plant systemic responses to stresses.170 Salinity stress results in a significant accumulation of ROS in plant roots.171,172 A major source of these ROS is NADPH oxidase, a plasma-membrane-bound enzyme complex from the NOX family.173 Osmotic stress-induced increase in H2O2 production requires NADPH oxidase stimulation by ABA.174 Also, inhibition of the NADPH oxidase-mediated H2O2 production in the root abolishes rapid stomatal closure, resulting in a salt-sensitive phenotype.172 Thus, salinity-induced increase in root ABA content may be critical for generating an “ROS wave” that triggers stomatal closure.
Increasing evidence has linked the JA pathway to salt stress responses in plants.175 Transcriptomic studies have revealed that many JA biosynthesis genes are upregulated under salt stress and that the JA signaling pathway is involved in the regulation of salt stress-responsive genes.176,177 In Arabidopsis, the JA signaling pathway is required for the inhibition of root elongation under high salinity.178 In rice, RICE SALT SENSITIVE3 (RSS3), a nuclear-localized protein, promotes root cell elongation under salt stress by physically interacting with class C basic-helix-loop-helix (bHLH) transcription factors and JASMONATE ZIM-DOMAIN (JAZ) proteins, the latter being the negative regulators of the JA pathway. Loss of function of RSS3 results in the upregulation of JA-responsive genes.179 These results support the notion that the JA pathway is required for root growth inhibition under high salinity. Overexpression of the OsCYP94C2b gene, which encodes an enzyme that catalyzes the conversion of bioactive JA-Ile to an inactive form, enhances the survival rate of rice under high salinity, demonstrating the negative role of JA in salt stress tolerance. However, opposite phenotypes have also been observed in studies showing that overexpression of the JA biosynthesis gene TaORP1 or application of exogenous JA enhances salt stress tolerance in wheat, rice, and soybean.180, 181, 182 Together, these results suggest that JA may act as a positive or negative regulator of salt stress response in a spatially and temporally dependent manner.
Coordination of growth and stress tolerance is critical for the survival of plants under unfavorable conditions. GA, as an important hormone that regulates plant growth, has been linked to the regulation of growth under abiotic stress.183 Treatment of Arabidopsis seedlings with salt reduces endogenous bioactive GAs and increases the accumulation of DELLA protein.184,185 In the quadruple-della mutant, salt stress-triggered growth inhibition and delayed flowering are attenuated, and salt stress-induced death is enhanced,184 suggesting that DELLA proteins promote the survival of plants by restricting growth under high salinity. Ethylene is also involved in salt stress tolerance in plants. Application of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) increases salt stress tolerance,186 while mutations in ethylene signaling pathway-associated genes, such as ETR1, EIN4, EIN2, and EIN3, lead to hypersensitivity to high salinity.186, 187, 188, 189 The positive effect of ethylene on salt stress tolerance is largely mediated via the modulation of ROS-generating and ROS-scavenging machineries.187,190 The role of SA in salt stress tolerance is represented by its ability to improve the accumulation of osmoprotectants, such as glycine betaine, proline, and polyamines,191 and to enhance antioxidant enzyme activities under high salinity.192 Pretreatment of plants with SA also reduces NaCl-induced K+ efflux and H+ influx and thus promotes the adaptation of plants to high salinity.193
Salt-Responsive Gene Expression and Epigenetic Regulation of Salt Stress Tolerance
Transcription is the first and the most critical step in the regulation of gene expression. Early transcriptome analyses in Arabidopsis indicated that salt stress causes differential expression in hundreds to thousands of genes depending on the strength or duration of the treatment.194,195 Salt stress, osmotic stress, and ABA treatment induce common sets of differentially expressed genes, especially during the early stages of stress treatment.194,195 The transcriptome is significantly different when salt stress is combined with other types of environmental stresses compared with when salt stress is applied alone, indicating extensive crosstalk between the salt stress signaling pathway and other stress signaling pathways.196 To date, hundreds if not thousands of salt stress-related transcriptomes have been conducted in many plant species. Members from all of the major transcription factor (TF) families (such as NAC, ERF/AP2, bZIP, MYB, and WRKY) have been found to be involved in the salt stress response. However, unlike cold acclimation, for which CBF (C-repeat binding factor)/DREB (dehydration responsive element binding protein) TFs function as master regulators, master transcriptional regulators for salt stress have not been identified. CBF1/2/3, belonging to the AP2/ERF (APETALA2/Ethylene-Responsive Factor) family, were first identified for their involvement in low-temperature responses.197,198 Overexpression of CBF3/DREB1A enhances plant tolerance to salinity,199 and mutation of all three CBF genes results in hypersensitivity to salt stress,200 indicating a positive role of CBFs in plant salt response. ABA mediates transcriptional regulation mainly through the AREB/ABF subfamily of bZIP TFs.201,202 For example, overexpression of ABF2 increases plant resistance to multiple stresses, including salt stress.203 A new class of ABA-responsive TFs named DIG (dynamic influencer of gene expression)/DIL (DIG-like) was also identified, and overexpression of these TFs results in hypersensitivity to high salt or ABA.204 TFs involved in other hormone signaling pathways also function in the salt stress response. MYC2, a master TF of jasmonate signaling, is a positive regulator of salt tolerance.181,205 EIN3, a TF that mediates core ethylene signaling, is stabilized by salt treatment and increases plant salt tolerance through the DELLA proteins.184,187 EIN3 increases salt tolerance partly through two downstream TFs, ERF1 (Ethylene Response Factor 1) and ESE1 (Ethylene and Salt Inducible 1), which directly bind to salt-responsive genes and activate their expression.189,206 Many other TFs from non-model plant species have also been identified mainly through transcriptome analyses. Although the specific mechanisms by which most of these TFs operate are not clear, their roles in salt tolerance have usually been validated using transgenic approaches in Arabidopsis or the original species.207 It is also important to keep in mind that TFs are highly dynamic and that the transcriptional network functions in a spatially and temporally specific manner. A recent study constructed an ABA-responsive network and defined the hierarchy among 21 ABA-related TFs by associating the in vivo binding dynamics of these TFs with time-series transcriptome data.204 The results revealed that dynamic binding of multiple TFs, compared with static binding, better predicts changes in gene expression over time.204 In summary, many TFs are involved in the regulation of salt-induced changes in gene expression. High-throughput sequencing technology facilitates the identification of TFs involved in the salt stress response. Additional research on the dynamics of transcriptional networks is needed, however, to increase systematic understanding and identify key players in transcriptional regulation of the salt stress response.
Genomic DNA in the eukaryotic nucleus is packed into the highly ordered structure of chromatin. Transcriptional regulation inevitably requires dynamic changes in chromatin conformation. Many chromatin modifiers have been identified as regulators of the salt stress response in plants. The nucleosome is the basic unit of chromatin and is typically composed of 147 bp of genomic DNA wrapped around a histone octamer containing two copies of histone H2A, H2B, H3, and H4. The structure of chromatin can be remodeled by ATP-dependent remodelers or covalent modifications. Histones contain flexible N-terminal tails, many residues of which are post-translationally modified by methylation, acetylation, phosphorylation, ubiquitination, etc. In addition, genomic DNA can be methylated. Specific modifications or a combination of modifications can directly change the chromatin conformation or can recruit specific binding proteins that promote changes. For example, acetylation of the histone lysine residue neutralizes the positive charge of the lysine side chain and reduces the interaction between the histones and the negatively charged DNA backbone, resulting in a less-compact chromatin conformation. As a consequence, histone acetylation is usually associated with transcriptional activation. Histone acetylation is catalyzed by histone acetyltransferases (HATs) and is removed by histone deacetylases (HDACs). Multiple HATs and HDACs are involved in the salt stress response. The plant-specific histone deacetylase HD2C interacts with HDA6 and reduces salt tolerance by repressing the expression of ABA-responsive genes such as ABI1 and ABI2.208 The class I (HDA6/HDA9/HDA19) and class II enzymes (HDA5/14/15/18) of the RPD3 histone deacetylase family play negative and positive roles in plant salinity tolerance, respectively.209, 210, 211 Furthermore, HUB2, a ubiquitin E3 ligase responsible for mono-ubiquitination of histone H2B, increases plant responses to drought and salt.212,213 In Arabidopsis, the NAP1 (Nucleosome Assembly Protein 1) protein, which functions as a histone chaperone for histone H2A and H2B, is a positive regulator of ABA signaling.214 In addition to factors that modify chromatin composition and/or conformation, various types of noncoding RNAs, including siRNAs (small interfering RNA), miRNAs (microRNA), or lncRNAs (long noncoding RNAs), also contribute to salt stress response. Additional details are provided in other reviews.215, 216, 217
Certain chromatin modifications such as DNA methylation and histone H3 lysine 27 trimethylation (H3K27me3) can be faithfully transmitted through meiosis or mitosis and are therefore considered epigenetics.218, 219, 220 Epigenetic regulatory mechanisms of salt stress tolerance or abiotic stress tolerance, in general, have drawn substantial attention in the past two decades because they represent attractive mechanisms for plant stress memory, which enables plants to enhance their stress response by remembering previous stresses. This effect is also termed “stress priming.” Priming of seeds using sodium chloride, hyperosmotic reagents, or BABA (β-aminobutyric acid) enhances the drought or salinity tolerance of the plants.221, 222, 223 In Arabidopsis, mild salt priming of seeds results in genome-wide alteration of H3K27me3, a repressive histone mark typically associated with developmental genes.223 In addition, multiple studies have reported salt stress-induced changes in DNA methylation across the genome.224, 225, 226 Natural epigenetic variations or DNA methylation defects resulting from mutations in the DNA methylation machinery have been linked to gene expression differences involved in salt tolerance.227, 228, 229 Despite these observations, convincing examples showing salt-induced epigenetic (i.e., heritable) changes that are important for salt stress response have yet to be reported. The stability of the epigenetic mark and the functional consequences of stress-induced epigenomic reprograming remain unclear. A recent study of DNA methylome changes in response to phosphate starvation in Arabidopsis found that most DNA methylation changes occur after transcriptional changes and are usually transient.230 Many studies on stress-induced epigenetic changes have been performed in Arabidopsis, which has an unusual plant genome with a very low amount of transposable elements. Salt-induced epigenetic changes in other plant species remain to be determined.
Ion Homeostasis under Salinity
Salinity stress is usually associated with too much NaCl. Because Na+ is considered to be toxic (see the section on physiological responses to salt stress for details), it is hardly surprising that most research concerning salinity stress has been aimed at revealing the mechanisms of Na+ transport and sequestration in plants. This was nicely summarized by Cheeseman,231 who stated “physiological folklore has elevated sodium toxicity to a belief in the almost paranoiac avoidance of cytoplasmic Na+”. Even though that paper was published more than 30 years ago, Cheeseman's statement still describes the current view.
Plants can use two major ways to prevent accumulation of high levels of Na+ in the cytosol of root cells: (1) Na+ exclusion from root uptake and (2) vacuolar Na+ sequestration. For photosynthetically active mesophyll cells, there are three additional ways: (3) control of xylem Na+ loading; (4) Na+ retrieval from the xylem; and (5) Na+ recirculation from the shoot via the phloem.
Na+ Exclusion from Uptake
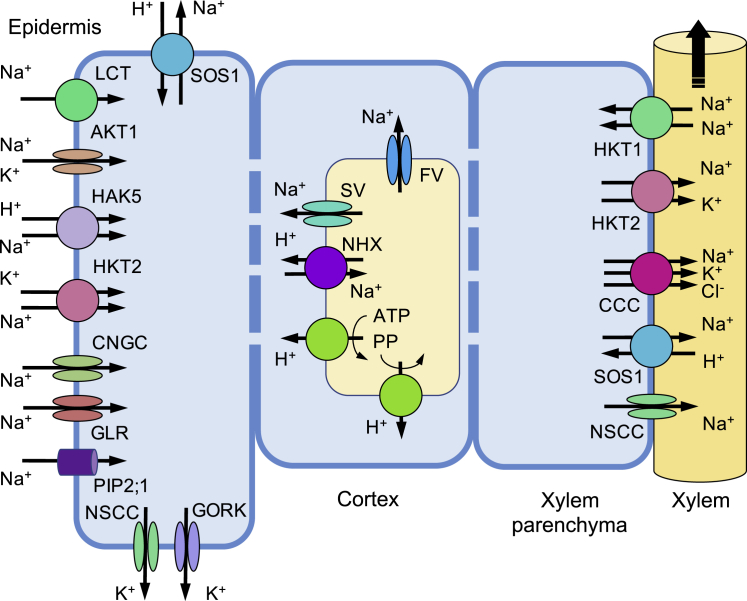
Symplastic root uptake of Na+ is potentially mediated by several types of ion transporters (Figure 3). Two major types are non-selective cation channels,52 either glutamate receptor-like (GLRs) or cyclic nucleotide-gated (CNGCs) channels; and HKT2 high-affinity K+ transporters.232 Other possible pathways for Na+ uptake may involve AKT1 Shaker-type K+ channels, HAK5 high-affinity K+ transporters,232,233 and the low-affinity cation transporter LCT1,234,235 although arguments against their involvement have been presented.233 PIP2;1 aquaporins (initially identified as plant water channels) were recently added to the list of candidates for root Na+ uptake.236 The uptake of Na+ is counterbalanced by active Na+ extrusion (Figure 3). The most prominent component in this process is the SOS1 Na+/H+ exchanger,237 although the involvement of vesicle-mediated Na+ transport cannot be ruled out.238 It is generally assumed that ∼95% of all Na+ taken up by the root is then exported back to the rhizosphere.26 The reasons for and the consequences of such a futile cycle are discussed in detail elsewhere.239,240
Figure 3.
