Abstract
New technologies and analysis methods are enabling genomic structural variants (SVs) to be detected with ever-increasing accuracy, resolution, and comprehensiveness. To help translate these methods to routine research and clinical practice, we developed the first sequence-resolved benchmark set for identification of both false negative and false positive germline large insertions and deletions. To create this benchmark for a broadly consented son in a Personal Genome Project trio with broadly available cells and DNA, the Genome in a Bottle (GIAB) Consortium integrated 19 sequence-resolved variant calling methods from diverse technologies. The final benchmark set contains 12745 isolated, sequence-resolved insertion (7281) and deletion (5464) calls ≥50 base pairs (bp). The Tier 1 benchmark regions, for which any extra calls are putative false positives, cover 2.51 Gbp and 5262 insertions and 4095 deletions supported by ≥1 diploid assembly. We demonstrate the benchmark set reliably identifies false negatives and false positives in high-quality SV callsets from short-, linked-, and long-read sequencing and optical mapping.
Introduction
Many diseases have been linked to structural variations (SVs), most often defined as genomic changes at least 50 base pairs (bp) in size, but SVs are challenging to detect accurately. Conditions linked to SVs include autism,1 schizophrenia, cardiovascular disease,2 Huntington’s Disease, and several other disorders.3 Far fewer SVs exist in germline genomes relative to small variants, but SVs affect more base pairs and each SV may be more likely to impact phenotype.4–6 While next generation sequencing technologies can detect many SVs, each technology and analysis method has different strengths and weaknesses. To enable the community to benchmark these methods, the Genome in a Bottle Consortium (GIAB) here developed benchmark SV calls and benchmark regions for the son (HG002/NA24385) in a broadly consented and available Ashkenazi Jewish trio from the Personal Genome Project,7 which are disseminated as National Institute of Standards and Technology (NIST) Reference Material 8392.8,9
Many approaches have been developed to detect SVs from different sequencing technologies. Microarrays can detect large deletions and duplications, but not with sequence-level resolution.10 Since short reads (<<1000bp) are often smaller than or similar to the SV size, bioinformaticians have developed a variety of methods to infer SVs, including using split reads, discordant read pairs, depth of coverage, and local de novo assembly. Linked reads add long-range (100kb+) information to short reads, enabling phasing of reads for haplotype-specific deletion detection, large SV detection,11–13 and diploid de novo assembly.14 Long reads (>>1000bp), which can fully traverse many more SVs, further enable SV detection, often sequence-resolved, using mapped reads,15,16 local assembly after phasing long reads,6,17 and global de novo assembly.18,19 Finally, optical mapping and electronic mapping provide an orthogonal approach capable of determining the approximate size and location of insertions, deletions, inversions, and translocations while spanning even very large SVs.20–22
GIAB recently published benchmark sets for small variants for seven genomes,9,23 and the Global Alliance for Genomics and Health Benchmarking Team established best practices for using these and other benchmark sets to benchmark germline variants.24 These benchmark sets are widely used in developing, optimizing, and demonstrating new technologies and bioinformatics methods, as well as part of clinical laboratory validation.12,15,25,26 Benchmarking tool development has also been critical to standardize definitions of performance metrics, robustly compare VCFs with different representations of complex variants, and enable stratification of performance by variant type and genome context. Benchmark set and benchmarking tool development is even more challenging and important for SVs given the wide spectrum of types and sizes of SVs, complexity of SVs (particularly in repetitive genome contexts), and that many SV callers output imprecise or imperfect breakpoints and sequence changes.
Several previous efforts have developed well-characterized SVs in human genomes. The 1000 Genomes Project catalogued copy-number variants (CNVs) and SVs in thousands of individuals.27,28 A subset of CNVs from NA12878 were confirmed and further refined to those with support from multiple technologies using SVClassify.29 The unique collection of Sanger sequencing from the HuRef sample has also been used to characterize SVs.30,31 Long reads were used to broadly characterize SVs in a haploid hydatidiform mole cell line.32 The Parliament framework was developed to integrate short and long reads for the HS1011 sample.33 Most recently, the Human Genome Structural Variation Consortium (HGSVC)6 and the Genome Reference Consortium34 used short, linked, and long reads to develop phased, sequence-resolved SV callsets, greatly expanding the number of SVs in three trios from 1000 Genomes, particularly in tandem repeats. Detection of somatic SVs in cancer genomes is a very active field, with numerous methods in development.35–37 While some of the problems are similar between germline and somatic SV detection, somatic detection is complicated by the need to distinguish somatic from germline events in the face of differential coverage, subclonal mutations and impure tumor samples, amongst others.38,39
We build on these efforts by enabling anyone to assess both false negatives (FNs) AND false positives (FPs) for a well-defined set of sequence-resolved insertions and deletions ≥50 bp in specified genomic regions. The HGSVC reports 27622 SVs per genome, but states in the discussion that “there is a pressing need to reduce the FDR of SV calling to below the current standard of 5%.”6 The Genome Reference Consortium developed SV calls in 15 individuals from de novo assembly, but these assemblies were not haplotype-resolved and therefore missed some heterozygous variants.34 In addition, neither of these studies define benchmark regions, which are critical in enabling reliable identification of false positives. HGSVC provides a very valuable resource, allowing the community to understand the spectrum of structural variation, but its lack of benchmark regions and its tradeoff of comprehensiveness for false positives limits its utility in benchmarking the performance of methods.
Our work in an open, public consortium is uniquely aimed at providing authoritative SVs and regions to enable technology and bioinformatics developers to benchmark and optimize their methods, and allow clinical laboratories to validate SV detection methods. We have developed methods and a benchmark set of SV calls and genomic regions that can be used to assess the performance of any sequencing and SV calling method. The ability to reliably identify false negative and false positives has been critical to the enduring success of our widely-adopted small variant benchmarks.9,23 We reach a similar goal for SVs by defining regions of the genome in which we are able to identify SVs with high precision and recall (here encompassing 2.51 Gb of the genome and 5262 insertions and 4095 deletions). While we include SVs only discovered by long reads, we exclude regions with more than one SV, mostly in tandem repeats, as these regions are not handled by current SV comparison and benchmarking tools. In SV calls for the Puerto Rican child HG00733 from HGSVC6 and de novo assembly34 in dbVar nstd152 and nstd162, respectively, we found that 24632 out of 33499 HGSVC calls and 10164 out of 22558 assembly-based calls were in clusters (within 1000 bp of another SV call in the same callset). We also cluster calls by their specific sequence, improving upon previous work that clustered loosely by position, overlap, or size; we address challenges in comparing calls with different representations in repetitive regions to enable the integration of a wide variety of sequence-resolved input callsets from different technologies. Most importantly, we show it correctly identifies false positives and false negatives across a diversity of technologies and SV callers. This is our principal goal: to make trustworthy assessment data and tools available as a common reference point for performance evaluation of SV calling.
Results
Candidate SV callsets differ by sequencing technology and analysis method
We generated 28 sequence-resolved candidate SV callsets from 19 variant calling methods from 4 sequencing technologies for the Ashkenazi son (HG002), as well as 20 callsets each from the parents HG003 and HG004 (Supplementary Table 1). We integrated a total of 68 callsets, where we define a “callset” as the result of a particular variant calling method using data from one or more technologies for an individual. The variant calling methods included 3 small variant callers, 9 alignment-based SV callers, and 7 global de novo assembly-based SV callers. The technologies included short-read (Illumina and Complete Genomics), linked-read (10x Genomics), and long-read (PacBio) sequencing technologies as well as SV size estimates from optical (Bionano) and electronic (Nabsys) mapping.
