Abstract
N6-Methyladenosine (m6A) RNA modification brings a new dawn for RNA modification researches in recent years. This posttranscriptional RNA modification is dynamic and reversible, and is regulated by methylases (“writers”), demethylases (“erasers”), and proteins that preferentially recognize m6A modifications (“readers”). The change of RNA m6A modification regulates RNA metabolism in eucaryon, including translation, splicing, exporting, decay, and processing. Thereby the dysregulation of m6A may lead to tumorigenesis and progression. Given the tumorigenic role of abnormal m6A expression, m6A regulators may function as potential clinical therapeutic targets for cancers. In this review, we emphasize on the underlying mechanisms of m6A modifications in tumorigenesis and further introduce the potential m6A regulators-associated therapeutic targets for tumor therapy.
Keywords: N6-methyladenosine, cancer, RNA methylation, therapy
Public Summary
-
•
N6-Methyladenosine (m6A) RNA modification brings a new dawn for RNA modification researches in recent years.
-
•
The dysregulation of m6A may lead to tumorigenesis and progression.
-
•
m6A regulators may function as potential clinical therapeutic targets for cancers.
Graphical Abstract
Main Text
From the 1950s, an increasing accumulated number of chemically modified nucleosides in RNA have been identified, which change the RNA structures, leading to different biological functions.1,2 Many RNA modifications have been identified in eukaryotic transcripts (Figure 1). N6-Methyladenosine (m6A) RNA modification defines as a methylation of adenosine at the N6 position, brings a new dawn for RNA modification research since its first discovery in 1974.3 m6A has been identified as the most abundant mRNA modification, widely distributing in the majority of eukaryotic species including mammals,3, 4, 5, 6 plants,7 insects,8 yeast,9 and certain viruses.10,11 The identifications and characterizations of “writer,” “eraser,” and “reader” proteins, and development of high-throughput sequencing provide new insight into RNA modification biology, especially m6A RNA modification.12, 13, 14
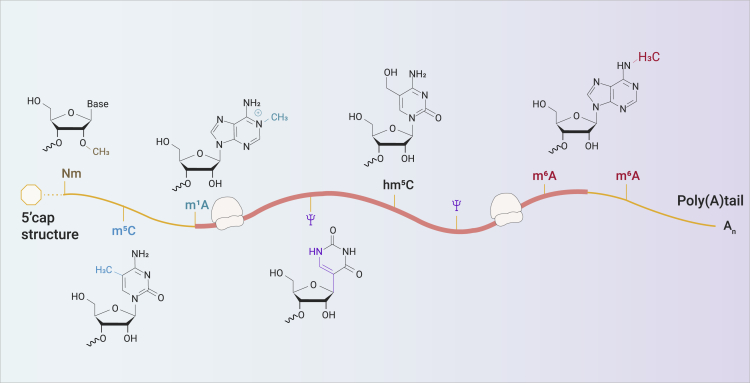
Figure 1.
RNA Modifications in Eucaryon
A schematic diagram of common RNA modifications in eukaryotic transcripts. m1A, N1-methyladenosine; hm5C, 5-hydroxymethylcytosine; m6Am, N6,2′-O-dimethyladenosine
m6A was identified in about one-third of the mRNAs in mammals while an average number of 3 to 5 m6A modifications were found in each mRNA. Of note, a lot of m6A sites are evolutionally conserved between humans and mice. The deposition of m6A modification in the transcriptome is not random.15 A characteristic DRACH sequence (D = G, A, or U; R = G or A; H = A, C, or U) often enriched in the 3′ untranslated region (3′ UTR) and the coding sequence (CDS) when m6A modification occurs.12,13 Besides, it has clearly known that m6A modifications exist on almost all types of coding and non-coding RNAs (ncRNAs) and dynamically regulate their relevant molecular processes, physiological and pathological functions.
In this review, we emphasize the underlying mechanisms of m6A modifications in tumorigenesis and further introduce the potential m6A regulator-associated therapeutic targets for tumor therapy.
Dynamic Regulation of m6A
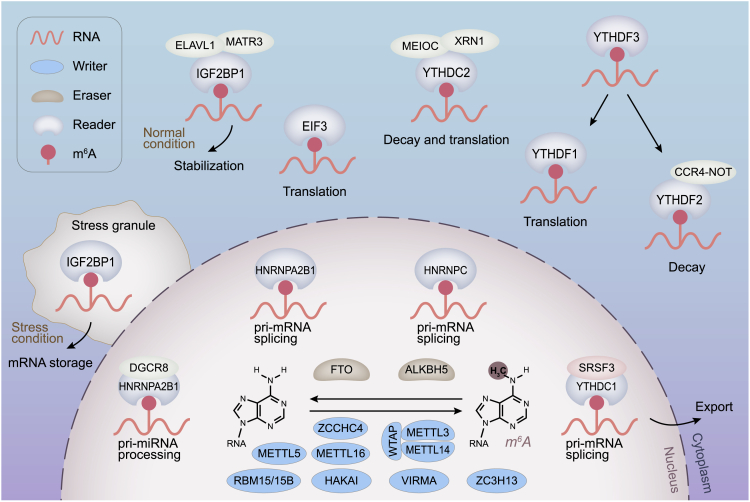
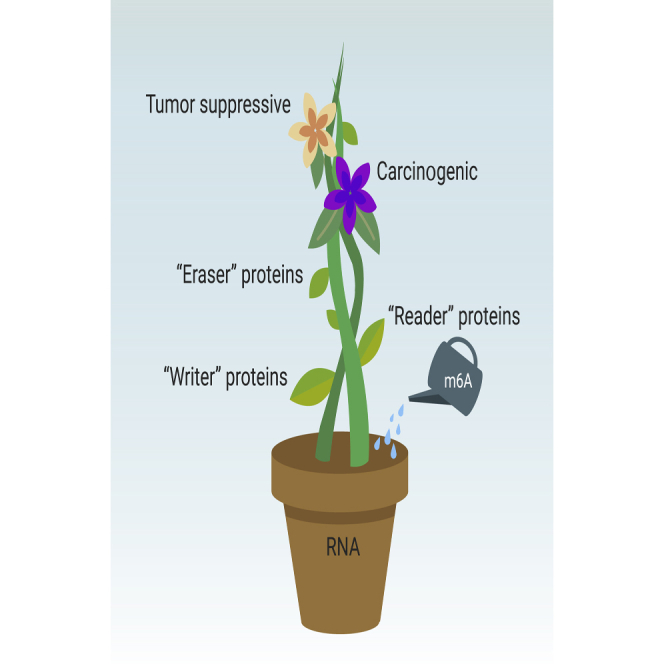
The RNA m6A modifications are dynamically and reversibly regulated by two important catalytic proteins, methyltransferases and demethylases, which are also recognized as “writers” and “erasers,” respectively. In addition, a set of binding proteins (“readers”) function as decoding the m6A modifications and recruiting downstream functional complexes (Figure 2).
Figure 2.
Mechanism of RNA m6A Modifications
The m6A modification is catalyzed by the “writer” proteins including METTL3, METTL14, WTAP, METTL5, METTL16, ZCCHC4, VIRMA, RBM15/15B, HAKAI, and ZC3H13. The m6A methylation is removed by “eraser” proteins FTO or ALKBH5. “Reader” proteins recognize m6A modifications and determine targeted RNA destiny.