Major Transporters Mediating Na+ Homeostasis in Salinized Root Tissues
The major pathways for Na+ uptake in the root epidermis are glutamate receptor-like (GLRs) channels or cyclic nucleotide-gated (CNGCs) non-selective cation channels and HKT2 high-affinity K+ transporters. Other possible pathways for Na+ uptake may involve AKT1 Shaker-type K+ channels, HAK5 high-affinity K+ transporters, the low-affinity cation transporter LCT1, and PIP2;1 aquaporins. The uptake of Na+ is counterbalanced by active Na+ extrusion via SOS1 Na+/H+ exchangers. Vacuolar Na+ sequestration is conferred by tonoplast-based Na+/H+ exchangers from the NHX family fueled by either H+-ATPase or H+-PPase pumps. Another component of vacuolar Na+ sequestration is efficient control over tonoplast slow- (SV) and fast- (FV) activating ion channels that may allow Na+ to leak back to the cytosol. Passive Na+ loading into the xylem is mediated by non-selective cation channels (NSCCs), and its active loading requires operation of cotransporters such as SOS1, CCC (cation-chloride cotransporters), and HKT2 (K+/Na+ symporter). Na+ withdrawal from the xylem is achieved by HKT1 high-affinity K+ transporters. Salinity-induced K+ loss from the root epidermis is mediated by NSCCs and depolarization-activated outward-rectifying GORK K+ channels.
Vacuolar Na+ Sequestration
Another way of avoiding excessive Na+ accumulation in the cytosol is to deposit it in the vacuole. The traditional view is that such vacuolar sequestration is conferred by operation of the tonoplast-based Na+/H+ exchangers in the NHX family.241 Both NHX activity and transcript levels are inducible by salt in glycophytes, and such tonoplast antiporters are constitutive in halophytes.238,242,243 More recent studies have suggested that NHX exchangers represent only part of the vacuolar Na+ sequestration mechanism. Another equally important sequestration mechanism involves the efficient control of tonoplast leak channels, which enables vacuolar Na+ retention in the tonoplast.244 Two types of Na+-permeable channels, namely slow- (SV) and fast- (FV) activating ion channels, are present at the tonoplast, and model calculations show that if each cell opened only one SV channel at a specific time, the back-leak fraction would range from 30% to 100%.244 Thus, to avoid the futile movement of Na+ into and out of the tonoplast, plants can afford to open only a very small percentage (about 0.1%) of all tonoplast channels; this is consistent with experimental observations in salt-tolerant species.245 Recent studies have reported additional complexity in the relationship between Na+/H+ antiporters and vacuolar Na+ sequestration; the studies showed that NHX antiporters have higher affinity to K+ than to Na+ and thus operate predominantly as K+/H+ exchangers.246,247 This prompted the suggestion that some other mechanisms and, specifically, enhanced vacuolar trafficking, may also deliver Na+ to the vacuole.248
Control of Xylem Na+ Loading
Despite its critical importance for salinity tolerance, whether Na+ loading into the xylem is an active or passive process is still debated. Both active and passive transport systems are probably involved, but their respective roles may differ depending on the length of time since salinity onset.249,250 Passive Na+ loading is likely to be mediated by non-selective cation channels (NSCC).251 Consideration of thermodynamics suggests, however, that under most physiologically relevant scenarios, xylem Na+ loading should be an active process (see Shabala250 for supporting arguments). One of the most likely candidates for such active loading is the Na+/H+ antiporter encoded by the SOS1 gene. Highly abundant in the xylem parenchyma,237 SOS1 belongs to the cation proton antiporter (CPA) subfamily of proteins. The SOS1 protein is a homodimer and consists of 10–12 transmembrane domains. Class 2 HKT transporters represent another pathway for active xylem Na+ loading. Functionally, HKT2 operates as a K+/Na+ symporter. HKT2 transporters are highly expressed in the stellar root tissues.252 The depolarization of parenchyma cells under saline conditions12 favors a passive outward movement of K+ into the xylem, thus creating a driving force for the loading of Na+ into the xylem.249 Finally, cation-chloride cotransporters (CCC) were found to be preferentially expressed at the xylem/symplast boundary in Arabidopsis.253 These transporters mediate symport of Cl−, Na+, and K+, and given that Cl− transport into the xylem is thermodynamically passive, they may provide a driving force for Na+ (secondary) active loading into the xylem.249 Supporting evidence for that view was provided by the pharmacological studies conducted by Zhu et al.,254 who demonstrated a significant reduction in the magnitude of Na+ efflux from barley root stellar tissue in the presence of bumetanide, a known inhibitor of mammalian CCC.
Retrieval of Na+ from the Xylem
Class I high-affinity K+ transporters have been firmly established to operate in the withdrawal of Na+ from the xylem sap in various species.255, 256, 257, 258, 259 HKT proteins belong to the HKT/Trk/Ktr-type superfamily of K+ transporters, which consists of four repeats of transmembrane/pore-loop/transmembrane motifs, similar to the ion-conducting pore-forming units of K+ channels. Members of class I (HKT1) contain a Ser residue at the first pore-loop domain and are highly selective for Na+ over K+.260 The Arabidopsis genome contains only a single copy of the AtHKT1;1 gene, but its halophytic relative Thellungiella salsuginea contains three copies of HKT1-type genes.261 When expressed in Xenopus laevis oocytes and yeast, HKT1 transporters show a highly specific Na+ influx.259,262 Arabidopsis hkt1;1 mutants were salt-sensitive compared with the wild type and hyperaccumulate Na+ in the shoot, but accumulate less Na+ in the root,263 and targeted overexpression of the Arabidopsis HKT1;5 homolog in Arabidopsis and rice increases Na+ exclusion from the shoot.264,265 In rice, the OsHKT1;5 locus has been narrowed down as a salt tolerance determinant by QTL (quantitative trait locus) analysis.257 In wheat, the TmHKT1; 5-A locus derived from a wild wheat relative T. monococcum corresponds to the Na+ exclusion 2 (Nax2) QTL that contributes to the lowering of the Na+ level in leaves.266,267
Na+ Recirculation via the Phloem
The Na+ load in the shoot may also be reduced by its recirculation back to the roots via the phloem.268 The molecular mechanisms of this process remain elusive, although the HKT1 class of transporters is thought to play a major role.263,269 The fate of the Na+ remobilized in the phloem is also unclear. The anatomical structure of the root favors a unidirectional, radial transport of Na+. Once it passes through the Casparian strip and is loaded into the stele, Na+ has very little chance of being transported back to the cortex. Hence, if a substantial quantity of Na+ is returned to the root, it presumably must remain in the rather limited number of parenchyma cells in the stele. Unless sequestered properly in vacuoles, this could cause phytotoxicity and compromise root functions. The export of Na+ in the phloem could also cause damage to growing leaves and meristematic regions of the shoot, assuming that they are connected to the phloem via sieve tubes.270 This may explain why salt-tolerant species have much lower rates of Na+ export in the phloem than salt-sensitive species; for example, the percentage of Na+ that is exported in the phloem is only ∼10% in salt-tolerant barley species271 but is 50% in salt-sensitive white lupin.272
Potassium Retention in the Cytosol
Over the last decade, researchers have found that cytosolic K+ homeostasis and the ability of various plant tissues to retain K+ under stress conditions are essential for salinity tolerance (reviewed by Shabala et al.14 and Shabala and Pottosin273). Reported initially for barley roots,274 a positive correlation between the overall salinity tolerance and the ability of a root tissue to retain K+ was later expanded to at least a dozen other plant species (reviewed in Wu et al.18). Efficient cytosolic K+ retention is also considered to be a hallmark of halophytes.275, 276, 277 There are at least four physiological reasons why K+ retention is important under saline conditions. First, a high level of K+ retention allows the plant to accumulate high amounts of Na+ in the cytosol without compromising the cytosolic K+/Na+ ratio, the ratio that determines cell metabolic competence and, ultimately, the ability of a plant to survive in saline environments. Second, depletion of the cytosolic K+ pool may activate caspase-like proteases and endonucleases, thus triggering PCD.34, 35, 36 While the physiological role of PCD under saline conditions is still debated,33,36 the causal relationship between K+ efflux and stress-induced PCD is beyond any doubt.18 Third, to maintain normal metabolic activity in the cytosol, plants rely on the vacuolar K+ pool to replenish the K+ lost from the cytosol. The vacuolar K+ buffering capacity in a typical plant cell is estimated to be between 1 and 7 h.18,244 Thus, unless the cell is able to activate high-affinity K+-uptake systems within this time frame, depletion of the vacuolar K+ pool may result in a loss of turgor and collapse of the cell.
Two major pathways mediate salinity-induced K+ loss from the cell.14,18 One pathway involves the GORK channel, which belongs to the Shaker family of K+ channels and consists of six transmembrane domains (TMDs), a pore helix, and a selectivity filter between the last two TMDs.278 The GORK channel is highly sensitive to changes in membrane potential (as occur under stress conditions) and is activated upon depolarization.279 To prevent depolarization-induced K+ leakage, plants must restore (otherwise depolarized) the membrane potential by more active H+ pumping.280 This comes at a significant ATP cost281 and may compromise the plant's ability to adapt and grow. The second pathway for salinity stress-induced K+ loss from the cell is via K+-permeable ROS-activated NSCCs.18 Differential sensitivity of K+-permeable NSCC to various ROS (e.g., H2O2 and ⋅OH) explains the intraspecific,275,282 genotypic,283,284 and tissue-specific14,49 differences in salinity stress tolerance.
Potassium as a Second Messenger
In addition to being an essential nutrient, K+ functions in signaling.285,286 When plants deal with energy crises (as they do under saline conditions240), they must use a large fraction of available ATP for defense, at the expense of “business as usual” metabolism. Given that many metabolic enzymes require K+,18,287 transient cytosolic K+ efflux is thought to operate as a “metabolic switch” that inhibits energy-consuming anabolic reactions and saves energy for adaptation and repair, which may give species a competitive advantage under the energy-limiting conditions imposed by salinity.35,286 At the same time, the amount of K+ lost for signaling purposes should not compromise the plant's nutritional demand for this element. This dilemma is resolved by the transient nature and high tissue specificity of stress-induced K+ efflux.14 In addition, different plant species may display distinct salt stress-induced K+ flux “signatures,”281 prompting analogies with cytosolic Ca2+ signaling.67 In the latter case, an array of protein kinases and calcium-binding proteins decode transient Ca2+ spikes.288 It remains to be determined whether a similar mechanism characterizes K+ signaling.
Salt Stress-Induced Metabolite Changes
A major strategy used by plants to maintain a low intracellular osmotic potential under high salinity is the accumulation of compatible osmolytes,26,103 including proline, hydroxyproline, glycine betaine, sugars, polyamines, and proteins from the late embryogenesis abundant (LEA) superfamily.289, 290, 291 Among these metabolites, proline plays a dominant role in osmotic adjustment under salt stress.292 The accumulation of proline under osmotic stress can be achieved both by activation of the proline biosynthesis pathway and by inactivation of the proline catabolic pathway.293 Proline is biosynthesized mainly via reductions of glutamic acid by two successive enzymes: P5C synthase (P5CS) and P5C reductase (P5CR).294,295 P5CS is a rate-limiting enzyme in proline biosynthesis, and its activity is mainly controlled at the transcriptional level.295 Both P5CS and P5CR genes are upregulated under high salinity, which enables the accumulation of proline and subsequent salt stress tolerance.295 In the catabolic pathway, proline can be converted to glutamate via the reactions catalyzed by proline dehydrogenase (PDH) and P5C dehydrogenase (P5CDH).296 The negative role of PDH in salt stress tolerance is supported by the fact that plants with lower transcript levels of PDH have higher salt stress tolerance.297,298 Apart from its role in osmotic adjustment, proline acts to stabilize proteins and membrane structures, as well as an ROS scavenger that attenuates oxidative stress under high salinity.299, 300, 301
GB is another major metabolite that is increased under salt stress and is associated with adjustment of osmotic potential under dehydrating conditions.302,303 Choline monooxygenase (CMO) and NAD+-dependent betaine aldehyde dehydrogenase (BADH) are required for the biosynthesis of glycine betaine from choline via two oxidation reactions. Glycine betaine is also important for protecting enzymes, stabilizing membranes, and reducing oxidative stress under stress conditions.304 Trehalose is a non-reducing disaccharide of glucose that helps protect plants against abiotic stress.305 Application of trehalose reduces ROS accumulation, which in turn alleviates oxidative stress under high salinity.305,306 Transgenic rice plants that overexpress OsTRE1 have enhanced salt stress tolerance.307 Other sugars, such as glucose, fructose, and fructans, are also involved in the osmotic adjustment under drought and salt stress.193 Polyamines are organic amines, which include putrescine, spermidine, and spermine.308,309 Application of exogenous polyamines or the engineering of plants with increased levels of polyamines increases tolerance to salinity and to other abiotic stresses.310
Myo-inositol metabolism is important for the relay of specific signals to the cell. Several myo-inositol-derived inositol phosphates have been shown that are required for the response to environmental stresses. Inositol trisphosphate (IP3) is rapidly induced upon salt stress and ABA treatment.311,312 In plants, IP3 is associated with the release of Ca2+ from vacuolar vesicles and thus relay of environmental stress signals. Inositol 1,3,4-trisphosphate 5/6-kinase (ITPK) catalyzes the conversion of IP3 to inositol tetrakisphopsphate (IP4). In rice, disruption and overexpression of the OsITPK2 gene both lead to increased sensitivity to salt and drought stress,313 which implies that the homeostasis between IP3 and IP4 is important for stress tolerance.
Salinity Tolerance in Halophytes
According to the Dictionary of Botany, halophytes are “plants that are adapted to live in soil containing a high concentration of salt;” many other definitions of halophytes are available in the literature. Despite these differences in definition, all authors agree that the physiological, anatomical, or genetic traits do not fundamentally differ between halophytic species and domesticated (non-halophytic) crop species.314 The major difference is that halophytes are simply much more efficient in implementing these traits.238 Some traits, however, can be considered as hallmarks of halophytic species, and these are briefly summarized in the following sections.