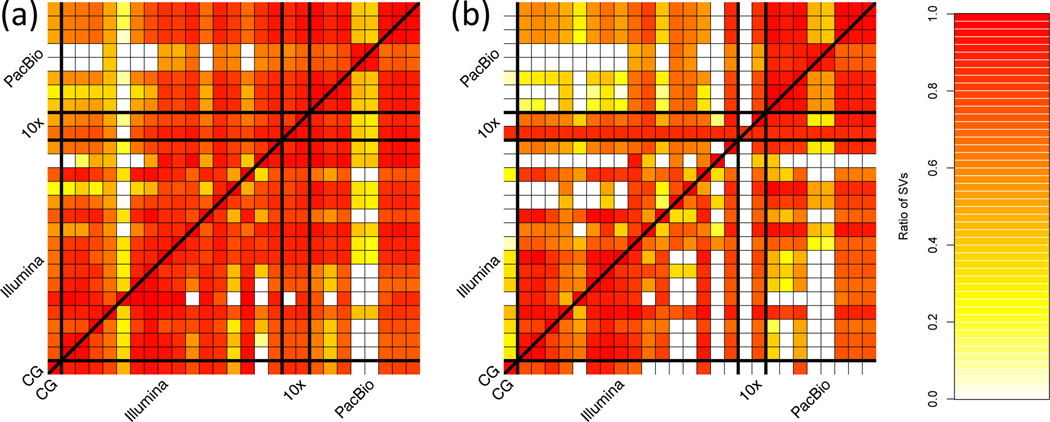
Figure 1 shows the number of SVs overlapping between our sequence-resolved callsets from different variant calling methods and technologies for HG002, with loose matching by SV type within 1 kbp using SURVIVOR.40 In general, the concordance for insertions is lower than the concordance for deletions, except among long-read callsets, mostly because current short read-based methods do not sequence-resolve large insertions. This highlights the importance of developing benchmark SV sets to identify which callset is correct when they disagree, and potentially when both are incorrect even when they agree.
Figure 1: Pairwise comparison of sequence-resolved SV callsets obtained from multiple technologies and SV callers for SVs ≥50bp from HG002.
Heatmap produced by SURVIVOR40 shows the fraction of SVs overlapping between the individual SV caller and technologies split between (a) deletions and (b) insertions. The color corresponds to the fraction of SVs in the caller on the x axis that overlap the caller on the y axis. Overall we obtained a quite diverse picture of SVs calls supported by each SV caller and technology, highlighting the need for benchmark sets.
Design objectives for our benchmark SV set
Our objective was that, when comparing any callset (the “test set” or “query set”) to the “benchmark set,” it reliably identifies FPs and FNs. In practice, we aimed to demonstrate that most (ideally approaching 100%) of conflicts (both FPs and FNs) between any given test set and the benchmark set were actually errors in the test set. This goal is typically challenging to meet across the wide spectrum of sequencing technologies and calling methods. Secondarily, to the extent possible, our goal was for the benchmark set to include a large, representative variety of SVs in the human genome. By integrating results from a large suite of high-throughput, whole genome methods, each with their own signatures of bias, biases from any particular method are minimized. We systematically establish the “benchmark regions” in this genome in which we are close to comprehensively characterizing SVs. We exclude regions from our benchmark if we could not reliably reach near-comprehensive characterization (e.g., in segmental duplications). Importantly, we demonstrate the benchmark set is fit for purpose for benchmarking by presenting examples of comparisons of SVs from multiple technologies and manual curation of discordant calls.
Benchmark set is formed by clustering and evaluating support for candidate SVs
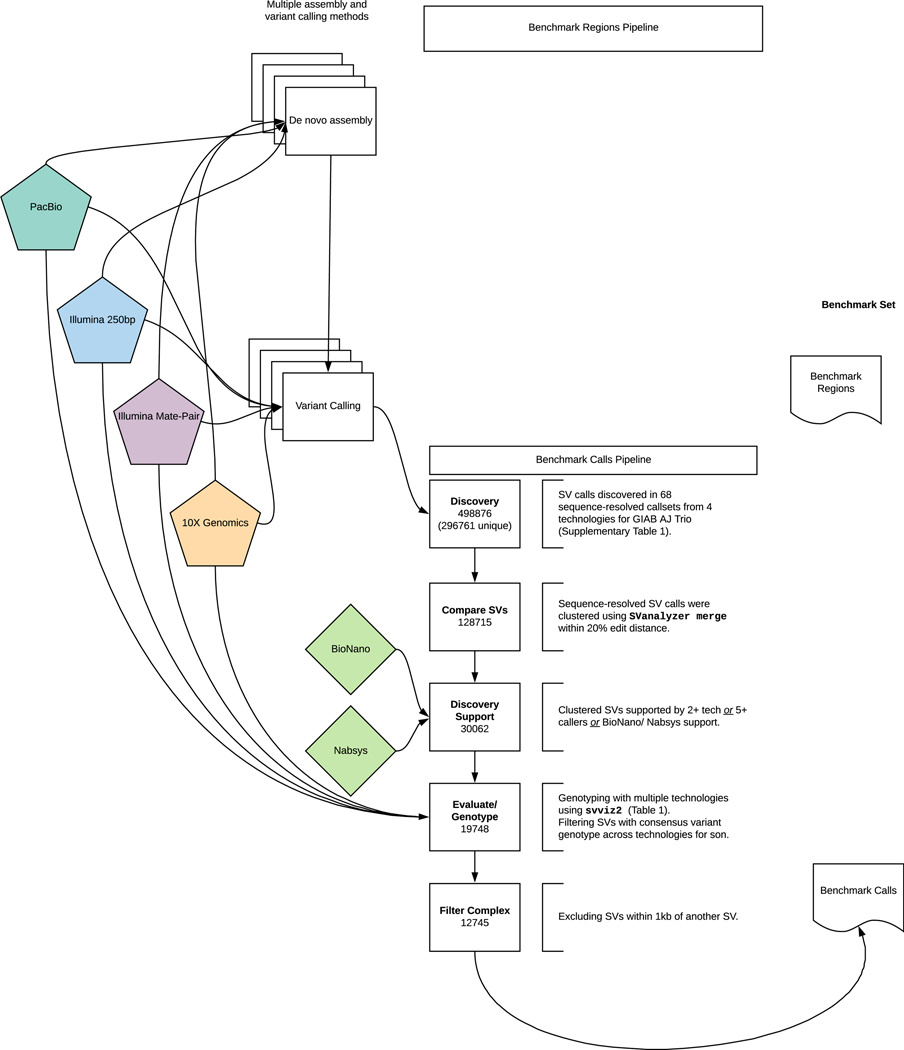
We integrated all sequence-resolved candidate SV callsets (“Discovery callsets” in Supplementary Table 1) to form the benchmark set, using the process described in Figure 2. Since candidate SV calls often differ in their exact breakpoints, size, and/or sequence change estimated, we used a new method called SVanalyzer (https://svanalyzer.readthedocs.io) to cluster calls estimating similar sequence changes. This new method was needed to account for both differences in SV representation (e.g., different alignments within a tandem repeat) and differences in the precise sequence change estimated. Of the 498876 candidate insertion and deletion calls ≥50 bp in the son-father-mother trio, 296761 were unique after removing duplicate calls and calls that were the same when taking into account representation differences (e.g., different alignment locations in a tandem repeat). When clustering variants for which the estimated sequence change was <20 % divergent, 128715 unique SVs remain. We then filtered to retain SV clusters supported by: more than one technology, ≥5 callsets from a single technology, Bionano, or Nabsys. The 30062 SVs remaining were then evaluated and genotyped in each member of the trio using svviz 41 to align reads to reference and alternate alleles from PCR-free Illumina, Illumina 6 kbp mate-pair, haplotype-partitioned 10x Genomics, and PacBio with and without haplotype partitioning. We further filtered for SVs covered in HG002 by 8 or more PacBio reads (mean coverage of about 60), with at least 25% of PacBio reads supporting the alternate allele and consistent genotypes from all technologies that could be confidently assessed with svviz. This left 19748 SVs. The number of PacBio reads supporting the SV allele and reference allele for each benchmark SV is in .Extended Data Figure 1.