m6A Writer Proteins
The RNA m6A modification writer proteins are a kind of methyltransferase, consist of methyltransferase-like 3 (METTL3), methyltransferase-like 5 (METTL5), methyltransferase-like 16 (METTL16), zinc finger CCHC-type containing 4 (ZCCHC4), and additional partner proteins such as methyltransferase-like 14 (METTL14), Wilms tumor 1-associating protein (WTAP), Vir like m6A methyltransferase associated (VIRMA), zinc finger CCCH-type containing 13 (ZC3H13), and RNA-binding motif 15/15B (RBM15/15B).16, 17, 18, 19, 20, 21 METTL3 was found acting as the key methyltransferase for m6A methylation in 1997 and abnormal expression of METTL3 often affects the total level of m6A methylation.22, 23, 24 Meanwhile, METTL14 functions as a synergistical partner for METTL3, which has structural support for METTL3 when they combine into a core methyltransferase complex, and therefore facilitates RNA binding.24,25 The main function of WTAP is binding to METTL3/METTL14 complex and is necessary for the recruitment of optimal substrate and the localization of METTL3/METTL14 complex.17,26 RBM15/15B has been confirmed, helping the combination process between METTL3 and WTAP by specific binding to U-rich regions.27 HAKAI, also known as Cbl Proto-Oncogene Like 1 (CBLL1), assists controlling nuclear m6A methylation. The function of ZC3H13 is similar to CBLL1, which helps nuclear localization.20 VIRMA is very important for m6A modification, especially locating at the 3′-UTR and around stop codon sequences.19
Of all the m6A methyltransferases, METTL5, METTL16, and ZCCHC4 could function independent of other methyltransferase proteins. As for METTL5, it has been identified as a kind of independent m6A methyltransferase, catalyzing m6A on some certain structured RNAs, including 18S rRNA, 28S rRNA, and U6 small nuclear (snRNA).21,28, 29, 30, 31 A special structure of heterodimeric complex with TRMT112 is formed by METTL5 and could enhance its own metabolic stability in cells.28 Meanwhile, TRMT112 functions as a coactivator of METTL5, and the relationship between METTL5 and TRMT112 is similar to the relationship between METTL3 and METTL14.28 Besides, the atomic resolution structure of METTL5-TRMT112 complex demonstrates the RNA-binding mode of the complex differs considerably from that of other m6A RNA methyltransferases.28 Similar to METTL5, METTL16 is also demonstrated as an independent m6A methyltransferase, whose binding sites have no overlap with METTL3/METTL14 complex, regulating the stability and splicing of mRNAs.31,32 Therefore, the overexpression of METTL16, with a mutation on catalytic domain, activates the splicing process.33 Besides, METTL16 has been reported binding to U6 snRNA and numerous ncRNAs (ncRNAs), long ncRNAs (lncRNAs), and pre-mRNAs.30,31 With respect to ZCCHC4, it has been identified as an RNA m6A methyltransferase, methylating human 28S rRNA. Besides, the rRNA m6A methylation mediated by ZCCHC4 makes a difference to the distribution of ribosome subunit, global translation, and cell proliferation, which may lead to tumorigenesis when aberrant ZCCHC4 expression occurs.21
m6A Eraser Proteins
The m6A eraser proteins, acting as demethylase, remove m6A modifications by raising ferrous iron as co-factor and α-ketoglutarate as co-substrate at the same time.34 Unlike the numerous types of m6A writer proteins, only two m6A demethylase, including fat mass and obesity-associated (FTO) protein and AlkB homolog 5 (ALKBH5) protein, have been identified so far. In addition, m6A eraser proteins often work in the nucleus where the process of demethylation occurs.
FTO, the first recognized m6A eraser protein, belongs to AlkB family, which can mediate m6A modification by demethylating not only internal m6A but also N6, 2ʹ-O-dimethyladenosine (5ʹ cap m6Am) mRNA.35,36 Besides, FTO is nucleus localized to a large extent and regulates −10% of total m6A modification in almost all the cell lines and mRNA internal m6A is the major substrate of FTO.37, 38, 39 Moreover, FTO is highly expressed in the brain, a metabolically active organ. The processes of RNA transcription and translation regulate metabolism, and of course reversible m6A modification plays an important role in these processes. A recent study showed that NADP directly binds FTO, independently increases FTO activity, promoting the demethylation of RNA m6A modifications and adipogenesis.40 The discovery of FTO proves the process of m6A methylation is reversed and controlled under physiological and pathological status.
Another eraser protein is ALKBH5, which has functional similarity with FTO in m6A demethylation. ALKBH5 is widely expressed in various tissues, especially in the testes.41 The differential expression of FTO and ALKBH5 among different tissues reveals that the two proteins each play their roles in different biological pathways. Specifically, m6A is the only known substrate of ALKBH5 to date.42 Even more encouraging is the crystal structures of ALKBH5 have been analyzed, providing new insights into the underlying mechanisms for the procedures of m6A recognition and demethylation.41,43,44 The above findings greatly accelerate the development of targeted drugs for m6A demethylation inhibitors.
m6A Reader Proteins
The m6A-mediated posttranscriptional gene regulation has been further understood by identification and characterization of m6A readers. The m6A reader proteins control the destinies of the modified RNAs. Therefore irregulation of the proteins may cause misconception of the binding modified RNAs and following RNA metabolic disturbance. The m6A reader proteins consist of the YT521-B homology (YTH) domain family proteins (YTHDF1-3), YTH domain containing proteins (YTHDC1-2), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and heterogeneous nuclear ribonucleoproteins (HNRNPs, including hnRNPA2B1, hnRNPC, and HNRNPG).
The CCR4-NOT deadenylase complex mediates deadenylation of m6A modified RNAs.45 Cytoplasmic YTHDF2, interacting with the SH domain of the CNOT1 subunit, recruits the CCR4-NOT complex and subsequently destabilizes m6A modified RNAs, leading to their deadenylation and decay.45,46 On the contrary, the other two YTH domain family proteins, YTHDF1 and YTHDF3, are reported to facilitate translation by recruiting translation initiation factors.47,48 In addition, YTHDF3 accelerates mRNA translation in conjunction with YTHDF1 and promotes the deadenylation and decay of m6A modified RNAs collaboratively with YTHDF2.48,49
In the cell nucleus, YTHDC1 regulates mRNA splicing through recruiting and combining a certain splicing factor serine/arginine-rich splicing factor 3 (SRSF3).50 Besides, YTHDC1 also helps mRNA exportation from nucleus to cytoplasm.51 Recently, YTHDC1 has been found to accelerate the decay of a subset of m6A modifications on chromosome-associated regulatory RNAs (carRNAs), especially the long interspersed element-1 (LINE1) elements, via the nuclear exosome targeting complex-mediated nuclear degradation.52 YTHDC2 enhances the translation efficacy of target RNAs while decreasing their abundance in the cytoplasm.53
Currently, IGF2BPs are demonstrated to maintain the stable structures of target mRNA by m6A-dependent approaches under both normal and stressed circumstances.54 HNRNPA2B1 recognizes the m6A signals of primary microRNA and interplays with DiGeorge syndrome critical region gene 8 (DGCR8), while selective recognition of m6A-dependent splicing in mRNA secondary structures was proven as the function of HNRNPC.55, 56, 57, 58, 59
m6A in Cancer
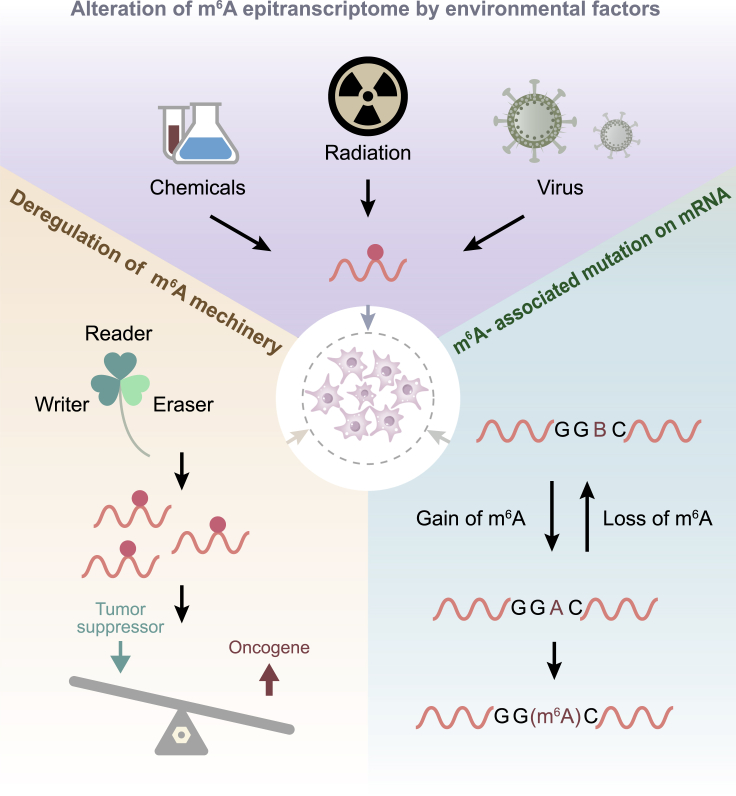
Previous studies have suggested the effects of m6A modification and its capacity to modulate and coordinate gene expression. The level of m6A may profoundly affect the characteristics of cancer. It is suggested that m6A may play an important role in carcinogenesis or inhibition in malignant tumor effect. Some proteins need to be modified by m6A to participate in the mechanism of carcinogenesis, but it is still not clear whether they take effect in the modification. The main causes of tumorigenesis in m6A-dependent manner (Figure 3) and the functions of the main m6A proteins in most of the tumor types (Figure 4) are shown.