Succulence
Succulence is an important adaptive strategy that accumulates excessive salt and conserves water in saline-grown plants. This trait is not unique to halophytes, because increased leaf succulence also occurs in glycophytes,315 although to a much lesser extent than in halophytes. In halophytes, a specialized parenchymatous tissue beneath the photosynthetically active chlorechymatous tissue functions in both salt storage316 and water storage.317 Succulence is mostly characteristic of dicot species belonging to the Chenopodioideae and Salicornioideae.318 The mechanistic basis of development of succulent structures in plants is linked to endopolyploidy or endoreduplication,319 although specific details are still emerging. An increase in the activity of aquaporins increases succulence in some halophyte species,320 suggesting that a turgor-driven mechanism may also be involved. The molecular mechanisms mediating salt deposition in succulent storage tissues remain elusive. A recent study found that storage parenchyma cells in the succulent halophyte Carpobrotus rosii act as Na+ sinks and have both a higher Na+ sequestration ability and a higher K+ retention ability than mesophyll cells;316 the latter trait was indicated by a higher rate of H+-ATPase operation and higher non-enzymatic antioxidant activity in this tissue. Also, storage parenchyma cells have a constitutively lower number of open SV vacuolar channels and the ability to downregulate activity of FV vacuolar channels. Because both SV and FV channels represent major pathways for Na+ leakage into the cytosol, the efficiency of salt load sequestration in parenchymatous tissues depends on the ability of the plant to control this process.
Salt Glands and Bladders
Salt glands are present in more than 50 halophyte species from 14 families321 and are believed to have evolved independently at least 12 times. At the functional level, two types of salt glands can be distinguished: those that directly secrete salts to the surface of the leaf (exo-recretohalophytes), and those that collect salt in the vacuole of a specialized bladder cell (endo-recretohalophytes).321,322 Structurally, salt glands are divided into four groups: (1) highly vacuolated unicellular secretary cells (e.g., those in Porteracia species); (2) two-celled excretory structures (those in graminoids); (3) multi-cellular (up to 40 cells) glands found in some graminoids and several dicot families; and (4) a structure consisting of a stalk cell and an epidermal bladder cell (e.g., those in quinoa or Atriplex species) (Figure 4).238,323 Salt glands in the first three groups, which are characteristic of exo-recretohalophytes, release salt periodically. NaCl is thought to be transported into the salt gland through both the plasmodesmata and the regions that are not covered by the cuticle.323 The salt secretion per se involves membrane-bound transport proteins. Several such transporters were postulated to exist in salt glands of various halophyte species.324 None of them, however, has been characterized at the molecular and functional level.
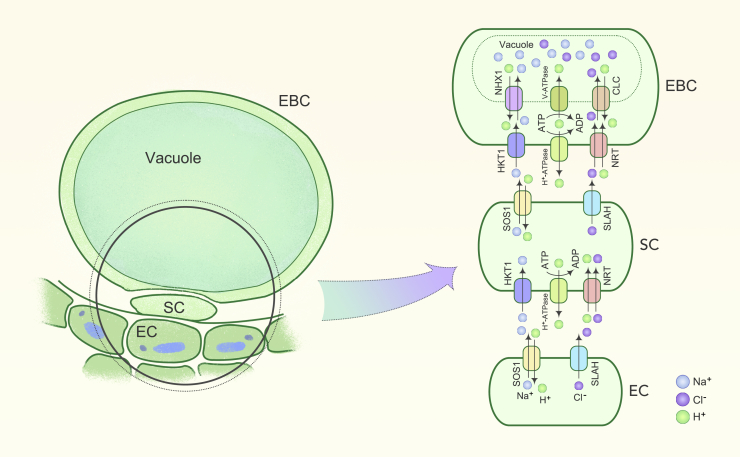
Figure 4.
A Proposed Model for the Mechanism of Salt Sequestration in Epidermal Bladder Cells
Left part of figure shows the morphology of an epidermal cell (EC), stalk cell (SC), and epidermal bladder cell (EBC). Right part of the figure presents ion transport systems in the EC, SC, and EBC. SOS1 (Na+/H+ plasma membrane exchanger) and HKT1 (high-affinity potassium transporter) transport Na+, while SLAH (Cl− permeable anion channel) and NRT (Cl−/H+ co-transporter) transport Cl− from EC to EBC. In the EBC, NHX1 (tonoplast-based Na+/H+ exchanger) and CLC (anion channel) are required for the sequestration of excessive Na+ and Cl− in the vacuole. Plasma membrane-localized H+-ATPase and vacuolar ATPase (V-ATPase) are essential for the generation of proton gradients and membrane potential that drive the transport of Na+ and Cl− from the EC to the vacuole of the EBC.
Salt glands in the fourth group, which are characteristic of endo-recretohalophytes, deposit the salt load in a bladder-like epidermal outgrowth called the epidermal bladder cell (EBC). Because EBCs are large (typically 100–200 μm in diameter),325,326 each EBC has a volume that is three orders of magnitude larger than that of a mesophyll cell or an epidermal cell; the large volume facilitates external salt storage, away from metabolically active photosynthetic tissues.327 EBC density is higher in younger leaves than in older leaves.326 During ontogeny, EBCs increase in size and operate as salt depots. Once an EBC accumulates a certain threshold quantity of salt, it ruptures and releases the salt into the external environment.328 The causal link between EBCs and salinity stress tolerance was demonstrated when the mechanical removal of bladder cells in quinoa resulted in a salt-sensitive phenotype.329 The molecular identities of key transporters involved in accumulation of Na+ and Cl− in EBCs in quinoa have recently been postulated330 and characterized at the functional level.331 Salt bladders may also function as a secondary epidermis to reduce water loss and prevent excessive UV damage in addition to functioning as reservoirs for the storage of water and various metabolic compounds.332,333
Osmotic Adjustment
Plants may achieve osmotic adjustment via two major avenues, i.e., by de novo synthesis of organic osmolytes and by increased uptake of inorganic ions. Because the production of organic osmolytes has a very high carbon cost,334,335 plants that use this strategy may be able to survive stress conditions but may grow poorly. The carbon cost of osmotic adjustment via inorganic ion uptake is an order of magnitude lower than that via organic osmolyte synthesis334,336 and is generally preferred, assuming that the problem of Na+ toxicity is resolved. This is the case for halophytes.250,314,337 Among inorganic osmolytes, K+ is the most abundant in the cell and non-toxic so is preferred. However, K+ uptake under saline conditions has an additional cost,281 while Na+ is present in the soil in high concentration and thus can be taken passively. Thus, halophytes rely on Na+ as a “cheap osmoticum” to maintain cell turgor pressure (hence, elongation growth and stomatal operation). This trait is complemented by the superior ability of halophytes to safely sequester toxic Na+ and Cl− ions in the vacuole and to thereby maintain their cytosolic and organellar concentrations below toxic levels (as discussed in previous sections). The osmotic potential of the cytosol is then adjusted by an increased accumulation of organic osmolytes to match that of the vacuole. As the volume of the cytosol is only ∼10% of the total cell volume, such a strategy is energetically more favorable than one in which the cell deposits large amounts of organic osmolytes into its vacuole in order to maintain its turgor.
Leaf Photochemistry
Both PSII and PSI are inactivated by increasing NaCl levels in the cytosol.276,338 The dark reactions of photosynthesis are also affected by salinity,339 with many key photosynthetically related enzymes being inactivated.134 Such inactivation, however, appears to differ between halophytic and glycophytic species. The transport of some key metabolites such as pyruvate, ascorbate, and phosphate into chloroplasts in halophytes requires the presence of Na+.134 Chloroplastic fructose 1,6-bisphosphatase (FruP2ase) from the halophyte Porteresia coarctata was less sensitive to NaCl in vitro compared with its domesticated relative Oryza sativa (cultivated rice);340 this difference was attributed to mutations in some amino acid residues in the FruP2ase gene.341 Some halophytes require high concentrations of Cl− to enhance electron transport and oxygen evolution during salt stress;134 they also demonstrate a higher extent of attachment of Rubisco activase to the thylakoid membrane.342,343 The sensitivity of grana unstacking to the ionic strength of the stroma also differs significantly between glycophytes and halophytes.344,345 Salinity also has a greater effect on rETR and maximal photochemical efficiency of PSII in isolated chloroplasts of glycophytes than of halophytes.276 There seems to be some important differences in the structure of the PSII complex between halophytes and glycophytes, with PsbQ protein (one of the extrinsic proteins mediating binding affinity of Cl− to the Mn-Ca-Cl complex) being completely absent in halophytes.345,346
Stomatal Patterning and Operation
The low soil-water potential imposed by salinity causes a marked decline in stomatal conductance (Gs); the physiological rationale behind this reduction is the plant's attempt to minimize water loss under the conditions of reduced water availability (“physiological drought”) imposed by salinity. This reduction in Gs comes, however, with a reduction in net CO2 assimilation, and therefore a reduction in plant growth. To understand how this problem is solved by halophytes, consider that Gs may be reduced by a decrease in stomatal aperture or stomatal density.250 Halophytes seem to be highly efficient in controlling both stomatal aperture and density. Stomatal aperture is controlled by many environmental and internal signals; among these, ABA and H2O2 are the most important. Salt stress-induced ABA production is highly dynamic, with peak ABA levels detected within 15 min of salinity exposure in plant roots and within 30 min in leaves.347 However, while all crop species respond to salinity by a rapid increase in the xylem sap ABA content,348, 349, 350 this seems not to be the case for halophytes.351,352 It also appears that the basal leaf ABA content (i.e., content under non-saline conditions) is much lower in halophytes than in glycophytes353 and is increased only slightly in salt-grown plants, while in glycophytes, salinity stress results in a 2- to 3-fold increase in leaf ABA content. It was suggested that modulation of the stomatal aperture by altering ABA levels operates over a much lower concentration range in halophytes than in glycophytes.353 Another difference concerns the stress-induced increases in ROS levels, which are accelerated in halophytes compared with glycophytes.354 Consequently, the ROS sensitivity of the guard cell channels and antioxidant systems should differ between halophytes and glycophytes.353 Another striking difference comes from the ability of halophytes to substitute K+ by Na+ in stomatal operations.355
Leaves can lose water even when stomata are fully closed. This process, which is termed residual transpiration, is controlled by several factors, one of which is stomatal density. Stomatal density is positively correlated with Gs,356 and it was argued that a reduction in stomatal density may represent a fundamental mechanism by which plants can optimize water-use efficiency (WUE).250 Consistent with this idea are observations by Franks et al.,357 who found that WUE was increased by ∼20% in Arabidopsis mutants that had a reduced stomatal density as a result of overexpressing the EPF2 (epidermal pattering factor) gene. Salinity causes a marked (about 30%) decrease in stomatal density in quinoa,250,326,358 and a negative correlation between stomatal density and salt tolerance has been reported for many other halophytic species (reviewed in Shabala250).The stomatal lineage is dynamic and flexible, altering stomatal production in response to environmental change, with numerous transcriptional regulators, cell-to-cell signaling, and polarity proteins involved.359,360 An increased understanding of this process in halophytes could suggest ways to increase WUE (and therefore overall salinity stress tolerance) in crops.
Salt Cress as Model Halophytic Organisms
More than 15 years ago, two close relatives of Arabidopsis from the genus of Thellungiella were proposed as model halophytes for molecular genetics studies, because of their ease of genetic transformation, short life cycle, and relatively small genome.361,362 One of them is Schrenkiella parvula (formerly known as Eutrema parvulum and before that as Thellungiella parvula), and the other one is Eutrema salsugineum (formerly known as T. salsuginea). Consistent with the notion that the same molecular machinery for salt tolerance operates in both glycophytes and halophytes, loss-of-function mutants of the SOS1 homolog in E. salsugineum lost halophytism.363 The genomic sequences for both E. salsugineum and S. parvula have been reported. Although both genomes are diploid and contain a similar number of genes, they differ significantly in their content of repeat/transposable elements (TEs).261,364,365 About 50% of the E. salsugineum genome is composed of TEs, and it remains to be investigated whether these TEs contribute to the evolution of its salt tolerance traits. Certain physiological traits are associated with high salt tolerance in salt cress, including high accumulation of the osmoprotectant proline, duplication or increased expression of salt stress-associated genes, swollen roots with additional layers of endodermis and cortex, and efficient management of the Na+/K+ ratio.366,367 In particular, the expression of SOS1 and the vacuolar H+-pyrophosphatase VP1 are increased in response to high salinity in E. salsugineum.363,368 EsSOS1 is under positive selection and operates more efficiently than SOS1 from Arabidopsis.369 Interestingly, a single amino acid substitution determines the ion selectivity of EsHKT1;2, which prefers K+ over Na+ and confers salt tolerance in Arabidopsis.370 These findings indicate that evolution acts on multiple levels to enhance the function of salinity-related genes in halophytes.
Future Perspective
Soil salinity will continue to threaten crop production and food security in the future. Cultivation of salt-tolerant crops is the most effective way to overcome this environmental problem. In the last three decades, extensive efforts have contributed to our understanding of the mechanisms of salt stress tolerance in plants. However, application of this fundamental knowledge to improve salt stress tolerance of crops in the field is a slow and challenging process. With the aid of gene editing technologies and advances in efficient genetic transformation in different species, improvement of salt stress tolerance of crops will become more feasible. Halophytes have developed special molecular mechanisms or special cellular structures to tolerate high concentration of salts. Growing halophytes in saline soils can greatly increase the area of arable land and also improve salt conditions in the soil. Knowledge of the molecular mechanisms underlying the formation and function of salt tolerance structures, such as EBCs, in halophytes can be potentially applied in crops to improve their capacity to accumulate high concentrations of ions.
Identification of salt stress sensor(s) is one of the most important questions concerning the molecular mechanisms of the salt stress response in plants. The discovery of GIPCs as potential monovalent-cation sensors7 opens a road to understand how plants sense salt stress at the plasma membrane. However, Na+ can accumulate both in the apoplast and in the cytosol, which implies that Na+ can be sensed not only at the plasma membrane but can also be perceived in different organelles, such as chloroplasts, mitochondria, and the ER. Most likely, the signals from different organelles need to be integrated and coordinated to achieve optimized cellular responses to salt stress. In the future, salt stress-induced organellar stress and the crosstalk between different organelles need to be further studied. The cell wall, as a front line that is directly exposed to stress, is involved in the sensing of salt stress. The status of cell wall integrity under salt stress is crucial for plants to determine the strength and duration of salt stress response. Currently little is known about the salt-induced cell wall changes and the mechanisms underlying the sensing and repair of cell wall integrity under salt stress. Engineering of crops with enhanced cell wall biosynthesis may not only increase the biomass but also improve the tolerance to a variety of environmental stresses. At the transcriptional level, it is critical to understand the expression of a specific gene in a strict tissue- and cell-based context and also study the gene regulatory network at the whole-plant level. The molecular mechanisms underlying root-to-shoot signaling need to be further elucidated. It has been shown that multiple phytohormones are involved in the response to high salinity. Systemic studies on the crosstalks between different hormones in response to salt stress need to be emphasized in the future.