Figure 2: Process to integrate SV callsets and diploid assemblies from different technologies and analysis methods and form the benchmark set.
The input datasets are depicted in the center of the figure with the benchmark calls and region pipelines to the left and right of the input data, respectively. The number of variants in each step of the benchmark calls integration pipeline is indicated in the white boxes. See the Methods section for additional description of the pipeline steps. Briefly, approximately 0.5 million input SV calls were locally clustered based on their estimated sequence change, and we kept only those discovered by at least two technologies or at least 5 callsets in the trio. We then used svviz with short, linked, and long reads to evaluate and genotype these calls, keeping only those with a consensus heterozygous or homozygous variant genotype in the son. We filtered potentially complex calls in regions with multiple discordant SV calls, as well as regions around 20 bp to 49 bp indels, and our final Tier 1 benchmark set included 12745 total insertions and deletions ≥50 with 9357 inside the 2.51 Gbp of the genome where diploid assemblies had no additional SVs beyond those in our benchmark set. We also define a Tier 2 set of 6007 additional regions where there was substantial support for one or more SVs but the precise SV was not yet determined.
In our evaluations of these well-supported SVs, we found that 12745 were isolated, while 7003 (35 %) were within 1000 bp of another well-supported SV call. Upon manual curation, we found that the variants within 1000 bp of another variant were mostly in tandem repeats and fell into several classes: (1) inferred complex variants with more than one SV call on the same haplotype, (2) inferred compound heterozygous variant with different SV calls on each haplotype, and (3) regions where some methods had the correct SV call and others had inaccurate sequence, size, or breakpoint estimates, but svviz still aligned reads to it because reads matched it better than the reference. We chose to exclude these clustered SVs from our benchmark set because methods do not exist to confidently distinguish between the above classes, nor do SV comparison tools for robust benchmarking of complex and compound structural variants.
Finally, to enable assessment of both FNs and FPs, benchmark regions were defined using diploid assemblies and candidate variants. These regions were designed such that our benchmark variant callset should contain almost all true SVs within these regions. These regions define our Tier 1 benchmark set, which spans 2.51 Gbp and includes 5262 insertions and 4095 deletions. These regions exclude 1837 of the 12745 SVs because they were within 50 bp of a 20 bp to 49 bp indel; they exclude an additional 856 SVs within 50 bp of a candidate SV for which no consensus genotype could be determined; and they exclude an additional 411 calls that were not fully supported by a diploid assembly as the only SV in the region. A large number of annotations are associated with the Tier 1 SV calls (e.g., number of discovery callsets from each technology, number of reads supporting reference and alternate alleles from each technology, number of callsets with exactly matching sequence estimates), which enable users to filter to a more specific callset. We also define Tier 2 regions that delineate 6007 additional regions in addition to the 12745 isolated SVs, which are regions with substantial evidence for one or more SVs but we could not precisely determine the SV. For the Tier 2 regions, multiple SVs within 1 kb or in the same or adjacent tandem repeats are counted as a single region, so many SV callers would be expected to call more than 6007 SVs in these regions.
Benchmark calls are well-supported
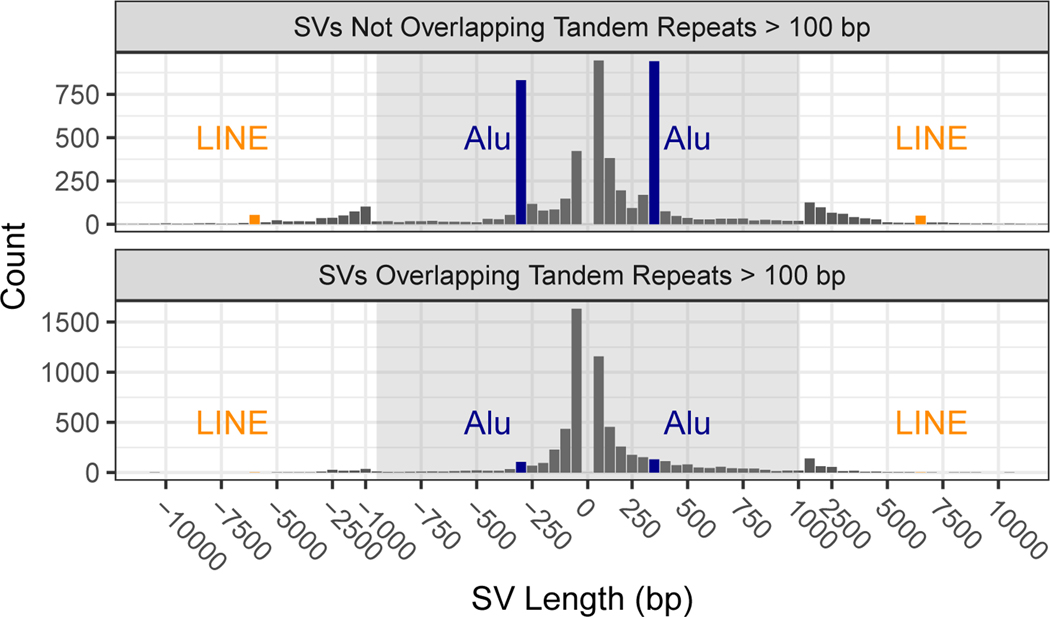
The 12745 isolated SV calls had size distributions consistent with previous work detecting SVs from long reads,6,15,17,26 with the clear, expected peaks for insertions and deletions near 300 bp related to Alu’s and for insertions and deletions near 6000 bp related to full-length LINE1’s (Figure 3). Note that deletion calls of Alu and LINE elements are most likely mobile element insertions in the GRCh37 sequence that are not in HG002. SVs have an exponentially decreasing abundance vs. size if they fall in tandem repeats longer than 100 bp in the reference. Interestingly, there are more large insertions than large deletions in tandem repeats, despite insertions being more challenging to detect. This is consistent with previous work detecting SVs from long read sequencing15,17 and may result from instability of tandem repeats in the BAC clones used to create the reference genome.42
Figure 3: Size distributions of deletions and insertions in the benchmark set.
Variants are split by SVs overlapping and not overlapping tandem repeats longer than 100bp in the reference. Deletions are indicated by negative SV lengths. The expected Alu mobile elements peaks near ± 300 bp are indicated in blue and LINE mobile elements peaks near ± 6000 bp indicated in orange.