Figure 3.
Precipitating Factors for Abnormal m6A Regulation in Cancer
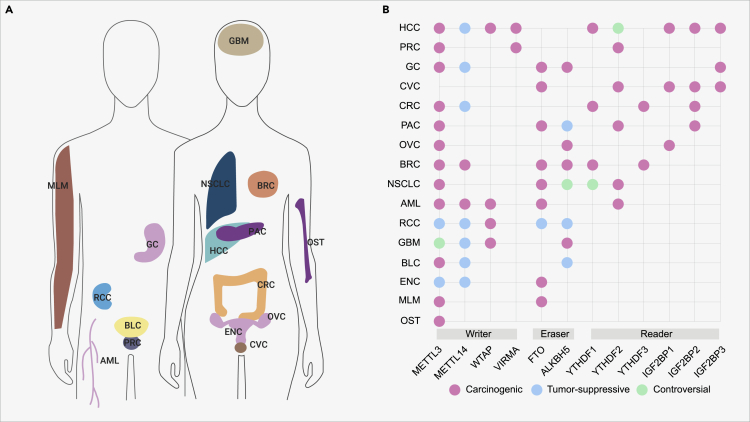
Figure 4.
m6A Modifiers in Human Cancers
The sites of different cancers (A) and the roles of m6A genes in different cancers (B). HCC, hepatocellular carcinoma; PRC, prostate cancer; GC, gastric cancer; CVC, cervical cancer; CRC, colorectal cancer; PAC, pancreatic cancer; OVC, ovarian cancer; BRC, breast cancer; NSCLC, non-small cell lung cancer; AML, acute myeloid leukemia; RCC, renal cell carcinoma; GBM, glioblastoma; BLC, bladder cancer; ENC, endometrial cancer; MLM, melanoma; OST, osteosarcoma.
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) has obvious genetic variation due to uncontrolled proliferation and cell differentiation defects of myeloid leukocytes. So far, the treatment for AML is still unsatisfactory. Previous studies have demonstrated that MELLT3 and METTL14 promoted the translation of MYC, MYB, BCL2, SP1, and PTEN, increasing the level of phosphorylated AKT.60,61 It has been found that FTO played an important role in the proliferation of AML cells.62, 63, 64, 65, 66 FTO can enhance leukemia oncogene-mediated cell transformation and leukemia, inhibit all trans retinoic acid–mediated differentiation of AML cells, and regulate the mRNA synthesis of target genes (such as ASB2 and RARA) by down-regulating the level of m6A.66 Su et al.39 found that R-2HG inhibited the proliferation of leukemia cells and resulted in cell-cycle arrest by inhibiting FTO activity, increasing m6A modification, reducing the stability of MYC/CEBPA transcription. Many studies have confirmed that YTHDF2 can promote tumor progression. Paris et al.67 suggested that YTHDF2 was highly expressed in AML and played a key role in the initiation and proliferation of AML. They found that targeting YTHDF2 could prolong the half-life of the m6A modified transcripts, thus selectively destroying the initiation and proliferation of AML without affecting normal hematopoiesis. Besides, WTAP is upregulated in AML and the high expression of WTAP predicts poor outcomes in patients with AML.68,69
Glioblastoma
Glioblastoma (GBM) is the deadliest primary brain tumor.70 Research on the role of METTL3 in GBM has produced conflicting conclusions. The different phenotype associated with METTL3 can be illustrated by the different dependence and genetic heterogeneity of m6A modified RNA in different types of GBM cells. Cui et al.71 demonstrated that METTL3 and METTL14 inhibit the growth and tumorigenesis of glioblastoma stem-like cells (GSCs) by the down-regulation of ADAM19/EPHA3/KLF4 pathway. ALKBH5 was also highly expressed in GSCs.72,73 Silencing ALKBH5 could inhibit the proliferation of GSCs. ALKBH5 can induce demethylation of FOXM, a transcription factor, and stimulate cell proliferation, resulting in increased FOXM1 expression.73 Inhibiting the expression of ALKBH5 provides a new direction for the treatment of glioma. Visvanathan et al.74 found that METTL3 was highly expressed in malignant GBM. METTL3 promoted tumor growth by targeting the 3′ UTR of SOX2 mRNA. Silencing METTL3 can inhibit tumor growth and enhance tumor radiosensitivity, which can be used as a potential molecular target for GBM therapy. Similar to AML, the high expression of WTAP also predicts poor prognosis in patients with GBM.75,76
Lung Cancer
Lung cancer is the leading cause of cancer-related mortality worldwide.77, 78, 79, 80, 81, 82, 83 METTL3 is the oncogene of lung cancer through different mechanisms. Lin et al.84 found that METTL3 can enhance RNA translation without the aid of methyltransferase and reader protein activity. METTL3 increases RNA translation by directly recruiting translation initiation factors. METTL3 increases the growth, survival and invasion of lung adenocarcinoma cells by increasing EGFR and TAZ.84 Choe et al.85 demonstrated that METTL3 could enhance translation, transform oncogenes, and form dense polyribosomes by interacting with EIF3H in primary lung cancer, which can be applied as a potential therapeutic target. METTL3 can also promote tumor progression by regulating some microRNAs (miRNAs). Du et al.86 found that mir-33a can inhibit the proliferation of non-small cell lung cancer (NSCLC) by targeting METTL3 mRNA. Wei et al.87 revealed that mir-600 can inhibit the migration and proliferation of lung cancer cells by down-regulating the expression of METTL3. Li et al.88 found that FTO promotes the proliferation of NSCLC by increasing the expression of USP7. Liu et al.89 found that overexpression of FTO can down-regulate the level of m6A in MZF1 mRNA transcripts, increase the stability of mRNA, and promote the expression of MZF1, leading to the proliferation and invasion of lung squamous cell carcinoma cells. In addition, m6A demethylase ALKBH5 was indicated to inhibit tumor growth and metastasis in patients with NSCLC by reducing the expression of YTHDFs-mediated YAP, whereas some studies suggested ALKBH5 promoted the progression of NSCLC.90,91 As for YTHDF1, it is also controversial for its role in NSCLC. On one hand, YTHDF1 deficiency was proved to suppress NSCLC cell proliferation and xenograft neoplasia via affecting the translational efficacy of cyclin D1, CDK2, and CDK4. On the other hand, it was observed that the high expression of YTHDF1 associated with better prognosis and the depletion of YTHDF1 helped tumor cells resistant to cisplatin.92
Endometrial Cancer
Approximately 70% of patients with endometrial cancer show decreased m6A expression level because of either reduced expression of METTL3 or mutation (loss-of-function) in METTL14 and the tumorigenicity is associated with activation of the AKT pathway.93 Down-regulation of METTL14 can reduce the level of m6A of RNA. Recent studies have shown that the decrease of m6A level is associated with endometrial cancer caused by METTL14 gene mutation.93 They found that the deletion of METTL14 increased cancer cell proliferation, clone formation, and metastasis. They observed that the level of m6A in tumor tissue was lower than that in adjacent normal tissue. They believe that the METTL14 mutation reduces the level of m6A, which plays a key role in the progression of endometrial cancer. Liu et al.93 also found that the down-regulation of METTL14 could lead to the decrease of the expression of PHLPP2, the negative regulator of AKT pathway, and increase the expression of mTORC2, the positive regulator of AKT pathway, which led to the proliferation of endometrial cancer cells. FTO can also participate in the occurrence and development of endometrial cancer through a variety of mechanisms. Zhu et al.94 have shown that estrogen induces FTO nuclear aggregation through the mammalian target of Rapamycin signaling pathway, thus enhancing the proliferation and activity of endometrial cancer cells and promoting tumor progression. Another study showed that estrogen induces overexpression of FTO gene by activating PI3K/AKT and MAPK signaling pathways, which leads to the proliferation and invasion of endometrial cancer cells.95
Cervical Cancer
The role of FTO in cervical cancer tumorigenesis has been uncovered, FTO functions as an oncogenic regulator for cervical cancer in terms of promoting tumor cell proliferation and migration by regulating E2F1 and Myc transcripts.96 Long ncRNA GAS5-AS1 was found having a lower expression in cervical cancer when compared with that of adjacent normal tissues and played its role on YTHDF2-dependent pathway.97 Besides, m6A “reader” IGF2BP family proteins were also reported serving as carcinogenic roles in CVC.98,99
Ovarian Cancer
