Abstract
CHROMagar Salmonella (CAS), a new chromogenic medium, was retrospectively compared to Hektoen enteric agar (HEA) with 501 Salmonella stock isolates and was then prospectively compared to HEA for the detection and presumptive identification of Salmonella spp. with 508 stool samples before and after enrichment. All stock cultures (100%), including cultures of H2S-negative isolates, yielded typical mauve colonies on CAS, while 497 (99%) isolates produced typical lactose-negative, black-centered colonies on HEA. Following overnight incubation at 37°C, a total of 20 Salmonella strains were isolated from the 508 clinical samples. Sensitivities for primary plating and after enrichment were 95% (19 isolates) and 100% (20 isolates), respectively, for CAS and 80% (16 isolates) and 100% (20 isolates), respectively, for HEA. The specificity of CAS (88.9%) was significantly higher than that of HEA (78.5%; P < 0.0001). On the basis of its good sensitivity and specificity, CAS medium can be recommended for use for primary plating when human stool samples are screened for Salmonella spp.
Infections due to Salmonella spp. continue to be a major health problem, and their diagnosis most often relies on direct detection of bacteria in stools by culture or, more recently, by PCR after enrichment (18, 19). Contrary to PCR, isolation of Salmonella on selective culture media allows further identification of the bacteria and antibiotic susceptibility testing, which are critical for disease control. Hektoen enteric agar (HEA) has been widely used for this purpose since its introduction in 1968 (11), and because of its good sensitivity, it remains the standard primary plating medium in our routine screening for Salmonella and Shigella spp. However, its specificity is poor (4, 5, 12) and the numerous false-positive results require time-consuming complementary testing to identify or, in most cases, to exclude the presence of Salmonella colonies. Recently, new media which allow the detection of Salmonella by the use of chromogenic substrates have been introduced (4, 14). When compared to conventional media like HEA, chromogenic media had higher specificities, i.e., fewer false-positive results, but lower sensitivities, i.e., more false-negative results. Consequently, they are not recommended for the screening of Salmonella on primary plating of stool specimens (4, 12, 13, 15), and there is still a need for more specific culture media that will at least retain the sensitivities of media such as HEA. CHROMagar Salmonella (CAS) is a new selective chromogenic medium that allows the detection of Salmonella as mauve colonies after 18 h of incubation, whereas other members of the family Enterobacteriaceae grow as blue or uncolored colonies. A preliminary study (7) showed that the sensitivity of CAS was similar to that of HEA on primary plating and after enrichment of human stool specimens. However, its specificity was hampered by the presence of false-positive colonies of Pseudomonas aeruginosa. We attempted to increase the selectivity of CAS by adding cefsulodin, a narrow-spectrum cephalosporin mostly active against P. aeruginosa. CAS was then compared to HEA (i) with 501 consecutive clinical Salmonella strains isolated from different laboratories in order to assess the performance of both media with a wide variety of isolates and (ii) for routine analysis of 508 consecutive clinical stool samples.
MATERIALS AND METHODS
Culture media.
CAS, a proprietary product, was provided for evaluation by CHROMagar Microbiology, Paris, France. The medium was supplied as a white powder in preweighed batches sufficient for 250 ml and was prepared according to the manufacturer’s instructions. Powdered CAS was added to distilled water and was dissolved by slow rotation. When it was dissolved, the medium was boiled with continuous stirring for about 2 min until the complete fusion of the agar grains was detected. After boiling, the medium was swirled gently while cooling to 48°C. Cefsulodin (Sigma, Saint-Quentin-Fallavier, France) was added at this stage to obtain a final concentration of 10 mg/liter. Then, 20 ml of the medium was dispensed into sterile petri dishes and was allowed to solidify and dry with the plate lids kept ajar. As indicated by the manufacturer, CAS plates were stored at room temperature in a dark container and were used within a week. Storage in a refrigerator was not recommended. HEA was purchased as commercially prepared plates (Becton Dickinson, Le Pont-de-Claix, France).
Stock isolates.