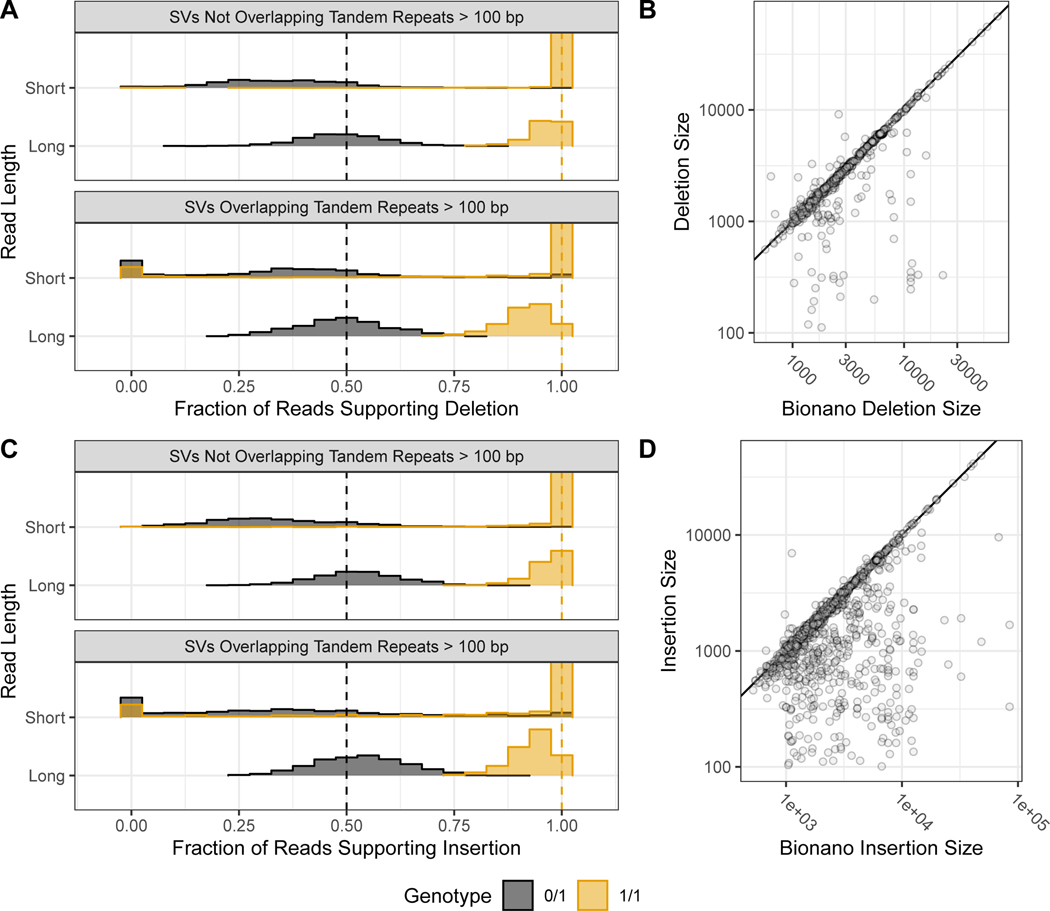
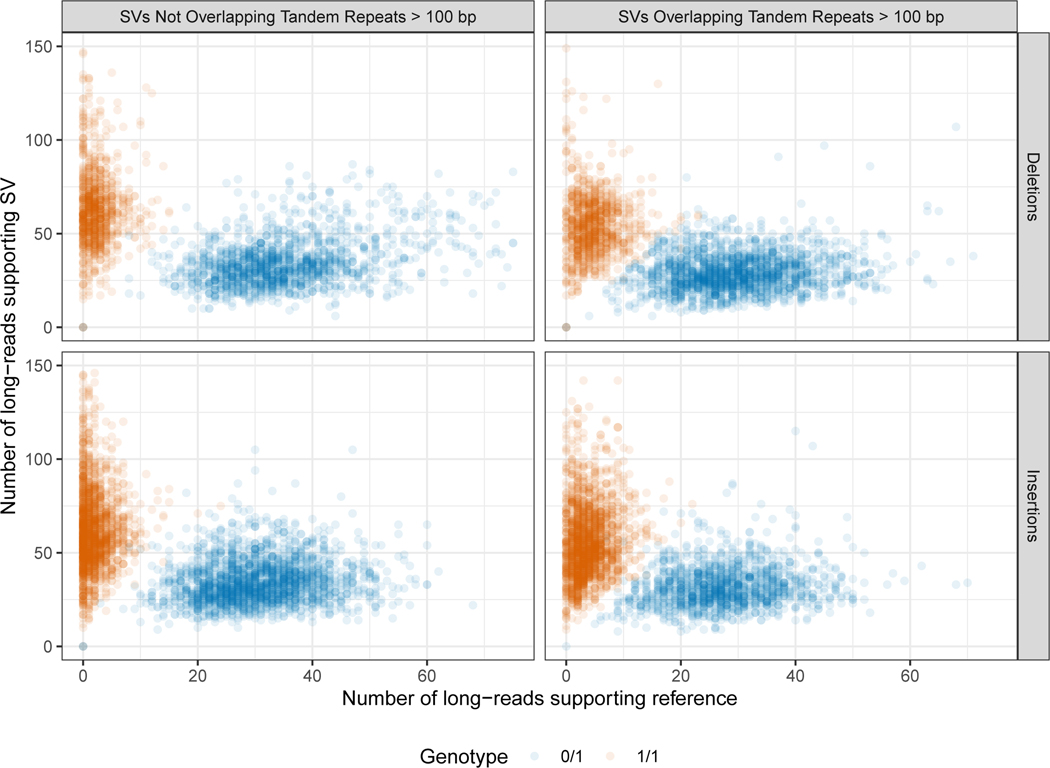
When evaluating the support for our benchmark SVs, approximately 50 % of long reads more closely matched the SV allele for heterozygous SVs, and approximately 100 % for homozygous SVs, as expected (Figure 4A and 4C). While short reads clearly supported and differentiated homozygous and heterozygous genotypes for many SVs, the support for heterozygous calls was less balanced, with a mode around 30%, and they did not definitively genotype 35 % of deletions and 47 % of insertions in tandem repeats because reads were not sufficiently long to traverse the repeat. These results highlight the difficulty in detecting SVs with short reads in long tandem repeats, as a sizeable fraction of reads containing the variant either map without showing the variant or fail to map at all. We also found high size concordance with Bionano (Figure 4B and 4D). Since the region between Bionano markers can contain multiple SVs, the Bionano estimate will be the sum of all SVs between the markers, which can cause apparent differences in size estimates. For example, for insertions > 300bp where the Bionano DLS size estimate is > 300 bp higher and > 30 % higher than the v0.6 insertion size, and where the entire region between Bionano markers is included in our benchmark bed, 23 out of the 40 Bionano insertions have multiple v0.6 insertions in the interval that sum to the Bionano size. In general, there was strong support from multiple technologies for the benchmark SVs, with 90 % of the Tier 1 SVs having support from more than one technology.
Figure 4: Support for benchmark SVs by long reads, short reads, and optical mapping.
Histograms show the fraction of PacBio (long-reads) and Illumina 150 bp (short-reads) reads that aligned better to the SV allele than to the reference allele using svviz, colored by v0.6 genotype, where blue is heterozygous and orange is homozygous. Variants are stratified into deletions (A) & and insertions (C), and into SVs overlapping and not overlapping tandem repeats longer than 100bp in the reference. Vertical dashed lines correspond to the expected fractions 0.5 for heterozygous (blue) and 1.0 for homozygous variants (dark orange). The v0.6 benchmark set sequence-revolved deletion (B) and insertion (D) SV size is plotted against the size estimated by BioNano in any overlapping intervals, where points below the diagonal (indicated by the black line) represent smaller sequence-resolved SVs in the overlapping interval.
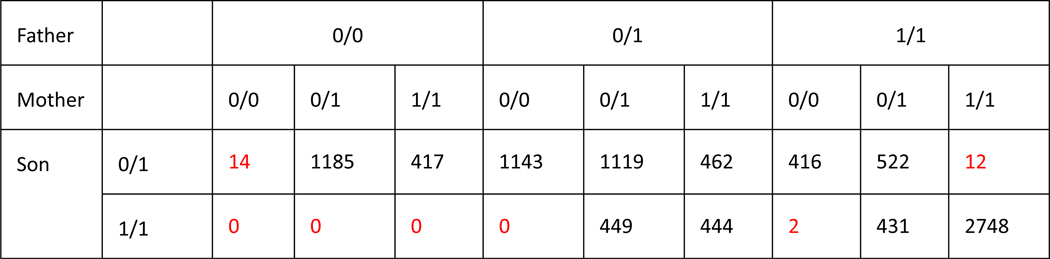
For SVs on autosomes, we also identified if genotypes were consistent with Mendelian inheritance. When limiting to 7973 autosomal SVs in the benchmark set for which a consensus genotype from svviz was determined for both of the parents, only 20 violated Mendelian inheritance. Upon manual curation of these 20 sites, 16 were correct in HG002 (mostly misidentified as homozygous reference in both parents due to lower long read sequencing coverage), 1 was a likely de novo deletion in HG002 (17:51417826–51417932), 1 was a deletion in the T cell receptor alpha locus known to undergo somatic rearrangement (14:22918114–22982920), and 2 were insertions mis-genotyped as heterozygous in HG002 when in fact they were likely homozygous variant or complex (2:232734665 and 8:43034905). Extended Data Figure 2 is a detailed contingency table of genotypes in the son, father, and mother.