Acknowledgments
We are grateful to Zichen Xu for assistance in the preparation of the figures. This work was supported by the Strategic Priority Research Program (grant no. XDB27040101) of the Chinese Academy of Sciences.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Chunzhao Zhao, Email: czzhao@psc.ac.cn.
Jian-Kang Zhu, Email: jkzhu@sibs.ac.cn.
Sergey Shabala, Email: sergey.shabala@utas.edu.au.
References
- 1.FAO . Food and Agriculture Organization of the United Nations and Earthscan; 2011. The State of the World’s Land and Water Resources for Food and Agriculture (SOLAW)—Managing Systems at Risk.http://www.fao.org/3/a-i1688e.pdf [Google Scholar]
- 2.Rengasamy P. World salinization with emphasis on Australia. J. Exp. Bot. 2006;57:1017–1023. doi: 10.1093/jxb/erj108. [DOI] [PubMed] [Google Scholar]
- 3.Flowers T.J., Munns R., Colmer T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015;115:419–431. doi: 10.1093/aob/mcu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 5.Halfter U., Ishitani M., Zhu J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U S A. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H., Ishitani M., Kim C., Zhu J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U S A. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Z., Zhou X., Tao M., Yuan F., Liu L., Wu F., Wu X., Xiang Y., Niu Y., Liu F., et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature. 2019;572:341–346. doi: 10.1038/s41586-019-1449-z. [DOI] [PubMed] [Google Scholar]
- 8.Munns R., Guo J., Passioura J.B., Cramer G.R. Leaf water status controls day-time but not daily rates of leaf expansion in salt-treated barley. Aust. J. Plant Physiol. 2000;27:949–957. [Google Scholar]
- 9.Fricke W., Akhiyarova G., Veselov D., Kudoyarova G. Rapid and tissue-specific changes in ABA and in growth rate in response to salinity in barley leaves. J. Exp. Bot. 2004;55:1115–1123. doi: 10.1093/jxb/erh117. [DOI] [PubMed] [Google Scholar]
- 10.Shabala S.N., Lew R.R. Turgor regulation in osmotically stressed Arabidopsis epidermal root cells. Direct support for the role of cell turgor measurements. Plant Physiol. 2014;129:290–299. doi: 10.1104/pp.020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrt C.S., Munns R., Burton R.A., Gilliham M., Wege S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018;269:47–55. doi: 10.1016/j.plantsci.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Wegner L.H., Stefano G., Shabala L., Rossi M., Mancuso S., Shabala S. Sequential depolarization of root cortical and stelar cells induced by an acute salt shock- implications for Na+ and K+ transport into xylem vessels. Plant Cell Environ. 2011;34:859–869. doi: 10.1111/j.1365-3040.2011.02291.x. [DOI] [PubMed] [Google Scholar]
- 13.Christmann A., Grill E., Huang J. Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 2013;16:293–300. doi: 10.1016/j.pbi.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Shabala L., Zhang J., Pottosin I., Bose J., Zhu M., Fuglsang A.T., Velarde-Buendia A., Massart A., Hill C.B., Roessner U., et al. Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity Stress. Plant Physiol. 2016;172:2445–2458. doi: 10.1104/pp.16.01347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosgrove D.J., Hedrich R. Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta. 1991;186:143–153. doi: 10.1007/BF00201510. [DOI] [PubMed] [Google Scholar]
- 16.Furuichi T., Tatsumi H., Sokabe M. Mechano-sensitive channels regulate the stomatal aperture in Vicia faba. Biochem. Biophys. Res. Commun. 2008;366:758–762. doi: 10.1016/j.bbrc.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 17.Cheeseman J.M. The integration of activity in saline environments: problems and perspectives. Funct. Plant Biol. 2013;40:759–774. doi: 10.1071/FP12285. [DOI] [PubMed] [Google Scholar]
- 18.Wu H.H., Zhang X.C., Giraldo J.P., Shabala S. It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil. 2018;431:1–17. [Google Scholar]
- 19.Benito B., Haro R., Amtmann A., Cuin T.A., Dreyer I. The twins K+ and Na+ in plants. J. Plant Physiol. 2014;171:723–731. doi: 10.1016/j.jplph.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Shabala S., Cuin T.A. Potassium transport and plant salt tolerance. Physiol. Plant. 2008;133:651–669. doi: 10.1111/j.1399-3054.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 21.Spitzer J., Poolman B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence. Microbiol. Mol. Biol. Rev. 2009;73:371–388. doi: 10.1128/MMBR.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geilfus C.M. Review on the significance of chlorine for crop yield and quality. Plant Sci. 2018;270:114–122. doi: 10.1016/j.plantsci.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Bazihizina N., Colmer T.D., Cuin T.A., Mancuso S., Shabala S. Friend or foe? Chloride patterning in halophytes. Trends Plant Sci. 2019;24:142–151. doi: 10.1016/j.tplants.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Teakle N.L., Tyerman S.D. Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 2010;33:566–589. doi: 10.1111/j.1365-3040.2009.02060.x. [DOI] [PubMed] [Google Scholar]
- 25.Moir-Barnetson L., Veneklaas E.J., Colmer T.D. Salinity tolerances of three succulent halophytes (Tecticornia spp.) differentially distributed along a salinity gradient. Funct. Plant Biol. 2016;43:739–750. doi: 10.1071/FP16025. [DOI] [PubMed] [Google Scholar]
- 26.Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- 27.Roy S.J., Negrão S., Tester M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Katsuhara M. Apoptosis-like cell death in barley roots under salt stress. Plant Cell Physiol. 1997;38:1091–1093. [Google Scholar]
- 29.Li J., Jiang A., Chen H., Wang Y., Zhang W. Lanthanum prevents salt stress-induced programmed cell death in rice root tip cells by controlling early induction events. J. Integr. Plant Biol. 2007;49:1024–1031. [Google Scholar]
- 30.Li J., Jiang A., Zhang W. Salt stress-induced programmed cell death in rice root tip cells. J. Integr. Plant Biol. 2007;49:481–486. [Google Scholar]
- 31.Luo L., Zhang P., Zhu R., Fu J., Su J., Zheng J., Wang Z., Wang D., Gong Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017;8:1–13. doi: 10.3389/fpls.2017.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Affenzeller M.J., Darehshouri A., Andosch A., Lütz C., Lütz-Meindl U. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 2009;60:939–954. doi: 10.1093/jxb/ern348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh G.H., Damsz B., Matsumoto T.K., Reddy M.P., Rus A.M., Ibeas J.I., Narasimhan M.L., Bressan R.A., Hasegawa P.M. Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J. 2002;29:649–659. doi: 10.1046/j.0960-7412.2001.01247.x. [DOI] [PubMed] [Google Scholar]
- 34.Demidchik V., Cuin T.A., Svistunenko D., Smith S.J., Miller A.J., Shabala S., Sokolik A., Yurin V. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 2010;123:1468–1479. doi: 10.1242/jcs.064352. [DOI] [PubMed] [Google Scholar]
- 35.Demidchik V. Mechanisms and physiological roles of K+ efflux from root cells. J. Plant Physiol. 2014;171:696–707. doi: 10.1016/j.jplph.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Shabala S., Pang J., Zhou M., Shabala L., Cuin T.A., Nick P., Wegner L.H. Electrical signalling and cytokinins mediate effects of light and root cutting on ion uptake in intact plants. Plant Cell Environ. 2009;32:194–207. doi: 10.1111/j.1365-3040.2008.01914.x. [DOI] [PubMed] [Google Scholar]
- 37.Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 38.Ma L., Zhang H., Sun L., Jiao Y., Zhang G., Miao C., Hao F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012;63:305–317. doi: 10.1093/jxb/err280. [DOI] [PubMed] [Google Scholar]
- 39.Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A.C., Harberd N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012;31:4359–4370. doi: 10.1038/emboj.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009;2:1–11. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 41.Ben Rejeb K., Benzarti M., Debez A., Bailly C., Savouré A., Abdelly C. NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. J. Plant Physiol. 2015;174:5–15. doi: 10.1016/j.jplph.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y., Rubio M.C., Alassimone J., Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153:402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 43.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allakhverdiev S.I., Nishiyama Y., Miyairi S., Yamamoto H., Inagaki N., Kanesaki Y., Murata N. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiol. 2002;130:1443–1453. doi: 10.1104/pp.011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pospisil P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta. 2009;1787:1151–1160. doi: 10.1016/j.bbabio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Del Río L.A., López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016;57:1364–1376. doi: 10.1093/pcp/pcw076. [DOI] [PubMed] [Google Scholar]
- 47.Wingler A., Lea P.J., Quick W.P., Leegood R.C. Photorespiration: metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. B Biol. Sci. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noctor G., Foyer C.H. Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 49.Demidchik V., Shabala S.N., Davies J.M. Spatial variation in H2O2 response of Arabidopsis thaliana root epidermal Ca2+ flux and plasma membrane Ca2+ channels. Plant J. 2007;49:377–386. doi: 10.1111/j.1365-313X.2006.02971.x. [DOI] [PubMed] [Google Scholar]
- 50.Pel Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 51.Köhler B., Hills A., Blatt M.R. Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol. 2003;131:385–388. doi: 10.1104/pp.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demidchik V. ROS-activated ion channels in plants: biophysical characteristics, physiological functions and molecular nature. Int. J. Mol. Sci. 2018;19:17–21. doi: 10.3390/ijms19041263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demidchik V., Shabala S.N., Coutts K.B., Tester M.A., Davies J.M. Free oxygen radicals regulate plasma membrane Ca2+- and K+-permeable channels in plant root cells. J. Cell Sci. 2003;116:81–88. doi: 10.1242/jcs.00201. [DOI] [PubMed] [Google Scholar]
- 54.Zepeda-Jazo I., Velarde-Buendía A.M., Enríquez-Figueroa R., Bose J., Shabala S., Muñiz-Murguía J., Pottosin I.I. Polyamines interact with hydroxyl radicals in activating Ca2+ and K+ transport across the root epidermal plasma membranes. Plant Physiol. 2011;157:2167–2180. doi: 10.1104/pp.111.179671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laohavisit A., Shang Z., Rubio L., Cuin T.A., Véry A.A., Wang A., Mortimer J.C., Macpherson N., Coxon K.M., Battey N.H., et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+-and K+-permeable conductance in root cells. Plant Cell. 2012;24:1522–1533. doi: 10.1105/tpc.112.097881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pottosin I., Shabala S. Transport across chloroplast membranes: optimizing photosynthesis for adverse environmental conditions. Mol. Plant. 2016;9:356–370. doi: 10.1016/j.molp.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Pottosin I., Wherrett T., Shabala S. SV channels dominate the vacuolar Ca2+ release during intracellular signaling. FEBS Lett. 2009;583:921–926. doi: 10.1016/j.febslet.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Barhoumi Z., Djebali W., Chaïbi W., Abdelly C., Smaoui A. Salt impact on photosynthesis and leaf ultrastructure of Aeluropus littoralis. J. Plant Res. 2007;120:529–537. doi: 10.1007/s10265-007-0094-z. [DOI] [PubMed] [Google Scholar]
- 59.Bose J., Rodrigo-Moreno A., Shabala S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014;65:1241–1257. doi: 10.1093/jxb/ert430. [DOI] [PubMed] [Google Scholar]
- 60.Hanin M., Ebel C., Ngom M., Laplaze L., Masmoudi K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016;7:1–17. doi: 10.3389/fpls.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mittova V., Volokita M., Guy M., Tal M. Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol. Plant. 2000;110:42–51. [Google Scholar]
- 62.Koffler B.E., Luschin-Ebengreuth N., Zechmann B. Compartment specific changes of the antioxidative status in Arabidopsis thaliana during salt stress. J. Plant Biol. 2015;58:8–16. [Google Scholar]
- 63.Badawi G.H., Yamauchi Y., Kawano N., Tanaka K., Tanaka K. Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit Ghazi. Physiol. Plant. 2004;121:231–238. doi: 10.1111/j.0031-9317.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 64.Li J., Liu J., Wang G., Cha J.Y., Li G., Chen S., Li Z., Guo J., Zhang C., Yang Y., et al. A chaperone function of NO CATALASE ACTIVITY1 Is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell. 2015;27:908–925. doi: 10.1105/tpc.114.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giraud E., Ho L.H.M., Clifton R., Carroll A., Estavillo G., Tan Y.F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., et al. The absence of Alternative Oxidase1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008;147:595–610. doi: 10.1104/pp.107.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mittova V., Tal M., Volokita M., Guy M. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environ. 2003;26:845–856. doi: 10.1046/j.1365-3040.2003.01016.x. [DOI] [PubMed] [Google Scholar]
- 67.Dodd A.N., Kudla J., Sanders D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 68.Knight H., Trewavas A.J., Knight M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 69.Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., Zhang J., Theprungsirikul L., Shrift T., Krichilsky B., et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014;514:367–371. doi: 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- 70.Feng W., Kita D., Peaucelle A., Cartwright H.N., Doan V., Duan Q., Liu M.-C., Maman J., Steinhorst L., Schmitz-Thom I., et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ Signaling. Curr. Biol. 2018;28:666–675. doi: 10.1016/j.cub.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laohavisit A., Richards S.L., Shabala L., Chen C., Colaço R.D.D.R., Swarbreck S.M., Shaw E., Dark A., Shabala S., Shang Z., et al. Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 2013;163:253–262. doi: 10.1104/pp.113.217810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephan A.B., Kunz H.H., Yang E., Schroeder J.I. Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U S A. 2016;113:E5242–E5249. doi: 10.1073/pnas.1519555113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamilton E.S., Schlegel A.M., Haswell E.S. United in diversity: mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015;66:113–137. doi: 10.1146/annurev-arplant-043014-114700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamilton E.S., Jensen G.S., Maksaev G., Katims A., Sherp A.M., Haswell E.S. Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science. 2015;350:438–441. doi: 10.1126/science.aac6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zanto T.P., Hennigan K., Östberg M., Clapp W.C., Gazzaley A. Mechanosensitive channels protect plastids from hypoosmotic stress during normal plant growth. Curr. Biol. 2011;46:564–574. doi: 10.1016/j.cub.2012.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolf S. Plant cell wall signalling and receptor-like kinases. Biochem. J. 2017;474:471–492. doi: 10.1042/BCJ20160238. [DOI] [PubMed] [Google Scholar]