The 501 stock Salmonella strains representing 38 serovars (Table 1) were isolated from human stool samples by different clinical laboratories in the northern region of France between October 1996 and September 1997. They were consecutively received for serological identification by the Microbiology Laboratory of Lille University Hospital. After typing, the isolates were stored frozen at −80°C. For the present study, the isolates were thawed, seeded on blood agar, and incubated overnight at 37°C. Suspensions were prepared from freshly grown colonies in saline solution and were adjusted to a turbidity equivalent to that of a 0.5 McFarland standard suspension. Dilutions were made from these suspensions in order to inoculate approximately 200 CFU of each strain onto HEA and CAS plates.
TABLE 1.
Distribution of S. enterica serovars among stock isolates and fresh stool isolatesa
| Serogroup | Serovar | No. of isolates tested
|
|
|---|---|---|---|
| Stock | Fresh | ||
| A | Paratyphi A | 1 | |
| B | Typhimurium | 197 | 7 |
| Derby | 5 | ||
| Brandenburg | 4 | ||
| Heidelberg | 4 | ||
| Indiana | 3 | 1 | |
| Saint-Paul | 3 | ||
| Paratyphi B | 2 | ||
| Agona | 1 | ||
| Bredeney | 1 | ||
| C1 | Virchow | 10 | 2 |
| Infantis | 6 | 1 | |
| Montevideo | 5 | ||
| Braenderup | 2 | ||
| Isangi | 2 | ||
| Livingstone | 1 | ||
| Mbandaka | 1 | ||
| Othmarschen | 1 | ||
| Orianenburg | 1 | ||
| C2 | Hadar | 23 | 2 |
| Goldcoast | 4 | ||
| Bovismorbificans | 3 | ||
| Blockley | 1 | ||
| Gatuni | 1 | ||
| Litchfield | 1 | ||
| Muenchen | 1 | ||
| Newport | 1 | ||
| C3 | Emek | 2 | |
| Kentucky | 1 | ||
| D | Enteritidis | 202 | 7 |
| Panama | 2 | ||
| Typhi | 1 | ||
| E | Anatum | 2 | |
| Senftenberg | 2 | ||
| Give | 1 | ||
| Munster | 1 | ||
| Oxford | 1 | ||
| Uganda | 1 | ||
A total of 501 stock isolates and 20 fresh stool isolates were tested.
Clinical samples.
The second arm of the study was carried out between October 1997 and August 1998 in the 380-bed Boucicaut Hospital with 508 consecutive stool samples from adult inpatients and outpatients with symptoms of gastroenteritis. The same amount of each sample (50 μl of liquid stool or stool liquefied in saline solution) was streaked onto CAS and HEA plates. Concurrently, enrichment was performed in Muller-Kauffmann broth (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) inoculated with 1 ml of stool specimen, and the mixture was incubated overnight at 41°C and then streaked onto CAS and HEA plates (50 μl per plate). All plates were incubated at 37°C in air.
Presumptive identification.
All plates were inspected by the same technologist. Colonies suspected of being Salmonella spp. were defined as follows: on CAS, mauve colonies after incubation for 18 to 24 h; on HEA, green (lactose-negative), black-centered (H2S-positive) colonies after incubation for 18 to 48 h. The green colonies on HEA plates (lactose and H2S negative) were identified as part of our routine search for Shigella spp., but they were not included as suspect Salmonella colonies in the evaluation of the specificity of the medium.
Confirmatory tests and final identification.
A maximum of 10 suspect colonies from each positive agar plate were selected for complete identification. Bacterial colonies were identified with the API 20E and/or API 32GN systems (bioMérieux, Marcy l’Etoile, France). The oxidase test was performed with oxidase disks (Sanofi Diagnostics Pasteur) according to the manufacturer’s instructions. Salmonella serovars were determined after overnight culture on Kligler’s iron agar (Sanofi Diagnostics Pasteur) by agglutination with anti-O and anti-H antisera (Sanofi Diagnostics Pasteur) by following the Kauffmann-White scheme (10). Any questionable identifications were confirmed by the Centre National de Référence des Salmonella et Shigella (Institut Pasteur, Paris, France). Yeast colonies were identified by a germ tube test in rabbit serum (bioMérieux) and with the Auxacolor system (Sanofi Diagnostics Pasteur).