The GIAB community also manually curated a random subset of SVs from different size ranges in the union of all discovered SVs.43 When comparing the consensus genotype from expert manual curation to our benchmark SV genotypes, 627/635 genotypes agreed. Most discordant genotypes were identified as complex by the curators, with a 20 bp to 49 bp indel near an SV in our benchmark set, because they were asked to include indels 20 bp to 49 bp in size in their curation, whereas our SV benchmark set focused on SVs >49 bp.
We compared the v0.6 Tier 1 deletion breakpoints to the deletion breakpoints from a different set of samples analyzed by HGSVC6 and GRC.34 Of the 5464 deletions in v0.6, (a) 45 % had breakpoints and 57 % had size matching an HGSVC call, (b) 49 % had breakpoints and 66 % had size matching a GRC call, and (c) 58 % had breakpoints and 73 % had size matching either an HGSVC call or a GRC call. This comparison permitted 1 bp differences in the left and right breakpoints or 1 bp difference in size for any overlap, which ignores slight imprecision and off-by-one file format errors, but does not account for all differences in representation within repeats. This high degree of overlap supports the base-level accuracy of our calls and previous findings that many SVs are shared between even small numbers of sequenced individuals.34
We also evaluated the sensitivity of v0.6 to 429 deletions from the population-based gnomAD-SV v2.1 callset 44 that were homozygous reference in less than 5 % of individuals of European ancestry and at least 1000 Europeans had the variant. Of these 429 deletions, 296 were in the v0.6 benchmark bed, and 286 of the 296 (97 %) overlapped a v0.6 deletion. We manually curated the 4 deletions that had size estimates > 30 % different between gnomAD-SV and v0.6, and all were in tandem repeats and the v0.6 breakpoints were clearly supported by long read alignments. We also manually curated the 10 deletions that did not overlap a v0.6 deletion, which had Homozygous Reference frequencies in Europeans between 1.8 % and 5 %, and all 10 were clearly homozygous reference in HG002, and 9 of the 10 were in our discovery callset and were genotyped as heterozygous in both parents but homozygous reference in HG002 (Supplementary Table 2). This demonstrates that even though population-based callsets were not included in our discovery methods, v0.6 does not miss many common SVs within the benchmark bed.
Benchmark set is useful for identifying false positives and false negatives across technologies
Our goal in designing this SV benchmark set was that, when comparing any callset to our benchmark VCF within the benchmark BED file, most putative FPs and FNs should be errors in the tested callset. To determine if we meet this goal, we benchmarked several callsets from assembly- and non-assembly-based methods that use short or long reads. Most of these callsets (“Evaluation callsets” in Supplementary Table 1) are different from the callsets used in the integration process by using different callers, new data types, or new tool versions. We developed a new benchmarking tool truvari (https://github.com/spiralgenetics/truvari) to perform these comparisons at different matching stringencies, since truvari enables users to specify matching stringency for size, sequence, and/or distance. We performed some comparisons requiring only that the variant size to be within 30 % of the benchmark size and the position to be within 2 kb, and some comparisons additionally requiring the sequence edit distance to be less than 30 % of the SV size. We compared at both stringencies because truvari sometimes could not match different representations of the same variant. An alternative benchmarking tool developed more recently, which has more sophisticated sequence matching, is SVanalyzer SVbenchmark (https://github.com/nhansen/SVanalyzer/blob/master/docs/svbenchmark.rst).
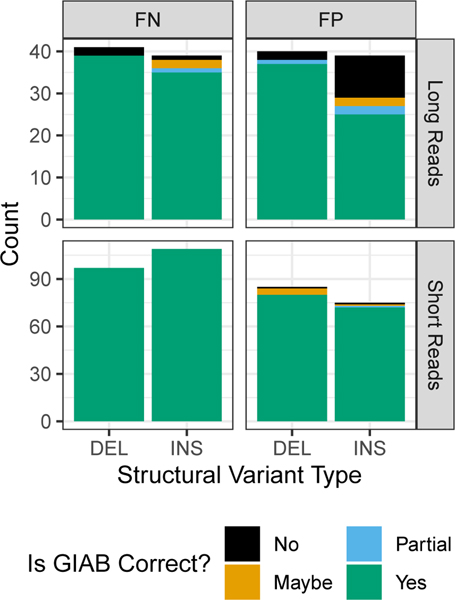
Upon manual curation of a random 10 FP and FN insertions and deletions (40 total SVs) from each callset being compared to the benchmark, nearly all of the FPs and FNs were errors in each of the tested callsets and not errors in the GIAB callset (Figure 5 and Supplementary Table 2). The version of the truvari tool we used could not always account for all differences in representation, so if manual curation determined both the benchmark and test sets were correct, they were counted as correct. The only notable exception to the high GIAB callset accuracy was for FP insertions from the PacBio caller pbsv (https://github.com/PacificBiosciences/pbsv), for which about half of the putative FP insertions were true insertions missed in the benchmark regions. This suggests the GIAB callset may be missing approximately 5 % of true insertions in the benchmark regions. When comparing BioNano calls to our benchmark, we also found one region with multiple insertions where our benchmark had a heterozygous 1412 bp insertion at chr6:65000859, but we incorrectly called a homozygous 101 bp insertion in a nearby tandem repeat at chr6:65005337, when in fact there is an insertion of approximately 5400 bp in this tandem repeat on the same haplotype as the 1412 bp insertion, and the 101 bp insertion is on the other haplotype.
Figure 5: Summary of manual curation of putative FPs and FNs when benchmarking short and long reads against the v0.6 benchmark set.
Most FP and FN SVs were determined to be correct in the v0.6 benchmark (green), but some were partially correct due to missing part of the SV in the region (blue), were incorrect in v0.6 (orange), or were in difficult locations where the evidence was unclear (black).
To evaluate the utility of v0.6 to benchmark genotypes, we also compared genotypes from two graph-based genotypers for short reads: vg45 and paragraph46. Of the 5293 heterozygous and 4245 homozygous variant v0.6 calls that had genotypes from both genotypers, 3642 heterozygous and 2970 homozygous calls had identical genotypes for vg, paragraph, and v0.6. 925 heterozygous and 496 homozygous variant v0.6 calls had genotypes that were different from both vg and paragraph. However, after filtering v0.6 calls annotated as overlapping tandem repeats, which are less accurately genotyped by short reads, only 326 heterozygous and 69 homozygous discordant genotypes remained. We manually curated 10 randomly-selected discordant heterozygous and homozygous genotype calls, and all 10 heterozygous and all 10 homozygous calls were correctly genotyped in v0.6, and were errors in short read genotyping mostly in short tandem repeats, transposable elements, or tandem duplications, demonstrating the utility of v0.6 for benchmarking genotypes. The ratio of heterozygous to homozygous sites in v0.6 is 3433 to 2031 for deletions and 3505 to 3776 for insertions, significantly lower than the ratio of approximately 2 for small variants, particularly for insertions. This difference likely results both from homozygous variants being easier to discover and from tandem repeats that are systematically compressed in GRCh37, which result in homozygous insertions in our calls.