METTL3 promotes the maturation of miR-126-5p and therefore accelerates ovarian cancer (OVC) progression.100 In addition, another study demonstrated that METTL3 facilitated OVC growth and invasion via activating epithelial to mesenchymal transition.101 The expression of ALKBH5 was found to be increased in epithelial OVC tissues when compared with the normal ovarian tissues and ALKBH5 was identified as a candidate oncogene in epithelial OVC.102 The “reader” protein IGF2BP1 enhanced SRC/MAPK-driven invasive growth of OVC cells and the high expression of IGF2BP1 was related to poor prognosis of patients with OVC.103,104 Most recently, recruitment of YTHDF1 to m6A-modified TRIM29 was participated in accelerating TRIM29 translation in the OVC cells with cisplatin-resistance, which would be a potential therapeutic target.105
Breast Cancer
There is now a compelling body of evidence demonstrating that epigenetic modifications including RNA m6A modification play a vital role in the tumorigenesis and progression of breast cancer.106 Cai et al.107 found that METTL3 interacts with hepatitis B X-interacting protein (HBXIP) in breast cancer cells, and HBXIP inhibits the expression of METTL3 by acting on 3′UTR of miRNA let-7g. At the same time, METTL3 promoted the expression of HBXIP through m6A modification. A positive feedback loop was formed between HBXIP/miRNA let-7g/METTL3, which promoted the proliferation of breast cancer cells. METTL14 was promoted by LINC00942, enhancing the initiation and progression of breast cancer.108 Niu et al.109 found that FTO is highly expressed in breast cancer tissues, and the higher the expression, the worse the prognosis of patients. FTO promotes the proliferation and metastasis of breast cancer cells by inhibiting BNIP3. In one study, Zhang et al.110 found that ALKBH5 mediated m6A demethylation of NANOG (an embryonic stem cell transcription factor) mRNA, which increased the expression of NANOG in breast cancer. Knockout of ALKBH5 can increase the demethylation of NANOG mRNA and decrease the expression level of NANOG, which can significantly inhibit lung metastasis of breast cancer. Subsequently, they found that HIFs promoted the invasion and metastasis of breast cancer cells by regulating ZNF217 to inhibit methylation and ALKBH5 induced demethylation.111
Colorectal Cancer
Epigenetic alterations exist in various aspects of colorectal tumorigenesis.112 In colorectal cancer, METTL3 acts as a functional oncogene in an m6A-IGF2BP2/3-dependent manner.112 Oncogene c-Myc can promote the expression of YTHDF1, induce the proliferation and metastasis of cancer cells, and increase their resistance to chemotherapy drugs.113 Knockdown of c-Myc can inhibit the expression of YTHDF1 in colorectal cancer through HIF-1α. Besides, oncogene c-Myc can promote the expression of YTHDF1, inducing the proliferation and metastasis of cancer cells, and increasing their resistance to chemotherapy drugs.113 YTHDF3 negatively regulated lncRNA GAS5 through GAS5-YAP-YTHDF3 axis both in vivo and in vitro.114 The expression of YTHDC2 is positively correlated with the stage and metastasis of colon cancer.115 Knockdown the expression of YTHDC2 can inhibit the metastasis of tumor cells in vivo and in vitro through HIF-1α. Except all the preceding m6A-associated oncogenes, METTL14 is demonstrated as a tumor suppresser, decreasing the proliferation and tumor metastasis of colorectal cancer via different molecular mechanisms.116, 117, 118, 119
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is the most common primary neoplasm of the liver.120 Chen et al.121 found that METTL3 can promote the progression of liver cancer cells through YTHDF2-dependent transcriptional regulation of SOCS2 silencing. Further study showed that METTL14 interacted with DGCR8 and positively regulated the expression of miRNA 126.122 Overexpression of METTL14 can inhibit the metastasis of liver cancer cells in mice. According to these results, they speculated that METTL14 may regulate the expression of miRNA 126 through the modification of m6A, and then regulate its downstream target to inhibit the metastasis of HCC. Therefore, METTL14 may be an important adverse prognostic factor for HCC.122,123 As for IGF2BP family, IGF2BP1,124, 125, 126 IGF2BP2,127 and IGF2BP3128,129 were all identified as oncogenes to promote the carcinogenesis of HCC. YTHDF2 not only acts as a tumor activating protein, but also acts as a tumor suppressor protein. Some studies have shown that, hypoxia can induce the decrease of the expression of YTHDF2 in hepatocellular carcinoma.130,131 They also found that overexpression of YTHDF2 inhibited the proliferation of HCC cells and activated MEK and ERK. YTHDF2 can directly act on the 3′UTR m6A modification site of EGFR mRNA, resulting in the degradation of EGFR mRNA. In addition, the phosphorylation of ERK induced by hypoxia was also blocked by YTHDF2, suggesting that hypoxia can down-regulate the phosphorylation of ERK induced by YTHDF2. YTHDF2 inhibits ERK/MAPK signal transduction by reducing the stability of EGFR mRNA in HCC, thus inhibiting the proliferation of hepatoma cells.131 WTAP was examined to determine whether it was related to clinicopathological factors of patients with HCC, and the results showed that the expression level of WTAP was increased more in HCC than in para-carcinoma tissues and associated with worse prognosis.132,133 Besides, VIRMA was also defined as an oncogene in HCC.134,135
Pancreas Cancer
Pancreas cancer is recognized as a kind of high-grade malignant neoplasm. He et al.54 revealed that ALKBH5 may be a potential therapeutic target for pancreatic cancer by down-regulating methylation of lncRNA KCNK15-AS1 in pancreatic cancer cells and inhibiting cell motility. Taketo et al.136 found that pancreatic cancer cells with low METTL3 expression are more sensitive to chemotherapy drugs such as gemcitabine, 5-fluorouracil, and cisplatin and radiotherapy, it provides a potential target for the treatment of pancreatic cancer. Many studies have proved that IGF2BP2 was overexpressed in pancreas cancer and promoted cancer proliferation.137,138 A case-control study demonstrated that variants in the FTO gene was associated with pancreatic cancer risk.139 YTHDF2 promoted proliferation while inhibiting migration and invasion in pancreatic cancer cells.140
Gastric Carcinoma
With improved diagnostic strategy, patients who diagnosed with early-stage gastric carcinoma (GC) is increasing. Previous studies have shown that epigenetics may play an important role in the genesis and growth of GC. Li et al.141 demonstrated the high expression of FTO and ALKBH1 mRNA indicates poor prognosis of GC through mining TCGA database. METTL3 promotes GC angiogenesis and glycolysis by increasing the stability of HDGF mRNA and activating the AKT signaling pathway.142 ALKBH5 promotes invasion and metastasis of GC by decreasing methylation of the lncRNA NEAT1.143 Some studies demonstrated that IGF2BP3 functioned as an oncogene to promote tumor progression in GC.144,145 Knockdown of METTL14 (m6A suppression) promotes GC development through activating the Wnt/PI3K-AKT signaling pathway, whereas increasing m6A levels reversed these phenotypical and molecular changes.146
Prostate Cancer
Prostate cancer (PRC) is one of the most common malignancies worldwide.147 Some studies have shown that down-regulation of YTHDF2 can inhibit the proliferation and invasion of PRC cells.148,149 There was a negative correlation between YTHDF2 and miR-493-3p. Knockout of YTHDF2 could increase the level of m6A and inhibit the proliferation and invasion of GC cell lines. Besides, METTL3 and VIRMA were also reported promoting the development and progression of PRC in different manners.150, 151, 152, 153
Renal Cell Carcinoma