Statistical analysis.
The differences in the sensitivities and specificities of the two media were evaluated by the Yates-corrected χ2 test.
RESULTS
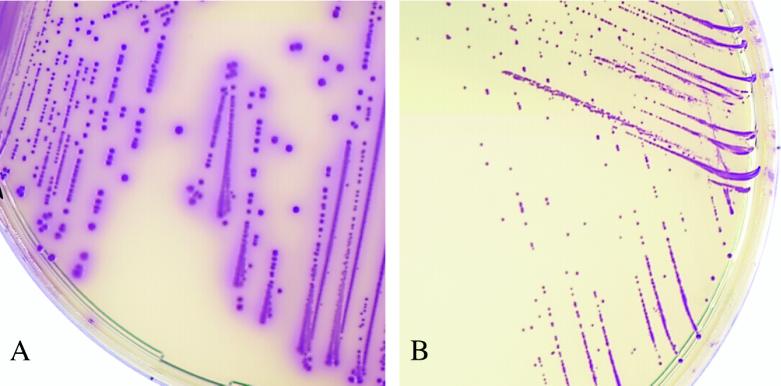
At first, we evaluated the colonial appearance of 501 Salmonella stock isolates on both media. All of them (100%) produced typical mauve colonies on CAS after 18 h of incubation (Fig. 1A). On HEA, 496 isolates (99%) yielded typical green, black-centered colonies after 24 to 36 h of incubation; the remaining 5 isolates belonged to serovars Heidelberg (n = 3), Typhi (n = 1), and Virchow (n = 1) and grew as green, H2S-negative colonies. For each isolate, the colony counts performed on both media were similar (data not shown).
FIG. 1.
Colonies plated out from saline suspensions of Salmonella serovar Enteritidis (A) and Candida albicans (B). C. albicans colonies are tiny and convex. CAS plates were incubated for 24 h at 37°C. Magnification, ×2.
We next evaluated the performance of the two media in a routine laboratory. A total of 20 salmonellae were isolated on at least one medium from the 508 fresh stool samples. The distribution of serovars is indicated in Table 1. On primary plating, 19 and 16 isolates were detected on CAS and HEA, respectively. At this step, the strain that was missed on CAS was also missed on HEA. After enrichment, all isolates grew on both media. The sensitivities of CAS and HEA were not significantly different on primary plating (95 and 80%, respectively; P ≥ 0.05) and were identical (100%) after enrichment (Table 2). The 20 isolates were lactose negative and H2S positive and yielded typical mauve colonies on CAS (Fig. 1A). Biochemical identification of all the isolates was satisfactorily performed with colonies picked from CAS plates.
TABLE 2.
Recovery of Salmonella spp. from 508 stool specimens before and after enrichment in Muller-Kauffmann brotha
| Medium (step) | No. of true-positive isolates | No. of false-negative isolates | Sensitivity (%) |
|---|---|---|---|
| CAS (primary plating) | 19 | 1 | 95 |
| HEA (primary plating) | 16 | 4 | 80 |
| CAS (after enrichment) | 20 | 0 | 100 |
| HEA (after enrichment) | 20 | 0 | 100 |
A total of 20 isolates were recovered on at least one medium (sensitivity, 100%).