Technologies and variant callers have different strengths and weaknesses
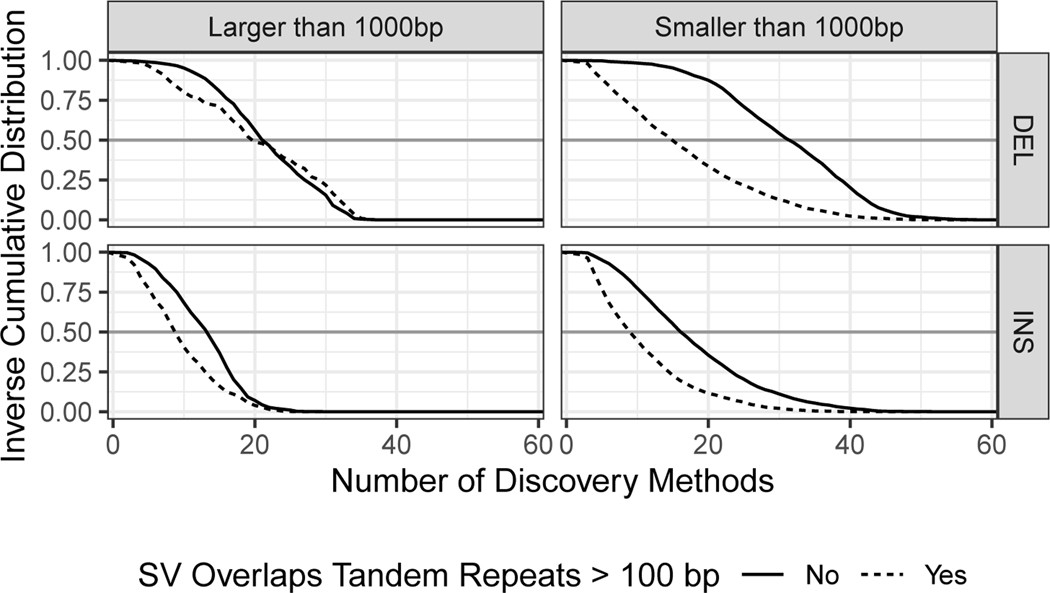
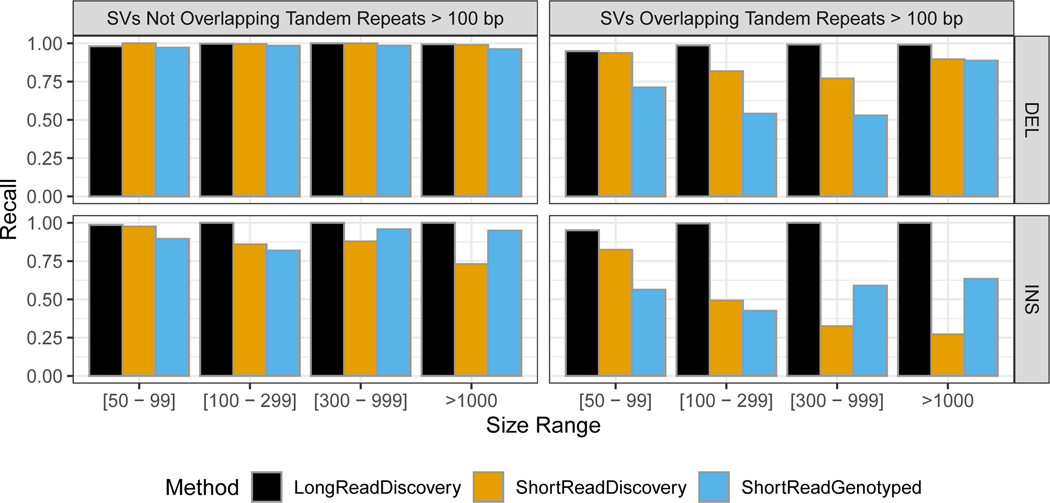
Amongst the extensive candidate SV callsets we collected from different technologies and analyses, we found that certain SV types and sizes in our benchmark set were discovered by fewer methods (Figure 6). In particular, more methods discovered sequence-resolved deletions than insertions, more methods discovered SVs not in tandem repeats, and the most methods discovered deletions smaller than 1000 bp not in tandem repeats. These results confirm the intuition that SV detection outside of repeats is simpler than within repeats, and that deletions are simpler to detect than insertions since deletions do not require mapping to new sequence. Extended Data Figure 3 further shows that the fewest SVs were missed by the union of all long read discovery methods. The only exception was (50 to 99) bp deletions, which were all found by at least one short read discovery method. Many insertions >300 bp that were not discovered by any short read method could be accurately genotyped in this sample by short reads. Interestingly, many deletions and insertions <300 bp that were not genotyped accurately by short reads were discovered by at least one short read-based method. This likely reflects a limitation of the heuristics we used for genotyping, which reduces the false positive rate but may increase the false negative rate. Both discovery and genotyping based on short reads had limitations for SVs in tandem repeats. These results confirm the importance of long read data for comprehensive SV detection.
Figure 6: Inverse cumulative distribution showing the number of discovery methods that supported each SV.
All 68 callsets from all variant calling methods and technologies in all three members of the trio are included in these distributions. SVs larger than 1000 bp (top) are displayed separately from SVs smaller than 1000 bp (bottom). Results are stratified into deletions (left) and insertions (right), and into SVs overlapping (black) and not overlapping (gold) tandem repeats longer than 100 bp in the reference. Grey horizontal line at 0.5 added to aid comparison between panels.
Sequence-resolved benchmark calls have annotations related to base-level accuracy
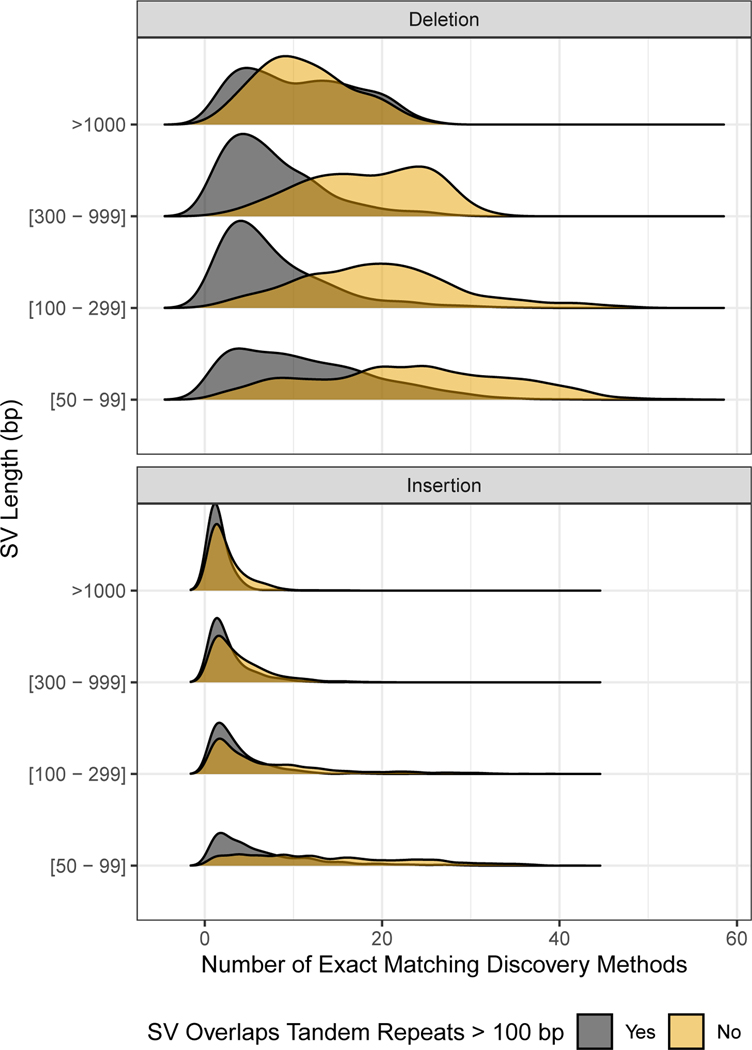
We provide sequence-resolved calls in our benchmark set to enable benchmarking of sequence change predictions, but importantly not all calls are perfect on a base-level. When discovered SVs from multiple callsets have exactly matching sequence changes, we output the sequence change from the largest number of callsets. However, as shown in Figure 7, not all benchmark SVs have calls that exactly matched between discovery callsets. For deletions not in tandem repeats, at least 99 % of the calls had exact matches, but there were no exact matches for ~5% of DELs in TRs, and for large insertions no exact matches existed for ~50% of the calls. This is likely because SVs in tandem repeats and larger insertions are more likely to be discovered only by methods using relatively noisy long reads.