- 77.Engelsdorf T., Hamann T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann. Bot. 2014;114:1339–1347. doi: 10.1093/aob/mcu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franck C.M., Westermann J. Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 2018;69:301–328. doi: 10.1146/annurev-arplant-042817-040557. [DOI] [PubMed] [Google Scholar]
- 79.Zhao C., Zayed O., Yu Z., Jiang W., Zhu P., Hsu C.-C., Zhang L., Tao W.A., Lozano-Durán R., Zhu J.-K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2018;115:13123–13128. doi: 10.1073/pnas.1816991115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Decreux A., Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46:268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- 81.Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343:408–411. doi: 10.1126/science.1244454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Basu D., Tian L., DeBrosse T., Poirier E., Emch K., Herock H., Travers A., Showalter A.M. Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One. 2016;11:1–27. doi: 10.1371/journal.pone.0145092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu S.L., Rahman A., Baskin T.I., Kieber J.J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell. 2008;20:3065–3079. doi: 10.1105/tpc.108.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K.S., Pardo J.M., Guo Y. Phosphorylation of SOS3-like calcium binding protein8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 2009;21:1607–1619. doi: 10.1105/tpc.109.066217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishitani M., Liu J., Halfter U., Kim C.S., Shi W., Zhu J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu J., Zhu J.K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 87.Kim W.Y., Ali Z., Park H.J., Park S.J., Cha J.Y., Perez-Hormaeche J., Quintero F.J., Shin G., Kim M.R., Qiang Z., et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013;4:1312–1357. doi: 10.1038/ncomms2357. [DOI] [PubMed] [Google Scholar]
- 88.Zhou H., Lin H., Chen S., Becker K., Yang Y., Zhao J., Kudla J., Schumaker K.S., Guo Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 2014;26:1166–1182. doi: 10.1105/tpc.113.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Z., Wang C., Xue Y., Liu X., Chen S., Song C.P., Yang Y., Guo Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019;10:1199. doi: 10.1038/s41467-019-09181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohta M., Guo Y., Halfter U., Zhu J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U S A. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quintero F.J., Martinez-Atienza J., Villalta I., Jiang X., Kim W.Y., Ali Z., Fujii H., Mendoza I., Yun D.J., Zhu J.K., et al. Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U S A. 2011;108:2611–2616. doi: 10.1073/pnas.1018921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren X.L., Qi G.N., Feng H.Q., Zhao S., Zhao S.S., Wang Y., Wu W.H. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013;74:258–266. doi: 10.1111/tpj.12123. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y., Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217:523–539. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- 94.Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 95.Wang F., Jing W., Zhang W. The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 2014;227:181–189. doi: 10.1016/j.plantsci.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 96.Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008;283:26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- 97.Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010;188:762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- 98.Bargmann B.O.R., Laxalt A.M., Riet B., Schooten B. Van, Merquiol E., Testerink C., Haring M.A., Bartels D., Munnik T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 2009;50:78–89. doi: 10.1093/pcp/pcn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hong Y., Pan X., Welti R., Wang X. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell. 2008;20:803–816. doi: 10.1105/tpc.107.056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katagiri T., Takahashi S., Shinozaki K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001;26:595–605. doi: 10.1046/j.1365-313x.2001.01060.x. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Y., Wang L., Liu Y., Zhang Q., Wei Q., Zhang W. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta. 2006;224:545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
- 102.Hong Y., Devaiah S.P., Bahn S.C., Thamasandra B.N., Li M., Welti R., Wang X. Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 2009;58:376–387. doi: 10.1111/j.1365-313X.2009.03788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Apse M.P., Blumwald E. Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 2002;13:146–150. doi: 10.1016/s0958-1669(02)00298-7. [DOI] [PubMed] [Google Scholar]
- 104.Boudsocq M., Barbier-Brygoo H., Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 105.McLoughlin F., Galvan-Ampudia C.S., Julkowska M.M., Caarls L., Van Der Does D., Laurière C., Munnik T., Haring M.A., Testerink C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012;72:436–449. doi: 10.1111/j.1365-313X.2012.05089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fujii H., Zhu J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U S A. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., Umezawa T., Fujita M., Maruyama K., Ishiyama K., et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 108.Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 110.Saruhashi M., Ghosh T.K., Arai K., Ishizaki Y., Hagiwara K., Komatsu K., Shiwa Y., Izumikawa K., Yoshikawa H., Umezawa T., et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U S A. 2015;112:E6388–E6396. doi: 10.1073/pnas.1511238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fujii H., Verslues P.E., Zhu J.-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U S A. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kawa D., Meyer A.J., Dekker H.L., Abd-El-Haliem A., Gevaert K., Van De Slijke E., Maszkowska J., Bucholc M., Dobrowolska G., De Jaeger G., et al. SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 2020;182:361–377. doi: 10.1104/pp.19.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim J.M., Woo D.H., Kim S.H., Lee S.Y., Park H.Y., Seok H.Y., Chung W.S., Moon Y.H. Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 2012;31:217–224. doi: 10.1007/s00299-011-1157-0. [DOI] [PubMed] [Google Scholar]
- 114.Kim S.H., Woo D.H., Kim J.M., Lee S.Y., Chung W.S., Moon Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011;412:150–154. doi: 10.1016/j.bbrc.2011.07.064. [DOI] [PubMed] [Google Scholar]
- 115.Zhu J.K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang S.S., Sun L., Dong X., Lu S.J., Tian W., Liu J.X. Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. J. Integr. Plant Biol. 2016;58:623–626. doi: 10.1111/jipb.12442. [DOI] [PubMed] [Google Scholar]
- 117.Shi H., Kim Y., Guo Y., Stevenson B., Zhu J.-K. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu J., Lee B., Dellinger M., Cui X., Zhang C., Wu S., Nothnagel E.A., Zhu J. A cellulose synthase like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2011;63:128–140. doi: 10.1111/j.1365-313X.2010.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Endler A., Kesten C., Schneider R., Zhang Y., Ivakov A., Froehlich A., Funke N., Persson S. A mechanism for sustained cellulose synthesis during salt stress. Cell. 2015;162:1353–1364. doi: 10.1016/j.cell.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 120.Zhao C., Zayed O., Zeng F., Liu C., Zhang L., Zhu P., Hsu C., Tuncil Y.E., Tao W.A., Carpita N.C., et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019;224:274–290. doi: 10.1111/nph.15867. [DOI] [PubMed] [Google Scholar]
- 121.Shomer I., Novacky A.J., Pike S.M., Yermiyahu U., Kinraide T.B. Electrical potentials of plant cell walls in response to the ionic environment. Plant Physiol. 2003;133:411–422. doi: 10.1104/pp.103.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 2004;55:109–139. doi: 10.1146/annurev.arplant.55.031903.141750. [DOI] [PubMed] [Google Scholar]
- 123.Mohnen D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 124.Wu Y., Cosgrove D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000;51:1543–1553. doi: 10.1093/jexbot/51.350.1543. [DOI] [PubMed] [Google Scholar]
- 125.Hocq L., Pelloux J., Lefebvre V. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 2017;22:20–29. doi: 10.1016/j.tplants.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 126.Proseus T.E., Boyer J.S. Pectate chemistry links cell expansion to wall deposition. J. Exp. Bot. 2012;63:3953–3958. doi: 10.4161/psb.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nari J., Noat G., Ricard J. Pectin methylesterase, metal ions and plant cell-wall extension. Hydrolysis of pectin by plant cell-wall pectin methylesterase. Biochem. J. 1991;279:343–350. doi: 10.1042/bj2790343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Olmos E., García De La Garma J., Gomez-Jimenez M.C., Fernandez-Garcia N. Arabinogalactan proteins are involved in salt-adaptation and vesicle trafficking in tobacco by-2 cell cultures. Front. Plant Sci. 2017;8:1092. doi: 10.3389/fpls.2017.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Griffiths J.S., Tsai A.Y.L., Xue H., Voiniciuc C., Šola K., Seifert G.J., Mansfield S.D., Haughn G.W. SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol. 2014;165:991–1004. doi: 10.1104/pp.114.239400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tryfona T., Theys T.E., Wagner T., Stott K., Keegstra K., Dupree P. Characterisation of FUT4 and FUT6 α-(1→2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS One. 2014;9:e93291. doi: 10.1371/journal.pone.0093291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geilfus C.M. The pH of the apoplast: dynamic factor with functional impact under stress. Mol. Plant. 2017;10:1371–1386. doi: 10.1016/j.molp.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 132.Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 133.Suo J., Zhao Q., David L., Chen S., Dai S. Salinity response in chloroplasts: insights from gene characterization. Int. J. Mol. Sci. 2017;18:1011. doi: 10.3390/ijms18051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bose J., Munns R., Shabala S., Gilliham M., Pogson B., Tyerman S.D. Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J. Exp. Bot. 2017;68:3129–3143. doi: 10.1093/jxb/erx142. [DOI] [PubMed] [Google Scholar]
- 135.Chaves M.M., Flexas J., Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Finazzi G., Petroutsos D., Tomizioli M., Flori S., Sautron E., Villanova V., Rolland N., Seigneurin-Berny D. Ions channels/transporters and chloroplast regulation. Cell Calcium. 2015;58:86–97. doi: 10.1016/j.ceca.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 137.Iuchi S., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;123:553–562. doi: 10.1104/pp.123.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xiong L., Lee H., Ishitani M., Zhu J.K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- 139.Kempa S., Rozhon W., Šamaj J., Erban A., Baluška F., Becker T., Haselmayer J., Schleiff E., Kopka J., Hirt H., et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007;49:1076–1090. doi: 10.1111/j.1365-313X.2006.03025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun Y.L., Li F., Su N., Sun X.L., Zhao S.J., Meng Q.W. The increase in unsaturation of fatty acids of phosphatidylglycerol in thylakoid membrane enhanced salt tolerance in tomato. Photosynthetica. 2010;48:400–408. [Google Scholar]
- 141.Zhang J.T., Zhu J.Q., Zhu Q., Liu H., Gao X.S., Zhang H.X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009;390:469–474. doi: 10.1016/j.bbrc.2009.09.095. [DOI] [PubMed] [Google Scholar]
- 142.Calderon R.H., García-Cerdán J.G., Malnoë A., Cook R., Russell J.J., Gaw C., Dent R.M., De Vitry C., Niyogi K.K. A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 2013;288:26688–26696. doi: 10.1074/jbc.M113.487629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khurana N., Chauhan H., Khurana P. Characterization of a chloroplast localized wheat membrane protein (TaRCI) and its role in heat, drought and salinity stress tolerance in Arabidopsis thaliana. Plant Gene. 2015;4:45–54. [Google Scholar]
- 144.Dogra V., Li M., Singh S., Li M., Kim C. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun. 2019;10:2834. doi: 10.1038/s41467-019-10760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wagner D., Przybyla D., Op Den Camp R., Kim C., Landgraf F., Keun P.L., Würsch M., Laloi C., Nater M., Hideg E., et al. The genetic basis of singlet oxygen-induced stress response of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 146.Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U S A. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xiao Y., Savchenko T., Baidoo E.E.K., Chehab W.E., Hayden D.M., Tolstikov V., Corwin J.A., Kliebenstein D.J., Keasling J.D., Dehesh K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149:1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 148.Estavillo G.M., Crisp P.A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu J.X., Howell S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- 150.Liu L., Cui F., Li Q., Yin B., Zhang H., Lin B., Wu Y., Xia R., Tang S., Xie Q. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011;21:957–969. doi: 10.1038/cr.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., Wu Y., Tang S., Xie Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell. 2012;24:233–244. doi: 10.1105/tpc.111.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu J.X., Srivastava R., Che P., Howell S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Che P., Bussell J.D., Zhou W., Estavillo G.M., Pogson B.J., Smith S.M. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 2010;3:1–13. doi: 10.1126/scisignal.2001140. [DOI] [PubMed] [Google Scholar]