As shown in Table 3, the specificity of CAS on primary plating (88.9%) was significantly higher than that of HEA (78.5%) (P < 0.0001), with a total of 54 and 105 false-positive strains isolated on CAS and HEA, respectively. They consisted of Candida albicans (n = 30), P. aeruginosa (n = 23), and Aeromonas hydrophila (n = 1) on CAS and Proteus mirabilis (n = 88), Citrobacter freundii (n = 14), Escherichia coli (n = 2), and Shewanella (previously Alteromonas) putrefaciens (n = 1) on HEA. After enrichment, the specificities increased to 96.7 and 88.7% for CAS and HEA, respectively (P < 0.0001). At this step, false-positive results were obtained for C. albicans (n = 2) and P. aeruginosa (n = 16) on CAS and P. mirabilis (n = 45) and C. freundii (n = 10) on HEA. It is noteworthy that no swarming of Proteus colonies was observed on either medium. C. albicans was the only yeast species that grew as mauve colonies on CAS. Other species including Candida glabrata, Candida krusei, and Candida parapsilosis were identified during the study and grew on CAS as tiny colorless colonies after a minimum of 48 h. The pinpoint and convex morphologies of the colonies of C. albicans on CAS allowed instant differentiation from other suspect colonies (Fig. 1B). The fungal nature of these colonies could be rapidly confirmed by microscopic examination of a wet preparation (magnification, ×400), showed which budding yeast cells instead of bacteria. Other false-positive organisms on CAS (P. aeruginosa and A. hydrophila) were macroscopically indistinguishable from the salmonella colonies (data not shown) but differed by a positive oxidase reaction and by their polar flagella. All 23 P. aeruginosa isolates that grew on CAS were resistant to cefsulodin and ticarcillin. A majority of them (n = 18) were isolated from patients undergoing prolonged hospitalization in intensive care units and were also resistant to aminoglycosides and/or to fluoroquinolones. The A. hydrophila strain was isolated in pure culture on both media and was resistant to cefsulodin (MIC, 32 μg/ml). All false-positive colonies on HEA were subcultured on CAS, and none of them grew as mauve colonies. Conversely, none of the false positive colonies on CAS yielded green, black-centered colonies on HEA.
TABLE 3.
Specificities of the two media on primary plating
| Medium | No. of true-negative results | No. of false-positive results | Specificity (%)a |
|---|---|---|---|
| CAS | 434 | 54b | 88.9 |
| HEA | 383 | 105c | 78.5 |
(Number of true-negative results on the medium/number of negative samples) × 100.
C. albicans, n = 30; P. aeruginosa, n = 23; A. hydrophila, n = 1.
P. mirabilis, n = 88; C. freundii, n = 14; E. coli, n = 2; S. putrefaciens, n = 1.
DISCUSSION
The first stage of this study was performed with stock cultures and demonstrated the ability of CAS and HEA to grow a variety of clinical Salmonella strains of human origin. However, five H2S-negative isolates were misidentified on HEA. Conversely, all isolates tested yielded typical positive (mauve) colonies on CAS, including serovars Typhi and Paratyphi A previously reported as being undetectable on Salmonella-specific media such as novobiocin-brilliant green-glycerol-lactose agar (5, 13, 15), modified semisolid Rappaport-Vassiliadis agar (1, 16), or Rambach agar (5, 8, 15). The identical outputs of the cultures on both media demonstrated the absence of an inhibitory effect of CAS on the growth of Salmonella compared to the effect of HEA. In addition to these results, CAS and HEA were then compared in a routine analysis of clinical stool specimens.