Figure 7: Fraction of SVs for each number of discovery callsets that estimated exactly matching sequence changes.
Variants are stratified into deletions (top) and insertions (bottom), and into SVs overlapping (black) and not overlapping (gold) tandem repeats longer than 100 bp in the reference. SVs are also stratified by size (y-axis) into 50 bp to 99 bp, 100 bp to 299 bp, 300 bp to 999 bp, and ≥1000 bp.
Discussion
We have integrated sequence-resolved SV calls from diverse technologies and SV calling approaches to produce a new benchmark set enabling anyone to assess both FN and FP rates. This benchmark is useful for evaluating accuracy of SVs from a variety of genomic technologies, including short, linked, and long read sequencing technologies, optical mapping and electronic mapping. This resource of benchmark SVs, data from a variety of technologies, and SVs from a variety of methods are all publicly available without embargo, and we encourage the community to give feedback and participate in GIAB to continue to improve and expand this benchmark set in the future.
When developing this benchmark set, several trade-offs were made. Most notably, we chose to exclude complex SVs and SVs for which we could not determine a consensus sequence. Limiting our set to isolated insertions and deletions removed approximately one half of SVs for which there was strong support that some SV occurred. However, by excluding these complex regions from our SV benchmark set, it enables anyone to use our sequence comparison-based benchmarking tools to confidently and automatically identify FPs and FNs at different matching stringencies (e.g., matching based on SV sequence, size, type, and/or genotype). Bionano also identified large heterozygous events outside the benchmark regions, and future work will be needed to sequence-resolve these large unresolved complex events, often near segmental duplications. In addition to our standard Tier 1 benchmark set, we also provide a set of Tier 2 regions in which we found substantial evidence for an SV but it was complex or we could not determine the precise SV. We also exclude regions from our benchmark set around putative indels (20 to 49) bp in size, which minimizes unreliable putative FP and FN SVs around clustered indels or variants just under or above 50 bp.
Our benchmark also currently does not include more complicated forms of structural variations including inversions, duplications (except for calls annotated as tandem duplications), very large copy number variants (only one deletion and one insertion >100 kb), calls in segmental duplications, calls in tandem repeats >10 kbp, or translocations. This benchmark does not enable performance assessment of inversion detection (e.g., with Strand-seq47) or in highly repetitive regions like segmental duplications, telomeres, and centromeres that are starting to be resolved by ultralong nanopore reads.48 We also do not explicitly call duplications, though in practice our insertions frequently are tandem duplications, and we have provisionally labeled them as such using SVanalyzer svwiden in the REPTYPE annotation in the benchmark VCF. Future work in GIAB will use new technologies and analysis methods to include new SV types and more challenging SVs. When using our current benchmark, it is critical to understand it does not enable performance assessment for all SV types nor the most challenging SVs.
GIAB is currently collecting new candidate SV callsets for GRCh37 and GRCh38 from new data types (e.g., Strand-seq,47 PacBio Circular Consensus Sequencing,26 and Oxford Nanopore ultra long reads49), new and updated SV callers, and new diploid de novo assemblies. We are also refining the integration methods (e.g., to include inversions), and developing an integration pipeline that is easier to reproduce. In the next several months, we plan to release improved benchmark sets for GRCh37 and GRCh38 using these new methods similar to how we have maintained and updated the small variant callsets for these samples over time. We will also use the reproducible integration pipeline developed here to benchmark SVs for all 7 GIAB genomes. We will continue to refine these methods to access more difficult SVs in more difficult regions of the genome. Finally, we plan to develop a manuscript describing best practices for using this benchmark set to benchmark any other SV callset, similar to our recent publication for small variants,24 with refined SV comparison tools and standardized definitions of performance metrics. We have summarized the limitations of the v0.6 benchmark in Extended Data Figure 4.
Methods
Cell Line and DNA availability
For the 10x Genomics and Oxford Nanopore sequencing and BioNano and Nabsys mapping, the following cell lines/DNA samples were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: GM24385. For the Illumina, Complete Genomics, and PacBio sequencing, NIST RM 8391 DNA was used, which was prepared from a large batch of GM24385.
Benchmark Integration process
The GIAB v0.6 Tier 1 and Tier 2 SV Benchmark Sets were generated (using methods summarized in Figure 2 and detailed in Supplementary Note 1) from the union vcf. The union vcf, generated from the discovery callsets described in Supplementary Note 2 and summarized in Supplementary Table 1 (68 callsets from 19 variant callers and 4 technologies for the GIAB Ashkenazi trio), is at ftp://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/data/AshkenazimTrio/analysis/NIST_UnionSVs_12122017/union_171212_refalt.sort.vcf.gz. Several draft SV benchmark sets were developed and evaluated by the GIAB community, and feedback from end users and new technologies and SV callers were used to improve each subsequent version. A description of each draft version is in Supplementary Note 3.
Evaluation of the Benchmark
GIAB asked for volunteers to compare their SV callsets to the v0.6 Tier 1 benchmark set with truvari as described in Supplementary Note 4. Each volunteer manually curated 10 randomly selected FPs and FNs each from insertions and deletions, subset to SVs overlapping and not overlapping tandem repeats longer than 100bp (80 total variants). Potential errors identified in GIAB were further examined by NIST and the final determination about whether v0.6 was correct was made in consultation between multiple curators.
Extended Data
Extended Data Fig. 1. Number of long reads supporting the SV allele vs. the reference allele in the benchmark set.
Variants are colored by heterozygous (blue) and homozygous (dark orange) genotype, and are stratified into deletions and insertions, and into SVs overlapping and not overlapping tandem repeats longer than 100 bp in the reference.
Extended Data Fig. 2. Mendelian contingency table for sites with consensus genotypes from svviz in the son, father, and mother.
SVs in boxes highlighted in red violate the expected Mendelian inheritance pattern. Variants on chromosomes X and Y are excluded.
Extended Data Fig. 3. Comparison of false negative rates for the union of all long read-based SV discovery methods, the union of all short read-based discovery methods, and paired-end and mate-pair short read genotyping of known SVs.
Variants are stratified into deletions (top) and insertions (bottom), and into SVs overlapping (right) and not overlapping (left) tandem repeats longer than 100 bp in the reference. SVs are also stratified by size into 50 bp to 99 bp, 100 bp to 299 bp, 300 bp to 999 bp, and ≥1000 bp.