METTL3 and METTL14 can not only promote the proliferation and metastasis of tumor cells, but also inhibit the progress of tumor.154,155 Some studies have shown that low expression of METTL3 is associated with larger tumors and higher histological grade in mice.156 The survival time of renal cell carcinoma patients with METTL3 overexpression was significantly prolonged.156 WTAP also plays a role in tumor growth by binding with mRNA and enhancing the stability of mRNA. WTAP can combine with cyclin dependent protein kinase (CDK) 2 transcripts, enhancing its stability, delaying cell apoptosis and promoting the proliferation of renal cell carcinoma.157 On the contrary, ALKBH5 and FTO was identified as a tumor suppresser for renal cell carcinoma.158, 159, 160
Potential Applications of Cancer Treatment Based on m6A
Many researchers have demonstrated that m6A has emerged as a widespread regulatory mechanism that controls gene abnormal expression in diverse pathological pathways, leading to tumorigenesis.161,162 Therefore, m6A regulators may function as potential clinical therapeutic targets for cancers. Since the first m6A demethylase FTO was identified, FTO has become the most striking target for developing targeted drugs against tumors.163 Several FTO-targeted inhibitors have already been developed, including MO-I-500, meclofenamic acid (MA), FB23, R-2HG, rhein, and so on. MO-I-500, a selective inhibitor, inhibits the enzyme activity of FTO and was reported to suppress the proliferation of breast cancer cell lines.109,164,165 MA also proved to be a selective inhibitor for treating GBM through inhibiting FTO over ALKBH5.71,166 Recently, FB23 and FB23-2 were designed as small-molecule FTO inhibitors, which have achieved remarkable inhibitory effect in AML models.64 As for nonselective inhibitors for FTO, rhein was the first to be uncovered and competitively binds to specific site of FTO.167 R-2HG, highly expressed by mutant isocitrate dehydrogenase 1/2 (IDH1/2) enzymes, was demonstrated to play important antitumor effect in glioma and leukemia cells.39
Multidisciplinary therapy, including neoadjuvant therapy, surgery, adjuvant chemoradiotherapy, targeted therapies, and immunotherapies, has been widely adopted in cancer treatment. However, drug resistance maintains a dominating hindrance to curative treatment, leading to treatment failure and tumor progression.168,169 Recently, the dysregulation of m6A regulators has been found related to the advent of treatment resistance.170, 171, 172 As for immunotherapy, it is an emerging way of dealing with cancer. YTHDF1 was proved to regulate antitumor immunity and had synergetic effect on immunotherapy by improving the therapeutic effect of PD-L1 inhibitor.173 In addition, Yang et al.174 found that m6A mRNA demethylation by FTO promoted melanoma growth and suppressed response to anti-PD-1 blockade, which indicated the combination of FTO inhibitors and anti-PD-1 blockade may help to promote sensibilization of immunotherapy in melanoma. Furthermore, the detailed information on drug resistance caused by m6A dysregulation is listed in Table 1. These studies indicated that m6A RNA modification signatures may provide important information for predicting drug resistance and help clinicians to adjust the treatment plan in time.
Table 1.
m6A Alteration in Drug-Resistant Cancer Cells
| Cancer Type | Therapeutic Agents | m6A Proteins Involved | Change of m6A Proteins | Reference |
|---|---|---|---|---|
| AML | R-2HG | FTO | Down-regulate | Su et al.39 |
| AML | TKI | FTO | Up-regulate | Yan et al.175 |
| GBM | γ-irradiation | METTL3 | Up-regulate | Visvanathan et al.74 |
| HCC | Sorafenib | METTL3 | Down-regulate | Lin et al.176 |
| CRC | Doxorubicin | METTL3 | Up-regulate | Uddin et al.172 |
| NSCLC | Cisplatin | METTL3 | Up-regulate | Jin et al.170 |
| NSCLC | Cisplatin | YTHDF1 | Down-regulate | Shi et al.92 |
| NSCLC | Afatinib | m6A | Up-regulate | Meng et al.177 |
| NSCLC | Crizotinib | METTL3, WTAP | Up-regulate | Ding et al.178 |
| PAC | cisplatin, 5-Fu and gemcitabine | METTL3 | Taketo et al.136 | |
| PRC | Cisplatin | VIRMA | – | Su et al.179 |
| CSCC | cisplatin, irradiation | FTO | Up-regulate | Zhou et al.180 |
| OVC | PARP inhibitor | FTO, ALKBH5 | Down-regulate | Fukumoto et al.181 |
| MLM | anti-PD-1 blockade | FTO, YTHDF2 | Up-regulate | Yang et al.174 |
AML, acute myeloid leukemia; GBM, glioblastoma; HCC, hepatocellular carcinoma; CRC, colorectal cancer; NSCLC, non-small cell lung cancer; PAC, pancreatic cancer; PRC, prostate cancer; CSCC, cervical squamous cell carcinoma; OVC, ovarian cancer; MLM, melanoma.
Conclusions and Future Perspectives
Different from DNA, RNA has much more intricate posttranscriptional processing: RNA splicing, RNA editing, and RNA chemical modifications. RNA chemical modifications, most of which have no influence on nucleoside sequence, are structurally diverse and functionally multiple, indicating their functional importance.
In recent years, epigenetics, especially m6A RNA modification, has been further understood and explored with the rapid advances in detection methods and high-throughput sequencing techniques. It has been widely illustrated that the dysregulation of m6A RNA modification is related to various types of cancers, as well as the drug resistance to antitumor therapy.182 However, as the evidence (m6A plays important role in tumors) began to accumulate, the underlying mechanisms of m6A in cancer still have not yet been fully realized. To be specific, in many kinds of cancers, such as NSCLC, breast cancer, acute myelocytic leukemia, and some gynecological tumors, overexpression of either writer or eraser proteins can play tumorigenesis roles, while controversies of the roles of m6A regulators still exist in other cancer types.183 Similar to DNA methylation signatures (hmc, 5hmc, and so on),184, 185, 186, 187, 188, 189 m6A RNA modifications of certain genetic loci could function as biomarkers associated with prognosis, molecular subtyping, and precise diagnosis.68,190 Many researchers focus on the identification of m6A-targeted mRNAs in diverse diseases, especially in cancers. Moreover, how m6A RNA modification affects the functions of ncRNAs still remains unclear and ncRNAs have been confirmed having a close link with a wide variety of tumors.15,191 carRNAs recently were found to have m6A modifications on them, which were deposited by METTL3, including repeat RNAs, promoter-associated RNAs, and enhancer RNAs.52 Thus, the direct crosstalk between the m6A carRNA modifications and chromatin state opens a new door on how m6A carRNA modifications regulate transcription. The Nobel Prize in Chemistry in 2020 was announced “for the development of a method for genome editing” (discovering the CRISPR/Cas9 genetic scissors). Therefore, whether a technology could developed for epitranscriptome editing is also worthy of expectation.
Considering that the dysregulation of m6A regulators is related to treatment resistance and cancer immunity, it is attractive to develop m6A-based targeted drugs for oncotherapy. Besides, m6A-based targeted small-molecule drugs could function as a kind of sensitizer, improving the therapeutic effects of chemotherapy, radiotherapy, and even immunotherapy in the future. In general, RNA m6A modification in cancers is an emerging field of cancer epigenetic research, providing not only new insights into the potential molecular mechanisms of tumorigenesis and cancer progression, but novel strategies for drug exploitation and clinical cancer therapies. At present, our understanding of RNA m6A modifications for cancers is still in its infancy and further studies are desperately required for the rosy scenario.