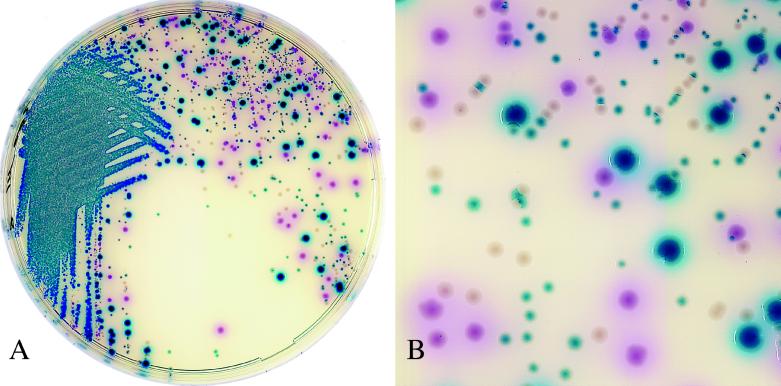
The number of Salmonella strains isolated from the 508 consecutive stool samples was relatively low (positivity rate, 3.9%) compared to the numbers isolated in similar studies that reported positivity rates ranging from 5.4 to 8.8% (4, 15). This could be explained by the fact that a significant proportion of samples originated from patients who had been hospitalized for a long time; such patients are less likely to carry Salmonella than patients with gastroenteritis in the community, as reported previously (6). The difference in the sensitivities between HEA and CAS observed on primary plating (Table 1) may have resulted from a better separation of colonies on CAS, whereas many colonies on HEA tended to be confluent. This was particularly noticeable when the number of positive colonies was small compared to the competing growth of other coliforms. Good separation added to the easy distinction of mauve from blue or uncolored colonies (Fig. 2) and was of considerable help for the further identification of suspect colonies. However, the sensitivities after enrichment in Muller-Kauffmann broth were identical for both media. When compared to our previous results (7), the addition of cefsulodin to the composition of CAS had no apparent effect on the sensitivity of the medium.
FIG. 2.
Colonies plated out from stool specimens containing Salmonella serovar Enteritidis. Salmonella strains appear as mauve colonies. Various blue colonies were identified as Escherichia coli and Citrobacter freundii, and uncolored colonies were identified as Morganella morganii. CAS plates were incubated for 20 h at 37°C. Magnifications: actual size (A) and ×3 (B).
In this study, CAS was shown to be more specific than HEA, despite the presence of false-positives colonies of P. aeruginosa, A. hydrophila, and C. albicans. However, the proportion of P. aeruginosa isolates which accounted for more than 70% of the false-positive colonies on CAS in our preliminary study (7) fell to less than 45% after the addition of cefsulodin to the medium. False-positive colonies on HEA included various H2S-producing members of the family Enterobacteriaceae and the less frequently isolated species S. putrefaciens. These organisms can be routinely differentiated from Salmonella spp. by combinations of various techniques that require at least 4 to 6 h of incubation (for examples, see references 2, 3, 9, and 17). In comparison, the presence of colonies of P. aeruginosa or A. hydrophila on CAS can be ruled out in a few minutes by the oxidase test.
With a little experience, the difference between C. albicans colonies and other suspect colonies was considered straightforward enough (Fig. 1A and B) such that further identification was not required, and we felt that the addition of an antifungal agent such as amphotericin B to CAS was not necessary. Furthermore, the detection of a heavy growth of C. albicans was of clinical interest in several instances, especially for intensive care unit patients receiving broad-spectrum antibiotics. Still, we recommend that workers who are using CAS for the first time include a mauve colony-producing C. albicans isolate to serve as a control.
Considering our results, we feel that the use of CAS provides a time-saving method for the detection and presumptive identification of salmonellae in the routine analysis of stool specimens. Interpretation of colors is easy, and all colonies of salmonellae tested displayed the same color and morphology. Compared to the number of false-positive colonies on HEA, we observed far fewer false-positive colonies on CAS, and all of them could be ruled out as Salmonella spp. with a minimum of rapid and inexpensive confirmatory testing (the disk oxidase test or direct microscopy). Furthermore, the good sensitivity of CAS qualifies this medium for use in the primary plating of stool specimens when searching for Salmonella spp.
ACKNOWLEDGMENTS
We thank Michel Simonet, Colin R. Tinsley, and Nicolas Fortineau for helpful comments and critical reading of the manuscript.