- 154.Wang Y., Berkowitz O., Selinski J., Xu Y., Hartmann A., Whelan J. Stress responsive mitochondrial proteins in Arabidopsis thaliana. Free Radic. Biol. Med. 2018;122:28–39. doi: 10.1016/j.freeradbiomed.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 155.Che-Othman M.H., Millar A.H., Taylor N.L. Connecting salt stress signalling pathways with salinity-induced changes in mitochondrial metabolic processes in C3 plants. Plant Cell Environ. 2017;40:2875–2905. doi: 10.1111/pce.13034. [DOI] [PubMed] [Google Scholar]
- 156.Vanderauwera S., Vandenbroucke K., Inzé A., Van De Cotte B., Mühlenbock P., De Rycke R., Naouar N., Van Gaever T., Van Montagu M.C.E., Van Breusegem F. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2012;109:20113–20118. doi: 10.1073/pnas.1217516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ng S., Giraud E., Duncan O., Law S.R., Wang Y., Xu L., Narsai R., Carrie C., Walker H., Day D.A., et al. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jia W. Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 2002;53:2201–2206. doi: 10.1093/jxb/erf079. [DOI] [PubMed] [Google Scholar]
- 159.Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014;202:35–49. doi: 10.1111/nph.12613. [DOI] [PubMed] [Google Scholar]
- 160.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1069. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 161.Park S.S.-Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL Family of START proteins. Science. 2009;324:1068–1069. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A.S., et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U S A. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jiang F., Hartung W. Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J. Exp. Bot. 2008;59:37–43. doi: 10.1093/jxb/erm127. [DOI] [PubMed] [Google Scholar]
- 165.Wilkinson S., Davies W.J. ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ. 2002;25:195–210. doi: 10.1046/j.0016-8025.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- 166.Buckley T.N. How do stomata respond to water status? New Phytol. 2019;224:21–36. doi: 10.1111/nph.15899. [DOI] [PubMed] [Google Scholar]
- 167.Christmann A., Weiler E.W., Steudle E., Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- 168.Endo A., Sawada Y., Takahashi H., Okamoto M., Ikegami K., Koiwai H., Seo M., Toyomasu T., Mitsuhashi W., Shinozaki K., et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008;147:1984–1993. doi: 10.1104/pp.108.116632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang F.P., Sussmilch F., Nichols D.S., Cardoso A.A., Brodribb T.J., McAdam S.A.M. Leaves, not roots or floral tissue, are the main site of rapid, external pressure-induced ABA biosynthesis in angiosperms. J. Exp. Bot. 2018;69:1261–1267. doi: 10.1093/jxb/erx480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mittler R., Blumwald E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell. 2015;27:64–70. doi: 10.1105/tpc.114.133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nath M., Bhatt D., Jain A., Saxena S.C., Saifi S.K., Yadav S., Negi M., Prasad R., Tuteja N. Salt stress triggers augmented levels of Na+, Ca2+ and ROS and alter stress-responsive gene expression in roots of CBL9 and CIPK23 knockout mutants of Arabidopsis thaliana. Environ. Exp. Bot. 2019;161:265–276. [Google Scholar]
- 172.Niu M., Huang Y., Sun S., Sun J., Cao H., Shabala S., Bie Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2018;69:3465–3476. doi: 10.1093/jxb/erx386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marino D., Dunand C., Puppo A., Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17:9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 174.Zhang Y., Tan J., Guo Z., Lu S., He S., Shu W., Zhou B. Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 2009;32:509–519. doi: 10.1111/j.1365-3040.2009.01945.x. [DOI] [PubMed] [Google Scholar]
- 175.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20:219–229. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 176.Ma S., Gong Q., Bohnert H.J. Dissecting salt stress pathways. J. Exp. Bot. 2006;57:1097–1107. doi: 10.1093/jxb/erj098. [DOI] [PubMed] [Google Scholar]
- 177.Yu Geng W., Wu R., Wei Wee C., Xie F., Wei X., Mei Yeen Chan P., Tham C., Duan L., Dinneny J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Valenzuela C.E., Acevedo-Acevedo O., Miranda G.S., Vergara-Barros P., Holuigue L., Figueroa C.R., Figueroa P.M., Murphy A. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016;67:4209–4220. doi: 10.1093/jxb/erw202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Toda Y., Tanaka M., Ogawa D., Kurata K., Kurotani K.I., Habu Y., Ando T., Sugimoto K., Mitsuda N., Katoh E., et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25:1709–1725. doi: 10.1105/tpc.113.112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kang D.J., Seo Y.J., Lee J.D., Ishii R., Kim K.U., Shin D.H., Park S.K., Jang S.W., Lee I.J. Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Sci. 2005;191:273–282. [Google Scholar]
- 181.Wei D., Wang M., Xu F., Quan T., Peng K., Xiao L., Xia G. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol. 2013;161:1217–1228. doi: 10.1104/pp.112.211854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yoon J.Y., Hamayun M., Lee S.-K., Lee I.-J. Methyl jasmonate alleviated salinity stress in soybean. J. Crop Sci. Biotechnol. 2009;12:63–68. [Google Scholar]
- 183.Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- 184.Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van D., Straeten D., Peng J., Harberd N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 185.Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008;56:613–626. doi: 10.1111/j.1365-313X.2008.03627.x. [DOI] [PubMed] [Google Scholar]
- 186.Cao W.H., Liu J., He X.J., Mu R.L., Zhou H.L., Chen S.Y., Zhang J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007;143:707–719. doi: 10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Peng J., Li Z., Wen X., Li W., Shi H., Yang L., Zhu H., Guo H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014;10:e1004664. doi: 10.1371/journal.pgen.1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang Y., Liu C., Li K., Sun F., Hu H., Li X., Zhao Y., Han C., Zhang W., Duan Y., et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007;64:633–644. doi: 10.1007/s11103-007-9182-7. [DOI] [PubMed] [Google Scholar]
- 189.Zhang L., Li Z., Quan R., Li G., Wang R., Huang R. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 2011;157:854–865. doi: 10.1104/pp.111.179028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jiang C., Belfield E.J., Cao Y., Smith J.A.C., Harberd N.P. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell. 2013;25:3535–3552. doi: 10.1105/tpc.113.115659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Misra N., Misra R. Salicylic acid changes plant growth parameters and proline metabolism in Rauwolfia serpentina leaves grown under salinity stress. Environ. Sci. 2012;12:1601–1609. [Google Scholar]
- 192.Szepesi Á., Csiszár J., Bajkán S., Gémes K., Horváth F., Erdei L., Deér A.K., Simon M.L., Tari I. Role of salicylic acid pre-treatment on the acclimation of tomato plants to salt- and osmotic stress. Acta Biol. Szeged. 2005;49:123–125. [Google Scholar]
- 193.Jayakannan M., Bose J., Babourina O., Rengel Z., Shabala S. Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 2013;64:2255–2268. doi: 10.1093/jxb/ert085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kreps J.A., Wu Y., Chang H.S., Zhu T., Wang X., Harper J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:1–21. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zeller G., Henz S.R., Widmer C.K., Sachsenberg T., Rätsch G., Weigel D., Laubinger S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
- 196.Rasmussen S., Barah P., Suarez-Rodriguez M.C., Bressendorff S., Friis P., Costantino P., Bones A.M., Nielsen H.B., Mundy J. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013;161:1783–1794. doi: 10.1104/pp.112.210773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stockinger E.J., Gilmour S.J., Thomashow M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U S A. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 200.Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.-K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016;171:2744–2759. doi: 10.1104/pp.16.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Choi H.I., Hong J.H., Ha J.O., Kang J.Y., Kim S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 202.Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U S A. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kim S., Kang J.Y., Cho D.I., Park J.H., Soo Y.K. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 204.Song L., Huang S.S.C., Wise A., Castanoz R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R. A transcription factor hierarchy defines an environmental stress response network. Science. 2016;354:aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhao Y., Dong W., Zhang N., Ai X., Wang M., Huang Z., Xiao L., Xia G. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014;164:1068–1076. doi: 10.1104/pp.113.227595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cheng M.C., Liao P.M., Kuo W.W., Lin T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baillo E.H., Kimotho R.N., Zhang Z., Xu P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (Basel) 2019;10:771. doi: 10.3390/genes10100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen L.T., Luo M., Wang Y.Y., Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ueda M., Matsui A., Tanaka M., Nakamura T., Abe T., Sako K., Sasaki T., Kim J.M., Ito A., Nishino N., et al. The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 2017;175:1760–1773. doi: 10.1104/pp.17.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zheng Y., Ding Y., Sun X., Xie S., Wang D., Liu X., Su L., Wei W., Pan L., Zhou D.X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016;67:1703–1713. doi: 10.1093/jxb/erv562. [DOI] [PubMed] [Google Scholar]
- 212.Chen H., Feng H., Zhang X., Zhang C., Wang T., Dong J. An Arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton. Plant Biotechnol. J. 2019;17:556–568. doi: 10.1111/pbi.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhou S., Chen Q., Sun Y., Li Y. Histone H2B monoubiquitination regulates salt stress-induced microtubule depolymerization in Arabidopsis. Plant Cell Environ. 2017;40:1512–1530. doi: 10.1111/pce.12950. [DOI] [PubMed] [Google Scholar]
- 214.Liu Z.Q., Gao J., Dong A.W., Shen W.H. A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the AtNAP1;3-2 mutant. Mol. Plant. 2009;2:688–699. doi: 10.1093/mp/ssp026. [DOI] [PubMed] [Google Scholar]
- 215.Sunkar R., Chinnusamy V., Zhu J., Zhu J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 216.Wang J., Meng X., Dobrovolskaya O.B., Orlov Y.L., Chen M. Non-coding RNAs and their roles in stress response in plants. Genomics Proteomics Bioinformatics. 2017;15:301–312. doi: 10.1016/j.gpb.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015;66:1749–1761. doi: 10.1093/jxb/erv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Eichten S.R., Schmitz R.J., Springer N.M. Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiol. 2014;165:933–947. doi: 10.1104/pp.113.234211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jiang D., Berger F. DNA replication–coupled histone modification maintains Polycomb gene silencing in plants. Science. 2017;357:1146–1149. doi: 10.1126/science.aan4965. [DOI] [PubMed] [Google Scholar]
- 220.Law J.A., Jacobsen S.E. Patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Cayuela E., Pérez-Alfocea F., Caro M., Bolarín M.C. Priming of seeds with NaCl induces physiological changes in tomato plants grown under salt stress. Physiol. Plant. 1996;96:231–236. [Google Scholar]
- 222.Jakab G., Ton J., Flors V., Zimmerli L., Metraux J.P., Mauch-Mani B. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 2005;139:267–274. doi: 10.1104/pp.105.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sani E., Herzyk P., Perrella G., Colot V., Amtmann A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013;14:R59. doi: 10.1186/gb-2013-14-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Karan R., DeLeon T., Biradar H., Subudhi P.K. Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS One. 2012;7:e40203. doi: 10.1371/journal.pone.0040203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Song Y., Ji D., Li S., Wang P., Li Q., Xiang F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS One. 2012;7:1–11. doi: 10.1371/journal.pone.0041274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang W., Huang F., Qin Q., Zhao X., Li Z., Fu B. Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochem. Biophys. Res. Commun. 2015;465:790–796. doi: 10.1016/j.bbrc.2015.08.089. [DOI] [PubMed] [Google Scholar]
- 227.Baek D., Jiang J., Chung J.S., Wang B., Chen J., Xin Z., Shi H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011;52:149–161. doi: 10.1093/pcp/pcq182. [DOI] [PubMed] [Google Scholar]
- 228.Huang C.F., Miki D., Tang K., Zhou H.R., Zheng Z., Chen W., Ma Z.Y., Yang L., Zhang H., Liu R., et al. A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013;9:e1003779. doi: 10.1371/journal.pgen.1003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang M., Qin L., Xie C., Li W., Yuan J., Kong L., Yu W., Xia G., Liu S. Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol. 2014;55:1354–1365. doi: 10.1093/pcp/pcu059. [DOI] [PubMed] [Google Scholar]
- 230.Secco D., Wang C., Shou H., Schultz M., Chiarenza S., Nussaume L., Ecker J., Whelan J., Lister R. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife. 2015;4:1–26. doi: 10.7554/eLife.09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cheeseman J.M. Mechanisms of salinity tolerance in plants. Plant Physiol. 1988;87:547–550. doi: 10.1104/pp.87.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mian A., Oomen R.J.F.J., Isayenkov S., Sentenac H., Maathuis F.J.M., Véry A.A. Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 2011;68:468–479. doi: 10.1111/j.1365-313X.2011.04701.x. [DOI] [PubMed] [Google Scholar]
- 233.Isayenkov S.V., Maathuis F.J.M. Plant salinity stress: many unanswered questions remain. Front. Plant Sci. 2019;10:80. doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kronzucker H.J., Britto D.T. Sodium transport in plants: a critical review. New Phytol. 2011;189:54–81. doi: 10.1111/j.1469-8137.2010.03540.x. [DOI] [PubMed] [Google Scholar]
- 235.Schachtman D.P., Kumar R., Schroeder J.I., Marsh E.L. Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc. Natl. Acad. Sci. U S A. 1997;94:11079–11084. doi: 10.1073/pnas.94.20.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Byrt C.S., Zhao M., Kourghi M., Bose J., Henderson S.W., Qiu J., Gilliham M., Schultz C., Schwarz M., Ramesh S.A., et al. Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 2017;40:802–815. doi: 10.1111/pce.12832. [DOI] [PubMed] [Google Scholar]