Acknowledgments
The work is supported by the projects from Shanghai Hospital Development Center (SHDC12015116), the Fundamental Research Funds for the Central Universities (22120180607), the National Natural Science Foundation of China (81802256), Science and Technology Commission of Shanghai Municipality (15411968400 and 14411962600), Shanghai Pujiang Program (15PJD034), the “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (18CG19), the “Outstanding young talent” project supported by Shanghai Pulmonary Hospital (FKYQ1907), and Shanghai Rising-Star Program (20QA1408300).
Declaration of Interests
Chuan He is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board.
Contributor Information
Chuan He, Email: chuanhe@uchicago.edu.
Tao Huang, Email: huangtao@sibs.ac.cn.
Chang Chen, Email: chenthoracic@126.com.
References
- 1.Cohn W.E., Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature. 1951;167:483–484. [Google Scholar]
- 2.Boccaletto P., Machnicka M.A., Purta E., et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams J.M., Cory S. Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA. Nature. 1975;255:28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 5.Perry R.P., Kelley D.E., Friderici K., et al. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus. Cell. 1975;4:387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- 6.Perry R.P., Kelley D.E. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1:37–42. [Google Scholar]
- 7.Nichols J.L. 'Cap' structures in maize poly(A)-containing RNA. Biochim. Biophys. Acta. 1979;563:490–495. doi: 10.1016/0005-2787(79)90067-4. [DOI] [PubMed] [Google Scholar]
- 8.Levis R., Penman S. 5'-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J. Mol. Biol. 1978;120:487–515. doi: 10.1016/0022-2836(78)90350-9. [DOI] [PubMed] [Google Scholar]
- 9.Clancy M.J., Shambaugh M.E., Timpte C.S., et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beemon K., Keith J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 1977;113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- 11.Aloni Y., Dhar R., Khoury G. Methylation of nuclear simian virus 40 RNAs. J. Virol. 1979;32:52–60. doi: 10.1128/jvi.32.1.52-60.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer K.D., Saletore Y., Zumbo P., et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 14.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of rna methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H., Weng H., Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knuckles P., Lence T., Haussmann I.U., et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ping X.L., Sun B.F., Wang L., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz S., Mumbach M.R., Jovanovic M., et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yue Y., Liu J., Cui X., et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen J., Lv R., Ma H., et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H., Wang X., Cai J., et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019;15:88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bokar J.A., Shambaugh M.E., Polayes D., et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Feng J., Xue Y., et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 24.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sledz P., Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong S., Li H., Bodi Z., et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil D.P., Chen C.K., Pickering B.F., et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Tran N., Ernst F.G.M., Hawley B.R., et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard E.M., Polla D.L., Assir M.Z., et al. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet. 2019;105:869–878. doi: 10.1016/j.ajhg.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pendleton K.E., Chen B., Liu K., et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warda A.S., Kretschmer J., Hackert P., et al. Human METTL16 is a N 6 -methyladenosine (m 6 A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shima H., Matsumoto M., Ishigami Y., et al. S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 33.Thomas J.M., Batista P.J., Meier J.L. Metabolic regulation of the epitranscriptome. ACS Chem. Biol. 2019;14:316–324. doi: 10.1021/acschembio.8b00951. [DOI] [PubMed] [Google Scholar]
- 34.Fedeles B.I., Singh V., Delaney J.C., et al. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linder B., Grozhik A.V., Olarerin-George A.O., et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei J., Liu F., Lu Z., et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Wei L.-H., Wang Y., et al. Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. USA. 2019;116:2919–2924. doi: 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su R., Dong L., Li C., et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105 e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L., Song C., Wang N., et al. NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 2020 doi: 10.1038/s41589-020-0601-2. [DOI] [PubMed] [Google Scholar]
- 41.Aik W., Scotti J.S., Choi H., et al. Structure of human RNA N⁶-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–4754. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng G., Dahl J.A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C., Liu K., Tempel W., et al. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 2014;289:17299–17311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W., Zhang L., Zheng G., et al. Crystal structure of the RNA demethylase ALKBH5 from zebrafish. FEBS Lett. 2014;588:892–898. doi: 10.1016/j.febslet.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du H., Zhao Y., He J., et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Lu Z., Gomez A., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Zhao B.S., Roundtree I.A., et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi H., Wang X., Lu Z., et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A., Chen Y.-S., Ping X.-L., et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao W., Adhikari S., Dahal U., et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 51.Roundtree I.A., Luo G.Z., Zhang Z., et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311 doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Dou X., Chen C., et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu P.J., Zhu Y., Ma H., et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H., Weng H., Sun W., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alarcon C.R., Goodarzi H., Lee H., et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N., Dai Q., Zheng G., et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu N., Zhou K.I., Parisien M., et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu B., Su S., Patil D.P., et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vu L.P., Pickering B.F., Cheng Y., et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng H., Huang H., Wu H., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen S.N., Armstrong S.A. It's not what you say but how you say it: targeting RNA methylation in AML. Mol. Cell. 2020;78:996–998. doi: 10.1016/j.molcel.2020.05.027. [DOI] [PubMed] [Google Scholar]
- 63.Weng H., Huang H., Chen J. RNA N (6)-methyladenosine modification in normal and malignant hematopoiesis. Adv. Exp. Med. Biol. 2019;1143:75–93. doi: 10.1007/978-981-13-7342-8_4. [DOI] [PubMed] [Google Scholar]
- 64.Huang Y., Su R., Sheng Y., et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Der Werf I., Jamieson C. The yin and yang of RNA methylation: an imbalance of erasers enhances sensitivity to FTO demethylase small-molecule targeting in leukemia stem cells. Cancer Cell. 2019;35:540–541. doi: 10.1016/j.ccell.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 66.Li Z., Weng H., Su R., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6 -methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paris J., Morgan M., Campos J., et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naren D., Yan T., Gong Y., et al. High Wilms' tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m(6)A methylation of MYC mRNA. J. Cancer Res. Clin. Oncol. 2020 doi: 10.1007/s00432-020-03373-w. [DOI] [PubMed] [Google Scholar]
- 69.Sorci M., Ianniello Z., Cruciani S., et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9:796. doi: 10.1038/s41419-018-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White K., Connor K., Clerkin J., et al. New hints towards a precision medicine strategy for IDH wild-type Glioblastoma. Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.08.2336. [DOI] [PubMed] [Google Scholar]
- 71.Cui Q., Shi H., Ye P., et al. ARRAY(0x6a0bdd4))A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong Z., Cui H. The emerging roles of RNA modifications in glioblastoma. Cancers (Basel) 2020;12 doi: 10.3390/cancers12030736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S., Zhao B.S., Zhou A., et al. ARRAY(0x6a102e0))A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Visvanathan A., Patil V., Arora A., et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 75.Xi Z., Xue Y., Zheng J., et al. WTAP expression predicts poor prognosis in malignant glioma patients. J. Mol. Neurosci. 2016;60:131–136. doi: 10.1007/s12031-016-0788-6. [DOI] [PubMed] [Google Scholar]
- 76.Jin D.I., Lee S.W., Han M.E., et al. Expression and roles of Wilms' tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103:2102–2109. doi: 10.1111/cas.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu C., Shi X., Huang Z., et al. A comprehensive study of construction and analysis of competitive endogenous RNA networks in lung adenocarcinoma. Biochim. Biophys. Acta Proteins Proteom. 2020;1868:140444. doi: 10.1016/j.bbapap.2020.140444. [DOI] [PubMed] [Google Scholar]