REFERENCES
- 1.Aspinall S T, Hindle M A, Hutchinson D N. Improved isolation of salmonellae from faeces using a semisolid Rappaport-Vassiliadis medium. Eur J Clin Microbiol Infect Dis. 1992;11:936–939. doi: 10.1007/BF01962379. [DOI] [PubMed] [Google Scholar]
- 2.Cherrington C A, Huis in’t Veld J H. Comparison of classical isolation protocols with a 24 h screen to detect viable salmonellas in faeces. J Appl Bacteriol. 1993;75:65–68. doi: 10.1111/j.1365-2672.1993.tb03409.x. [DOI] [PubMed] [Google Scholar]
- 3.De Ryck R, Struelens M J, Serruys E. Rapid biochemical screening for Salmonella, Shigella, Yersinia, and Aeromonas isolates from stool specimens. J Clin Microbiol. 1994;32:1583–1585. doi: 10.1128/jcm.32.6.1583-1585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dusch H, Altwegg M. Comparison of Rambach agar, SM-ID medium, and Hektoen enteric agar for primary isolation of non-typhi salmonellae from stool samples. J Clin Microbiol. 1993;31:410–412. doi: 10.1128/jcm.31.2.410-412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dusch H, Altwegg M. Evaluation of five new plating media for isolation of Salmonella species. J Clin Microbiol. 1995;33:802–804. doi: 10.1128/jcm.33.4.802-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan K, Morris A J, Reller L B. Application of rejection criteria for stool cultures for bacterial enteric pathogens. J Clin Microbiol. 1993;31:2233–2235. doi: 10.1128/jcm.31.8.2233-2235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillot O, Maruéjouls C, Di Camillo P, Fortineau N, Courcol R, Savage C. Program and abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Comparison of CHROMagar Salmonella medium and Hektoen enteric agar for detection of Salmonella in human stool samples, abstr. C-445; p. 205. [Google Scholar]
- 8.Gruenewald R, Henderson R W, Yappow S. Use of Rambach propylene glycol-containing agar for identification of Salmonella spp. J Clin Microbiol. 1991;29:2354–2356. doi: 10.1128/jcm.29.10.2354-2356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imperatrice C A, Nachamkin I. Evaluation of the Vitek EPS enteric pathogen screen card for detecting Salmonella, Shigella, and Yersinia spp. J Clin Microbiol. 1993;31:433–435. doi: 10.1128/jcm.31.2.433-435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauffmann F. Serological diagnosis of Salmonella-species. Kauffmann-White-Schema. Copenhagen, Denmark: Munksgaard; 1972. [Google Scholar]
- 11.King S, Metzger W I. A new plating medium for the isolation of enteric pathogen. Appl Microbiol. 1968;16:577–578. doi: 10.1128/am.16.4.577-578.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monnery I, Freydière A M, Baron C, Rousset A M, Tigaud S, Boude-Chevalier M, de Montclos H, Gille Y. Evaluation of two new chromogenic media for detection of Salmonella in stools. Eur J Clin Microbiol Infect Dis. 1994;13:257–261. doi: 10.1007/BF01974547. [DOI] [PubMed] [Google Scholar]
- 13.Poisson D M, Nugier J P, Rousseau P. Study of Rambach and NBGL agar on 4037 stools of human origin and 584 veterinary samples submitted for isolation of Salmonellae. Pathol Biol (Paris) 1993;41:543–546. [PubMed] [Google Scholar]
- 14.Rambach A. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl Environ Microbiol. 1990;56:301–303. doi: 10.1128/aem.56.1.301-303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz J, Nunez M L, Diaz J, Lorente I, Perez J, Gomez J. Comparison of five plating media for isolation of Salmonella species from human stools. J Clin Microbiol. 1996;34:686–688. doi: 10.1128/jcm.34.3.686-688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz J, Nunez M L, Lorente I, Perez J, Simarro E, Gomez J. Performance of six culture media for isolation of Salmonella species from stool samples. Eur J Clin Microbiol Infect Dis. 1996;15:922–926. doi: 10.1007/BF01690509. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz J, Sempere M A, Varela M C, Gomez J. Modification of the methodology of stool culture for Salmonella detection. J Clin Microbiol. 1992;30:525–526. doi: 10.1128/jcm.30.2.525-526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone G G, Oberst R D, Hays M P, McVey S, Chengappa M M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J Clin Microbiol. 1994;32:1742–1749. doi: 10.1128/jcm.32.7.1742-1749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]