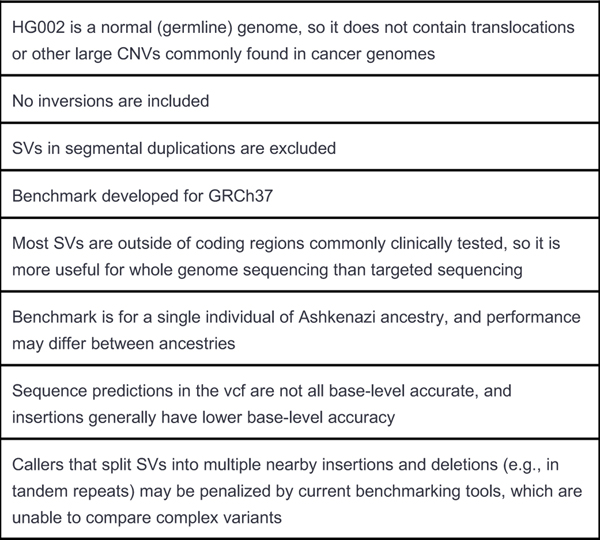
Extended Data Fig. 4. Known limitations of the v0.6 benchmark.
It is important to understand the limitations of any benchmark, such as the limitations below for v0.6, when interpreting the resulting performance metrics.
Supplementary Material
Acknowledgments
We thank many Genome in a Bottle Consortium Analysis Team members for helpful discussions about the design of this benchmark. We thank Jean Monlong and Glenn Hickey for sharing genotypes for HG002 from vg and paragraph. We thank Timothy Hefferon at NIH/NCBI for assistance with the dbVar submission. Certain commercial equipment, instruments, or materials are identified to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose. Chunlin Xiao and Steve Sherry were supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. NFH, JCM, SK, and AMP were supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health
Footnotes
Competing Interests Statement
A.M.W. is an employee and shareholder of Pacific Biosciences. AMB and ITF are employees and shareholders of 10X Genomics. GMC is the founder and holds leadership positions of many companies described at http://arep.med.harvard.edu/gmc/tech.html. FJS has received sponsored travel from Oxford Nanopore and Pacific Biosciences, and received a 2018 sequencing grant from Pacific Biosciences. J.L. is an employee and shareholder of Bionano Genomics. AC is an employee of Google Inc. and is a former employee of DNAnexus. JMS, JRD, MDK, JSO, and APC are employees of Nabsys 2.0, LLC. ACE is an employee and shareholder of Spiral Genetics. SMES is an employee of Roche.
Data availability
Raw sequence data were previously published in Scientific Data (DOI: 10.1038/sdata.2016.25), and were deposited in the NCBI SRA with the accession codes SRX847862 to SRX848317, SRX1388732 to SRX1388743, SRX852933, SRX5527202, SRX5327410, and SRX1033793-SRX1033798. 10x Genomics Chromium bam files used are at ftp://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/data/AshkenazimTrio/analysis/10XGenomics_ChromiumGenome_LongRanger2.2_Supernova2.0.1_04122018/. The data used in this manuscript and other datasets for these genomes are available in ftp://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/data/, and in the NCBI BioProject PRJNA200694.
The v0.6 SV benchmark set (only compare to variants in the Tier 1 vcf inside the Tier 1 bed with the FILTER “PASS”) for HG002 on GRCh37 is available in dbVar accession nstd175 and at: ftp://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/data/AshkenazimTrio/analysis/NIST_SVs_Integration_v0.6/
Input SV callsets, assemblies, and other analyses for this trio are available under: ftp://ftp-trace.ncbi.nlm.nih.gov/ReferenceSamples/giab/data/AshkenazimTrio/analysis/
Code availability
Scripts for integrating candidate structural variants to form the benchmark set in this manuscript are available in a GitHub repository at https://github.com/jzook/genome-data-integration/tree/master/StructuralVariants/NISTv0.6. This repository includes jupyter notebooks for the comparisons to HGSVC, GRC, vg, paragraph, and Bionano. Publicly available software used to generate input callsets is described below in the methods.
References
- 1.Sebat J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merker JD et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med 20, 159–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantere T, Kersten S. & Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet 10, 426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roses AD et al. Structural variants can be more informative for disease diagnostics, prognostics and translation than current SNP mapping and exon sequencing. Expert Opin. Drug Metab. Toxicol 12, 135–147 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Chiang C. et al. The impact of structural variation on human gene expression. Nat. Genet 49, 692–699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball MP et al. A public resource facilitating clinical use of genomes. Proc. Natl. Acad. Sci. U. S. A 109, 11920–11927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Scientific Data 3, 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zook JM et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol 37, 561–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebat J. et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Spies N. et al. Genome-wide reconstruction of complex structural variants using read clouds. Nat. Methods 14, 915–920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marks P. et al. Resolving the full spectrum of human genome variation using Linked-Reads. Genome Res. 29, 635–645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karaoglanoglu F. et al. Characterization of segmental duplications and large inversions using Linked-Reads. bioRxiv 394528 (2018). [Google Scholar]
- 14.Weisenfeld NI, Kumar V, Shah P, Church DM & Jaffe DB Direct determination of diploid genome sequences. Genome Res. 27, 757–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sedlazeck FJ et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cretu Stancu M. et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun 8, 1326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaisson MJP et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin C-S et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koren S. et al. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol (2018) doi: 10.1038/nbt.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser MD et al. Automated Structural Variant Verification in Human Genomes using Single-Molecule Electronic DNA Mapping. bioRxiv 140699 (2017) doi: 10.1101/140699. [DOI] [Google Scholar]
- 21.Lam ET et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol 30, 771–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barseghyan H. et al. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 9, 90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zook JM et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol 32, 246–251 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Krusche P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol 37, 555–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleveland MH, Zook JM, Salit M. & Vallone PM Determining Performance Metrics for Targeted Next-Generation Sequencing Panels Using Reference Materials. J. Mol. Diagn 20, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenger AM et al. Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. bioRxiv 519025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudmant PH et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad DF et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh H. et al. svclassify: a method to establish benchmark structural variant calls. BMC Genomics 17, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang AW et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11, R52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mu JC et al. Leveraging long read sequencing from a single individual to provide a comprehensive resource for benchmarking variant calling methods. Sci. Rep 5, 14493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huddleston J. et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.English AC et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics 16, 286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Audano PA et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell 176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wala JA et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 28, 581–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron DL et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 27, 2050–2060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nattestad M. et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 28, 1126–1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee AY et al. Combining accurate tumor genome simulation with crowdsourcing to benchmark somatic structural variant detection. Genome Biol. 19, 188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia LC et al. SVEngine: an efficient and versatile simulator of genome structural variations with features of cancer clonal evolution. Gigascience 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffares DC et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun 8, 14061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spies N, Zook JM, Salit M. & Sidow A. Svviz: A read viewer for validating structural variants. Bioinformatics 31, (2015). [DOI] [PubMed] [Google Scholar]
- 42.Song JHT, Lowe CB & Kingsley DM Characterization of a Human-Specific Tandem Repeat Associated with Bipolar Disorder and Schizophrenia. Am. J. Hum. Genet 103, 421–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman LM et al. SVCurator: A Crowdsourcing app to visualize evidence of structural variants for the human genome. bioRxiv 581264 (2019) doi: 10.1101/581264. [DOI] [Google Scholar]
- 44.Collins RL et al. An open resource of structural variation for medical and population genetics. bioRxiv 578674 (2019) doi: 10.1101/578674. [DOI] [Google Scholar]
- 45.Hickey G. et al. Genotyping structural variants in pangenome graphs using the vg toolkit. bioRxiv 654566 (2019) doi: 10.1101/654566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S. et al. Paragraph: A graph-based structural variant genotyper for short-read sequence data. bioRxiv 635011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falconer E. et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 9, 1107–1112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miga KH et al. Telomere-to-telomere assembly of a complete human X chromosome. bioRxiv 735928 (2019) doi: 10.1101/735928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.