- 78.Gu C., Huang Z., Chen X., et al. TEAD4 promotes tumor development in patients with lung adenocarcinoma via ERK signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165921. doi: 10.1016/j.bbadis.2020.165921. [DOI] [PubMed] [Google Scholar]
- 79.Gu C., Pan X., Chen Y., et al. Short-term and mid-term survival in bronchial sleeve resection by robotic system versus thoracotomy for centrally located lung cancer. Eur. J. Cardiothorac. Surg. 2018;53:648–655. doi: 10.1093/ejcts/ezx355. [DOI] [PubMed] [Google Scholar]
- 80.Gu C., Huang Z., Dai C., et al. Prognostic analysis of limited resection versus lobectomy in stage IA small cell lung cancer patients based on the surveillance, epidemiology, and end results registry database. Front. Genet. 2018;9:568. doi: 10.3389/fgene.2018.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu C., Wang R., Pan X., et al. Sublobar resection versus lobectomy in patients aged ≤35 years with stage IA non-small cell lung cancer: a SEER database analysis. J. Cancer Res. Clin. Oncol. 2017;143:2375–2382. doi: 10.1007/s00432-017-2499-y. [DOI] [PubMed] [Google Scholar]
- 82.Gu C., Pan X., Wang R., et al. Analysis of mutational and clinicopathologic characteristics of lung adenocarcinoma with clear cell component. Oncotarget. 2016;7:24596–24603. doi: 10.18632/oncotarget.8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gu C., Wang R., Pan X., et al. Comprehensive study of prognostic risk factors of patients underwent pneumonectomy. J. Cancer. 2017;8:2097–2103. doi: 10.7150/jca.19454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin S., Choe J., Du P., et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choe J., Lin S., Zhang W., et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Du M., Zhang Y., Mao Y., et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 2017;482:582–589. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 87.Wei W., Huo B., Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag. Res. 2019;11:1177–1187. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J., Han Y., Zhang H., et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 89.Liu J., Ren D., Du Z., et al. m)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 90.Jin D., Guo J., Wu Y., et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol. Cancer. 2020;19:40. doi: 10.1186/s12943-020-01161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu Z., Qian Q., Zhao X., et al. N(6)-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020;731:144348. doi: 10.1016/j.gene.2020.144348. [DOI] [PubMed] [Google Scholar]
- 92.Shi Y., Fan S., Wu M., et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu J., Eckert M.A., Harada B.T., et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Y., Shen J., Gao L., et al. Estrogen promotes fat mass and obesity-associated protein nuclear localization and enhances endometrial cancer cell proliferation via the mTOR signaling pathway. Oncol. Rep. 2016;35:2391–2397. doi: 10.3892/or.2016.4613. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Z., Zhou D., Lai Y., et al. Estrogen induces endometrial cancer cell proliferation and invasion by regulating the fat mass and obesity-associated gene via PI3K/AKT and MAPK signaling pathways. Cancer Lett. 2012;319:89–97. doi: 10.1016/j.canlet.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 96.Zou D., Dong L., Li C., et al. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019;19:321. doi: 10.1186/s12935-019-1045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X., Zhang J., Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am. J. Transl. Res. 2019;11:4909–4921. [PMC free article] [PubMed] [Google Scholar]
- 98.Wang P., Zhang L., Zhang J., et al. MicroRNA-124-3p inhibits cell growth and metastasis in cervical cancer by targeting IGF2BP1. Exp. Ther. Med. 2018;15:1385–1393. doi: 10.3892/etm.2017.5528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Zhang Y., Wang D., Wu D., et al. Long noncoding RNA KCNMB2-AS1 stabilized by N(6)-methyladenosine modification promotes cervical cancer growth through acting as a competing endogenous RNA. Cell Transpl. 2020;29 doi: 10.1177/0963689720964382. 963689720964382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bi X., Lv X., Liu D., et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2020 doi: 10.1038/s41417-020-00222-3. [DOI] [PubMed] [Google Scholar]
- 101.Hua W., Zhao Y., Jin X., et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol. Oncol. 2018;151:356–365. doi: 10.1016/j.ygyno.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 102.Zhu H., Gan X., Jiang X., et al. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muller S., Bley N., Glaß M., et al. IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 2018;46:6285–6303. doi: 10.1093/nar/gky229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bley N., Schott A., Muller S., et al. IGF2BP1 is a targetable SRC/MAPK-dependent driver of invasive growth in ovarian cancer. RNA Biol. 2020:1–13. doi: 10.1080/15476286.2020.1812894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao L., Wang J.M., Liu B.Q., et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2020:118878. doi: 10.1016/j.bbamcr.2020.118878. [DOI] [PubMed] [Google Scholar]
- 106.Pasculli B., Barbano R., Parrella P. Epigenetics of breast cancer: biology and clinical implication in the era of precision medicine. Semin. Cancer Biol. 2018;51:22–35. doi: 10.1016/j.semcancer.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 107.Cai X., Wang X., Cao C., et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 108.Sun T., Wu Z., Wang X., et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–5372. doi: 10.1038/s41388-020-1338-9. [DOI] [PubMed] [Google Scholar]
- 109.Niu Y., Lin Z., Wan A., et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang C., Samanta D., Lu H., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U S A. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang C., Zhi W.I., Lu H., et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen C., Xuan B., Yan T., et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nishizawa Y., Konno M., et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ni W., Yao S., Zhou Y., et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu T., Li C., Jin L., et al. The prognostic value of m6A RNA methylation regulators in colon adenocarcinoma. Med. Sci. Monit. 2019;25:9435–9445. doi: 10.12659/MSM.920381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen X., Xu M., Xu X., et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer. 2020;19:106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang X., Zhang S., He C., et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen X., Xu M., Xu X., et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol. Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Liu X., Liu L., Dong Z., et al. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am. J. Transl Res. 2019;11:3972–3991. [PMC free article] [PubMed] [Google Scholar]
- 120.Dhayat S.A., Yang Z. Impact of circulating tumor DNA in hepatocellular and pancreatic carcinomas. J. Cancer Res. Clin. Oncol. 2020;146:1625–1645. doi: 10.1007/s00432-020-03219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 122.Ma J.Z., Yang F., Zhou C.C., et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 123.Li Z., Li F., Peng Y., et al. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 2020;9:1877–1889. doi: 10.1002/cam4.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J., Hu K., Yang Y.Q., et al. LIN28B-AS1-IGF2BP1 binding promotes hepatocellular carcinoma cell progression. Cell Death Dis. 2020;11:741. doi: 10.1038/s41419-020-02967-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ding H., Liu J., Wang C., et al. NONO promotes hepatocellular carcinoma progression by enhancing fatty acids biosynthesis through interacting with ACLY mRNA. Cancer Cell Int. 2020;20:425. doi: 10.1186/s12935-020-01520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu F., Li C.H., Wong C.H., et al. Genome-wide screening and functional analysis identifies tumor suppressor long noncoding RNAs epigenetically silenced in hepatocellular carcinoma. Cancer Res. 2019;79:1305–1317. doi: 10.1158/0008-5472.CAN-18-1659. [DOI] [PubMed] [Google Scholar]
- 127.Liu F.Y., Zhou S.J., Deng Y.L., et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2015;6:e1670. doi: 10.1038/cddis.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang W., Cheng X., Wang T., et al. LINC00467 promotes cell proliferation and metastasis by binding with IGF2BP3 to enhance the mRNA stability of TRAF5 in hepatocellular carcinoma. J. Gene Med. 2020;22:e3134. doi: 10.1002/jgm.3134. [DOI] [PubMed] [Google Scholar]
- 129.Gao Y., Luo T., Ouyang X., et al. IGF2BP3 and miR191-5p synergistically increase HCC cell invasiveness by altering ZO-1 expression. Oncol. Lett. 2020;20:1423–1431. doi: 10.3892/ol.2020.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhong L., Liao D., Zhang M., et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 131.Hou J., Zhang H., Liu J., et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer. 2019;18:163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sera T., Hiasa Y., Mashiba T., et al. Wilms' tumour 1 gene expression is increased in hepatocellular carcinoma and associated with poor prognosis. Eur. J. Cancer. 2008;44:600–608. doi: 10.1016/j.ejca.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 133.Chen Y., Peng C., Chen J., et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lan T., Li H., Zhang D., et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer. 2019;18:186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang M., Yang Y., Yang J., et al. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020;257:118082. doi: 10.1016/j.lfs.2020.118082. [DOI] [PubMed] [Google Scholar]