- 237.Shi H., Quintero F.J., Pardo J.M., Zhu J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shabala S., Mackay A. Ion transport in halophytes. Adv. Bot. Res. 2011;57:151–199. [Google Scholar]
- 239.Malagoli P., Britto D.T., Schulze L.M., Kronzucker H.J. Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetics, energetics, and relationship to salinity tolerance. J. Exp. Bot. 2008;59:4109–4117. doi: 10.1093/jxb/ern249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Munns R., Passioura J.B., Colmer T.D., Byrt C.S. Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 2019;225:1091–1096. doi: 10.1111/nph.15862. [DOI] [PubMed] [Google Scholar]
- 241.Bassil E., Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 242.Barkla B.J., Zingarelli L., Blumwald E., Smith J.A.C. Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophytic plant Mesembryanthemum crystallinum L. Plant Physiol. 1995;109:549–556. doi: 10.1104/pp.109.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang H.X., Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 2001;19:765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- 244.Shabala S., Chen G., Chen Z.H., Pottosin I. The energy cost of the tonoplast futile sodium leak. New Phytol. 2019;225:1105–1110. doi: 10.1111/nph.15758. [DOI] [PubMed] [Google Scholar]
- 245.Pérez V., Wherrett T., Shabala S., Muñiz J., Dobrovinskaya O., Pottosin I. Homeostatic control of slow vacuolar channels by luminal cations and evaluation of the channel-mediated tonoplast Ca2+ fluxes in situ. J. Exp. Bot. 2008;59:3845–3855. doi: 10.1093/jxb/ern225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bassil E., Tajima H., Liang Y.C., Ohto M.A., Ushijima K., Nakano R., Esumi T., Coku A., Belmonte M., Blumwald E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23:3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Leidi E.O., Barragán V., Rubio L., El-Hamdaoui A., Ruiz M.T., Cubero B., Fernández J.A., Bressan R.A., Hasegawa P.M., Quintero F.J., et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010;61:495–506. doi: 10.1111/j.1365-313X.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- 248.Baral A., Shruthi K.S., Mathew M.K. Vesicular trafficking and salinity responses in plants. IUBMB Life. 2015;67:677–686. doi: 10.1002/iub.1425. [DOI] [PubMed] [Google Scholar]
- 249.Ishikawa T., Cuin T.A., Bazihizina N., Shabala S. Xylem ion loading and its implications for plant abiotic stress tolerance. Membr. Transp. Plants. 2018;87:267–301. [Google Scholar]
- 250.Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013;112:1209–1221. doi: 10.1093/aob/mct205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Guo K.M., Babourina O., Christopher D.A., Borsic T., Rengel Z. The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in Arabidopsis. Physiol. Plant. 2010;139:303–312. doi: 10.1111/j.1399-3054.2010.01366.x. [DOI] [PubMed] [Google Scholar]
- 252.Jabnoune M., Espeout S., Mieulet D., Fizames C., Verdeil J.L., Conéjéro G., Rodríguez-Navarro A., Sentenac H., Guiderdoni E., Abdelly C., et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009;150:1955–1971. doi: 10.1104/pp.109.138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Colmenero-Flores J.M., Martínez G., Gamba G., Vázquez N., Iglesias D.J., Brumós J., Talón M. Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 2007;50:278–292. doi: 10.1111/j.1365-313X.2007.03048.x. [DOI] [PubMed] [Google Scholar]
- 254.Zhu M., Zhou M., Shabala L., Shabala S. Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ. 2017;40:1009–1020. doi: 10.1111/pce.12727. [DOI] [PubMed] [Google Scholar]
- 255.Mäser P., Eckelman B., Vaidyanathan R., Horie T., Fairbairn D.J., Kubo M., Yamagami M., Yamaguchi K., Nishimura M., Uozumi N., et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- 256.Munns R., James R.A., Xu B., Athman A., Conn S.J., Jordans C., Byrt C.S., Hare R.A., Tyerman S.D., Tester M., et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012;30:360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- 257.Ren Z.H., Gao J.P., Li L.G., Cai X.L., Huang W., Chao D.Y., Zhu M.Z., Wang Z.Y., Luan S., Lin H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 258.Sunarpi, Horie T., Motoda J., Kubo M., Yang H., Yoda K., Horie R., Chan W.Y., Leung H.Y., Hattori K., et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- 259.Uozumi N., Kim E.J., Rubio F., Yamaguchi T., Muto S., Tsuboi A., Bakker E.P., Nakamura T., Schroeder J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;122:1249–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Horie T., Yoshida K., Nakayama H., Yamada K., Oiki S., Shinmyo A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- 261.Wu H.J., Zhang Z., Wang J.Y., Oh D.H., Dassanayake M., Liu B., Huang Q., Sun H.X., Xia R., Wu Y., et al. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U S A. 2012;109:12219–12224. doi: 10.1073/pnas.1209954109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xue S., Yao X., Luo W., Jha D., Tester M., Horie T., Schroeder J.I. AtHKT1;1 mediates nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS One. 2011;6:e24725. doi: 10.1371/journal.pone.0024725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Berthomieu P., Conéjéro G., Nublat A., Brackenbury W.J., Lambert C., Savio C., Uozumi N., Oiki S., Yamada K., Cellier F., et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Møller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type- Specific alteration of Na+ transport in Arabidopsis. Plant Cell. 2009;21:2163–2178. doi: 10.1105/tpc.108.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Plett D., Safwat G., Gilliham M., Møller I.S., Roy S., Shirley N., Jacobs A., Johnson A., Tester M. Improved salinity tolerance of rice through cell type-specific expression of ATHKT1;1. PLoS One. 2010;5:1–8. doi: 10.1371/journal.pone.0012571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Byrt C.S., Platten J.D., Spielmeyer W., James R.A., Lagudah E.S., Dennis E.S., Tester M., Munns R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007;143:1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.James R.A., Davenport R.J., Munns R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006;142:1537–1547. doi: 10.1104/pp.106.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Blom-Zandstra M., Vogelzang S.A., Veen B.W. Sodium fluxes in sweet pepper exposed to varying sodium concentrations. J. Exp. Bot. 1998;49:1863–1868. [Google Scholar]
- 269.Kobayashi N.I., Yamaji N., Yamamoto H., Okubo K., Ueno H., Costa A., Tanoi K., Matsumura H., Fujii-Kashino M., Horiuchi T., et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017;91:657–670. doi: 10.1111/tpj.13595. [DOI] [PubMed] [Google Scholar]
- 270.Shabala S., Munns R. In: Plant Stress Physiology. Shalala S., editor. CABI Publishing; 2017. Salinity stress: physiological constraints and adaptive mechanisms; pp. 24–64. [Google Scholar]
- 271.Wolf O., Munns R., Tonnet M.L., Jeschke W.D. Concentrations and transport of solutes in xylem and phloem along the leaf axis of NaCl-treated Hordeum vulgare. J. Exp. Bot. 1990;41:1133–1141. [Google Scholar]
- 272.Jeschke W.D., Wolf O., Hartung W. Effect of NaCl salinity on flows and partitioning of C, N, and mineral ions in whole plants of white lupin, Lupinus albus L. J. Exp. Bot. 1992;43:777–788. [Google Scholar]
- 273.Shabala S., Pottosin I. Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014;151:257–279. doi: 10.1111/ppl.12165. [DOI] [PubMed] [Google Scholar]
- 274.Chen Z., Newman I., Zhou M., Mendham N., Zhang G., Shabala S. Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant Cell Environ. 2005;28:1230–1246. [Google Scholar]
- 275.Bose J., Rodrigo-Moreno A., Lai D., Xie Y., Shen W., Shabala S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2015;115:481–494. doi: 10.1093/aob/mcu219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Percey W.J., Shabala L., Wu Q., Su N., Breadmore M.C., Guijt R.M., Bose J., Shabala S. Potassium retention in leaf mesophyll as an element of salinity tissue tolerance in halophytes. Plant Physiol. Biochem. 2016;109:346–354. doi: 10.1016/j.plaphy.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 277.Volkov V., Wang B., Dominy P.J., Fricke W., Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ. 2004;27:1–14. [Google Scholar]
- 278.Riedelsberger J., Dreyer I., Gonzalez W. Outward rectification of voltage-gated K+ channels evolved at least twice in life history. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0137600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Véry A.A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 2014;171:748–769. doi: 10.1016/j.jplph.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 280.Falhof J., Pedersen J.T., Fuglsang A.T., Palmgren M. Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant. 2016;9:323–337. doi: 10.1016/j.molp.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 281.Rubio F., Nieves-Cordones M., Horie T., Shabala S. Doing ‘business as usual’ comes with a cost: evaluating energy cost of maintaining plant intracellular K+ homeostasis under saline conditions. New Phytol. 2019;225:1097–1104. doi: 10.1111/nph.15852. [DOI] [PubMed] [Google Scholar]
- 282.Chakraborty K., Bose J., Shabala L., Shabala S. Difference in root K+ retention ability and reduced sensitivity of K+-permeable channels to reactive oxygen species confer differential salt tolerance in three Brassica species. J. Exp. Bot. 2016;67:4611–4625. doi: 10.1093/jxb/erw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chen Z., Cuin T.A., Zhou M., Twomey A., Naidu B.P., Shiabala S. Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. J. Exp. Bot. 2007;58:4245–4255. doi: 10.1093/jxb/erm284. [DOI] [PubMed] [Google Scholar]
- 284.Wang H., Shabala L., Zhou M., Shabala S. Hydrogen peroxide-induced root Ca2+ and K+ fluxes correlate with salt tolerance in cereals: towards the cell-based phenotyping. Int. J. Mol. Sci. 2018;19:702. doi: 10.3390/ijms19030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Anschütz U., Becker D., Shabala S. Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014;171:670–687. doi: 10.1016/j.jplph.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 286.Shabala S. Signalling by potassium: another second messenger to add to the list? J. Exp. Bot. 2017;68:4003–4007. doi: 10.1093/jxb/erx238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Dreyer I., Uozumi N. Potassium channels in plant cells. FEBS J. 2011;278:4293–4303. doi: 10.1111/j.1742-4658.2011.08371.x. [DOI] [PubMed] [Google Scholar]
- 288.Kudla J., Becker D., Grill E., Hedrich R., Hippler M., Kummer U., Parniske M., Romeis T., Schumacher K. Advances and current challenges in calcium signaling. New Phytol. 2018;218:414–431. doi: 10.1111/nph.14966. [DOI] [PubMed] [Google Scholar]
- 289.Henry C., Bledsoe S.W., Griffiths C.A., Kollman A., Paul M.J., Sakr S., Lagrimini L.M. Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol. 2015;169:1072–1089. doi: 10.1104/pp.15.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pommerrenig B., Papini-Terzi F.S., Sauer N. Differential regulation of sorbitol and sucrose loading into the phloem of Plantago major in response to salt stress. Plant Physiol. 2007;144:1029–1038. doi: 10.1104/pp.106.089151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 292.Mansour M.M.F., Ali E.F. Evaluation of proline functions in saline conditions. Phytochemistry. 2017;140:52–68. doi: 10.1016/j.phytochem.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 293.Kiyosue T., Yoshiba Y., Yamaguchi-Shinozaki K., Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hayat S., Hayat Q., Alyemeni M.N., Wani A.S., Pichtel J., Ahmad A. Role of proline under changing environments: a review. Plant Signal. Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Székely G., Ábrahám E., Cséplo Á., Rigó G., Zsigmond L., Csiszár J., Ayaydin F., Strizhov N., Jásik J., Schmelzer E., et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- 296.Peng Z., Lu Q., Verma D.P.S. Reciprocal regulation of Δ1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet. 1996;253:334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- 297.Kubala S., Wojtyla L., Quinet M., Lechowska K., Lutts S., Garnczarska M. Enhanced expression of the proline synthesis gene P5CSA in relation to seed osmopriming improvement of Brassica napus germination under salinity stress. J. Plant Physiol. 2015;183:1–12. doi: 10.1016/j.jplph.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 298.Taji T., Seki M., Satou M., Sakurai T., Kobayashi M., Ishiyama K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ashraf M., Foolad M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007;59:206–216. [Google Scholar]
- 300.Ben Rejeb K., Abdelly C., Savouré A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014;80:278–284. doi: 10.1016/j.plaphy.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 301.Verbruggen N., Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- 302.Hasegawa P.M., Bressan R.A. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Mol. Plant Physiol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 303.Sakamoto A., Murata N. Genetic engineering of glycinebetaine synthesis in plants: current status and implications for enhancement of stress tolerance. J. Exp. Bot. 2000;51:81–88. [PubMed] [Google Scholar]
- 304.Chen T.H.H., Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 2011;34:1–20. doi: 10.1111/j.1365-3040.2010.02232.x. [DOI] [PubMed] [Google Scholar]
- 305.Mostofa M.G., Hossain M.A., Fujita M. Trehalose pretreatment induces salt tolerance in rice (Oryza sativa L.) seedlings: oxidative damage and co-induction of antioxidant defense and glyoxalase systems. Protoplasma. 2015;252:461–475. doi: 10.1007/s00709-014-0691-3. [DOI] [PubMed] [Google Scholar]
- 306.Iordachescu M., Imai R. Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 2008;50:1223–1229. doi: 10.1111/j.1744-7909.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- 307.Islam M.O., Kato H., Shima S., Tezuka D., Matsui H., Imai R. Functional identification of a rice trehalase gene involved in salt stress tolerance. Gene. 2019;685:42–49. doi: 10.1016/j.gene.2018.10.071. [DOI] [PubMed] [Google Scholar]
- 308.Ke Q., Ye J., Wang B., Ren J., Yin L., Deng X., Wang S. Melatonin mitigates salt stress in wheat seedlings by modulating polyamine metabolism. Front. Plant Sci. 2018;9:1–11. doi: 10.3389/fpls.2018.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Zarza X., Atanasov K.E., Marco F., Arbona V., Carrasco P., Kopka J., Fotopoulos V., Munnik T., Gómez-Cadenas A., Tiburcio A.F., et al. Polyamine oxidase 5 loss-of-function mutations in Arabidopsis thaliana trigger metabolic and transcriptional reprogramming and promote salt stress tolerance. Plant Cell Environ. 2017;40:527–542. doi: 10.1111/pce.12714. [DOI] [PubMed] [Google Scholar]