- 136.Taketo K., Konno M., Asai A., et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 137.Xu X., Yu Y., Zong K., et al. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:497. doi: 10.1186/s13046-019-1470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dahlem C., Barghash A., Puchas P., et al. The insulin-like growth factor 2 mRNA binding protein IMP2/IGF2BP2 is overexpressed and correlates with poor survival in pancreatic cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20133204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin Y., Ueda J., Yagyu K., et al. Association between variations in the fat mass and obesity-associated gene and pancreatic cancer risk: a case-control study in Japan. BMC Cancer. 2013;13:337. doi: 10.1186/1471-2407-13-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen J., Sun Y., Xu X., et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:1–13. doi: 10.1080/15384101.2017.1380125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li Y., Zheng D., Wang F., et al. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig. Dis. Sci. 2019;64:1503–1513. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Q., Chen C., Ding Q., et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 143.Zhang J., Guo S., Piao H.Y., et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee D., Yu E.J., Ham I.H., et al. Clinicopathological implication of insulin-like growth factor-II mRNA-binding protein 3 (IMP3) expression in gastric cancer. Anticancer Res. 2017;37:135–142. doi: 10.21873/anticanres.11298. [DOI] [PubMed] [Google Scholar]
- 145.Kim H.J., Kim G.E., Lee J.S., et al. Insulin-like growth factor-II mRNA-binding protein 3 expression in effusion cytology: a marker for metastatic adenocarcinoma cells and a potential prognostic indicator in gastric adenocarcinoma. Acta Cytol. 2014;58:167–173. doi: 10.1159/000357199. [DOI] [PubMed] [Google Scholar]
- 146.Zhang C., Zhang M., Ge S., et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766–4781. doi: 10.1002/cam4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu N., Wu Y.P., Yin H.B., et al. SHCBP1 promotes tumor cell proliferation, migration, and invasion, and is associated with poor prostate cancer prognosis. J. Cancer Res. Clin. Oncol. 2020;146:1953–1969. doi: 10.1007/s00432-020-03247-1. [DOI] [PubMed] [Google Scholar]
- 148.Li J., Meng S., Xu M., et al. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget. 2018;9:3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang J., Lin H., Zhou M., et al. The m6A methylation regulator-based signature for predicting the prognosis of prostate cancer. Future Oncol. 2020 doi: 10.2217/fon-2020-0330. [DOI] [PubMed] [Google Scholar]
- 150.Yuan Y., Du Y., Wang L., et al. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer. 2020;11:3588–3595. doi: 10.7150/jca.42338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li E., Wei B., Wang X., et al. METTL3 enhances cell adhesion through stabilizing integrin β1 mRNA via an m6A-HuR-dependent mechanism in prostatic carcinoma. Am. J. Cancer Res. 2020;10:1012–1025. [PMC free article] [PubMed] [Google Scholar]
- 152.Ma X.X., Cao Z.G., Zhao S.L. m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3565–3571. doi: 10.26355/eurrev_202004_20817. [DOI] [PubMed] [Google Scholar]
- 153.Barros-Silva D., Lobo J., Guimaraes-Teixeira C., et al. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers (Basel) 2020;12 doi: 10.3390/cancers12040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Q., Zhang H., Chen Q., et al. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Dis. Markers. 2019;2019:5648783. doi: 10.1155/2019/5648783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gong D., Zhang J., Chen Y., et al. The m(6)A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca(2+) influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:233. doi: 10.1186/s13046-019-1223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li X., Tang J., Huang W., et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017;8:96103–96116. doi: 10.18632/oncotarget.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tang J., Wang F., Cheng G., et al. Wilms' tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J. Exp. Clin. Cancer Res. 2018;37:40. doi: 10.1186/s13046-018-0706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhuang C., Zhuang C., Luo X., et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell Mol. Med. 2019;23:2163–2173. doi: 10.1111/jcmm.14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiao Y., Thakkar K.N., Zhao H., et al. The m(6)A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc. Natl. Acad. Sci. U S A. 2020;117:21441–21449. doi: 10.1073/pnas.2000516117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Strick A., von Hagen F., Gundert L., et al. The N(6)-methyladenosine (m(6) A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020;125:617–624. doi: 10.1111/bju.15019. [DOI] [PubMed] [Google Scholar]
- 161.Lan Q., Liu P.Y., Haase J., et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 162.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 163.Deng X., Su R., Stanford S., et al. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol. (Lausanne) 2018;9:396. doi: 10.3389/fendo.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh B., Kinne H.E., Milligan R.D., et al. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS One. 2016;11:e0159072. doi: 10.1371/journal.pone.0159072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zheng G., Cox T., Tribbey L., et al. Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem. Neurosci. 2014;5:658–665. doi: 10.1021/cn500042t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang Y., Yan J., Li Q., et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen B., Ye F., Yu L., et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 2012;134:17963–17971. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 168.Yuan Y., Huang Q., Gu C., et al. Disease-free survival improved by use of adjuvant EGFR tyrosine kinase inhibitors in resectable non-small cell lung cancer: an updated meta-analysis. J. Thorac. Dis. 2017;9:5314–5321. doi: 10.21037/jtd.2017.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Song H., Liu D., Dong S., et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther. 2020;5:193. doi: 10.1038/s41392-020-00300-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jin D., Guo J., Wu Y., et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 171.Li B., Jiang J., Assaraf Y.G., et al. Surmounting cancer drug resistance: new insights from the perspective of N(6)-methyladenosine RNA modification. Drug Resist. Updates. 2020;53:100720. doi: 10.1016/j.drup.2020.100720. [DOI] [PubMed] [Google Scholar]
- 172.Uddin M.B., Roy K.R., Hosain S.B., et al. An N(6)-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem. Pharmacol. 2019;160:134–145. doi: 10.1016/j.bcp.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han D., Liu J., Chen C., et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang S., Wei J., Cui Y.H., et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yan F., Al-Kali A., Zhang Z., et al. A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28:1062–1076. doi: 10.1038/s41422-018-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lin Z., Niu Y., Wan A., et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Meng Q., Wang S., Zhou S., et al. Dissecting the m(6)A methylation affection on afatinib resistance in non-small cell lung cancer. Pharmacogenomics J. 2020;20:227–234. doi: 10.1038/s41397-019-0110-4. [DOI] [PubMed] [Google Scholar]
- 178.Ding N., You A., Tian W., et al. Chidamide increases the sensitivity of non-small cell lung cancer to Crizotinib by decreasing c-MET mRNA methylation. Int. J. Biol. Sci. 2020;16:2595–2611. doi: 10.7150/ijbs.45886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Su Y., Zhong G., Lin T. MP17-06 CIRCRIP2 accelerates bladder cancer progression via MIR-1305/TGF-β2/SMAD3 pathway. J. Urol. 2020;203:e227–e228. [Google Scholar]
- 180.Zhou S., Bai Z.L., Xia D., et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol. Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 181.Fukumoto T., Zhu H., Nacarelli T., et al. N(6)-methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019;79:2812–2820. doi: 10.1158/0008-5472.CAN-18-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Frye M., Harada B.T., Behm M., et al. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Deng X., Su R., Weng H., et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gu C., Chen C. Methylation in lung cancer: a brief review. Methods Mol. Biol. 2020;2204:91–97. doi: 10.1007/978-1-0716-0904-0_8. [DOI] [PubMed] [Google Scholar]
- 185.Wang Z., Du M., Yuan Q., et al. Epigenomic analysis of 5-hydroxymethylcytosine (5hmC) reveals novel DNA methylation markers for lung cancers. Neoplasia. 2020;22:154–161. doi: 10.1016/j.neo.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Applebaum M.A., Barr E.K., Karpus J., et al. 5-Hydroxymethylcytosine profiles in circulating cell-free DNA associate with disease burden in children with neuroblastoma. Clin. Cancer Res. 2020;26:1309–1317. doi: 10.1158/1078-0432.CCR-19-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cai J., Chen L., Zhang Z., et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu C., Cui X., Zhao B.S., et al. DNA 5-methylcytosine-specific amplification and sequencing. J. Am. Chem. Soc. 2020;142:4539–4543. doi: 10.1021/jacs.9b12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li W., Zhang X., Lu X., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Anita R., Paramasivam A., Priyadharsini J.V., et al. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am. J. Cancer Res. 2020;10:2546–2554. [PMC free article] [PubMed] [Google Scholar]
- 191.Yang X., Liu M., Li M., et al. Epigenetic modulations of noncoding RNA: a novel dimension of Cancer biology. Mol. Cancer. 2020;19:64. doi: 10.1186/s12943-020-01159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]