- 310.Minocha R., Majumdar R., Minocha S.C. Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 2014;5:1–17. doi: 10.3389/fpls.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.DeWald D.B., Torabinejad J., Jones C.A., Shope J.C., Cangelosi A.R., Thompson J.E., Prestwich G.D., Hama H. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed arabidopsis. Plant Physiol. 2001;126:759–769. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Xiong L., Lee B.H., Ishitani M., Lee H., Zhang C., Zhu J.K. FIERY1 encoding an inositol polyphosphate 1-phosphatase is negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Du H., Liu L., You L., Yang M., He Y., Li X., Xiong L. Characterization of an inositol 1,3,4-trisphosphate 5/6-kinase gene that is essential for drought and salt stress responses in rice. Plant Mol. Biol. 2011;77:547–563. doi: 10.1007/s11103-011-9830-9. [DOI] [PubMed] [Google Scholar]
- 314.Flowers T.J., Colmer T.D. Salinity tolerance in halophytes. New Phytol. 2008;179:945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- 315.Gorham J., Wyn Jones R.G., McDonnell E. Some mechanisms of salt tolerance in crop plants. Plant Soil. 1985;89:15–40. [Google Scholar]
- 316.Zeng F., Shabala S., Maksimovic J.D., Maksimovic V., Bonales-Alatorre E., Shabala L., Yu M., Zhang G., Zivanovic B.D. Revealing mechanisms of salinity tissue tolerance in succulent halophytes: a case study for Carpobrotus rossi. Plant Cell Environ. 2018;41:2654–2667. doi: 10.1111/pce.13391. [DOI] [PubMed] [Google Scholar]
- 317.Chiang C.P., Li C.H., Jou Y., Chen Y.C., Lin Y.C., Yang F.Y., Huang N.C., Yen H.E. Suppressor of K+ transport growth defect 1 (SKD1) interacts with RING-type ubiquitin ligase and sucrose non-fermenting 1-related protein kinase (SnRK1) in the halophyte ice plant. J. Exp. Bot. 2013;64:2385–2400. doi: 10.1093/jxb/ert097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Flowers T.J., Colmer T.D. Plant salt tolerance: adaptations in halophytes. Ann. Bot. 2015;115:327–331. doi: 10.1093/aob/mcu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Braun P., Winkelmann T. Flow cytometric analyses of somatic and pollen nuclei in midday flowers (Aizoaceae) Caryologia. 2016;69:303–314. [Google Scholar]
- 320.Qi C.H., Chen M., Song J., Wang B.S. Increase in aquaporin activity is involved in leaf succulence of the euhalophyte Suaeda salsa, under salinity. Plant Sci. 2009;176:200–205. [Google Scholar]
- 321.Dassanayake M., Larkin J.C. Making plants break a sweat: the structure, function, and evolution of plant salt glands. Front. Plant Sci. 2017;8:1–20. doi: 10.3389/fpls.2017.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ding F., Chen M., Sui N., Wang B.S. Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S. Afr. J. Bot. 2010;76:95–101. [Google Scholar]
- 323.Yuan F., Leng B., Wang B. Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front. Plant Sci. 2016;7:1–12. doi: 10.3389/fpls.2016.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Yuan F., Lyu M.J.A., Leng B.Y., Zheng G.Y., Feng Z.T., Li P.H., Zhu X.G., Wang B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015;38:1637–1657. doi: 10.1111/pce.12514. [DOI] [PubMed] [Google Scholar]
- 325.Barkla B.J., Rhodes T., Tran K.N.T., Wijesinghege C., Larkin J.C., Dassanayake M. Making epidermal bladder cells bigger: developmental- and salinity-induced endopolyploidy in a model halophyte. Plant Physiol. 2018;177:615–632. doi: 10.1104/pp.18.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Shabala L., Mackay A., Tian Y., Jacobsen S.E., Zhou D., Shabala S. Oxidative stress protection and stomatal patterning as components of salinity tolerance mechanism in quinoa (Chenopodium quinoa) Physiol. Plant. 2012;146:26–38. doi: 10.1111/j.1399-3054.2012.01599.x. [DOI] [PubMed] [Google Scholar]
- 327.Kiani-Pouya A., Rasouli F., Bazihizina N., Zhang H., Hedrich R., Shabala S. A large-scale screening of quinoa accessions reveals an important role of epidermal bladder cells and stomatal patterning in salinity tolerance. Environ. Exp. Bot. 2019;168:103885. [Google Scholar]
- 328.Smaoui A., Barhoumi Z., Rabhi M., Abdelly C. Localization of potential ion transport pathways in vesicular trichome cells of Atriplex halimus L. Protoplasma. 2011;248:363–372. doi: 10.1007/s00709-010-0179-8. [DOI] [PubMed] [Google Scholar]
- 329.Kiani-Pouya A., Roessner U., Jayasinghe N.S., Lutz A., Rupasinghe T., Bazihizina N., Bohm J., Alharbi S., Hedrich R., Shabala S. Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 2017;40:1900–1915. doi: 10.1111/pce.12995. [DOI] [PubMed] [Google Scholar]
- 330.Shabala S., Bose J., Hedrich R. Salt bladders: do they matter? Trends Plant Sci. 2014;19:687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 331.Böhm J., Messerer M., Müller H.M., Scholz-Starke J., Gradogna A., Scherzer S., Maierhofer T., Bazihizina N., Zhang H., Stigloher C., et al. Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr. Biol. 2018;28:3075–3085.e7. doi: 10.1016/j.cub.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 332.Barkla B.J., Vera-Estrella R. Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte mesembryanthemum crystallinum. Front. Plant Sci. 2015;6:1–10. doi: 10.3389/fpls.2015.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Oh D.H., Barkla B.J., Vera-Estrella R., Pantoja O., Lee S.Y., Bohnert H.J., Dassanayake M. Cell type-specific responses to salinity–the epidermal bladder cell transcriptome of Mesembryanthemum crystallinum. New Phytol. 2015;207:627–644. doi: 10.1111/nph.13414. [DOI] [PubMed] [Google Scholar]
- 334.Munns R., Gilliham M. Salinity tolerance of crops - what is the cost? New Phytol. 2015;208:668–673. doi: 10.1111/nph.13519. [DOI] [PubMed] [Google Scholar]
- 335.Munns R., Day D.A., Fricke W., Watt M., Arsova B., Barkla B.J., Bose J., Byrt C.S., Chen Z., Foster K.J., et al. Energy costs of salt tolerance in crop plants. New Phytol. 2019;225:1072–1090. doi: 10.1111/nph.15864. [DOI] [PubMed] [Google Scholar]
- 336.Shabala S., Shabala L. Ion transport and osmotic adjustment in plants and bacteria. Biomol. Concepts. 2011;2:407–419. doi: 10.1515/BMC.2011.032. [DOI] [PubMed] [Google Scholar]
- 337.Hariadi Y., Marandon K., Tian Y., Jacobsen S.E., Shabala S. Ionic and osmotic relations in quinoa (Chenopodium quinoa Willd.) plants grown at various salinity levels. J. Exp. Bot. 2011;62:185–193. doi: 10.1093/jxb/erq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Zhang T., Gong H., Wen X., Lu C. Salt stress induces a decrease in excitation energy transfer from phycobilisomes to photosystem II but an increase to photosystem I in the cyanobacterium Spirulina platensis. J. Plant Physiol. 2010;167:951–958. doi: 10.1016/j.jplph.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 339.Bahmani K., Sadat-Noori S.A., Izadi A., Akbari A. Molecular mechanisms of plant salinity tolerance: a review. Aust. J. Crop Sci. 2015;9:321–336. [Google Scholar]
- 340.Ghosh S., Bagchi S., Lahiri Majumder A. Chloroplast fructose-1,6-bisphosphatase from Oryza differs in salt tolerance property from the Porteresia enzyme and is protected by osmolytes. Plant Sci. 2001;160:1171–1181. doi: 10.1016/s0168-9452(01)00361-2. [DOI] [PubMed] [Google Scholar]
- 341.Chatterjee J., Patra B., Mukherjee R., Basak P., Mukherjee S., Ray S., Bhattacharyya S., Maitra S., GhoshDastidar K., Ghosh S., et al. Cloning, characterization and expression of a chloroplastic fructose-1,6-bisphosphatase from Porteresia coarctata conferring salt-tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. 2013;114:395–409. [Google Scholar]
- 342.Sengupta S., Majumder A.L. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: a physiological and proteomic approach. Planta. 2009;229:911–929. doi: 10.1007/s00425-008-0878-y. [DOI] [PubMed] [Google Scholar]
- 343.Wiciarz M., Gubernator B., Kruk J., Niewiadomska E. Enhanced chloroplastic generation of H2O2 in stress-resistant Thellungiella salsuginea in comparison to Arabidopsis thaliana. Physiol. Plant. 2015;153:467–476. doi: 10.1111/ppl.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Redondo-Gómez S., Mateos-Naranjo E., Figueroa M.E., Davy A.J. Salt stimulation of growth and photosynthesis in an extreme halophyte, Arthrocnemum macrostachyum. Plant Biol. 2010;12:79–87. doi: 10.1111/j.1438-8677.2009.00207.x. [DOI] [PubMed] [Google Scholar]
- 345.Trotta A., Redondo-Gómez S., Pagliano C., Clemente M.E.F., Rascio N., La Rocca N., Antonacci A., Andreucci F., Barbato R. Chloroplast ultrastructure and thylakoid polypeptide composition are affected by different salt concentrations in the halophytic plant Arthrocnemum macrostachyum. J. Plant Physiol. 2012;169:111–116. doi: 10.1016/j.jplph.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 346.Pagliano C., La Rocca N., Andreucci F., Deák Z., Vass I., Rascio N., Barbato R. The extreme halophyte Salicornia veneta is depleted of the extrinsic PsbQ and PsbP proteins of the oxygen-evolving complex without loss of functional activity. Ann. Bot. 2009;103:505–515. doi: 10.1093/aob/mcn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Geilfus C.M., Mithöfer A., Ludwig-Müller J., Zörb C., Muehling K.H. Chloride-inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytol. 2015;208:803–816. doi: 10.1111/nph.13507. [DOI] [PubMed] [Google Scholar]
- 348.Albacete A., Ghanem M.E., Martínez-Andújar C., Acosta M., Sánchez-Bravo J., Martínez V., Lutts S., Dodd I.C., Pérez-Alfocea F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008;59:4119–4131. doi: 10.1093/jxb/ern251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Gómez-Cadenas A., Tadeo F.R., Primo-Millo E., Talon M. Involvement of abscisic acid and ethylene in the responses of citrus seedlings to salt shock. Physiol. Plant. 1998;103:475–484. [Google Scholar]
- 350.Kefu Z., Munns R., King R.W. Abscisic-acid levels in NaCl-treated barley, cotton and saltbush. Aust. J. Plant Physiol. 1991;18:17–24. [Google Scholar]
- 351.Reef R., Schmitz N., Rogers B.A., Ball M.C., Lovelock C.E. Differential responses of the mangrove Avicennia marina to salinity and abscisic acid. Funct. Plant Biol. 2012;39:1038–1046. doi: 10.1071/FP12178. [DOI] [PubMed] [Google Scholar]
- 352.Taybi T., Cushman J.C. Abscisic acid signaling and protein synthesis requirements for phosphoenolpyruvate carboxylase transcript induction in the common ice plant. J. Plant Physiol. 2002;159:1235–1243. [Google Scholar]
- 353.Hedrich R., Shabala S. Stomata in a saline world. Curr. Opin. Plant Biol. 2018;46:87–95. doi: 10.1016/j.pbi.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 354.Ellouzi H., Ben Hamed K., Cela J., Munné-Bosch S., Abdelly C. Early effects of salt stress on the physiological and oxidative status of Cakile maritima (halophyte) and Arabidopsis thaliana (glycophyte) Physiol. Plant. 2011;142:128–143. doi: 10.1111/j.1399-3054.2011.01450.x. [DOI] [PubMed] [Google Scholar]
- 355.Chiang C.P., Yim W.C., Sun Y.H., Ohnishi M., Mimura T., Cushman J.C., Yen H.E. Identification of ice plant (Mesembryanthemum crystallinum L.) microRNAs using RNA-seq and their putative roles in high salinity responses in seedlings. Front. Plant Sci. 2016;7:1–18. doi: 10.3389/fpls.2016.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Franks P.J., Drake P.L., Beerling D.J. Plasticity in maximum stomatal conductance constrained by negative correlation between stomatal size and density: an analysis using Eucalyptus globulus. Plant Cell Environ. 2009;32:1737–1748. doi: 10.1111/j.1365-3040.2009.02031.x. [DOI] [PubMed] [Google Scholar]
- 357.Franks P.J., W Doheny-Adams T., Britton-Harper Z.J., Gray J.E. Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol. 2015;207:188–195. doi: 10.1111/nph.13347. [DOI] [PubMed] [Google Scholar]
- 358.Orsini F., Accorsi M., Gianquinto G., Dinelli G., Antognoni F., Carrasco K.B.R., Martinez E.A., Alnayef M., Marotti I., Bosi S., et al. Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: functional elements of successful halophytism. Funct. Plant Biol. 2011;38:818–831. doi: 10.1071/FP11088. [DOI] [PubMed] [Google Scholar]
- 359.Adrian J., Chang J., Ballenger C.E., Bargmann B.O.R., Alassimone J., Davies K.A., Lau O.S., Matos J.L., Hachez C., Lanctot A., et al. Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell. 2015;33:107–118. doi: 10.1016/j.devcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Lee L.R., Bergmann D.C. The plant stomatal lineage at a glance. J. Cell Sci. 2019;132:jcs228551. doi: 10.1242/jcs.228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Bressan R.A., Zhang C., Zhang H., Hasegawa P.M., Bohnert H.J., Zhu J.K. Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiol. 2001;127:1354–1360. [PMC free article] [PubMed] [Google Scholar]
- 362.Wang G., Pantha P., Tran K.N., Oh D.H., Dassanayake M. Plant growth and agrobacterium-mediated floral-dip transformation of the extremophyte Schrenkiella parvula. J. Vis. Exp. 2019;143:e58544. doi: 10.3791/58544. [DOI] [PubMed] [Google Scholar]
- 363.Oh D.H., Leidi E., Zhang Q., Hwang S.M., Li Y., Quintero F.J., Jiang X., D’urzo M.P., Lee S.Y., Zhao Y., et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009;151:210–222. doi: 10.1104/pp.109.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Dassanayake M., Oh D.H., Haas J.S., Hernandez A., Hong H., Ali S., Yun D.J., Bressan R.A., Zhu J.K., Bohnert H.J., et al. The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 2011;43:913–918. doi: 10.1038/ng.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Yang R., Jarvis D.E., Chen H., Beilstein M.A., Grimwood J., Jenkins J., Shu S.Q., Prochnik S., Xin M., Ma C., et al. The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 2013;4:1–14. doi: 10.3389/fpls.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Amtmann A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2009;2:3–12. doi: 10.1093/mp/ssn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Kazachkova Y., Eshel G., Pantha P., Cheeseman J.M., Dassanayake M., Barak S. Halophytism: what have we learnt from Arabidopsis thaliana relative model systems? Plant Physiol. 2018;178:972–988. doi: 10.1104/pp.18.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sun Q., Gao F., Zhao L., Li K., Zhang J. Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol. 2010;10:90. doi: 10.1186/1471-2229-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Jarvis D.E., Ryu C.H., Beilstein M.A., Schumaker K.S. Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula. Mol. Biol. Evol. 2014;31:2094–2107. doi: 10.1093/molbev/msu152. [DOI] [PubMed] [Google Scholar]
- 370.Ali A., Raddatz N., Aman R., Kim S., Park H.C., Jan M., Baek D., Khan I.U., Oh D.H., Lee S.Y., et al. A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol. 2016;171:2112–2126. doi: 10.1104/pp.16.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]